Abstract
The functions of the vast majority of genes encoding R2R3 MYB domain proteins remain unknown. The closely related MYB33 and MYB65 genes of Arabidopsis thaliana have high sequence similarity to the barley (Hordeum vulgare) GAMYB gene. T-DNA insertional mutants were isolated for both genes, and a myb33 myb65 double mutant was defective in anther development. In myb33 myb65 anthers, the tapetum undergoes hypertrophy at the pollen mother cell stage, resulting in premeiotic abortion of pollen development. However, myb33 myb65 sterility was conditional, where fertility increased both under higher light or lower temperature conditions. Thus, MYB33/MYB65 facilitate, but are not essential for, anther development. Neither single mutant displayed a phenotype, implying that MYB33 and MYB65 are functionally redundant. Consistent with functional redundancy, promoter–β-glucuronidase (GUS) fusions of MYB33 and MYB65 gave identical expression patterns in flowers (sepals, style, receptacle, anther filaments, and connective but not in anthers themselves), shoot apices, and root tips. By contrast, expression of a MYB33:GUS translational fusion in flowers was solely in young anthers (consistent with the male sterile phenotype), and no staining was seen in shoot meristems or root tips. A microRNA target sequence is present in the MYB genes, and mutating this sequence in the MYB33:GUS fusion results in an expanded expression pattern, in tissues similar to that observed in the promoter-GUS lines, implying that the microRNA target sequence is restricting MYB33 expression. Arabidopsis transformed with MYB33 containing the mutated microRNA target had dramatic pleiotrophic developmental defects, suggesting that restricting MYB33 expression, especially in the shoot apices, is essential for proper plant development.
INTRODUCTION
With >120 genes in the Arabidopsis thaliana genome, the R2R3-MYB gene family has been identified as one of the most abundant classes of transcription factors in plants (Stracke et al., 2001). They are involved in a diverse range of processes, including controlling cell shape, disease resistance, regulating secondary metabolism, and hormone signal transduction (Jin and Martin, 1999). One MYB gene from barley (Hordeum vulgare), HvGAMYB, is involved in gibberellin (GA) signaling in the aleurone (Gubler et al., 1995). Here, expression of HvGAMYB is upregulated by GA, where it then binds to the TAACAAA motif of a barley high-pI α-amylase promoter, a motif that plays an important role in the GA-regulated expression of the α-amylase gene (Skriver et al., 1991; Gubler and Jacobsen, 1992; Lanahan et al., 1992). In transient assays, constitutive expression of HvGAMYB is sufficient to activate the α-amylase promoter (Gubler et al., 1995) and the promoters of other aleurone GA-regulated genes that are required for the mobilization of endosperm reserves (Cercós et al., 1999; Gubler et al., 1999). Thus, in regards to expression of these genes in aleurone layers, transient expression of HvGAMYB has the same effect as GA application. From this, it has been concluded that HvGAMYB is a positive regulator of the GA signal transduction pathway in the barley aleurone.
Recently, several studies have examined the role of GAMYB outside the aleurone. In rice (Oryza sativa), three independent Tos17 insertion alleles in the GAMYB gene have been isolated (Kaneko et al., 2004). As predicted, no α-amylase expression was induced in gamyb seeds that had been treated with GA, confirming the role of GAMYB in induction of α-amylase expression. Although no vegetative phenotype was observed in gamyb plants, after phase transition to the reproductive stage, shortened internodes and defects in floral organs were observed, especially defects in anthers (Kaneko et al., 2004). These phenotypic deviations from wild-type plants were consistent with the pattern of expression of the GAMYB gene. Similar results were found in barley, where HvGAMYB was found to be strongly expressed in the anthers, and transgenic barley overexpressing HvGAMYB was male sterile (Murray et al., 2003). However, whereas the male sterility in barley was due to the failure of anthers to dehisce, the block in the rice gamyb plants occurred before meiosis. Other studies have implicated a role for GAMYB in endosperm development in barley (Diaz et al., 2002), elongation of the first internode in wheat (Triticum aestivum) (Chen et al., 2001), and flowering in Lolium temulentum (Gocal et al., 1999).
In Arabidopsis, there is a small family of GAMYB-like genes (Gocal et al., 2001). Construction of a phylogenetic tree with the entire MYB gene family of Arabidopsis groups five genes (MYB33, MYB65, MYB97, MYB101, and MYB120) with HvGAMYB (Stracke et al., 2001). Furthermore, these five genes contain a unique intron located at the 3′ end of the open reading frame, implying that they belong to a distinct subclass in the MYB superfamily of transcription factors. Previously, MYB33, MYB65, and MYB101 have been shown to be able to substitute for HvGAMYB in transactivating the α-amylase promoter in barley aleurone layer assays (Gocal et al., 2001). The expression of these genes was consistent with roles in GA-mediated processes, with expression of MYB33 at the shoot apex coinciding with the onset of flowering, either when endogenous GA levels increased or when GA was applied to the plants (Gocal et al., 2001).
Recent evidence suggests that the GAMYB-like genes are subjected to posttranscriptional regulation by microRNAs (miRNAs). Initially, three Arabidopsis miRNAs, miR159a, b, and c, were identified that have complementarity to a highly conserved motif in the coding region of the GAMYB-like genes (Park et al., 2002; Rhoades et al., 2002). The isolation of miRNA-guided cleavage products for both the MYB33 and MYB65 genes further suggests these genes are being regulated by miRNAs (Palatnik et al., 2003), and using an Agrobacterium tumefaciens–mediated delivery system, it has been shown that miR159a can cleave MYB33 mRNA in planta (Achard et al., 2004). Furthermore, Arabidopsis plants with mutations in genes involved in miRNA-mediated gene regulation, such as hua enhancer 1 or hyponastic leaves 1, have much higher steady state levels of MYB33 transcripts when compared with wild-type plants (Han et al., 2004), whereas plants overexpressing miR159a have decreased levels of MYB33, are male sterile, and have delayed flowering time (Achard et al., 2004). Finally, this miRNA target motif has been shown to be functionally important, for 35S expression of a MYB33 gene with a mutated miRNA target sequence (mMYB33) results in plants with an altered leaf morphology, which is in contrast with 35S expression of a wild-type MYB33 gene that results in no observable phenotype (Palatnik et al., 2003).
In this article, we describe T-DNA–tagged mutants in two of the Arabidopsis GAMYB-like genes, MYB33 and MYB65, and show that these genes act redundantly in anther development. Using a series of β-glucuronidase (GUS) reporter constructs, we demonstrate that the miR159 target motif restricts MYB33 expression, and when this motif is absent, aberrant MYB33 gene expression occurs, resulting in gross defects in plant growth and development.
RESULTS
The Duplicated Genes MYB33 and MYB65 Are Putative GAMYB Orthologs
Based on several different phylogenetic trees, MYB33 and MYB65 are the Arabidopsis genes with the highest similarity to the GAMYB genes of cereals (Stracke et al., 2001; Yang et al., 2001). Furthermore, MYB33 and MYB65 have high sequence similarity to one another. Overall, they are 58.4% identical at the amino acid level, with >90% identity shared between their R2R3 domains but only 51.0% identity in their C-terminal domains. Analysis of their chromosomal locations reveal that they lie within segmental duplicated regions of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000), for the genes immediately upstream of the MYB genes encode proteins that have high sequence identity with one another (81.2% amino acid identity) as do the genes immediately downstream (69.7% amino acid identity). In addition to being duplicated genes, Gocal et al. (2001) found that they have similar expression patterns. These two facts make them strong candidates for being functionally redundant in the plant.
Isolation and Characterization of T-DNA Insertional Mutants in the MYB33 and MYB65 Genes
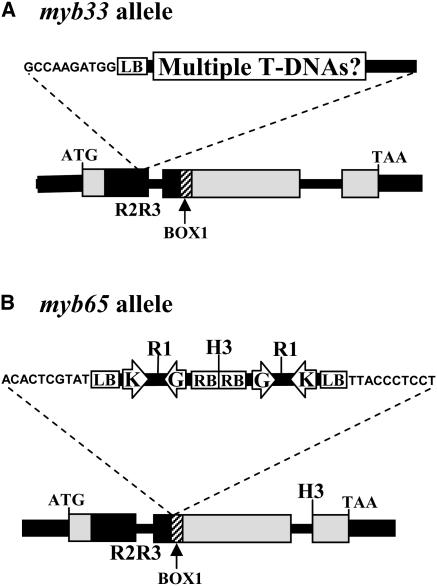
To determine the function of the Arabidopsis GAMYB-like genes, we undertook a reverse genetic approach and isolated T-DNA mutant alleles for both MYB33 and MYB65. Using a PCR-based screen on populations of T-DNA tagged lines, we identified one mutant that contained a T-DNA insert in the region encoding the R2R3 domain of the MYB33 gene, 15 amino acids into the R3 domain (Figure 1A). This insertion is likely to result in a null allele of MYB33 and was designated myb33. To help determine the number of T-DNA inserts, we performed DNA gel blot analysis on DNA from homozygous myb33 plants using the uidA (GUS) gene as a probe. Each lane had two to three hybridizing bands (data not shown), indicating that multiple T-DNA copies are present in the myb33 mutant line. In addition, segregation analysis for the nptII gene was performed by selecting for growth on medium containing kanamycin (50 μg/mL) with seeds from a heterozygous myb33 plant. There were 203 resistant plants and 76 sensitive plants that approximates to a 3:1 ratio (χ2 = 0.746; P > 0.25), implying that the T-DNAs segregated as one locus. This suggests that either multiple T-DNAs have inserted into one site as a concatamer, or T-DNAs have inserted into multiple linked sites. Plants homozygous for the myb33 allele were isolated; however, there were no obvious morphological alterations to the plants.
Figure 1.
Structure of the T-DNA Insertional Mutants.
(A) Structure of the myb33 allele.
(B) Structure of the myb65 allele.
Arrows indicate the location of the neomycin-phosphotransferase II gene (K) and the uidA gene driven by the −60 35S Cauliflower mosaic virus promoter (G). LB, left border; RB, right border; R1, EcoRI; H3, HindIII. As determined by sequencing the T-DNA/plant DNA junctions, the 10 nucleotides of Arabidopsis DNA flanking the T-DNAs are shown.
One mutant was found in which a T-DNA had inserted into the coding region of the MYB65 gene. In this allele, designated myb65, the T-DNA had inserted into the region of the gene that encodes Box 1 (Gocal et al., 2001), a conserved motif amongst the GAMYB-like genes that is immediately C-terminal to the R2R3 domain (Figure 1B). Both T-DNA/plant junctions could be amplified with the left border primer, JL202, implying that tandem T-DNAs had inserted into this locus in an inverted fashion. Restriction fragment patterns of a genomic DNA gel blot of myb65 hybridized to the GUS gene were consistent with this (data not shown). DNA digested with EcoRI gave a single expected band of 4.9 kb, whereas a HindIII digest gave two bands of >10 and 6.8 kb, which are consistent with the predicted restriction map (Figure 1B) and implying that there is only one T-DNA locus present in the plant. This is supported by segregation analysis of progeny of a MYB65/myb65 heterozygote on growth medium containing kanamycin (50 μg/mL). Here, 425 resistant plants were obtained compared with 135 sensitive plants, again an approximate 3:1 ratio (χ2 = 0.237; P > 0.5). In addition to the T-DNA insertion, 20 bp of the wild-type gene had been deleted. Thus, like the myb33 allele, it is assumed a nonfunctional protein is produced. Plants homozygous for the myb65 allele were isolated; however, there was no visible phenotype in these plants.
Double Mutant myb33 myb65 Plants Are Male Sterile
Although both single mutants have predicted loss-of-function alleles, neither myb33 nor myb65 plants show any detectable differences from wild-type plants. Thus, the generation of a myb33 myb65 double mutant may be necessary to elucidate their function(s). A cross was made between homozygous myb33 and homozygous myb65 plants. Of 173 F2 plants, nine plants failed to set seed. Using both PCR genotyping and genomic DNA gel blot analysis, it was found that only these plants were myb33 myb65 double mutants (data not shown). A 164:9 ratio is consistent with a 15:1 ratio (χ2 = 0.323; P > 0.5) expected with two segregating recessive loci. This implies that only plants homozygous for both myb33 and myb65 are sterile and that the presence of only one copy of MYB33 or MYB65 is enough to result in fertile plants.
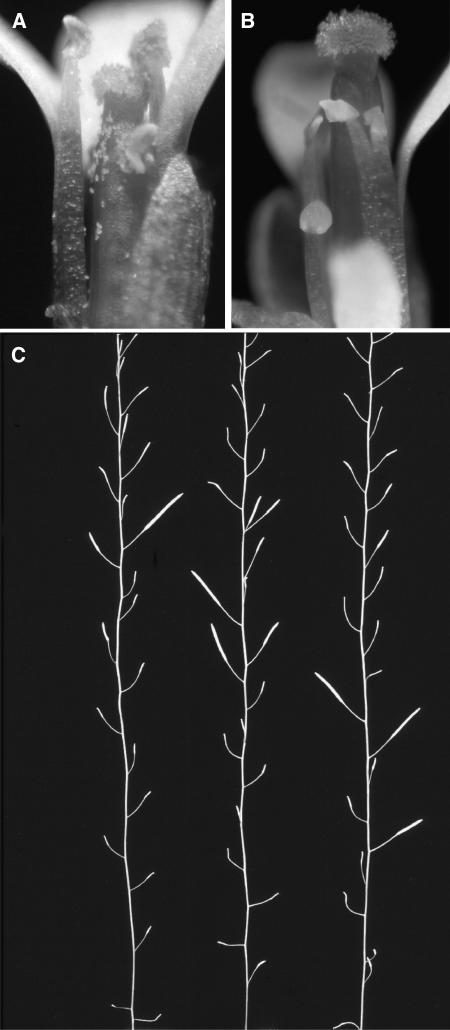
Stamens of myb33 myb65 were typically shorter than their wild-type counterparts (Figure 2A) and failed to fully extend to the pistil (Figure 2B). Furthermore, anthers from the mutant plants were smaller than the wild type and failed to produce pollen, indicating that the mutant is male sterile. Reciprocal crosses between myb33 myb65 and wild-type plants demonstrated that the female parts of the myb33 myb65 plant were fully fertile and confirmed the male sterile phenotype of myb33 myb65. Other than sterility and the associated characteristics of sterile plants, such as the increased numbers of flowers per inflorescence, myb33 myb65 shows no obvious morphological differences from wild-type plants. These observations were on plants grown under continuous light at ∼80 μmol·m−2s−1 light intensity and 21°C.
Figure 2.
Characteristics of the Male Sterile Phenotype of myb33 myb65.
(A) Flowers of the Columbia-0 ecotype photographed under bright-field microscopy.
(B) Flowers of the myb33 myb65 double mutant, photographed under bright-field microscopy.
(C) Three different bolts of myb33 myb65 that exhibit the sporadic setting of siliques, where the majority of siliques fail to set any seeds, but occasionally fully or partially filled siliques are set. Bolts are from plants grown at 22°C in a 12-h-day/12-h-night cycle and ∼90 μmol·m−2s−1 light intensity.
Of the nine myb33 myb65 plants obtained, two plants eventually set seed. From one plant, one silique was present, from which five seeds were obtained, and in the second plant, three consecutive siliques set seed, from which 44 seeds were obtained. PCR genotyping was performed on the F3 plants derived from these seed, and they were all shown to be myb33 myb65 double mutants, implying that these seeds arose through a self-crossing event. Like their F2 parents, these F3 plants were also male sterile, but again would sporadically produce partially or completely filled siliques. Along the bolt, sterility could cycle, with filled siliques followed by a few empty siliques (unfertilized carpels) and then again more filled siliques (Figure 2C). The unusual characteristic of this mutant is that a plant may only produce several siliques containing seeds and that these siliques may contain close to the same number of seeds as siliques from a wild-type plant. Thus, occasionally during the plant's life it is able to produce pollen and set seed. This is different from the known reduced fertility mutants of Arabidopsis that have been classified as having consistently short siliques (Sanders et al., 1999). Thus, the myb33 my65 double mutant appears to represent a new class of male sterile mutant in Arabidopsis.
Because of the presence of multiple T-DNAs in the myb33 allele, molecular complementation was needed to confirm that the male sterile phenotype observed in myb33 myb65 was in fact due to a mutant MYB33 gene. A genomic clone of MYB33 was transformed into myb33 myb65, and 25 of 30 T1 plants had fertility entirely restored. Fertile and sterile plants were found to be segregating in the progeny, and the transgene was found by PCR to be segregating with the fertile plants (data not shown), confirming that the MYB33 gene is able to complement the male sterile phenotype. For MYB65, an additional allele was obtained from the SALK collection (SALK_063552), which we designated myb65-2. We constructed a myb33 myb65-2 double mutant and found again the plants were male sterile. This second allele confirms that MYB65 is required for proper anther development in Arabidopsis.
The Block in Pollen Development Occurs Premeiotically Where the Tapetum Undergoes Hypertrophy
Transverse sections of wild-type and double mutant Arabidopsis anthers were prepared and stained with toluidine blue to identify the stage at which anther development is blocked. Sanders et al. (1999) have divided anther development into 14 stages, at which distinctive cellular events can be visualized at the level of the light microscope.
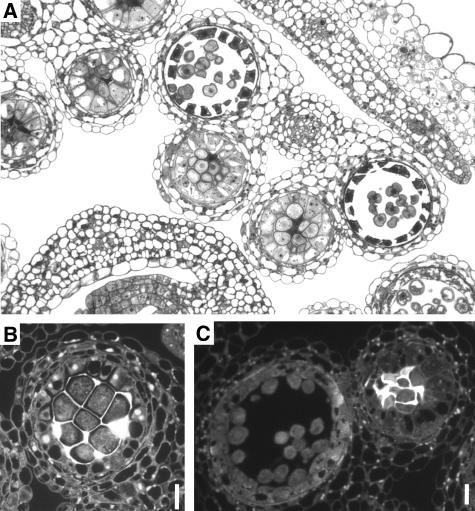
In myb33 myb65, no visible defects were found in early anther development up to stage five (Sanders et al., 1999). Here, the division of the archesporial cells to give rise to the pollen mother cells (PMCs), tapetum, middle layer, and endothecium (Figure 3F) appears indistinguishable from the wild type (Figure 3A). However, during stage six when the PMCs begin to separate in a clearly defined locule and the tapetum begins to vacuolate (Figure 3B), the mutant is similar, except that the tapetum begins to enlarge (Figure 3G). Furthermore, whereas the products of meiosis, the tetrads, become clearly visible in the wild-type anthers (Figure 3C), these structures are not visible in myb33 myb65, but rather the tapetum has expanded to such an extent that there is no locule, and the PMCs have an irregular shape (Figure 3H). Thus, the block in pollen development appears to be premeiotic, occurring between anther stages 5 and 6 (Sanders et al., 1999) or floral stages 9 and 10 (Smyth et al., 1990). Whereas microspores form in the locule of wild-type anthers (Figure 3D) and eventually form mature pollen (Figure 3E), the tapetum of the myb33 myb65 mutant continues to expand (Figure 3I) until the contents collapse and degenerate (Figure 3J). The expansion of the tapetum appears to be due to an increase in cell size, not in cell number.
Figure 3.
Comparison of the Development of Locules from Wild-Type and myb33 myb65 Plants Using Toluidine Blue Staining.
(A) to (E) Development in wild-type anthers.
(A) Early microsporocyte stage, with the pollen mother cells (pmc) surrounded by the four distinct cell layers, the epidermis (ep), endothecium (en), middle layer (m), and tapetum (t).
(B) PMCs begin to separate from one another and the locular space beginning to form.
(C) Tetrad stage, with the individual microspores separated by callose.
(D) Microspore stage. The tapetum shows signs of degradation and has a granular appearance.
(E) Mature pollen stage (tricellular pollen) before release from the anther. The tapetum has degenerated.
(F) to (J) Development in myb33 myb65 anthers.
(F) Development at the early microsporocyte stage appears indistinguishable from the wild type.
(G) At PMC stage, the tapetum (t) begins to enlarge.
(H) The locule becomes crowded, and the PMCs have an irregular shape.
(I) The tapetum continues to expand, and the PMCs begin to degrade.
(J) The tapetum occupies the majority of the locule, and the PMCs have degenerated.
Anthers are from plants grown at 22°C in a 12-h-day/12-h-night cycle and ∼90 μmol·m−2s−1 light intensity. All bars = 10 μm.
Such an early and comprehensive block in pollen development in the myb33 myb65 mutant appears inconsistent with the ability of the mutant to sporadically set partially or completely filled siliques. This apparent paradox was explained by the finding that some anthers from the mutant appear to undergo a wild-type developmental program. Strikingly, individual normal locules can be adjacent to locules in which pollen development has aborted through a hypertrophic tapetum. Figure 4A shows an example of this, where development in the two outer locules is at an early microspore stage, with the tapetum degrading and producing as many microspores as a locule from a wild-type plant (Figure 3D). The wild-type development explains the sporadic fertility of myb33 myb65. Thus, although there is some variation in anther development, locules appeared to undertake either one of two predominant courses of development; a completely wild-type developmental program or the tapetum undergoes hypertrophy before meiosis, resulting in the abortion of pollen development. In anthers in which both types of locules were present, there was no consistency in which of the locules of the anther developed normally. This alternative development of locules in the mutant anthers suggests that there are additional factors (environmental or genetic) that are influencing the development of the tapetum and that these factors can have a very localized affect.
Figure 4.
Further Microscopic Analysis of myb33 myb65 Anthers.
(A) Toluidine blue staining of a section showing the alternative developmental pathways within locules from the same myb33 myb65 anther.
(B) Alaline blue staining for callose in myb33 myb65 at the PMC stage under dark-field microscopy. Fluorescence indicates the presence of callose.
(C) Staining for callose in myb33 myb65 at the microspore stage. Wild-type development has proceeded in the locule on the left (no callose), whereas development has aborted in the locule on the right (callose present).
Anthers are from plants grown at 22°C in a 12-h-day/12-h-night cycle and ∼90 μmol·m−2s−1 light intensity. All bars = 10 μm.
To further characterize the mutant, analine blue staining was performed to detect the presence of callose. It was found that myb33 myb65 could in fact make callose, and similar to the wild type, it accumulates around the PMC at the expected developmental stage (Figure 4B). However, the callose persisted in locules where pollen development had aborted. In Figure 4C, callose is present in the locule that has a hypertrophic tapetum, whereas an adjacent locule that is undergoing a wild-type developmental program has no callose present. The fact that callose is persisting in the defective locule implies that callase has not been secreted from the tapetum and that the tapetum, in addition to undergoing hypertrophy, has ceased performing its normal functions. However, the fact that the other locule has been able to degrade its callose (Figure 4C) implies that the persistence of callose in the aborted locule is a secondary effect, rather than the primary cause of the locule aborting pollen production. The production of callose and its persistence are consistent with the block in development occurring before meiosis, for in the wild type, only after the tetrads are formed is the callose degraded.
Fertility Is Restored in myb33 myb65 Plants Grown under Higher Light Intensities or at a Lower Growth Temperature
The fact that the double mutant could sporadically set seed and the extent to which it could was highly variable suggested that environmental factors could be influencing the plant's fertility. Light intensity, daylength, and growth temperature can all influence the fertility of a plant (Kaul, 1988). To assess the affect of light intensity on fertility, myb33 myb65 plants were grown alongside wild-type controls at a light intensity of 95 μmol·m−2s−1, in a 12-h-day/12-h-night cycle at 22°C. When the plants began to bolt, plants were moved to either 140, 210, or 330 μmol·m−2s−1 photo intensities as well as being kept at 95 μmol·m−2s−1. Plants were then allowed to grow for 5 weeks, and fertility was scored by determining the total number of siliques that set seed of each plant, divided by the total number of siliques of the plant, including the empty siliques.
The increased light intensity had a dramatic effect on the number of siliques that wild-type plants produce; at 330 μmol·m−2s−1, plants produce more than twice as many siliques than at 95 μmol·m−2s−1 (Table 1). However, if fertility is defined as the percentage of siliques that set seed, there was no significance different in wild-type plants grown under the four different light intensities. By contrast, the double mutant plants produced more than four times as many siliques as Wassilewskija (Ws) at 95 μmol·m−2s−1, for similar to other male sterile mutants, they go into a phase of continuous flowering because of the inability to set seed. However, the total number of siliques that set seeds, along with the percentage of siliques that set seed, dramatically increased with increased light intensity. Less than four percent of myb33 myb65 siliques set seed at 95 μmol·m−2s−1, in comparison with 75% at 330 μmol·m−2s−1 (Table 1). Thus, myb33 myb65 plants are almost as fertile as wild-type plants when grown at a light intensity of 330 μmol·m−2s−1.
Table 1.
Fertility in myb33 myb65 Increases with Light Intensity
| Ecotype/Mutant | Light Intensity (μmol·m−2s−1)a | Total Number of Siliques/Plant | Total Number of Filled Siliques/Plant | Percentage of Filled Siliques |
|---|---|---|---|---|
| Ws | ||||
| 95 | 55.0 ± 4.7 | 52.6 ± 4.7 | 96.2 ± 1.1 | |
| 140 | 51.7 ± 12.2 | 49.4 ± 11.7 | 96.2 ± 1.3 | |
| 210 | 99.3 ± 11.1 | 95.5 ± 10.5 | 96.2 ± 0.6 | |
| 330 | 152.7 ± 15.3 | 152.6 ± 15.3 | 97.6 ± 0.5 | |
| Col | ||||
| 95 | 39.8 ± 6.7 | 35.0 ± 5.3 | 89.3 ± 1.5 | |
| 140 | 61.8 ± 10.0 | 57.8 ± 9.4 | 93.3 ± 1.9 | |
| 210 | 103.7 ± 11.2 | 97.5 ± 11.0 | 93.3 ± 1.7 | |
| 330 | 83.0 ± 10.9 | 75.1 ± 10.9 | 89.9 ± 3.4 | |
| myb33 myb65 | ||||
| 95 | 241.6 ± 37.7 | 7.1 ± 1.2 | 3.6 ± 0.9 | |
| 140 | 129.6 ± 16.8 | 46.3 ± 7.13 | 35.5 ± 3.7 | |
| 210 | 203.0 ± 39.2 | 107.3 ± 29.2 | 50.3 ± 3.5 | |
| 330 | 176.1 ± 23.0 | 136.0 ± 21.1 | 75.1 ± 2.6 |
Light intensity at which the plants were grown.
The fertility of myb33 myb65 is also influenced by temperature. Again, plants were grown at 22°C until they began to bolt. Plants were then either left at 22°C or shifted to 16°C at approximately the same light intensity of 86 μmol·m−2s−1 (86.9 μmol·m−2s−1 at 22°C and 85.5 μmol·m−2s−1 at 16°C). Fertility was scored after 5 weeks for the plants grown at 22°C and at 7 weeks for the plants grown at 16°C because of the slower growing conditions. At 22°C after 5 weeks, the myb33 myb65 plants had produced more than four times as many siliques as Columbia (Col) or Ws, but only 2.5% of these siliques set seed. At 16°C after 7 weeks, the myb33 myb65 plants were producing similar amounts of siliques as the wild-type controls, Col and Ws, and this reflected the fact that fertility had increased in the myb33 myb65 plants, for >30% of their siliques had set seed (Table 2). Thus, fertility was >10-fold higher at 16°C relative to 22°C, implying that temperature has a dramatic effect on fertility of the myb33 myb65 plants.
Table 2.
Lower Growth Temperature Partially Restores Fertility in myb33 myb65
| Growth Temperaturea | Ecotype/Mutant | Total Number of Siliques/Plant | Total Number of Filled Siliques/Plant | Percentage of Filled Siliques |
|---|---|---|---|---|
| 22°C | Col | 51.6 ± 5.5 | 45.9 ± 3.9 | 91.2 ± 2.0 |
| Ws | 49.7 ± 5.0 | 45.8 ± 4.3 | 92.8 ± 1.4 | |
| myb33 myb65 | 217.1 ± 14.8 | 5.0 ± 1.9 | 2.5 ± 0.7 | |
| 16°C | Col | 101.3 ± 29.0 | 87.8 ± 24.2 | 89.6 ± 2.5 |
| Ws | 154.4 ± 13.1 | 152.4 ± 13.2 | 98.4 ± 0.8 | |
| myb33 myb65 | 112.3 ± 10.0 | 36.4 ± 5.4 | 32.5 ± 3.4 |
Temperature at which the plants were grown.
MYB33 and MYB65 Promoter-GUS Fusions Direct Identical Expression Patterns, but Not in Anthers
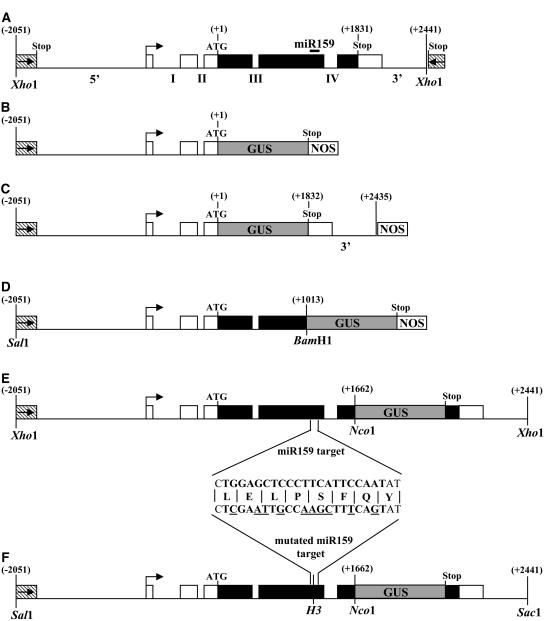
Previously, Gocal et al. (2001) examined the expression patterns of MYB33 and MYB65 using RNase protection assays and RNA in situ analyses. To extend these studies, promoter-GUS fusions, Pro33:GUS and Pro65:GUS (Figure 5B; see Methods), were transformed into Arabidopsis, multiple transgenic plants were obtained, and >12 independent lines were examined for each construct at various stages of development.
Figure 5.
Structure of the Constructs Used for Molecular Complementation, for MYB33 Expression Analysis, and for Overexpresion of MYB33 and mMYB33 Genes.
(A) Structure of the Landsberg erecta MYB33 genomic clone used for molecular complementation. This clone contains 1390 bp upstream of the putative transcription start site (arrow), which includes all the 5′ untranscribed regions of MYB33 and into the coding region of the upstream gene (At5g06090; the stop codon being −1833 relative to MYB33 start codon), 660 bp of 5′ untranslated region, including the first two introns (denoted I and II), 1833 bp of open reading frame, including two introns (denoted III and IV), and 611 bp of sequences 3′ of the MYB33 stop codon, which extends downstream 12 bp before the stop codon of the next gene (At5g060110; stop codon is +624 bp relative to MYB33 stop codon). Untranscribed regions and introns are shown as thin lines, untranslated regions as open boxes, and coding regions as closed boxes. Numbers shown are relative to the MYB33 start codon. The location of the miR159 target is indicated by a solid line. This construct was also used to overexpress MYB33 in wild-type plants.
(B) The Pro33:GUS transgene in the vector pBI101.1.
(C) The Pro33:GUS:3′end transgene in the vector backbone pBI101.1
(D) The 5′-MYB33:GUS transgene in the vector backbone pBI101.1.
(E) The MYB33:GUS transgene made in the vector pPZP200-Hygro.
(F) The mMYB33:GUS transgene made in the vector pPZP200-Hygro. The alteration to the nucleotide sequence in the miRNA target motif is shown, and nucleotides underlined are those differing from the wild type. The unaltered amino acid sequence is shown. The mMYB33 gene is identical but lacks the GUS reporter gene. H3, HindIII.
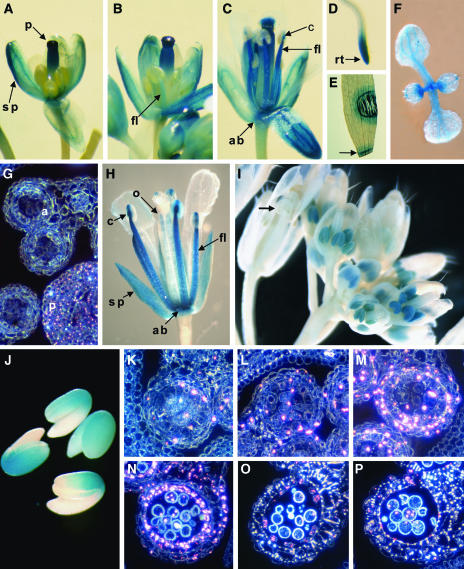
GUS staining in Pro33:GUS plants was strongest in the inflorescence and root tips (Figures 6A to 6D). At approximately floral stage 12 (petals level with long stamens; Smyth et al., 1990), the upper half of the pistil stained strongly, with weaker staining in the sepals and the anther filament (Figure 6A). However, no obvious GUS staining was observed in the anther tissues (Figures 6A and 6G). As the flower develops (Figures 6B and 6C), not only does the GUS staining persist in the sepals and upper half of the pistil, but it becomes much stronger in the anther filament and connective. Furthermore, there was strong staining in the receptacle (abscission zone). Thus, the MYB33 promoter directs GUS expression in the inflorescence in a very complex pattern, and no variation was seen amongst the transgenic lines. Although no GUS staining could be found in mature leaves or stems of plants, GUS staining could be seen in newly emerging leaves, predominantly at the proximal side (Figure 6F). Furthermore, wounding could induce GUS expression, either by crushing cells with a pair of forceps or slicing with a razor blade (Figure 6E, arrow). The response was very local, where GUS was expressed as a narrow band that encircled the wounded site, several cell layers away from the actual wound (Figure 6E). Furthermore, this induction of GUS occurs immediately after wounding, implying that the gene is rapidly induced.
Figure 6.
Expression of MYB33 and MYB65; GUS Staining of Transgenic Pro33:GUS, myb65 Enhancer Trap, and MYB33:GUS Arabidopsis Plants.
(A) GUS activity as determined by histochemical analysis in a Pro33:GUS Arabidopsis flower at floral stage 12.
(B) Pro33:GUS flower at floral stage 13.
(C) Pro33:GUS flower at floral stage 15.
(D) Pro33:GUS root of a 5-d-old plant.
(E) Wounded leaf of a Pro33:GUS plant.
(F) Seven-day-old Pro33:GUS seedling
(G) Transverse section of young anthers at the PMC stage of Pro33:GUS shown under dark-field microscopy. Pink crystals indicate GUS expression.
(H) A GUS-stained flower (at floral stage 15) from the myb65 enhancer trap line.
(I) A GUS-stained inflorescence of a MYB33:GUS plant.
(J) GUS-stained imbibed seeds from a MYB33:GUS plant.
(K) Transverse section of MYB33:GUS anthers at the archesporial cell stage.
(L) Early PMC stage.
(M) Meiosis.
(N) Young microspore stage.
(O) Stage of tapetal degeneration.
(P) Dehiscence.
a, anther; ab, abscission zone (receptacle); c, connective; fl, anther filament; o, ovule; p, pistil; rt, root tip; sp, sepal.
For the Pro65:GUS construct, 2145 bp upstream of the MYB65 ATG start codon was used. Sequence comparisons between this 2145-bp region to the 2051-bp MYB33 5′ region revealed that the first 73 bp immediately 5′ of the ATG of MYB33 shares significant homology to the corresponding region of the MYB65 gene. However, beyond this region, the sequences share little homology with one another, with only eight stretches of sequence with six or more base pairs that are identical. However, despite these highly divergent promoter regions, the pattern of GUS expression in Pro65:GUS transgenic plants was identical to that of the complex expression pattern of Pro33:GUS (data not shown) and was again not expressed in anthers.
The 3′ Region of MYB33 Does Not Direct Expression in Anthers
For correct expression, several genes have been shown to require their 3′ flanking sequences, for regulatory elements can reside in these regions (e.g., Larkin et al., 1993; Chen et al., 1998). Thus, we subcloned the 3′ noncoding sequences of MYB33, downstream of the GUS gene into the Pro33:GUS construct, resulting in the construct Pro33:GUS:3′end (Figure 5C). This fragment contains all sequences 3′ of the coding region of MYB33 to the next open reading frame downstream. Transgenic plants displayed a GUS expression pattern that was identical, both spatially and temporally, to Pro33:GUS and Pro65:GUS, and again no expression was seen in anthers, suggesting that sequences 3′ of the MYB33 coding region do not contain regulatory elements that control tissue-specific expression.
The myb65 Allele Contains an Enhancer Trap That Expresses GUS Similar to the Promoter-GUS Lines
The myb65 allele was generated using the binary vector pD991 that contains a minimal promoter fused to the GUS reporter gene (Figure 1B), and this can act as an enhancer trap (Campisi et al., 1999). The myb65 plants only had one T-DNA locus; thus, any GUS gene expression will be driven by regulatory elements of the MYB65 gene. When myb65 mutant plants were stained for GUS activity, a similar pattern of spatial and temporal expression was observed as the Pro33:GUS and Pro65:GUS lines. Staining was strongest in mature flowers, with sepals, receptacle, anther filaments, and connective all staining for GUS activity (Figure 6H). The fact that both the enhancer trap line and the Pro33:GUS and Pro65:GUS lines stain identically in these tissues would argue that MYB33/MYB65 are genuinely expressed in these locations. However, there were several differences between the enhancer trap and promoter-GUS lines. First, in female tissues, GUS staining was absent from the stigma/style, but some staining did appear in the ovules (Figure 6H). Furthermore, expression in the enhancer trap line was not wound inducible.
In Floral Tissues, a MYB33:GUS Translational Fusion Is Expressed Exclusively in Young Anthers
Neither the Pro33:GUS/Pro65:GUS lines or the enhancer trap line had any GUS staining in anthers, despite the phenotype of myb33 myb65 and the RNA in situ data suggesting MYB33/MYB65 was expressed in young anthers (Gocal et al., 2001). Thus, we constructed a MYB33:GUS translational fusion (Figure 5E) using the whole MYB33 gene (pPZP-MYB33, Figure 5A) that could fully complement the myb33 myb65 mutant, implying all the regulatory sequences required for anther development are present.
In stark contrast with the promoter-GUS lines and enhancer trap line, expression in flowers was found to only be located in young anthers (Figure 6I) in >24 independent lines examined. No expression could be seen in the rest of the flower, including sepals, receptacle, anther filaments, and stigma/style at any stage, and expression was not wound inducible. Expression was strongest in anthers before floral stage 12 (Smyth et al., 1990); however, after this developmental stage, expression appeared to be absent (arrow, Figure 6I). Expression from the transgene was generally low, and most lines required >30 h of incubation for strong staining. Expression was also found in imbibed seeds, being strongest in the region where the cotyledons met the hypocotyl (Figure 6J).
To analyze the spatial and temporal pattern of MYB33:GUS expression in anthers, inflorescences were fixed and sectioned transversely. Expression of GUS is shown as pink crystals, with the level of expression reflected in the number and size of the crystals. Through all stages of development, GUS crystals were only observed in anthers. GUS crystals were visible at the archesporial cell stage; however, their number was low and was mainly confined to the four anther cell layers (Figure 6K). Expression increased slightly at the PMC stage (Figure 6L), and at meiosis when the tetrads became visible, expression increased dramatically in the four anther cell layers (Figure 6M) and was also found in the connective region (data not shown) and the tetrads themselves. After this developmental stage, GUS expression remained strong but is mainly confined to the tapetum (Figure 6N), and expression persists in this cell layer until it degrades (Figure 6O). At dehiscence, low levels of expression are found in the pollen grains and in the epidermis, adjacent to the stomium (Figure 6P).
Posttranscriptional Regulation of MYB33/MYB65: An miR159 Target Sequence Restricts Expression to Anthers
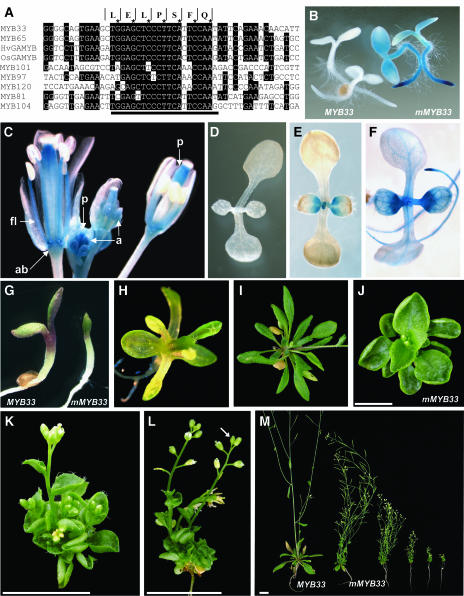
The promoter-GUS constructs and the MYB33:GUS translational fusion generate dramatically contrasting expression patterns. A possible explanation for this is that sequences are present downstream of the ATG (+1) that repress expression in tissues outside the anther (i.e., sepals, anther filaments, receptacle, stigma/style, roots, and wounding). One potential sequence is the miR159 target motif that is fully conserved between MYB33 and MYB65 (Figure 7A). Furthermore, this motif is highly conserved amongst the other closely related Arabidopsis MYB genes and in all cereal GAMYB genes cloned to date (Figure 7A). This high level of conservation implies that this motif plays an important role in the function of these MYB genes.
Figure 7.
The miR159 Target Motif Is Highly Conserved and Is Required to Restrict MYB33 Expression for Normal Plant Development.
(A) Alignment of the miR159 target sequence (black line) and surrounding sequences of the GAMYB-like genes of Arabidopsis (AtMYB33, AtMYB65, AtMYB101, AtMYB97, and AtMYB120), as well as two other closely related genes (AtMYB81 and AtMYB104), and the barley (HvGAMYB) and rice (OsGAMYB) genes. Amino acid sequence above corresponds to the sequence encoded by MYB33 and MYB65. The asterisk indicates the third position in each codon.
(B) GUS staining of 3-d-old MYB33:GUS and mMYB33:GUS seedlings.
(C) GUS staining of an inflorescence from a mMYB33:GUS transgenic line.
(D) GUS staining of a 7-d-old MYB33:GUS seedling (48 h of staining).
(E) GUS staining of a 7-d-old mMYB33:GUS seedling (16 h of staining).
(F) GUS staining of a 7-d-old 5′-MYB33:GUS seedling (6 h of staining).
(G) Comparison of 5-d-old transgenic plants transformed with the wild-type MYB33 gene (left) and mMYB33 (right).
(H) Ten-day-old mMYB33 transformant that is failing to form proper leaves.
(I) Rosette of a 4-week-old plant transformed with the MYB33 gene.
(J) Rosette of a 4-week-old plant transformed with the mMYB33 gene.
(K) Side view of a 3-week-old mMYB33 plant exhibiting extreme leaf curling.
(L) Side view of a 4-week-old mMYB33 plant exhibiting a short primary bolt with a terminal flower.
(M) Comparison of a 4-week-old MYB33 transformant with mMYB33 transformants with phenotypic defects of increasing degrees.
ab, abscission zone; a, anther; fl, anther filament; p, pistal. Bars = 1 cm.
To test the importance of the miR159 target sequence for MYB33 expression, or whether other regulatory sequences are present downstream of the ATG (+1), two different constructs were made. First, a construct was made, 5′-MYB33:GUS, where the MYB33 promoter, along with the first 1013 bp of the MYB33 gene, was fused in frame with the GUS reporter gene (Figure 5D). This includes all the sequences to 10 bp 5′ of the miR159 target motif. Multiple 5′-MYB33:GUS transgenic plants were generated, and >16 independent primary transformants were analyzed. All lines had a pattern of GUS expression that was indistinguishable from that of Pro33:GUS, Pro65:GUS, and Pro33:GUS:3′end. Furthermore, the strength of GUS activity is also similar; thus, it appears that there are no regulatory motifs, downstream of the ATG and upstream of the miR159 target sequence, that are regulating expression of the gene.
The second construct, mMYB33:GUS, is identical to MYB33:GUS, except that the miR159 target site has been mutated, altering the sequence of 10 nucleotides, but not the amino acid sequence the region encodes (Figure 5F; Palatnik et al., 2003). Multiple transgenic plants harboring this mMYB33:GUS construct were generated, and inflorescences from 16 different primary transformants were stained for GUS expression. There was variation in the pattern of GUS expression in the different primary transformants. For seven lines, weak GUS expression was found to be specifically in young anthers identical to MYB33:GUS. For seven other lines, much stronger expression was found, and in addition to expression being in young anthers, the expression pattern had increased in tissues outside the anther (Figure 7C), similar to the spatial and temporal expression of the promoter-GUS lines (Figures 6A to 6F). This included the upper half of the pistal throughout flower development, the receptacle, anther filaments, and connective, but a lack of expression in anther themselves after floral stage 12, with some expression in stems (Figure 7C). Furthermore, wounding induced GUS expression. Thus, the expression pattern of mMYB33:GUS resembled a combination of Pro33:GUS and MYB33:GUS, demonstrating that these two expression patterns were not mutually exclusive.
Expression was also examined in seedlings. Seeds from 16 different mMYB33:GUS lines were planted alongside eight MYB33:GUS lines and eight 5′-MYB33:GUS lines and were stained for GUS activity after 3 and 7 d of germination. After 3 d of germination, all mMYB33:GUS lines had strong GUS expression in root tips in all but two lines, and expression was also seen in the cotyledons of the strongest expressing lines (Figure 7B). GUS expression was seen in all eight 5′-MYB33:GUS lines examined, and typically stained stronger than mMYB33:GUS lines. By contrast, there was no GUS staining in any of the eight MYB33:GUS lines. In 7-d-old mMYB33:GUS seedlings, expression was still found in root tips, in both primary and secondary roots. Furthermore, expression was seen in and around the shoot meristem and in the proximal regions of the young emerging true leaves (Figure 7E). A similar pattern of expression was seen in the 5′-MYB33:GUS lines (Figure 7F). In contrast with the mMYB33:GUS and 5′-MYB33:GUS lines, no GUS was seen in any of eight MYB33:GUS lines examined (Figure 7D), even after 48 h of staining.
Thus, in conclusion, mutation of the miR159 target sequence results in increased expression of MYB33 in tissues outside the anther, and these are tissues in which the promoter-GUS lines were seen to be directing expression (Figure 6). This suggests that the miR159 target sequence restricts the expression pattern of MYB33.
Introduction of mMYB33 into Plants Results in Dramatic Pleiotrophic Developmental Defects
The altered expression pattern of mMYB33:GUS suggests that the miR159 target sequence is important for restricting the domain of MYB33 expression. To investigate whether expression of MYB33 in these tissues outside the anther have consequences for plant growth and development, we transformed Arabidopsis with the mMYB33 construct, which is identical to mMYB33:GUS, but lacking the GUS reporter gene (see Methods). For comparison, Arabidopsis transformants were also generated with the wild-type MYB33 gene (the pPZP-MYB33 construct).
For the MYB33 gene, 82 primary transformants were obtained and transferred to soil. Of these, two plants had a spindly/dwarf stature, and four others appeared male sterile. The phenotype of the other 76 appeared completely wild-type. For the mMYB33 construct, 74 primary transformants were obtained and transferred to soil. Many of these 5-d-old seedlings appeared to have developmental defects, with the cotyledons failing to expand (Figure 7G), and in 10-d-old seedlings, many plants had first true leaves that had unusual morphology (Figure 7H). Of the 74 plants, 62 survived, with only four plants having a phenotype resembling wild-type plants. The 12 plants that died indicated that expression of this gene results in seedling arrest. The remainder had mutant phenotypes with common developmental characteristics, but to differing degrees (Figure 7M). All the 58 mutant plants had a reduced size, with 11 plants having a rosette of <1 cm in diameter, and 44 plants having a diameter of <2 cm. The smaller the rosette, the shorter the bolts (shorter internode lengths) and the more reduced the apical dominance, indicating there was a general negative effect on plant growth. This was also the case with fertility, with the most severe transformants failing to set seed, whereas the less severe transformants had fully extended siliques. In all mMYB33 plants, leaves were profoundly more rounded than the wild type (cf. Figure 7I with 7J) and upturned at the sides (Figures 7J to 7L), petiole lengths were dramatically reduced (Figure 7J), and many had short bolts with terminal flowers (Figure 7L, arrow). Again these phenotypic characteristics were more pronounced in plants with the most stunted phenotype. All these characteristics were different in the two spindly/dwarf plants obtained with the MYB33 gene. Thus, it appears expression of MYB33, in which the miR159 target sequence has been mutated, has dramatic consequences for plant growth and development.
DISCUSSION
MYB33 and MYB65 Redundantly Facilitate Anther Development
Using a reverse genetic approach, we have shown a role for MYB33 and MYB65 in anther development. The fact that neither single mutant displayed a phenotype, whereas the myb33 myb65 double mutant was male sterile, implies that MYB33 and MYB65 are functionally redundant, which was not surprising considering the high homology they share and the fact that these genes lie within segmental chromosomal duplications. MYB33 and MYB65 have been previously coined the GAMYB-like genes, for they have high homology to HvGAMYB and evidence suggested that they were mediating the GA signal in plants (Gocal et al., 2001). A role in anther development is consistent with transducing a GA signal, for GA is known to control anther development (Jacobsen and Olszewski, 1991; Goto and Pharis, 1999) and HvGAMYB expression in barley anther increases after GA3 application (Murray et al., 2003).
The premeiotic block in myb33 myb65 anther development results from tapetal hypertrophy. The tapetum is a critical tissue, mediating between the gametophyte and sporophyte, and this is highlighted across the plant kingdom in that tapetal malfunction is regarded as the prime cause of male sterility, from either environmental or genetic causes (Kaul, 1988). In Arabidopsis, four male sterile mutants in which the tapetum undergoes hypertrophy have been described; ms3 (Chaudhury et al., 1994), fat tapetum (Sanders et al., 1999), gne1 and gne4 (Sorensen et al., 2002). In each case, hypertrophy occurs at the onset of meiosis similar to myb33 myb65. However, unlike myb33 myb65, in all these mutants, middle layer hypertrophy also occurs, and this along with the enlarged tapetum appears to crush the microsporocytes, resulting in their degeneration. Thus, myb33 myb65 is different with respect that only one microsporangial cell layer is affected, the tapetum, with the size of the middle layer remaining the same as the wild type. Also, in these previous Arabidopsis mutants, the tapetum degenerates leaving an empty locule that eventually results in a collapsed anther wall. In myb33 myb65 plants, the tapetum shows no sign of degeneration.
Consistent with the phenotype of myb33 myb65, the expression of a MYB33:GUS transgene was confined in the floral tissues to anthers, predominantly in the tapetum at meiosis, the developmental stage at which hypertrophy begins. However, this expression is not essential for anther development, for in myb33 myb65, adjacent locules of the same anther can undergo different developmental programs; either the tapetum undergoes hypertrophy just before meiosis or the anther develops normally. This stage-specific developmental decision is greatly influenced by environmental factors, where under conditions of favorable growth (16°C [low temperature] or 300 μmol·m−2s−1 [high light]), fertility of myb33 myb65 can be restored to almost wild-type levels. When we consider our standard growth conditions of 90 to 100 μmol light intensity, this would be regarded as low light intensity, whereas 22°C for a temperate plant as Arabidopsis could be considered as moderately high. Thus, the conditions that restore fertility in myb33 myb65 would probably be closer to the conditions Arabidopsis encounters in its natural environment. In this respect, the expression of MYB33/MYB65 could be considered as limiting in the anther under poor environmental conditions. Curiously, both the conditions that restore fertility, high light and lower growth temperature, result in higher levels of soluble carbohydrates in the plant (Hurry et al., 1995), and the MYB33 gene is being expressed in the tapetum at the time when tapetal carbohydrate (starch) reserves are being mobilized, just before meiosis (Clement et al., 1994). Although it is tempting to speculate that these MYB genes may play a role similar to HvGAMYB in barley aleurone (starch mobilization), further investigation is needed to elucidate the conditional nature of MYB33/MYB65 in the anther and alternative possibilities, such as further redundancy with other GAMYB-like homologs.
Rice gamyb mutants are blocked at a similar stage in anther development as myb33 myb65, where just before meiosis, the tapetum undergoes hypertrophy (Kaneko et al., 2004). Furthermore, the extent of sterility is variable, where it has been noted that greater sterility occurs at higher temperatures (Kaneko et al., 2004). The major difference in gamyb plants appears to be that other floral organs are also affected by this mutation. In barley, overexpression of HvGAMYB in transgenic plants also leads to a male sterile phenotype; however, the pollen developed normally, but dehiscence fails to occur, with the septum failing to rupture (Murray et al., 2003). Lastly, similar to Arabidopsis, GAMYB in both rice and barley is expressed in anthers (Murray et al., 2003; Kaneko et al., 2004). So both the expression and function of GAMYB in anther development appears to be highly conserved between Arabidopsis and cereals.
Analysis of the Control of MYB33 Expression
The dramatically contrasting expression patterns we obtained with our different reporter gene constructs highlights the complexities underlying the regulation of MYB33. The presence of the miR159 binding site within the MYB33 gene can explain why expression in MYB33:GUS plants is repressed in certain tissues, but it cannot explain the lack of expression in anthers of the promoter:GUS lines. Our experiments have enabled us to propose a model for the tissue-specific control of MYB33 expression (Figure 8). First, the 5′ upsteam region (Pro33:GUS) directs expression in flowers (receptacle, anther filaments/connective, pistal, and sepals), shoot apices, and root tips. The addition of region A (5′-MYB33:GUS) or region C (Pro33:GUS:3′end) alone does not alter the expression pattern (Figure 8). However, when they are present along with region B (mMYB33:GUS), they are sufficient for anther expression. Thus, either region B contains specific sequences required for anther expression, or a combination of all regions or subset of regions is required for anther expression. The only sequence we can exclude from being required for anther development is the miR159 binding site, for in mMYB33:GUS and another construct in which the miR159 binding site had been deleted (A.A. Millar and F. Gubler, unpublished data), expression still occurs in anthers. The fact that expression in the myb65 enhancer trap lacks expression in anthers and contains all these corresponding regions suggests that these anther-required sequences are not simply cis-acting enhancers, but may have to be transcribed or translated to exert their regulatory effect. Recently, evidence was presented that the SUPERMAN gene of Arabidopsis has both positive and negative regulatory elements in its protein-coding region (Ito et al., 2003), and this may be the case for MYB33. A second possibility is that some higher-order structure is required, such as a specific chromatin configuration (Lomvardas and Thanos, 2002) and that having any significant disruption, such as a large deletion or insertion of a T-DNA (e.g., the MYB65 enhancer trap), interferes with the required configuration for anther expression.
Figure 8.
Model for the Control of MYB33 Expression.
The 5′ upstream region strongly promotes expression in flowers (receptacle, pistal, sepals, and anther filaments/connective), meristems, and root tips. Addition of region A or region C alone does not alter this expression pattern. Addition of these two regions together, along with region B, enables expression in anthers. The miR159 target sequence (miR) represses expression in meristems, root tips, and flowers but has no apparent effect on anthers. Region A, contains sequences from the ATG (+1) to +1024; region B, +1043 to the stop codon (+1832); region C, from the stop codon to the open reading frame of the gene downstream of MYB33.
MYB33 Is Regulated by miRNAs
By mutating the miRNA target sequence within the MYB33:GUS gene (mMYB33:GUS), we have demonstrated visually how the expression pattern of MYB33 is restricted by miRNAs. They downregulate or silence MYB33 expression in shoot apices, root tips, and flowers, but not anthers, the only tissue in which we see a phenotype in the myb33 myb65 mutant. The most likely mechanism of regulation is mRNA cleavage, for miRNA-guided cleavage products have been found both for MYB33 and MYB65 (Palatnik et al., 2003), which have been shown to be the result of miR159 expression (Achard et al., 2004). In situ analysis (Gocal et al., 2001) found MYB33 mRNA in tissues in which MYB33 is being repressed by miRNAs, such as root tips and shoot apices. Therefore, we would predict that the signal detected in these tissues would predominantly represent cleaved transcript for this technique is unable to distinguish between intact and cleaved mRNA. Thus, in this instance, the MYB33:GUS transgene may be a more accurate reflection of where MYB33 protein accumulates rather than MYB33 RNA in situ hybridization.
The strong expression in germinating mMYB33:GUS seedlings in and around the shoot meristem region and on the proximal periphery of young developing leaves provides a rationale for the phenotype exhibited by mMYB33 transgenic plants. miR159 must be refining or abolishing the expression of MYB33 in shoot apices, the tissues from which leaf primordia arise, and gene expression programs are determining leaf size and shape (Kessler and Sinha, 2004). MYB33 has been hypothesized to be an activator of LEAFY (Gocal et al., 2001), and interestingly, mMYB33 plants do have some characteristics of 35S::LEAFY plants, such as short bolts with terminal flowers (Weigel and Nilsson, 1995). Thus, expression of MYB33 in the incorrect cells at a crucial time early on in development may be triggering incorrect developmental pathways that culminate in the gross abnormalities observed in the mMYB33 plants. The seedling arrest in mMYB33 plants may provide the rationale for the absence of mutants in the MYB33 miRNA target motif, similar to those leaf developmental mutants recovered for the PHABULOSA family (Emery et al., 2003).
The growth retarding effect of the mMYB33 transgene appears contradictory to the proposed role in mediating a GA signal. Furthermore, the mMYB33 phenotype appears unrelated to myb33 myb65 plants or plants overexpressing miR159 under the control of the 35S promoter (Achard et al., 2004). Transgenic 35S:miR159a Arabidopsis was found to have delayed flowering in short-day photoperiods and to be male sterile (Achard et al., 2004). Presumably in wild-type plants, miR159 is not expressed or weaky expressed in anthers, thus allowing MYB33 expression in these tissues, and this scenario has been changed in the 35S:miR159a plants. However, 35S:miR159a anthers appeared to be blocked at a different stage of development compared with myb33 myb65, for 35S:miR159a anthers increased in size and they darkened in color. This raises the possibility that other members of the Arabidopsis GAMYB-like family are targeted by miR159a and they too play roles in anther development.
We designed the mutations in the miR159 target to match those of Palatnik et al. (2003), who also transformed mMYB33 into Arabidopsis, but under the 35S promoter. Our results confirm some of their findings, such as upwardly curling leaves (Palatnik et al., 2003). However, although a detailed description of the 35S:mMYB33 plants was not given, there does appear to be significant differences between mMYB33 and 35S:mMYB33 plants. For instance, the petiole lengths of mMYB33 leaves are dramatically reduced when compared with the petioles of 35S:mMYB33 plants (Palatnik et al., 2003), and such dramatic reductions in plant size were not reported for mMYB33 plants. Seedling arrest was not reported for the 35S:mMYB33 gene, and the frequency of aberration was much higher in mMYB33 transformants (58 from 62) than in 35S:mMYB33 transformants (39 from 63; Palatnik et al., 2003). All these data indicate that a much stronger phenotype was produced with the endogenous MYB33 promoter than the 35S promoter.
One striking observation from comparison of MYB33:GUS and mMYB33:GUS plants is the apparent complete abolishment of expression of MYB33 in certain tissues, most notably in the root tips and the shoot apical meristem. It raises the interesting conundrum of why the gene is expressed in these tissues, but ultimately this expression is silenced or downregulated by miRNAs. Furthermore, from the phenotype of the mMYB33 plants, if these genes were not regulated correctly, they would be a liability to the plant. The fact that RNA in situ analyses have found GAMYB expression not only in meristems of Arabidopsis (Gocal et al., 2001), but also in the meristems of cereals such as Lolium (Gocal et al., 1999) and rice (Kaneko et al., 2004), would argue that GAMYB may have a highly conserved function within the meristem. It remains to be seen whether these genes are not silenced by miRNAs in a small group of cells within the meristem. And, as hypothesized by Rhoades et al. (2002), if MYB33 is a long-lived transcript, one way to abolish its expression in rapidly dividing cells (to allow differentiation) is through miRNA regulation, thus providing a rationale for our observations. However, with the loss-of-function myb33 myb65 mutant, the only phenotypic alteration we have found is a conditional role in anther development. If MYB33/MYB65 are performing additional roles in Arabidopsis and these roles are masked by further redundancy that may exist within the Arabidopsis GAMYB-like gene family, the generation of additional combinations of mutants may be needed to uncover further phenotypes.
METHODS
Isolation of T-DNA Insertional Mutants
Three pools of T-DNA–tagged mutants were screened, the collection at the Arabidopsis Knockout Facility at the University of Wisconsin, which was done as a service, and the Jack and Feldmann collection, which were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The Madison collection was only screened with the recommended left border primer, JL202. Both the Jack and Feldmann collections were screened with the recommended left and right border primers. The following primers were used to screen the T-DNA–tagged libraries for insertional mutants in the MYB65 gene, 65-1 (5′-TACCTCAGCTAGGGTTCGTCTTTGTTGTA-3′) and 65-2 (5′-ACCGTTACTTTGCGAGAAGCAAGACCTAA-3′), or the MYB33 gene, 33-1 (5′-TGTCGTATTTGTCGTTTCTCGATC-3′) and 33-2 (5′-CTAGTCCATGACCATGAGAAGTGAGAACT-3′). After the identification of initial positive, individual plants were isolated through the procedures outlined at the Arabidopsis Knockout Facility at the University of Wisconsin Web site.
Plant Lines and Growth Conditions
The myb65 mutant described in this article was isolated from the Madison collection (Krysan et al., 1999) and thus was in the ecotype Ws. The myb33 mutant described in this article was isolated from the Jack collection; thus, it was in the ecotype Col, with a glaborous1 background mutation. All plants were grown in a mix of 50% compost and 50% gravel. When growing plants for seed, plants were grown in a 21°C temperature growth room under continuous fluorescent illumination that varied from 60 to 100 μmol·m−2s−1.
Mutant Genotyping
Amplification with the oligonucleotide 33-1 and the T-DNA–specific primer, JL-202, yielded a 2222-bp product in plants containing the myb33 allele. Amplification using the oligonucleotides 33-1 and 33-2 yielded a 3644-bp product in plants with a wild-type allele of MYB33 and no product in plants homozygous for the myb33 allele. Amplification with the oligonucleotide 65-1 and the T-DNA–specific primer, JL-202, yielded a 1084-bp product in plants containing the myb65 allele. Amplification using the oligonucleotides 65-1 and 65-2 yielded a 2887-bp product in plants with a wild-type allele of MYB65 and no product in plants homozygous for the myb65 allele. All PCR reactions were performed in an Eppendorf Mastercycler gradient PCR machine (Hamburg, Germany), using 35 cycles of 94°C for 15 s, 65°C for 30 s, and 72°C for 2 min. PCR reactions were performed using AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA) according to the specifications of the supplier.
When required, the genotype of lines was verified using DNA gel blot analysis. For the myb33 allele, a 1893-bp EcoRI genomic clone fragment isolated from Col that spanned the T-DNA insertion site in myb33 would only give one hybridizing band in the wild-type of 1893 bp; however, in the mutant, it would yield two bands, one at ∼2.4 kb and the other at 0.8 kb. For genotyping of the myb65 allele, the probe used was the PCR product derived from the 65-1 and 65-2 primers. For the wild-type gene, it would hybridize to a fragment of 2643 bp, and for the myb65 mutant allele, two bands would be present, at ∼3.0 and 1.0 kb.
Determination of Fertility under Varying Growth Conditions
Seed was germinated on synthetic media and grown for 5 d at 22 °C under continuous light. Twenty seedlings per 10-cm pot were then transplanted into soil and grown under at 22°C in a 12-h-day/12-h-night cycle at a light intensity of ∼90 μmol·m−2s−1. At the onset of bolting, plants were transferred into the various treatments (higher light intensities; 140, 210, or 330 μmol·m−2s−1) or lower growth temperature. The higher light intensities were performed in the same growth cabinet but were done by moving the plants closer to the lights. Because of the air circulation, there is no temperature gradient within the cabinet. For the lower growth temperature, treatment plants were moved to a different growth cabinet compared with control plants, and light intensities of these two cabinets were almost identical (86.9 μmol·m−2s−1 at 22°C and 85.5 μmol·m−2s−1 at 16°C). After a period of growth, fertility was assessed by scoring each silique with respect to whether it set any seeds. Those siliques with no seeds were scored as sterile and those with at least one seed scored as fertile. Fertility was then defined by dividing the number of fertile siliques (per plant) by the total number of siliques (per plant).
The Generation of Binary Vectors and Transgenic Plants
All the following binary vectors were transformed into the Agrobacterium tumefaciens strain GV3101 and used to transform Arabidopsis thaliana ecotype Col using the floral dip method (Clough and Bent, 1998). For complementation of the myb33 myb65 mutant, plants were grown under conditions that promoted male fertility (300 μmol·m−2s−1, 16 h day, 16°C). All PCR amplifications for the following vectors were performed using high-fidelity PlatiniumR Pfx DNA polymerase (Invitrogen, Carlsbad, CA).
For complementation, a 4492-bp XhoI genomic fragment from Landsberg erecta containing the entire MYB33 gene (Figure 5; Gocal et al., 2001) was subcloned into the SalI site of the binary vector pPZP200-hygro (which was pPZP200 [Hajdukiewicz et al., 1994] with a 35S-hygromycin resistance gene inserted into an end-filled EcoRI site of the vector), resulting in construct pPZP-MYB33. The primers 2G (5′-CTGAAGGCGGGAAACGACAATCTGATCCA-3′) and MYB33-2 g (5′-GCAGCTTATGAAGACAATCCTTTTGGT-3′) were used to confirm the presence of the pPZP-MYB33 transgene in transgenic plants.
For the pPro33:GUS construct, the 2051 bp immediately upstream of the ATG of MYB33 was PCR amplified from MYB33 genomic clone using the oligonucleotides 5′-TAAGGATCCTCTTTTCTAATTAAACCAC-3′ and 5′-ACTCGAGGTGGAGGCGAC-3′. This includes all the sequences to the next gene upstream of MYB33. The amplified DNA was then cloned into BamHI/XhoI-cleaved pBluescript II SK−, and its sequence was determined to be correct. The BamHI/XhoI fragment was then subcloned into BamHI/SalI-cleaved pBI 101.1, resulting in the construct pPro33:GUS.
For pPro65:GUS, the oligonucleotides 5′-TAAAGCTTGAGTCTCAAAACATAAGCCAAAAAGCCG-3′ and 5′-TAGTCGACTCTTTCTACTTAAAGCAAGACAGACTCC-3′ were used to amplify the 2145 bp immediately upstream of the ATG of the MYB65 from Arabidopsis genomic DNA, ecotype Col. The sequences of the primers were based on the Arabidopsis BAC sequence AC008153 (which contains the MYB65 gene). This PCR product was subcloned into HindIII/SalI-cleaved pBluescript II SK−, and its sequenced was determined to be correct. This HindIII/SalI fragment was then cloned into HindIII/SalI-cleaved pBI 101.1, resulting in the plasmid pPro65:GUS.
For the pPro33:GUS:3′end construct, the 3′ end of the MYB33 gene was amplified from the 4492-bp XhoI genomic clone using the primers 5′-ATGAGCTCCGAGCCCGGGAAATCCTCCACT-3′ and 5′-ATGAGCTCCTGCACGCTTTGAGATTTT-G-3′, which amplifies the 603 bp immediately downstream of the stop codon of the MYB33 gene (Figure 5). The PCR fragment was ligated into the SmaI site of pBluescript II SK+, and its sequence was determined to be correct. This fragment was then cleaved out with SacI and ligated into the SacI site of pPro33:GUS. A plasmid with insert in the correct orientation was determined with a SmaI digest and called pPro33:GUS:3′end.
For the pMYB33:GUS translational fusion, the GUS gene was amplified with the primers 5′-GACCATGGTATGTTACGTCCTGTAGAAACCCCA-3′ and 5′-GACCATGGTTCATTGTTTGCCTCCCTGCTGCGG-3′ from the vector pBI 101.1. The product was digested with NcoI and ligated into the NcoI site of the 4492-bp XhoI genomic MYB33 clone. A clone was identified with the GUS gene in the correct orientation, and its sequence was determined to be correct. The MYB33:GUS gene fusion was then cleaved out with XhoI, and the resulting 6.3-kb fragment was subcloned into the SalI site of the binary vector pPZP200-hygro (Hajdukiewicz et al., 1994), resulting in the plasmid pMYB33:GUS. Thus, GUS has been fused in frame to the coding region of MYB33, 55 amino acids from the end of the gene.
For the p5′-MYB33:GUS translational fusion, the 5′ end of the MYB33 gene, including all upstream sequences (2044 bp upstream of the start codon), exons and introns up to amino acid 309 in the coding region (1013 bp downstream of the start codon), 10 bp before the potential miRNA target, was amplified from the 4492 bp Xho1 genomic clone with the primers 5′-ATGTCGACGTGGAGGCGACGTGTTCGTCAGG-3′ and 5′-TAGGATCCATATAAGGGCTTAGAATAAGGAACA-3′. This resulted in a 3071 bp amplification product, which was cloned into the Sma I site of pBluescript II SK+. The sequence of the amplified fragment was determined to be correct, it was then cleaved out with Sal I and BamH1, and ligated into the Sal I/BamH1sites of pBI 101.1, resulting in the plasmid p5′-MYB33:GUS.
For the construct pMYB33ΔmiR, we first amplified the 1386 bp of the 3′ end of the MYB33 gene with the primers 5′-TAGGATCCGAAACAACATTTGACCAGTGGAAG-3′ and 5′-TAGAGCTCCTGCACGCTTTGAGATTTTGTTTTGAC-3′ and ligated the PCR fragment into the SmaI site of pBluescript II SK+. These sequences contain the last intron in the gene, the last 198 amino acids of the MYB33 coding region, and 3′ sequences all the way to the gene downstream of MYB33 (Figure 5). The fragment was sequenced and determined to be correct. The fragment was cleaved out with BamHI and SacI and subcloned into the BamHI/SacI sites of pPZP200-hygro, creating the vector pPZP200-3′end. The 5′ end fragment used in the 5′-MYB33:GUS was then subcloned into the SalI/BamHI sites of pPZP200-3′end, resulting in the construct pMYB33ΔmiR.
For the constructs pmMYB33 and pmMYB33-GUS, a 988-bp KpnI/NcoI fragment containing the miR159 binding site was subcloned into pBluescript II SK+. The primers 5′-GGCAGTGAAGCTCGAATTGCCAAGCTTTCAGTATTCAGAAACAAC-3′ and 5′-GTTGTTTCTGAATACTGAAAGCTTGGCAATTCGAGCTTCACTGCC-3′ were used to introduce 10 nucleotide substitutions in the miR159 binding site, using PCR and a QuikChange site directed mutagenesis kit (Stratagene, La Jolla, CA), and this was verified by sequencing. The 988-bp KpnI/NcoI fragment was then used to replace the corresponding sequences in pMYB33ΔmiR, by subcloning into the KpnI/NcoI sites, resulting in the construct pmMYB33. The GUS gene was then subcloned into the NcoI site of pmMYB33 to result in the plasmid pmMYB33-GUS.
All transgenic plants were confirmed to be carrying the correct transgene through PCR analysis and primers that were specific for each construct.
Histochemical Localization of GUS Activity
In situ GUS staining was performed using the method of Jefferson (1987). Tissues were transferred to microfuge tubes containing a solution of 100 mM Na phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide at 37°C overnight or else as otherwise stated. Stained tissues were cleared with an ethanol series.
Light Microscopy
Arabidopsis inflorescences were fixed in 3% glutaraldehyde in 25 mM phosphate buffer, pH 7.2, at room temperature for several days before being rinsed in 25 mM phosphate buffer and dehydrated through a graded ethanol series. The inflorescences were then embedded in LR White resin and sliced into 1- to 2-μm transverse sections. Anther transverse sections were stained in 1% toluidine blue O and viewed under a Leica DMR compound microscope (Wetzlar, Germany) and images taken with a Leica DC200 digital camera. For examination of callose, sections were stained with 0.05% (w/v) analine blue in 0.067 M phosphate buffer, pH 8.5, and viewed under UV illumination. All GUS-stained tissues were viewed under dark-field conditions. Bright-field photographs of individual flowers or GUS-stained tissues were taken using a Wild Heerbrugg dissecting microscope and a Wild Leitz camera (Wetzlar, Germany).
Acknowledgments
We thank M. Keys for technical assistance, R. King for many helpful comments and experimental suggestions, R. White and C. Miller for sectioning and microscopy of anthers, C. Davies for photography, and C. Andersson for the pPZP200-hygro binary vector. We also thank E. Dennis, B. Furbank, and R. Dolferus for critically reading the manuscript and J. Jacobsen, M. Robertson, and C. MacMillan for many useful comments. A.A.M. was funded by Graingene1, a joint venture between Australian Wheat Board Limited, the Commonwealth Scientific and Industrial Research Organization, and the Grains Research and Development Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Anthony A. Millar (tony.millar@csiro.au).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027920.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Cercós, M., Gómez-Cadenas, A., and Ho, T.-H.D. (1999). Hormonal regulation of a cysteine proteinase gene, EPB1, in barley aleurone layers: Cis- and trans-acting elements involved in the coordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 19, 107–118. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Lavithis, M., Taylor, P.E., Craig, S., Singh, M.B., Signer, E.R., Knox, R.B., and Dennis, E.S. (1994). Genetic control of male fertility in Arabidopsis thaliana: Structural analysis of premeiotic developmental mutants. Sex. Plant Reprod. 7, 17–28. [DOI] [PubMed] [Google Scholar]
- Chen, L., Nishizawa, T., Higashitani, A., Suge, H., Wakeui, H., Takeda, K., and Takahashi, H. (2001). A variety of wheat tolerant to deep-seeding conditions: Elongation of the first internode depends on the response to gibberellin and potassium. Plant Cell Environ. 24, 469–476. [Google Scholar]
- Chen, R., Silver, D.L., and de Bruijn, F.J. (1998). Nodule parenchyma-specific expression of the sesbania rostrata early nodulin gene SrEnod2 is mediated by its 3′ untranslated region. Plant Cell 10, 1585–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, C., Chavant, L., Burrus, M., and Audran, J.C. (1994). Anther starch variations in Lilium during pollen development. Sex. Plant Reprod. 7, 347–356. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Matínez, M., Moneda, I.I.-L., and Carbonero, P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29, 453–464. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Gocal, G.F., Poole, A.T., Gubler, F., Watts, R.J., Blundell, C., and King, R.W. (1999). Long-day up-regulation of a GAMYB gene during Lolium temulentum inflorescence formation. Plant Physiol. 119, 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal, G.F., Sheldon, C.C., Gubler, F., Moritz, T., Bagnall, D.J., MacMillan, C.P., Li, S.F., Parish, R.W., Dennis, E.S., Weigel, D., and King, R.W. (2001). GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 127, 1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Goto, N., and Pharis, R.P. (1999). Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant ga1-1 of Arabidopsis thaliana. Can. J. Bot. 77, 944–954. [Google Scholar]
- Gubler, F., and Jacobsen, J.V. (1992). Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell 4, 1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Raventos, D., Keys, M., Watts, R., Mundy, J., and Jacobsen, J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17, 1–9. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, S., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Han, M.-H., Goud, S., Song, L., and Fedoroff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurry, V.M., Strand, A., Tobiaeson, M., Gardestrom, P., and Oquist, G. (1995). Cold hardening of spring and winter wheat and rape results in differential effects on growth, carbon metabolism, and carbohydrate content. Plant Physiol. 109, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Sakai, H., and Meyerowitz, E.M. (2003). Whorl-specific expression of the SUPERMAN gene of Arabidopsis is mediated by cis elements in the transcribed region. Curr. Biol. 13, 1524–1530. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1991). Characterization of the arrest in anther development associated with gibberellin deficiency of the gib-1 mutant of tomato. Plant Physiol. 97, 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., Inukai, Y., Ueguchi-Tanaka, M., Itoh, H., Izawa, T., Kobayashi, Y., Hattori, T., Miyao, A., Hirochika, H., Ashikari, M., and Matsuoka, M. (2004). Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul, M.L.H. (1988). Male Sterility in Higher Plants. (Berlin: Springer Verlag).
- Kessler, S., and Sinha, N. (2004). Shaping up: The genetic control of leaf shape. Curr. Opin. Plant Biol. 7, 65–72. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan, M.B., Ho, T.-H.D., Rogers, S.W., and Rogers, J.C. (1992). A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell 4, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C., Oppenheimer, D.G., Pollock, S., and Marks, M.D. (1993). Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5, 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas, S., and Thanos, D. (2002). Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110, 261–271. [DOI] [PubMed] [Google Scholar]
- Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2003). A role for HvGAMYB in anther development. Plant J. 33, 481–491. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.-C., Lee, P.Y., Truong, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- Sorensen, A., Guerineau, F., Canales-Holzeis, C., Dickinson, H.G., and Scott, R.J. (2002). A novel extinction screen in Arabidopsis thaliana identifies mutant plants defective in early microsporangial development. Plant J. 29, 581–594. [DOI] [PubMed] [Google Scholar]
- Skriver, K., Olsen, F.L., Rogers, J.C., and Mundy, J. (1991). cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 88, 7266–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500. [DOI] [PubMed] [Google Scholar]
- Yang, S., Sweetman, J.P., Amirsadeghi, S., Barghchi, M., Huttly, A.K., Chung, W., and Twell, D. (2001). Novel anther-specific myb genes from tobacco as putative regulators of phenylalanine ammonia-lyase expression. Plant Physiol. 126, 1738–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]