Abstract
We have characterized a rice (Oryza sativa) dwarf mutant, dwarf11 (d11), that bears seeds of reduced length. To understand the mechanism by which seed length is regulated, the D11 gene was isolated by a map-based cloning method. The gene was found to encode a novel cytochrome P450 (CYP724B1), which showed homology to enzymes involved in brassinosteroid (BR) biosynthesis. The dwarf phenotype of d11 mutants was restored by the application of the brassinolide (BL). Compared with wild-type plants, the aberrant D11 mRNA accumulated at higher levels in d11 mutants and was dramatically reduced by treatment with BL, implying that the gene is feedback-regulated by BL. Precise determination of the defective step(s) in BR synthesis in d11 mutants proved intractable because of tissue specificity and the complex control of BR accumulation in plants. However, 6-deoxotyphasterol (6-DeoxoTY) and typhasterol (TY), but not any upstream intermediates before these compounds, effectively restored BR response in d11 mutants in a lamina joint bending assay. Multiple lines of evidence together suggest that the D11/CYP724B1 gene plays a role in BR synthesis and may be involved in the supply of 6-DeoxoTY and TY in the BR biosynthesis network in rice.
INTRODUCTION
Dwarf plant mutants are very useful materials for elucidating the regulatory mechanisms of plant growth and development. Dwarfism in plants is also widely known to be an attractive characteristic in crop breeding. Dwarf mutants of many plant species have been isolated and characterized; in particular, they have been extensively analyzed for their modes of inheritance and their responses to plant hormones, such as gibberellins (GAs) (Ashikari et al., 1999; Ueguchi-Tanaka et al., 2000) and brassinosteroids (BRs) (Clouse and Sasse, 1998). A variety of causative agents for dwarfism in plants have been identified, GAs being the most investigated of these agents in rice (Oryza sativa) plants. Both of the semidwarf cultivars of rice and wheat (Triticum aestivum) varieties used in the green revolution were generated by changes in the production of GAs or responses to GAs, with rice being altered in GA metabolism (Sasaki et al., 2002; Spielmeyer et al., 2002) and wheat becoming GA insensitive (Peng et al., 1999). Recent molecular genetic investigations have revealed that BRs are also important agents for determining plant height (Fujioka and Yokota, 2003). In Arabidopsis thaliana, for example, several mutants are BR related and have a distinctive dwarf phenotype with dark green, rugose leaves (Szekeres et al., 1996; Azpiroz et al., 1998; Choe et al., 1999a, 1999b, 2000; Jang et al., 2000). When grown in the dark, these mutants show a deetiolated phenotype with reduced hypocotyl elongation (Chory et al., 1991; Chory, 1993). Through the characterization of these Arabidopsis mutants, BRs were shown to play important roles in morphogenesis during normal growth and also during development in light and darkness (Clouse and Sasse, 1998). Yamamuro et al. recently characterized a rice dwarf mutant, dwarf61 (d61), which shows pleiotropic phenotypes, including dwarfism and erection of leaves (Yamamuro et al., 2000). They showed that d61 was caused by loss of function of the O. sativa BR-insensitive 1 (OsBRI1) gene, which encodes a putative protein kinase very similar to the Arabidopsis BRI1 protein, a putative BR receptor (Li and Chory, 1997). Yamamuro et al. were the first to report that BRs seem to contribute to stem elongation in monocot plants (Yamamuro et al., 2000). Detailed studies of the BR-insensitive mutant, d61, should be helpful in understanding the functional roles of BRs in Gramineous plants. Phenotypic analysis of the mutant has shown that BRs seem to be important for elongation of the second internode, bending of the lamina joint, and skotomorphogenesis. Studies of other BR-deficient mutants, brd1 (Hong et al., 2002) and d2 (Hong et al., 2003), have confirmed that the phenotypic characteristics observed in d61 are not specific to BR-insensitive mutants but are common among both BR-insensitive and BR-deficient mutants.
So far, >60 rice dwarf mutants have been identified, some of which bear small round seeds (Iwata et al., 1984). To understand the mechanisms of production of seeds in a given form, we sought to identify the mutated gene of the rice dwarf mutant, dwarf11 (d11), which bears small round grains. In this article, we report that the D11 gene encodes a novel cytochrome P450 implicated in BR biosynthesis. On the basis of feeding experiments, we suggest that the D11 protein participates in the supply of 6-deoxotyphasterol (6-DeoxoTY) and typhasterol (TY) in the pathway of BR biosynthesis. The importance of BRs in the formation of normal seeds in rice plants is discussed in the light of our new information.
RESULTS
Characterization of Rice d11 Mutants
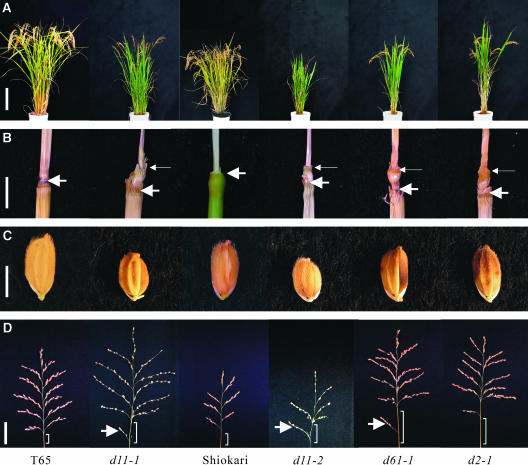
The gross morphologies of d11 mutants (d11-1 and d11-2 alleles), a BR-insensitive mutant (d61-1) (Yamamuro et al., 2000), and a BR-deficient mutant (d2-1) (Hong et al., 2003) are compared with those of Taichung 65 (T65) and Shiokari as wild-type plants are shown in Figure 1. The characteristic phenotypes of d11 mutants were the erection of leaves in mature stages (Figure 1A), the shortening of the second internode in culm, and the grain length (Figures 1B and 1C, Table 1). In particular, the second internode of d11 mutants was too short to be precisely measured. The d11 mutants also had a longer basal rachis internode than did the wild-type plants (Figure 1D, Table 1). Shortening of the second internode and overelongation of the basal rachis internode were also observed with d61-1 and d2-1 mutants.
Figure 1.
Pleiotropic Abnormalities Displayed by d11 Mutants.
(A) Gross morphology. From left to right: T65, d11-1, Shiokari, d11-2, d61-1, and d2-1 plants grown in the fields at heading stages. T65 and Shiokari, the recurrent parents for d11-1 and d11-2, respectively, were used as wild-type plants. Bar = 20 cm.
(B) Elongation patterns of the second internode. See (A) for rice plants. The second internode was extremely shortened in d11, d61-1, and d2-1 mutants. Thin and thick arrows indicate the upper and lower ends of the second internode, respectively. The second internodes of the wild-type plants were too long (see Table 1 for the length) to show their whole figures in this photograph. Bar = 1 cm.
(C) Grain morphology. See (A) for rice plants. d11 mutants have shortened grains. Bar = 0.5 cm.
(D) Panicle morphology. See (A) for rice plants. Brackets indicate the basal rachis internodes. Arrows show degenerated primary branches. The lengths of the basal rachis internodes of d11, d61-1, and d2-1 mutants are longer than those of wild-type plants. Bar = 5 cm.
Table 1.
Morphological Measurements of Wild-Type (T65 and Shiokari) and Mutant (d11-1, d11-2, d61-1, and d2-1) Rice Plants
| Internodes | T65 | d11-1 | Shiokari | d11-2 | d61-1 | d2-1 |
|---|---|---|---|---|---|---|
| Basal rachis internode | 23.1 ± 3.5 | 48.4 ± 3.1 | 30.2 ± 2.2 | 52.6 ± 2.5 | 73.7 ± 2.0 | 58.0 ± 1.7 |
| First internode | 375.2 ± 9.0 | 224.0 ± 2.4 | 303.3 ± 3.7 | 182.4 ± 1.9 | 349.2 ± 2.1 | 354.8 ± 2.1 |
| Second internode | 214.2 ± 8.8 | 5.0 ± 0.6 | 196.9 ± 1.9 | 4.6 ± 0.7 | 5.7 ± 1.0 | 5.2 ± 0.9 |
| Third internode | 99.3 ± 4.4 | 131.6 ± 2.3 | 74.7 ± 3.0 | 102.4 ± 2.7 | 58.2 ± 1.4 | 76.8 ± 1.4 |
| Fourth internode | 50.1 ± 4.8 | 57.4 ± 1.7 | 35.5 ± 2.7 | 25.2 ± 1.9 | 8.4 ± 2.0 | 38.8 ± 1.7 |
| Total | 761.9 ± 22.2 | 466.4 ± 4.6 | 640.6 ± 6.5 | 367.2 ± 4.0 | 495.2 ± 5.3 | 533.6 ± 5.1 |
The length is shown in millimeters. Data are averages of 10 plants (± sd). T65 and Shiokari were recurrent parents for d11-1 and d11-2, respectively.
Photomorphogenetic Phenotypes of the d11 Mutant in the Dark
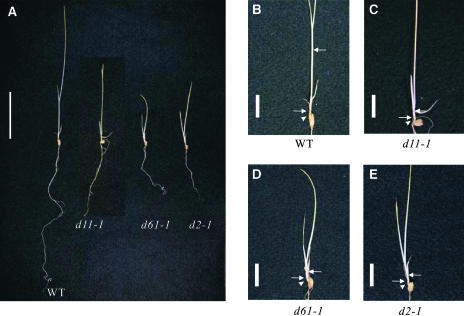
The abnormal phenotypes of the d11 mutants described above are similar to those of BR-deficient d2-1 and BR-insensitive d61-1 mutants; consequently, we suspected that d11 is deficient in BR biosynthesis or sensitivity. When wild-type (T65), d11-1, d61-1, and d2-1 plants were grown in the dark, the internodes (the parts between upper and lower nodes indicated by arrows in Figure 2B) of the wild-type (T65) plants elongated, whereas those of d11-1, d61-1, and d2-1 plants did not (Figures 2C to 2E). The mesocotyls of the wild-type (T65), d11-1, d61-1, and d2-1 plants (the parts indicated by arrowheads) were 3.0, 1.0, 0.9, and 0.8 mm long, respectively (Figures 2B to 2E). The absence of mesocotyl and internode elongation in the dark also suggests the possibility that d11 is a BR-related mutant.
Figure 2.
Aberrant Skotomorphogenesis of d11 Mutants.
Plants were germinated and grown in darkness.
(A) From left to right: wild-type, d11-1, d61-1, and d2-1 plants. T65, the recurrent parent for d11-1, d61-1, and d2-1, was used as wild-type plants. Bar = 5 cm.
(B) to (E) The node positions of the wild-type, d11-1, d61-1, and d2-1 plants are scaled up in (B) to (E), respectively, and indicated by arrows. The positions of the mesocotyls in these plants are indicated by arrowheads. Bar = 1 cm.
Restoration of the Dwarf Phenotype of d11 by BL Treatment
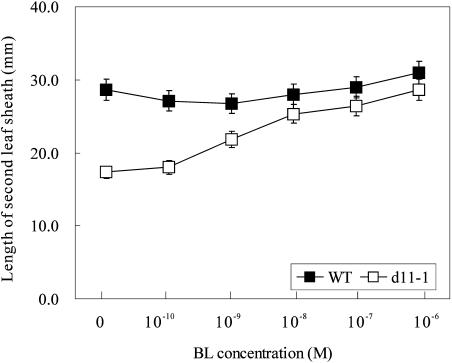
The phenotypic analyses of d11 suggested that d11 is a BR-deficient or BR-insensitive mutant. To determine whether d11 is BR deficient or BR insensitive, we treated d11 mutants with the most bioactive BR compound, brassinolide (BL). Although castasterone (CS), and not BL, seems to be the bioactive BR in rice, we chose BL for the biochemical complementation experiment because it was more active than CS in a rice lamina inclination bioassay (Fujioka et al., 1998). When d11 mutants were treated with 10−10 M to10−6 M BL, the length of the second leaf sheath of d11 mutants was elongated and was almost the same as that of wild-type plants (Figure 3). This result indicates that d11 can respond to exogenous BL to recover the dwarf phenotype. Therefore, d11 is unlikely to be a BR-insensitive mutant but is probably deficient in active BRs.
Figure 3.
Elongation of the Second Leaf Sheath in Wild-Type Plants and d11 Mutants after Treatment with BL.
The length of the second leaf sheath was measured 2 weeks after wild-type (closed squares) and d11-1 (open squares) seeds were germinated on half-strength MS medium containing various concentrations of BL. T65, the recurrent parent for d11-1, was used as wild-type plants. n = 30. Error bars indicate standard deviations.
The D11 Gene Encodes a Novel Cytochrome P450
Linkage mapping places the d11 locus on the long arm of chromosome 4 (Yoshimura et al., 1997). Identification of the D11 gene was performed by map-based cloning using F2 plants from a cross between d11, a japonica mutant, and SL14. The SL14 plant basically contains the japonica genome except for the long arm of chromosome 4, where the indica genome from Kasalath is present. The F2 plants segregated into two groups in a 3:1 ratio; one group showing the normal culm length of the japonica cultivar and the other the dwarf phenotype of d11. Three thousand and twenty F2 seeds were used for positional mapping of the d11 locus. The site of the d11 mutation was narrowed down to the region between two restriction fragment length polymorphism makers, G7008 and L353 (Figure 4A). To isolate the genomic clone, we performed PCR screening of a PAC library using two sequence tagged site primers (Baba et al., 2000). A PAC contig was constructed by PCR screening. Two PAC clones, P424E6 and P471B10, contained the entire coding region and the 5′ and 3′ flanking regions of the D11 gene. To identify the mutation sites of the d11 alleles, we sequenced DNA fragments corresponding to the open reading frames predicted by the GENSCAN program between markers T and K (Figure 4); two PAC clones, P424E6 and P471B10, were sequenced.
Figure 4.
Physical Map of the d11 Locus and Mutation Sites in d11 Alleles.
(A) High-resolution linkage and physical map of the d11 locus. Vertical lines represent the positions of molecular markers (C559, G7008, W390, and L353), and the numbers of recombinants are indicated above the linkage map. Genetic distances (centimorgan [cM]) between adjacent markers are shown in parentheses. The physical map of the d11 locus was constructed using three PAC clones, and the candidate region of the d11 mutation was present between markers T and K.
(B) Genomic structure of the D11 gene and positions of mutations in d11 alleles. Black boxes indicate exons. The D11 gene consists of nine exons and eight introns. The mutated DNA sequences of d11 alleles were shown in the bottom. d11-1 has a one-base deletion in the second exon, and d11-2 has a one-base insertion in the seventh exon. d11-3 has a one-base substitution in the fourth exon, and d11-4 has a one-base substitution of guanine for thymine at the last position of intron 3. A mutated mRNA with a five-base deletion in exon 4 (GCCAG) is produced in d11-4. The DNA sequences shown in the lowest part of the figure indicate those of mutated exons (d11-1, d11-2, and d11-3) and of the splicing site containing the mutated intron (d11-4).
Two markers, T and K, were designed with single nucleotide polymorphisms. The distance between the markers was 98 kb (Figure 4A). Nineteen open reading frames were identified using the GENSCAN program in the 98-kb segment of wild-type plants. We sequenced these open reading frames in d11-1, d11-2, d11-3, and d11-4; this sequence suggested that the candidate region of d11 mutants was likely to be a cytochrome P450 gene. A full-length cDNA clone encompassing the entire putative coding sequence of the D11 gene was isolated by RT-PCR using total RNA isolated from young seedlings. The cDNA clone contained a large open reading frame encoding 480 amino acid residues. Sequence analysis of cDNA for the cytochrome P450 in four d11 alleles was performed using the RT-PCR method; the mutated sequences in each allele were identified (Figure 4B). The allele d11-1 had a one-base deletion in exon 2, d11-2 had a one-base insertion in exon 7, d11-3 had a one-base substitution in exon 4, and d11-4 had a one-base substitution in the last base of intron 3, a base necessary for splicing. These mutations generated either a premature stop codon in exon 2 (d11-1), exon 4 (d11-4), or exon 7 (d11-2) or an amino acid substitution (Thr to Ile) in exon 4 (d11-3) (Figure 4B). The mutation sites of two d11-1 genotypes, HO556 and T65d11, and of two d11-2 genotypes, N58 and ID11, were identical. The results of our analyses of d11 alleles strongly support our suggestion that the D11 gene encodes a member of the cytochrome P450 superfamily.
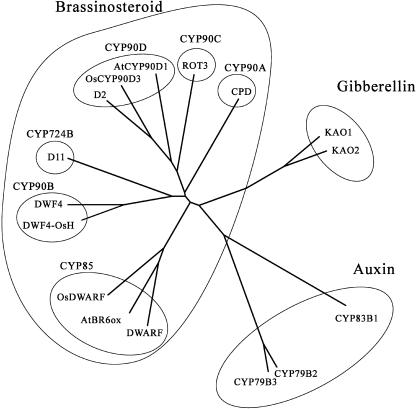
BLAST searches revealed that the deduced amino acid sequence of the D11 protein showed considerable similarity to those of previously reported proteins: 43% of identity and 62% similarity for DWF4/CYP90B protein from Arabidopsis (Choe et al., 1998); 42% of identity and 64% similarity for DWF4-OsH protein from rice (registered in GenBank) over almost the entire sequence (Figure 5). The sequence of the D11 protein also showed similarity to those of known BR-biosynthetic enzymes: 39% of identity for CPD/CYP90A1 protein (Szekeres et al., 1996); 36% of identity for D2/CYP90D2 protein (Hong et al., 2003); 33% of identity for OsDWARF/CYP85A4 protein (Hong et al., 2002; Mori et al., 2002) (Figure 6). This similarity raises the possibility that the D11 protein may be involved in BR biosynthesis. Positional cloning of the d11 locus revealed that the D11 gene encoded a putative cytochrome P450 protein named CYP724B1 (Nelson et al., 1996).
Figure 5.
Structure of the D11 Protein.
Comparisons of amino acid sequences among D11, DWF4-OsH, and DWF4. In accordance with the nomenclature of the P450 superfamily (Nelson et al., 1996), the D11 protein was named CYP724B1. Because the amino acid sequence of the D11 protein showed similarity to the sequences of BR-biosynthetic P450 proteins belonging to the CYP90B subfamily, the sequences of D11 (this study), DWF4-OsH, and DWF4 (Choe et al., 1998) were compared. Multiple sequence alignment was performed using the ClustalW analysis tool in the DNA Data Bank of Japan. Dashes (-) indicate gaps introduced to maximize alignment. Identical amino acid residues are represented by white-on-black letters. The triangles (d11-1), open circles (d11-4), closed circles (d11-3), and closed squares (d11-2) indicate the positions of deletion, splicing, amino acid substitution, and insertion mutations, respectively. Various domain regions, such as the anchor region, the Pro-rich region, Domain A, Domain B, Domain C, and the heme binding domain, are labeled (Kalb and Loper, 1988).
Figure 6.
Phylogenic Relationship in the Plant P450 Superfamily.
BR biosynthesis: D11 (this study), DWARF (Bishop et al., 1996), AtBR6ox (Shimada et al., 2001), OsDWARF (Hong et al., 2002), CPD (Szekeres et al., 1996), AtCYP90D1 (Hong et al., 2003), OsCYP90D3 (Hong et al., 2003), D2 (Hong et al., 2003), DWF4 (Choe et al., 1998), and DWF4-OsH (registered in GenBank). GA biosynthesis: AtKAO1 and AtKAO2 (Helliwell et al., 2001). Auxin biosynthesis: CYP79B2 (Hull et al., 2000), CYP79B3 (Hull et al., 2000), and CYP83B1 (Bak and Feyereisen, 2001). Other: ROT3 (Kim et al., 1998). The structural relationship was calculated by ClustalW and illustrated by Treeview.
Complementation Test
To confirm that the D11 gene is the mutated gene in d11, we performed a complementation experiment using d11-1. A DNA fragment ∼6.2 kb in size, including the entire sequence of the putative D11 gene, was introduced into d11-1 using the Agrobacterium tumefaciens–mediated transformation method (Hiei et al., 1994). Transformants with the D11 gene vector were identified by their resistance to the transformation selection marker, hygromycin; the transformants showed wild type–like phenotypes in the length of seeds and internodes and the whole form of leaves and ears (Figure 7, Transformant-D11). Transformants with a control vector that contained no insert had no apparent effect on the dwarf phenotype (Figure 7, Transformant-control vector). In a DNA gel blot analysis, we detected the cosegregation of T-DNA with the introduced D11 gene in the rice genome (data not shown). These results confirm that d11 was caused by a loss-of-function mutation of the D11 gene, a new member of the cytochrome P450 gene family.
Figure 7.
Phenotypic Complementation by Introduction of the D11 Gene.
The entire D11 gene, excised at the restriction sites XbaI and EcoRI (Figure 4B), was subcloned into a binary vector, pPZP2H-lac, to generate pPZP2H-lac-D11. The pPZP2H-lac and pPZP2H-lac-D11 were introduced into the Agrobacterium, EHA101. Transformants were generated from d11-1 using the Agrobacterium-mediated transformation method. Transformant-D11 and transformant-control vector were obtained by the infections of Agrobacterium containing pPZP2H-lac-D11 and pPZP2H-lac as the control vector, respectively. T65 was the recurrent parent for d11-1 and used as the wild-type plant.
(A) Gross morphology. Left, the wild type (T65); center, transformant-D11; right, transformant-control vector. Arrows and arrowheads indicate mature leaves and ears, respectively. Bar = 20 cm.
(B) Seeds. See (A) for rice plants. Seed form of transformant-D11 was the same as that of the wild-type plant. Bar = 0.5 cm.
Expression Analysis of the D11 Gene
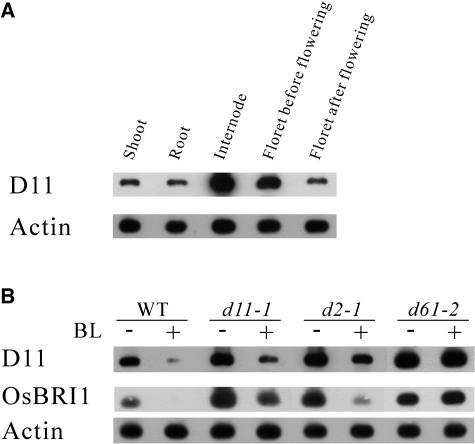
We examined the expression pattern of the D11 gene in various organs of rice plants. RNA gel blot analysis, using the entire length of the cDNA fragment as a probe, resulted in no detection of any bands, indicating that the level of the D11 gene expression was very low in the organs we tested (data not shown). Next, we performed a semiquantitative RT-PCR analysis to estimate the level of the D11 transcript. The highest concentrations of D11 mRNA were found in the internodes and the florets before flowering (Figure 8A). There was also expression of D11 in the shoots, roots, and florets after flowering. We suggest that the preferential expression of the D11 gene in the shoots, internodes, and florets before flowering of wild-type plants is correlated with the abnormal phenotypes of the shortened leaf sheath, stem, and grain in d11 plants.
Figure 8.
Comparison of D11 Gene Expression in Various Organs and the Negative Feedback Effect of BL on Gene Expression.
T65, the recurrent parent for d11-1, d2-1, and d61-2, was used as wild-type plants.
(A) Organ-specific expression of the D11 gene in the wild-type plants. Total RNAs were isolated from shoots, roots, internodes, and florets before flowering and florets after flowering, and RT-PCR was conducted to amplify a fragment of the D11 cDNA with specific primers. The expression of the Actin gene was used as a control.
(B) Negative feedback regulation of the D11 and OsBRI1 genes by BL. Total RNA was prepared from 2-week-old wild-type, d11-1, d2-1, and d61-2 seedlings with (+) or without (−) exogenous supplement of 10−6 M BL. RT-PCR was conducted as described in (A). The expression of the Actin gene was used as a control.
We also studied the effect of BL on the expression of D11 and OsBRI1 genes. When d11-1, d2-1, and d61-2 mutants and the wild-type (T65) plants were grown on half-strength MS medium without BL, the expression levels of the D11 and OsBRI1 genes were higher in d11-1, d2-1, and d61-2 mutants than in the wild-type (T65) plants (Figure 8B, lanes indicated as −). The expression of the D11 and OsBRI1 genes in d11-1 and d2-1 mutants, as well as that in the wild-type plants, was largely reduced by treatment with BL, whereas the expression was not affected by BL treatment in the d61-2 mutant, a BR-insensitive mutant (Figure 8B, lanes indicated as +).
Quantification of Endogenous BR Intermediates in d11 Mutants
To investigate the possibility that d11 has a defect in BR biosynthesis, we measured the amounts of BRs in d11-1, d11-2, and their respective recurrent parents by gas chromatography–mass spectrometry analysis. First, we analyzed the amounts of BR intermediates in the shoots of two d11 alleles and their respective recurrent parents (wild-type plants) (Figure 9A). No cathasterone (CT), 3-dehydroteasterone (3DT), or BL was detected in either the d11 mutants or the wild-type plants. The levels of 24-methylenecholesterol, campestanol (CN), 6-oxocampestanol (6-OxoCN), 6-deoxocathasterone (6-DeoxoCT), 6-DeoxoTY, TY, 6-deoxocastasterone (6-DeoxoCS), and CS in the mutants were lower than those in the wild-type plants (Figure 9A). The levels of 6-deoxoteasterone (6-DeoxoTE) in both mutants were slightly higher than those in wild-type plants, but those of teasterone (TE) were the same in d11-1 and reduced in d11-2 compared with the recurrent parents. The level of 3-dehydro-6-deoxoteasterone (6-Deoxo3DT) was somewhat higher in d11-2, but not in d11-1, than that of wild-type plants. One possible explanation for the observed differences between the mutants and wild-type plants in the levels of 6-DeoxoTE, TE, and 6-Deoxo3DT, as well as in those of the downstream intermediates after 6-Deoxo3DT, may be that the D11 protein participates in the supply of 6-DeoxoTY and TY in the BR biosynthesis network in rice.
Figure 9.
Quantitative Analysis of Endogenous BR Intermediates in Wild-Type and d11 Plants.
BR intermediates were isolated from shoots (A) and florets before flowering (B) as described in Methods. Sterol and BR levels (ng/g fresh weight) of two d11 alleles and their respective recurrent parents are shown below each product. The levels in d11-1 and its recurrent parent, T65, are indicated right and left, respectively, in the top parts marked with open circles. The levels in d11-2 and its recurrent parent, Shiokari, are indicated right and left, respectively, in the bottom parts marked with closed squares. The biosynthetic steps, in which D11 protein may participate, are indicated as “possible defect step 1” in (A) and “possible defect step 2” in (B). The steps catalyzed by D2 and OsDWARF (BRD1) proteins are shown as D2 and OsDWARF, respectively. nd, not detected; na, not analyzed.
Because D11 mRNA is strongly expressed in florets before flowering, we suspected there would also be marked differences in the levels of BR intermediates in the florets of mutant and wild-type plants. We tested this supposition by assaying the levels of BR intermediates in the florets before flowering of the two d11 alleles and their recurrent parents (Figure 9B). The levels of 24-methylenecholesterol, campesterol, CN, and 6-OxoCN in d11-1 and d11-2 mutants were slightly higher than those in the wild-type plants (Figure 9B). The results suggest that the D11 protein may be concerned with the conversion steps from CN to 6-DeoxoCT and from 6-OxoCN to CT (shown as the possible defect step 2 in Figure 9B).
Our analyses of the levels of BR intermediates in shoot and flowers before flowering both provided evidence for a defect in BR biosynthesis. However, the two analyses suggest different potential defects. We are therefore forced to conclude that this analytic approach has not provided an unambiguous identification of the defective step(s) in BR biosynthesis in d11 mutants.
The d11 Mutation Lies Downstream in the BR-Biosynthetic Pathway
As described previously, the addition of exogenous BL promoted the elongation of the leaf sheath of d11 mutants, and the amino acid sequence of the D11 protein was similar to the sequences of BR-biosynthetic enzymes such as CPD, DWF4, and DWARF. Because the quantitative analysis was unable to identify the reaction step(s) in which the D11 protein is involved, we undertook an alternative approach of using a series of feeding experiments with BR intermediates in an attempt to determine the reaction step(s) catalyzed by the D11 protein in the BR-biosynthetic pathway. In these experiments, we used the lamina joint test (i.e., the degree of bending between the rice leaf sheath and blade), which is highly sensitive to bioactive BRs exogenously applied. We applied BR intermediates to two d11 mutants (d11-1 and d11-2) and their respective recurrent parents (T65 and Shiokari) according to the method described previously (Fujioka et al., 1998). All the intermediates increased lamina joint bending in wild-type plants (Figures 10A to 10V, closed squares). By contrast, only some of the compounds were effective in the d11 mutants: 6-DeoxoTY, TY, and 6-DeoxoCS, and the active BRs, CS and BL, increased lamina joint bending (Figures 10D, 10E, 10I, 10K, 10O, 10P, and 10T to 10V); however, 6-DeoxoCT, CT, 6-DeoxoTE, TE, 6-Deoxo3DT, and 3DT induced no or little positive response (Figures 10A to 10C, 10F to 10H, 10L to 10N, and 10Q to 10S). The results suggest that D11 protein participates in the supply of 6-DeoxoTY in the late C-6 oxidation pathway and TY in the early C-6 oxidation pathway. The d11 mutants were slightly more responsive to BL in the test compared with wild-type plants (Figures 10K and 10V, respectively).
Figure 10.
Effects of BR Intermediates on the Degree of Inclination of the Leaf Lamina in Wild-Type and d11 Plants.
(A) to (K) T65 and d11-1.
(L) to (V) Shiokari and d11-2.
The effect of BR intermediates (ng/plant) on the degree of inclination of the leaf lamina in wild-type plants (closed squares) and d11 mutants (open squares). Data presented are means from 10 plants. Error bars indicate standard deviations.
DISCUSSION
This article reports the molecular characterization of a rice dwarf mutant, d11, first described in 1984 (Iwata et al., 1984). The mutant has several phenotypic characteristics, including the erection of leaves in mature stages, the inability to elongate the second internode, and the fruition of small round seeds. Seed lengths are shorter in d11 than in wild-type plants, although widths are the same. Dwarf mutants with a phenotypic effect on seed size are rare in rice. Because we are very interested in the molecular mechanisms of both dwarfism and the control of seed size and shape, we were particularly keen to isolate the mutated gene in d11.
Arabidopsis mutants defective in BR biosynthesis or signal transduction have shortened hypocotyls and open cotyledons even when grown in the dark (Chory et al., 1991; Szekeres et al., 1996; Li and Chory, 1997). They also develop primary leaves in the dark, although wild-type plants never develop the leaves under the same conditions. Arabidopsis wild-type plants develop long hypocotyls and small, closed cotyledons when grown in the dark. In rice, the BR-insensitive mutant, d61-1, and the BR-deficient mutants, brd1 and d2-1, show photomorphogenic phenotypes: in the dark, their mesocotyls and internodes do not elongate, unlike those of wild-type plants (Figure 2). In similar fashion to these rice mutants, d11 failed to elongate mesocotyls and internodes in the dark. When exogenous BL was applied to d11, the second leaf sheath was elongated and the length was almost the same as that of the wild-type plants, suggesting that d11 may be a BR-related mutant (Figure 3).
The D11 Gene Encodes a Novel Cytochrome P450
The D11 gene was identified by map-based cloning and found to encode a novel cytochrome P450. We studied four alleles of d11, d11-1, d11-2, d11-3, and d11-4 (Figure 4), and, using a complementation test, confirmed that the mutation was caused by a single gene, D11 (Figure 7). Cytochrome P450 proteins are well known to be heme binding enzymes, with mono-oxygenase activity such as oxidation, hydroxylation, isomerization, and dehydration of various kinds of compounds (Nelson et al., 1996). The phylogenic tree of rice and Arabidopsis cytochrome P450 proteins shows that the D11/CYP724B1 protein appears to be included in the CYP90B group, although the intervening relationship between rice and Arabidopsis proteins in the CYP90B group is not very close. There are four CYP90 groups, CPD/CYP90A (Szekeres et al., 1996), DWF4/CYP90B (Choe et al., 1998), ROT3/CYP90C (Kim et al., 1998), and D2/CYP90D (Hong et al., 2003); three of them have been identified as BR-biosynthetic enzymes, which catalyze the hydroxylation of C-23 (CPD/CYP90A1) and C-22 (DWF4/CYP90B1) and the oxidation of C-3 (D2/CYP90D2) in BRs (Figure 6). The function of ROT3/CYP90C1 is unknown. Another cytochrome P450 group, the CYP85 group (DWARF in Figure 6), catalyzes the oxidation of C-6 in BRs. Our study has shown that the D11 gene encodes a novel cytochrome P450 that seems to be involved in BR biosynthesis. To the best of our knowledge, no homolog of the D11 gene has been reported in other plant species.
Feedback Regulation of BR-Biosynthetic Genes by BL
It has been shown that the feedback regulation of BR-biosynthetic genes is regulated by the BR-signaling pathway. In plants, a complex of BRI1 and BRI1-associated receptor kinase 1 triggers BRI1 autophosphorylation after reception of exogenous BL (Li et al., 2002; Nam and Li, 2002). The signal is transmitted by molecules such as the protein kinase, BR-insensitive 2 (BIN2) (Li and Nam, 2002), and the transcription factors, brassinazole resistant 1 (BZR1) (He et al., 2002; Wang et al., 2002) and bri1 EMS suppressor 1 (Yin et al., 2002). The BZR1 protein is involved in a negative feedback pathway of the BR-biosynthetic genes and, in the presence of exogenous BL, inhibits the transcription of BR-biosynthetic genes, such as CPD and DWARF4 genes.
We asked the question of whether the D11 gene was regulated by exogenous BL. Treatment of the d11 mutant with BL was found to downregulate expression of the mutated D11 gene, in the same manner as that seen in a BR-deficient mutant, d2-1 (Figure 8B). We also observed that gene expression in d61-2 (defective in the BR receptor kinase gene, OsBRI1 gene) was not suppressed by exogenous BL (Figure 8B). This indicates that the OsBRI1 protein is necessary for the suppression of the D11 gene with BL. In d61-1, a leaky mutant for BL signaling, expression of the D11 gene was slightly suppressed by exogenous BL (data not shown). Hong and colleagues demonstrated that expression of the rice OsDWARF and D2 genes, which respectively encode oxidases of C-6 and C-3 of BRs, were regulated in a feedback manner by BL (Hong et al., 2002, 2003). Similarly, expression of the Arabidopsis cytochrome P450 genes, DWF4/CYP90B1, CPD/CYP90A1, DWARF/CYP85A1, ROT3/CYP90C1, and CYP90D1, were suppressed by BL (Bancos et al., 2002; Goda et al., 2002). These observations indicate that the negative feedback regulation of genes related to BR biosynthesis by BL is a common phenomenon in both dicot and monocot plants. Our finding here of negative feedback by BL of D11 gene expression is therefore consistent with our proposition that the gene is involved in BR biosynthesis. We also found that D11 mRNA accumulated at a higher level in non-BL-treated d11 mutants than in wild-type plants. This raises the possibility that the negative feedback regulation of the D11 gene expression is mediated through a BR-sensing pathway containing BR-signaling molecules, such as the BRI1, BIN2, and BZR1 proteins.
In the lamina joint inclination test, two d11 alleles, d11-1 and d11-2, showed a hypersensitive response to treatment with BL, the final product of the pathway of BR biosynthesis (open squares in Figures 10K and 10V). We compared the pattern of expression of the BR receptor kinase, OsBRI1, in d11-1 with those in the BR-deficient mutant, d2-1, and wild-type plants. In the absence of BL, OsBRI1 mRNA accumulated in the d11-1 and d2-1 mutants compared with wild-type plants (Figure 8B). Accordingly, we propose that the higher sensitivity of d11 and d2 mutants to BL may be due to enrichment of the BR receptor kinase. Because OsBRI1 mRNA was feedback-regulated by BL in d11-1 and d2-1 mutants, the BL-sensing pathways in these mutants are considered to be intact. Thus, the hypersensitive response of d11 mutants to BL suggests that the mutant may have a defect in the pathway of BR biosynthesis.
d11 Is a BR-Deficient Mutant
For a variety of reasons, we conclude that d11 is a BR-deficient mutant. First, the abnormal phenotypes of d11 are similar to those of BR-deficient (d2-1) and BR-insensitive (d61-1) mutants (Figures 1 and 2). Second, the treatment of d11 mutants with BL rescues the dwarf phenotype (Figure 3). Third, the D11 gene (defective gene in d11 mutants) encodes a novel cytochrome P450 with an amino acid sequence showing similarity to those of known BR-biosynthetic enzymes (Figures 5 and 6). Fourth, the expression of the D11 gene is regulated in a negative feedback manner by BL (Figure 8B). Fifth, in the lamina joint inclination test, only some intermediates of the BR-biosynthetic pathway (6-DeoxoTY, TY, and 6-DeoxoCS and the active BRs, CS and BL) had positive effects on inclination in d11 mutants (Figure 10); in wild-type plants, exogenous BL and all its intermediates increased inclination. This difference in response suggests that d11 mutants have a defective reaction step(s) in a late stage of the BR-biosynthetic pathway.
In rice mutants, a breakdown of one step in BR biosynthesis has been shown to cause increases in the level of upstream intermediates; for example, some upstream intermediates increased 6.6-fold (Hong et al., 2002), as compared with wild-type plants. In this study, measurement of BR intermediates identified different candidates for the defective step of biosynthesis in shoots and florets. The inconsistent results may be caused by a complex control in BR accumulation in plants. At present, we are unable to give a suitable explanation for this inconsistency between the results with shoots and florets. BR intermediates are unlikely to be transported long distances in plants (Symons and Reid, 2004). The complex control of BR accumulation may thus be a consequence of tissue-specific expression of the genes required for synthesis of BR intermediates. Another possible cause for the different candidates for the defective step of biosynthesis in shoots and florets may be the presence of a D11 homolog in the rice genome. The homolog may have differential activities in different tissues. The differential activities could occur on the transcriptional, translational, posttranslational, or enzymatic levels. As a result, the homolog may compensate defects of the d11 mutant differentially in different tissues. If this is the case, then the d11 mutant would effectively be leaky.
BR intermediates upstream from 6-Deoxo3DT and 3DT did not increase the lamina joint inclination in d11 mutants. On the basis of results from feeding experiments, we suggest that the D11 protein participates in the supply of 6-DeoxoTY and TY (possible defect step 1 in Figure 9A). At present, we cannot completely rule out the possibility that the D11 protein participates in possible defect step 2 (Figure 9B) because the D11 gene expressed largely in the internodes and the florets before flowering of wild-type plants (Figure 8A). If the D11 protein participates in possible defect step 2, however, we are unable to explain the results of feeding experiments (Figure 10); namely, we cannot logically explain why 6-DeoxoCT, CT, 6-DeoxoTE, TE, 6-Deoxo3DT, and 3DT could not rescue the increase of the lamina joint inclination. If the D11 protein should participate in possible defect step 2, we would suppose the existence of other pathways for BR biosynthesis in rice. After all, the concrete enzymatic function of the D11 protein remains ambiguous. The enzymatic functions of BR-related P450s have been studied using the yeast system on brassinosteroid-6-oxidases from tomato (Lycopersicon esculentum) (Bishop et al., 1999), Arabidopsis (Shimada et al., 2001), and rice (Hong et al., 2002) and on CYP72B1 from Arabidopsis (Turk et al., 2003). To address the function of D11 protein, biochemical studies using in vitro–synthesized D11 protein may be necessary.
Determinants of Seed Form
The dwarf rice mutant, d11, bears small seeds that are shorter in length but equal in width to wild-type seeds. Other rice mutants defective in BR biosynthesis, d2 (Hong et al., 2003) and brd1 (Hong et al., 2002), also bear small seeds, but their seeds are shorter in both length and width than wild-type seeds. A dwarf rice mutant, d1, which is a deficient mutant for the gene of the heterotrimeric G protein α subunit, bears small seeds similar to those of the d11 mutant (Ashikari et al., 1999; Fujisawa et al., 1999). The BR-signaling pathway in plants shares some similarity to the Wnt-signaling pathway in higher vertebrates (Huelsken and Birchmeier, 2001). The Wnt receptor, Frizzled, is a G protein–coupled receptor. However, the plant BR receptor, BRI1, is a Leu-rich repeat receptor kinase and does not contain the seven transmembrane spanning domain structures that are found in G protein–coupled receptors. On the basis of the comparison of receptor structure, BR signaling may be independent of the plant G protein signaling. Accordingly, we consider that both BR-signaling and G protein–coupled pathways are necessary for the formation of seeds of normal length. At present, it is unclear how the actions of active BRs and G protein–mediated signaling are related to each other in the formation of normal rice seeds.
METHODS
Plant Materials and Growth Conditions
We analyzed four d11 mutants, d11-1 (HO556 and T65d11), d11-2 (N58 and ID11), d11-3 (TCM2410), and d11-4 (Tos3220) in this study; three rice cultivars (Oryza sativa cv Nipponbare, Taichung 65, and Shiokari) were used as wild-type plants. All d11 mutants have been proven to be allelic. HO556 and N58 were first named d11, and physiological characteristics of these mutants were reported by Iwata et al. (1984). HO556 was backcrossed to Taichung 65 (T65) as the recurrent parent eight times to obtain T65d11, which was used as a near-isogenic line of T65. Thus, HO556 and T65d11 were the same allele in two different genotypes, and both HO556 and T65d11 were named d11-1 in this article. N58 was backcrossed to Shiokari as the recurrent parent nine times, and the resultant ID11, a near-isogenic line of Shiokari, was obtained (Mitsunaga et al., 1994). Thus, N58 and ID11 were the same allele in two different genotypes, and both N58 and ID11 were named d11-2 in this article. TCM2410 was a d11 allele obtained from a T65 library mutagenized with N-methyl-N-nitrosourea treatment, and TCM2410 was named d11-3 in this article. Tos3220 was a d11 allele obtained from a Nipponbare library with tissue culture–induced mutations (Hirochika et al., 1996), and Tos3220 was named d11-4. Two d11 mutants, T65d11 as d11-1 and ID11 as d11-2, were used for physiological experiments and the measurement of BR intermediates. All rice plants were usually grown in a greenhouse at 30°C.
The wild type (T65), d11-1, d61-1 (BR-insensitive mutants), and d2-1 (a BR-deficient mutant) were used in the analyses of deetiolation phenotypes and BL induction in shoot elongation. The recurrent parent of d61-1 and d2-1 was T65. Rice seeds were sterilized with 1% NaClO before use in the following experiments. (1) Rice seeds were grown on 1% agar medium containing half-strength MS medium in complete darkness at 30°C for 4 weeks. (2) For BL induction of shoot elongation, rice seeds were grown on 1% agar medium containing half-strength MS medium and various concentrations of BL and incubated at 30°C under continuous light. After 2 weeks, the length of the second leaf sheath was measured. A total 30 plants were used for each treatment. For the negative feedback analysis, d61-2 (Yamamuro et al., 2000) was used in place of d61-1 because d61-1 is a leaky mutant with regard to BR signaling. The recurrent parent of d61-2 was T65. (3) Rice seeds were grown on 1% agar medium containing half-strength MS medium with or without 10−6 M BL at 30°C under the continuous light for 2 weeks.
Mapping of the D11 Gene
The d11 locus has previously been shown to map to the long arm of chromosome 4 (Yoshimura et al., 1997). For approximate positioning of the d11 mutants, we used the indica strain, Kasalath. For fine mapping of d11 mutants, we used a substitution line, SL14, in which chromosome 4 of Kasalath was substituted for that of Nipponbare, the remaining chromosomes of the genome being of Nipponbare origin. Approximately 15,000 F3 seeds were sown in nurseries. At the seedling stages, 3020 dwarf plants were selected and transplanted into a paddy field. These dwarf mutants were screened using four cleaved and amplified, polymorphic sequence markers, C559, G7008, W390, and L353. Genomic DNA was extracted from rice leaves using the cetyltrimethylammonium bromide method, and DNA gel blot hybridization analysis was performed with several restriction fragment length polymorphism markers (Harushima et al., 1998) according to the protocol for the ECL direct nucleic acid labeling and detection system (Amersham Biosciences, Tokyo, Japan). To isolate a genomic D11 gene, we performed PCR screening using sequence tagged site primers of the PAC library produced by the Rice Genome Research Program (Baba et al., 2000). A contig was constructed from PAC clones obtained by the PCR screening. Two PAC clones, P424E6 and P471B10, contained the entire coding region and the 5′ and 3′ flanking regions of the D11 gene. To identify the mutation sites of the d11 alleles, we sequenced DNA fragments corresponding to the open reading frames predicted by GENSCAN program between markers T and K (Figure 4). The genomic DNA of the four d11 alleles was used as templates, and PCR was performed using >70 sets of PCR primers, using sequences between markers T and K. The PCR conditions were 94°C for 2 min, 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min in 25 cycles. The amplified DNA fragments were directly sequenced, without cloning, using the same primer as for amplification.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted with the RNeasy mini kit (Qiagen, Tokyo, Japan) from whole seedlings and various organs of d11 mutants and wild-type plants. The extracted RNA was treated with DNase during RNA purification to eliminate genomic DNA contamination: the DNase treatment was performed by the protocols recommended by the manufacture. The first strand of cDNA was synthesized from 1 μg of total RNA using the Super Script first-strand synthesis system for RT-PCR (Invitrogen, Tokyo, Japan). For the analysis of the mutation sites of d11 alleles, two primers, 5′-GAAACTTGGTAGAATAGACAGA-3′ and 5′-TTCACTGACTAATCCTCTCAGT-3′, were used to amplify the full-length fragments of D11 cDNA, and the amplified cDNA fragments were directly sequenced, without cloning, using appropriate primers. The conditions used during PCR were 94°C for 2 min, 94°C for 15 s, 55°C for 30 s, and 68°C for 1 min in 25 cycles.
For the analysis of the expression patterns of the D11, OsBRI1, and Actin genes, a semiquantitative RT-PCR analysis was performed. Two primers, 5′-TCACTGCCATGATACTTGAAGA-3′ and 5′-TTCACTGACTAATCCTCTCAGT-3′, were used to amplify 938-bp cDNA fragments covering the third exon to the ninth exon of the D11 gene. Two primers, 5′-GGCCAAACAGTCTGGGAAGATG-3′ and 5′-ACTGGTTATACGACAGGTCAAG-3′, were used to amplify OsBRI1 cDNA fragments. Two PCR primers, 5′-CGTCAGCAACTGGGATGATATG-3′ and 5′-GTGTGGCTGACACCATCACCAG-3′, were used to amplify a 262-bp cDNA fragment covering the third exon to the fourth exon of the Actin gene as a control. The conditions for semiquantitative RT-PCR were 94°C for 2 min, 94°C for 15 s, 55°C for 30 s, and 68°C for 1 min in 25 cycles using KOD plus (TOYOBO, Tokyo, Japan). PCR products for the D11, OsBRI1, and Actin genes were amplified in a linear range up to 28 cycles of PCR. The OsBRI1 gene has no intron, and so amplification of the OsBRI1 gene along with contaminating genomic DNA could not be completely excluded. When cDNA prepared from DNase-treated RNA was used as template for RT-PCR, the 938-bp cDNA fragment from D11 was free of contaminating genomic DNA. On the basis of this result, we consider it likely that the OsBRI1 gene amplification had little to no genomic DNA contamination. The amplified PCR products were loaded on a 1.5% agarose gel and run for 15 h at 20 V, then blotted onto a nylon membrane. The membranes were hybridized with enhanced chemiluminescent–labeled cDNA fragments for the D11, OsBRI1, and Actin genes as probes. DNA gel blot hybridization conditions were the same as described above.
Complementation Test
A DNA fragment containing a full-length genomic D11 gene was obtained by digesting the PAC clone, P424E6, with EcoRI and XbaI. The digested fragment was inserted into a binary vector pPZP2H-lac harboring a hygromycin-resistant gene (Fuse et al., 2001). The fragment was introduced into d11-1 by an Agrobacterium tumefaciens–mediated transformation method. An insert-free pPZP2H-lac vector harboring a hygromycin-resistant gene was transformed into d11-1 as a control.
Quantification of Endogenous BRs
Shoots and florets before flowering of two wild-type plants (T65 and Shiokari) and two d11 mutants (d11-1 and d11-2) were harvested at the sixth week and ∼4 months after germination, respectively. These were immediately lyophilized at −80°C. To analyze endogenous BRs, lyophilized shoots (equivalent to 20 g of fresh weight) were extracted twice with 250 mL of methanol:chloroform (4:1 [v:v]), and the BRs were purified and quantified according to the previously described method.
Lamina Joint Inclination for Feeding Experiments
Germinated seeds were selected for uniformity of coleoptile length, transplanted onto 1% agar medium, and grown at 30°C for 3 d. One microliter of ethanol containing 0, 10, 100, or 1000 ng of BR intermediates was spotted at the top of the lamina of two wild-type plants (T65 and Shiokari) and two d11 mutants (d11-1 and d11-2). After incubation for 3 d, the angle between the lamina and its leaf sheath was measured (Fujioka et al., 1998). Ten plants were used for each measurement.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries. The accession number of D11/CYP724B1 is AB158759, and those for the sequences shown in Figures 5 and 6 are as follows: DWF4-OsH (AAN60994); DWF4/CYP90B1 (AF044216); CPD/CYP90A1 (X87367); ROT3/CYP90C1 (AB008097); D2/CYP90D2 (AP003244); CYP90D3 (AC130732); CYP90D1 (AB066286); AtBR6ox/CYP85A2 (AB035868); DWARF/CYP85A1 (U54770); OsDWARF/CYP85A4 (AB084385); AtKAO1/CYP88A3 (AF318500); AtKAO2/CYP88A4 (AF318501); CYP83B1 (BAA28531); CYP79B2 (ATT5J17.120); CYP79B3 (AC006592). The accession numbers of OsBRI1 and Actin genes are NM191780 and X16280, respectively.
Acknowledgments
We thank Hikaru Satoh for the gift of TCM2410; Yoshio Sano for the gift of ID11 and N58; Takuji Sasaki and Takashi Matsumoto for the sequencing of two PAC clones, P424E6 and P471B10; and Masayo Sekimoto for her technical assistance. We thank Tadashi Asahi for his critical review of this manuscript. Part of the work was performed at the Biological Resource Research and Development Center, Fukui Prefectural University (Fukui, Japan). We acknowledge funding from three sources: a Grant-in-Aid for Scientific Research on Priority Areas (15031223) from the Ministry of Education, Science, and Culture of Japan for Y.I., a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Functional analysis of genes relevant to agriculturally important traits in rice genome, IP-1002) for Y.I., and Special Coordination Funds for Promoting Science and Technology from a Research Fellowship of the Japan Society for the Promotion of Young Scientists for S.T.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yukimoto Iwasaki (iwasaki@fpu.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024950.
References
- Ashikari, M., Wu, J., Yano, M., Sasaki, T., and Yoshimura, A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 96, 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz, R., Wu, Y., LoCascio, J.C., and Feldmann, K.A. (1998). An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, T., et al. (2000). Construction and characterization of rice genomic libraries: PAC library of japonica variety, Nipponbare and BAC library of indica variety. Kasalath. Bull. Natl. Inst. Agrobiol. Resour. 14, 41–49. [Google Scholar]
- Bak, S., and Feyereisen, R. (2001). The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127, 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancos, S., Nomura, T., Sato, T., Molnar, G., Bishop, G.J., Koncz, C., Yokota, T., Nagy, F., and Szekeres, M. (2002). Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130, 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Harrison, K., and Jones, J.D. (1996). The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., Takatsuto, S., Jones, J.D., and Kamiya, Y. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96, 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Gregory, B.D., Ross, A.S., Yuan, H., Noguchi, T., Fujioka, S., Takatsuto, S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. b). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C.P., Gregory, B.D., Ross, A.S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. a). The Arabidopsis dwf7/ste1 mutant is defective in the Δ 7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11, 207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Tanaka, A., Noguchi, T., Fujioka, S., Takatsuto, S., Ross, A.S., Tax, F.E., Yoshida, S., and Feldmann, K.A. (2000). Lesions in the sterol Δ reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 21, 431–443. [DOI] [PubMed] [Google Scholar]
- Chory, J. (1993). Out of darkness: Mutants reveal pathways controlling light-regulated development in plants. Trends Genet. 9, 167–172. [DOI] [PubMed] [Google Scholar]
- Chory, J., Nagpal, P., and Peto, C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse, S.D., and Sasse, J.M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. [DOI] [PubMed] [Google Scholar]
- Fujioka, S., Noguchi, T., Takatsuto, S., and Yoshida, S. (1998). Activity of brassinosteroid in the dwarf rice lamina inclination bioassay. Phytochemistry 49, 1841–1848. [Google Scholar]
- Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164. [DOI] [PubMed] [Google Scholar]
- Fujisawa, Y., Kato, T., Ohki, S., Ishikawa, A., Kitano, H., Sasaki, T., Asahi, T., and Iwasaki, Y. (1999). Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 96, 7575–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse, T., Sasaki, T., and Yano, M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18, 219–222. [Google Scholar]
- Goda, H., Shimada, Y., Asami, T., Fujioka, S., and Yoshida, S. (2002). Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J.X., Gendron, J.M., Yang, Y., Li, J., and Wang, Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Chandler, P.M., Poole, A., Dennis, E.S., and Peacock, W.J. (2001). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 98, 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., Yoshida, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken, J., and Birchmeier, W. (2001). New aspects of Wnt signaling pathways in higher vertebrates. Curr. Opin. Genet. Dev. 11, 547–553. [DOI] [PubMed] [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, N., Satoh, H., and Omura, T. (1984). The relationships between chromosomes identified cytologically and linkage groups. Rice Genet. Newsl. 1, 128–132. [Google Scholar]
- Jang, J.C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14, 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Kalb, V.F., and Loper, J.C. (1988). Proteins from eight eukaryotic cytochrome P-450 families share a segmented region of sequence similarity. Proc. Natl. Acad. Sci. USA 85, 7221–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12, 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., and Nam, K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., and Walker, J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Mitsunaga, S., Tashiro, T., and Yamaguchi, J. (1994). Identification and characterization of gibberellin-insensitive mutants selected from among dwarf mutants of rice. Theor. Appl. Genet. 87, 705–712. [DOI] [PubMed] [Google Scholar]
- Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T., Sugimoto, K., Okabe, K., Kajiwara, H., Satoh, K., Yamamoto, K., Hirochika, H., and Kikuchi, S. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130, 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Nelson, D.R., Koymans, L., Kamataki, T., Stegeman, J.J., Feyereisen, R., Waxman, D.J., Waterman, M.R., Gotoh, O., Coon, M.J., Estabrook, R.W., Gunsalus, I.C., and Nebert, D.W. (1996). P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42. [DOI] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., Ishiyama, K., Saito, T., Kobayashi, M., Khush, G.S., Kitano, H., and Matsuoka, M. (2002). Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. [DOI] [PubMed] [Google Scholar]
- Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yoshida, S. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126, 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer, W., Ellis, M.H., and Chandler, P.M. (2002). Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99, 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons, G.M., and Reid, J.B. (2004). Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 135, 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Turk, E.M., Fujioka, S., Seto, H., Shimada, Y., Takatsuto, S., Yoshida, S., Denzel, M.A., Torres, Q.I., and Neff, M.M. (2003). CYP72B1 inactivates brassinosteroid hormones: An intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 133, 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H., and Matsuoka, M. (2000). Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 97, 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., Yang, Y., Fujioka, S., Yoshida, S., Asami, T., and Chory, J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Wang, Z.Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., and Chory, J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yoshimura, A., Ideta, O., and Iwata, N. (1997). Linkage map of phenotype and RFLP markers in rice. Plant Mol. Biol. 35, 49–60. [PubMed] [Google Scholar]