Abstract
Aurora-like kinases play key roles in chromosome segregation and cytokinesis in yeast, plant, and animal systems. Here, we characterize three Arabidopsis thaliana protein kinases, designated AtAurora1, AtAurora2, and AtAurora3, which share high amino acid identities with the Ser/Thr kinase domain of yeast Ipl1 and animal Auroras. Structure and expression of AtAurora1 and AtAurora2 suggest that these genes arose by a recent gene duplication, whereas the diversification of plant α and β Aurora kinases predates the origin of land plants. The transcripts and proteins of all three kinases are most abundant in tissues containing dividing cells. Intracellular localization of green fluorescent protein–tagged AtAuroras revealed an AtAurora-type specific association mainly with dynamic mitotic structures, such as microtubule spindles and centromeres, and with the emerging cell plate of dividing tobacco (Nicotiana tabacum) BY-2 cells. Immunolabeling using AtAurora antibodies yielded specific signals at the centromeres that are coincident with histone H3 that is phosphorylated at Ser position10 during mitosis. An in vitro kinase assay demonstrated that AtAurora1 preferentially phosphorylates histone H3 at Ser 10 but not at Ser 28 or Thr 3, 11, and 32. The phylogenetic analysis of available Aurora sequences from different eukaryotic origins suggests that, although a plant Aurora gene has been duplicated early in the evolution of plants, the paralogs nevertheless maintained a role in cell cycle–related signal transduction pathways.
INTRODUCTION
A crucial process in cell division concerns the dynamic restructuring and segregation of chromosomes, which is necessary for the correct transmission of genetic information to daughter cells. Studies of cell cycle regulation in eukaryotes, including plants, have revealed that the molecular mechanisms that control cell cycle events are highly conserved (Burssens et al., 1998).
Reversible protein phosphorylation plays a major role in the regulation of cell division. Checkpoint regulation of cell-cycle progression relies on the cyclin-dependent kinases. Although these kinases cover the different cell cycle phases, including mitosis, recent studies have brought to light additional mitotic kinases termed Aurora. Many Aurora kinases have been shown to play key roles in regulating chromosome segregation and cytokinesis (Bischoff and Plowman, 1999; Giet and Prigent, 1999; Adams et al., 2001a; Nigg, 2001; Shannon and Salmon, 2002; Andrews et al., 2003; Carmena and Earnshaw, 2003; Kufer et al., 2003). In addition, in the biflagellated alga Chlamydomonas reinhardtii, the Aurora protein kinase CALK plays a key role in the cellular morphogenetic system that regulates the development of the flagellum (Pan et al., 2004).
The Aurora kinases are a family of Ser/Thr kinases whose activities peak during cell division. Studies of the intracellular localization of Aurora kinases in mitotic cells revealed an association with mitotic structures. Members of this family are overexpressed in a variety of cancers (Bischoff et al., 1998; Tatsuka et al., 1998; Sen et al., 2002), suggesting a crucial role in cell proliferation. Aurora kinases are likely to function downstream of the cell cycle–regulated signaling pathway and are activated by phosphorylation. Cdc37, an upstream regulatory protein regulates Aurora B in Drosophila melanogaster (Lange et al., 2002), and the association of Aurora A with the spindle assembly factor TPX2 leads to activation of the kinase in vertebrates (Eyers and Maller, 2004). Ubiquitin-dependent proteolysis of Aurora A depends on recognition by the anaphase-promoting complex Cdh1-APC/C (Castro et al., 2002).
Metazoans have evolved up to three Aurora kinase genes with three family members in human and mouse and two in Drosophila and Caenorhabditis elegans. Saccharomyces cerevisiae and Schizosaccharomyces pombe carry a single Aurora kinase gene (Meraldi et al., 2004). Based on function and subcellular localization, these Aurora genes are subdivided into three classes: Auroras A, B, and C (Adams et al., 2001a). The founding members of the Aurora family are budding yeast Ipl1p (Tung et al., 1995) and Drosophila Aurora A (Glover et al., 1995). All Aurora kinases share similar structures, with catalytic domains flanked by short C-terminal tails and N-terminal domains of variable lengths.
The precise function of the Aurora A kinases is not known, but roles related to centrosome separation, spindle assembly, and spindle maintenance have been suggested (Bischoff et al., 1998; Zhou et al., 1998; Giet et al., 2002). Overexpression of Aurora A in mammals gives rise to extra centrosomes, defects in cell division, and consequent tetraploidization (Meraldi et al., 2002). Drosophila Aurora A is required for centrosome separation (Glover et al., 1995) and for actin-dependent asymmetric protein localization (Berdnik and Knoblich, 2002).
Aurora B plays multiple roles in structural and regulatory events of mitosis and displays the dynamic properties of a chromosomal passenger protein (Bischoff et al., 1998; Bischoff and Plowman, 1999). In mammals it first associates with the centromere/kinetochore, where it forms a complex with the inner centromere protein INCENP and Survivin (Adams et al., 2001a, 2001c). Then it relocalizes to the midzone of the central spindle and finally concentrates at the midbody between dividing cells (Crosio et al., 2002). High expression of a mammalian Aurora B kinase-dead mutant gene caused multiple defects, including microtubule-dependent loss of the motor protein dynein and CENP-E from kinetochores, suggesting that Aurora B plays a role in kinetochore assembly (Murata-Hori and Wang, 2002). In mammals, CENP-A, a kinetochore-localized histone H3 variant, is the target for the Aurora B kinase (Zeitlin et al., 2001). Microinjection of function-blocking anti-X Aurora B antibody into mitotic Xenopus laevis cells induced aberrant anaphases and cytokinesis (Kallio et al., 2002). The single budding yeast Aurora ortholog Ipl1 corrects misorientation of kinetochores by destabilizing microtubule attachment at kinetochores under low tension (Biggins and Murray, 2001). In addition to its roles in bipolar chromosome orientation, kinetochore assembly, spindle checkpoint, and microtubule dynamics, Aurora B kinase is involved in chromosome condensation and cohesion in C. elegans (Kaitna et al., 2002). Disruption of AIR-2 expression by RNA interference is lethal (Rogers et al., 2002). Aurora kinases are also important for the cohesion of homologous chromosomes in meiosis I. Here, Aurora B is restricted to the region distal to chiasmata when crossover occurs (Kaitna et al., 2002; Rogers et al., 2002). The third type of Aurora-related kinases, Aurora C, has been described only in mammals, where it is expressed in testis tissue, with the highest level of transcription in pachytene cells (Tseng et al., 1998; Kimura et al., 1999) and certain tumor cell lines. These kinases also localize to spindle poles during late mitosis (Katayama et al., 2003).
Among the Aurora substrates identified to date are the kinetochore protein Ndc10p of yeast (Biggins and Murray, 2001), the kinesin-related protein XlEg5 (Giet et al., 1999a), and the kinetochore-localized histone H3 variant CENP-A (Zeitlin et al., 2001). Genetic and biochemical data indicate that Aurora family members, in particular Ipl1p of S. cerevisiae and the B-type Aurora of C. elegans, Drosophila, and mammals, can control cell cycle–regulated histone H3 phosphorylation at Ser positions 10 and 28 in opposition to a type 1 phosphatase (Glc7p in S. cerevisiae) (Hsu et al., 2000; Giet and Glover, 2001; Crosio et al., 2002; Goto et al., 2002). A similar posttranslational modification of histone H3 has also been demonstrated for higher plants (Houben et al., 1999b; Kaszas and Cande, 2000; Manzanero et al., 2000; Fass et al., 2002), but characterization of the kinase responsible is, as yet, lacking. In this report, we characterize three members of the Arabidopsis thaliana Aurora (AtAurora) gene family, encoding Ser/Thr kinases very similar in structure to those of nonplant species. Expression patterns and subcellular localization suggest that plant Aurora kinases are involved in cell cycle–related signal transduction pathways.
RESULTS
AtAurora Proteins Are Encoded by a Multigene Family in Arabidopsis
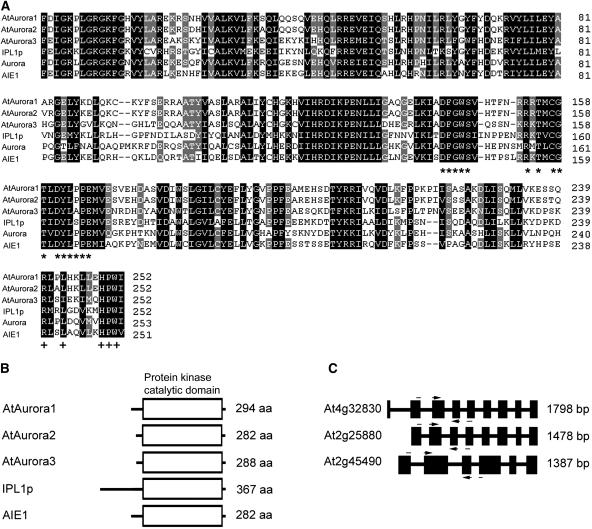
Aurora Ser/Thr kinases have a characteristic catalytic domain (Giet and Prigent, 1999) that is conserved in three Arabidopsis Aurora-like genes: AtAurora1 (At4g32830), AtAurora2 (At2g25880), and AtAurora3 (At2g45490). The cDNAs corresponding to the Aurora-like genes were isolated by RT-PCR after the corresponding exonic sequences were identified using gene prediction programs and published EST sequences. The exon/intron structures of the Arabidopsis Aurora genes were deduced by comparison of cDNA and genomic sequences. AtAurora1 to 3 encode predicted proteins of 294, 282, and 288 amino acids (Figure 1B). Alignment (Figure 1A) reveals the conserved catalytic domain of Arabidopsis Aurora-like kinases, the budding yeast Aurora kinase Ipl1, Drosophila Aurora, and mouse Aurora (AIE1). The catalytic domain is highly conserved, showing 64 to 95% identity between the different AtAuroras. It contains a motif in the activation loop (DFGWSxxxxxxxRxTxCGTxDYLPPE, see Figure 1A, asterisks), in which is imbedded a residue that in animal systems is phosphorylated by cAMP-dependent kinase PKA (Walter et al., 2000). Downstream of the activation loop, a D2-type destruction box [RXX(L/I)XXVXX HPW, see Figure 1A, indicated by plus signs] is present that may be subject to proteasome-dependent targeting and protein degradation (Arlot-Bonnemains et al., 2001). At a similar position, other Aurora proteins also carry a degradation box that has been shown to be essential for human Aurora protein degradation (Honda et al., 2000; Taguchi et al., 2002). The N- and C-terminal domains are more divergent and are presumably responsible for substrate specificity (Figure 1B).
Figure 1.
Sequence, Structure, and Similarity of AtAurora1, 2, and 3.
(A) Comparison of AtAuroras' kinase catalytic domains, deduced from AtAurora cDNAs with the domains of the Aurora/Ipl1p protein related kinases, IPL1p of Saccaromyces cerevisiae (accession number U07163), Aurora of Drosophila melanogaster (accession number X83465), and AIE1 of Mus musculus (accession number AF054620). Residues that are similar (conservative substitutions mode) in all six proteins are shaded black; those 80% similar are shaded gray. Dashes indicate gaps introduced to maximize alignment. The asterisks indicate the consensus sequence of the proposed activation motif that is a conserved signature for Aurora/Ipl1p family members (Giet and Prigent, 1999; Giet et al., 1999b). The plus signs indicate the conserved degradation box in the C-terminal domain RXXLXX(V)XXHPW (Arlot-Bonnemains et al., 2001).
(B) Diagrammatic representation of AtAuroras and other Aurora/IPL1p protein kinases. aa, amino acids.
(C) Intron–exon structure of AtAuroras. Introns are shown as black boxes. Nucleotide positions of exons were deduced by comparing genomic and cDNA sequences. Positions of primers employed in subsequent experiments are arrowed.
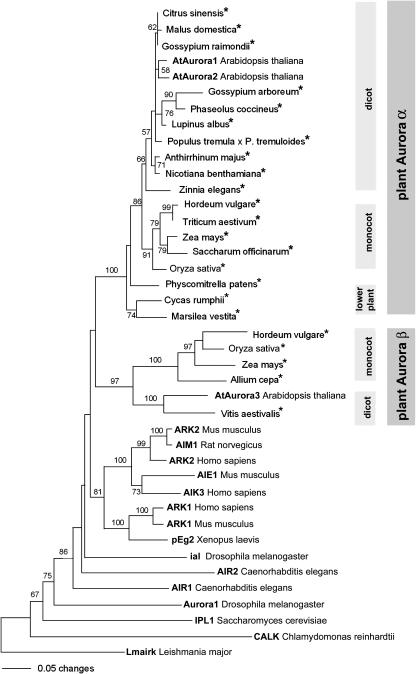
To investigate the evolutionary relationship of the Aurora family, the conserved catalytic domain was used to build a phylogenetic tree (Figure 2). We searched the EMBL plant EST database, querying with the predicted protein sequences of AtAuroras. We identified ESTs from putative Aurora-like genes in 15 angiosperm species (six monocots and nine dicots) and one member each in cycads, ferns, and mosses. All plant Auroras are grouped in a plant-specific clade divided in two major subgroups that we called plant Aurora α and plant Aurora β. The monocots Zea mays, Oryza sativa, and Hordeum vulgare and the dicot Arabidopsis had representatives in both subclades, whereas lower plants contained only plant Aurora α. Arabidopsis has two Aurora α-type copies that are highly similar at the nucleotide sequence level and in intron/exon structure (Figure 1C), suggesting that these originated via a relatively recent duplication event.
Figure 2.
The Relationships between the Kinase Domains of the Aurora Kinases Revealed by Neighbor-Joining Analysis.
Numbers depict bootstrap values (above 50%) derived from 500 bootstrap resamples. Nearly the same tree topology resulted from parsimony analysis. Auroras derived from EST sequences are indicated by asterisks. The accession numbers of the proteins in the tree are as follows: C. reinhardtii (AAF97501), S. cerevisiae (U07163), D. melanogaster (A56220 and AAF53026), L. major (AAD00707), C. elegans (AF071206 and AF071207), X. laevis (Z17207), H. sapiens (AF008551, AF054621, and AF008552), M. musculus (U69106, AF054620, and U69107), R. norvegicus (D89731), A. thaliana (At4g32830, At2g25880, and At2g45490), C. sinensis (CF263820), M. domestica (CN879930), G. raimondii (CO130728), G. arboreum (BQ411481), P. coccineus (CA908888), L. albus (CA411358), P. tremula × P. tremuloides (BU8303), A. majus (AJ795958), N. benthamiana (CK289306), Z. elegans (AU308986), H. vulgare (BF263829 and BF260084), T. aestivum (BJ305786), Z. mays (BI319148 and AY106302), S. officinarum (CA114185), O. sativa (BAA99439 and BE40040), P. patens (AJ566700), C. rumphii (CB091521), M. vestita (CD651861), A. cepa (CF444489), and V. aestivalis (CF355370).
The Three AtAuroras Are Expressed Similarly in Dividing Cells
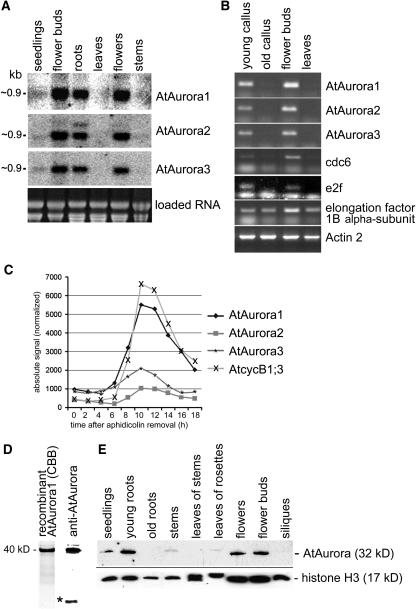
The tissue distribution of AtAurora expression was determined by RNA gel blot hybridization of total RNA isolated from young roots, stems, flower buds, flowers, fully expanded leaves, and 4-d-old seedlings possessing small apical meristems (Figure 3A) and by semiquantitative RT-PCR (see Supplemental Figure 1 online). Specific Aurora probes hybridized to RNA of ∼0.9 kb. High abundance of Aurora transcripts was detected in root and flower tissue. Remarkably, fully expanded leaves and stems contained little or no Aurora RNA. Similarly, transcript levels were high in flower buds as well as in extracts from cultured cells (Figure 3B) when analyzed by semiquantitative RT-PCR. The Aurora transcript levels paralleled those from cdc6 (Ramos et al., 2001) and transcription factor e2f (Mariconti et al., 2002), suggesting that these samples were enriched for cycling transcripts. The elongation factor 1B α-subunit (Becher et al., 2004) and actin 3 were included as loading standards. Because Aurora RNA was most abundant in extracts from tissue rich in dividing cells, we plotted the RNA profiles of the AtAuroras in synchronized Arabidopsis tissue culture cells using publicly available Affymetrix microarray datasets (Menges et al., 2002). The transcription profiles of AtAurora1, AtAurora2, AtAurora3, and the strongly mitotically induced cyclinB1;3 show that these four genes are activated together 8 h after release from the aphidicolin block (Figure 3C). The increase in mRNA coincides with entry into mitosis when expression of B-type cyclins is upregulated (Shaul et al., 1996; Ito et al., 1998). Expression of the Auroras peaks at mitosis, after which they are downregulated along with cyclinB1;3. AtAurora1 shows the strongest induction level increasing approximately fivefold over the RNA level before the onset of mitosis (Figure 3C). The downregulation of AtAuroras overlaps with the end of mitosis 12 to 13 h after aphidicolin removal.
Figure 3.
Transcriptional Analysis of AtAurora1, 2, and 3 and Expression Pattern of AtAurora Proteins.
(A) RNA gel blot analysis of AtAurora1, 2, and 3 transcripts isolated from Arabidopsis seedlings, flower buds, roots, leaves, flowers, and stems. Ethidium bromide staining of the gel is shown in the bottom panel.
(B) AtAurora mRNA abundance assessed by semiquantitative RT-PCR in undifferentiated tissue culture cells undergoing rapid (young callus) or stationary growth phase (old callus) in differentiated tissues of fast-growing flower buds and mature leaves. In parallel, to correlate AtAurora expression with active cell division, the activity of the cell cycle–specific genes CDC6 and AtE2F was monitored. Each sample contained approximately the same amount of RNA as revealed by the nearly homogeneous amplification of constitutive actin and elongation factor 1B α-subunit transcripts.
(C) mRNA profiles of the AtAuroras during the mitotic cell cycle deduced from publicly available microarray data of synchronized Arabidopsis tissue culture cells (Menges et al., 2003). AtAuroras are coactivated with cycB1;3 at the onset of mitosis, 8 h after release from the aphidicolin block.
(D) SDS-PAGE separation of purified recombinant AtAurora1 protein used for immunization after Coomassie blue (CBB) staining (first lane) or after immunoblotting using the antisera produced against recombinant AtAurora1 (second lane). The position of the recombinant AtAurora1 (40 kD) protein is indicated. The smaller extra band (asterisk) is background specific to bacteria.
(E) Forty micrograms of total protein isolated from seedlings, young and old roots, stems, leaves of stems and rosettes, flowers, flower buds, and siliques of Arabidopsis were separated by SDS-PAGE and probed by immunoblotting with antirecombinant AtAurora1. To demonstrate the uniformity of loading, the various protein samples were probed by immunoblotting with antihistone H3.
AtAurora Protein Accumulates in Dividing Cells
To quantify the AtAurora protein, a polyclonal antibody against recombinant AtAurora1 expressed in Escherichia coli was generated. The reactivity of the antiserum was tested with the same mixture of the recombinant AtAurora1 protein that was used for immunization of rabbits (Figure 3D). The antiserum recognized a 32-kD protein corresponding to the predicted molecular mass of AtAuroras. The smaller extra band (indicated with an asterisk in Figure 3D) is background specific to bacteria. In view of the high similarity (64 to 95%) of the AtAurora proteins and because the antibodies were prepared to a full-length AtAurora1, we anticipated a cross-reaction with all three members of the protein family and will therefore refer to the antiserum as anti-AtAurora. High protein levels were detected in young roots, flowers, and flower buds of Arabidopsis, in accordance with the transcript abundance (Figure 3E). A weaker immunosignal was detected in the protein extracts from seedlings and even less in extracts of mature leaves and old roots. No signal was obtained from siliques. The protein mobility in stems and leaves of rosettes was shifted, suggesting posttranslational modification of AtAurora in these tissues. To establish the uniformity of loading of the various protein samples, histone H3-specific antibody staining is shown in Figure 3D.
Dynamics of Green Fluorescent Protein–Tagged AtAuroras in Dividing Cells
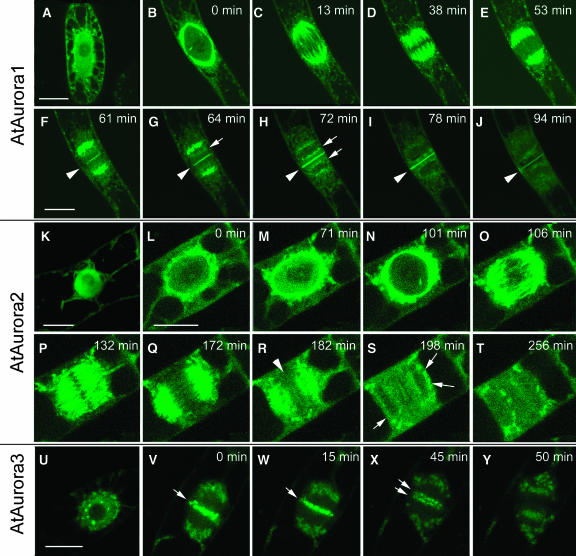
To analyze the cellular localization of plant Auroras, the open reading frames of AtAurora1, 2, and 3 were fused upstream and downstream of the green fluorescent protein (GFP) coding sequence, and the resulting constructs were introduced into tobacco (Nicotiana tabacum) BY-2 suspension cultures. Fusions with GFP at the N-terminal side produced more stable and intense fluorescence in BY-2 cells than fusions at the C-terminal end. In interphase cells, AtAurora1, 2, and 3 were predominantly located in the nucleus (Figures 4A, 4K, and 4U). To study the distribution of the fusion proteins during cell division, time-lapse photographs were recorded. At the onset of prophase, before nuclear envelope breakdown, GFP-AtAurora1 (Figure 4B) and GFP-AtAurora2 (Figures 4L to 4N) were located on the cytoplasmic side of the nuclear membrane, gradually migrating to the poles of the nascent spindle as mitosis progressed. Upon formation of the spindle microtubular basket, GFP-AtAurora1 (Figures 4C to 4F) and GFP-AtAurora2 (Figures 4O to 4R) associated with the microtubule fibrils and moved toward the midzone of the spindle. Later, both moved away from the spindle midzone toward the spindle poles (Figures 4E and 4Q). A weaker concentration of GFP-AtAurora1 was detectable in regions that colocalize with the position of the centromeres directed toward the spindle poles (Figures 4G and 4H, arrows). GFP-AtAurora1 and GFP-AtAurora2 behavior was remarkably similar until the end of anaphase when GFP-AtAurora1 concentrated at the midline of the phragmoplast (Figures 4F to 4I). It progressively followed the expansion of the cell plate until the mother wall was reached. GFP-AtAurora2 (Figure 4R, arrowhead) was much less abundant at the cell plate, and it disappeared at the onset of expansion.
Figure 4.
Subcellular Localization of GFP-Tagged AtAurora1, AtAurora2, and AtAurora3 in Transgenic Mitotic Tobacco BY-2 Cells.
AtAurora1, 2, and 3 are present in the cytoplasm and in the nucleus during interphase ([A], [K], and [U]). Upon entry into the mitotic cell cycle phase, AtAurora1 and 2 concentrated at the perinuclear region ([B] and [L]), whereas AtAurora3 concentrated at dots around the nucleolus (U). AtAurora1 and 2 associated with the spindle microtubules and moved toward the polar sides during later stages of the metaphase ([C] to [F] and [O] to [Q]). AtAurora3 moved to the midzone of the spindle, where it appeared as discrete spots, presumably corresponding to the centromeric region of chromosomes ([V] and [W]). At the start of anaphase, AtAurora3 was dispersed or destroyed ([X] and [Y]). At the end of mitosis, AtAurora1 and 2 localized at the cell plate (arrowheads; [G] to [J] and [R] to [T]). This was most pronounced for AtAurora1. In addition, some accumulation occurred at the nuclear rim ([H] to [J], [S], and [T]; arrows). Bars = 20 μm.
In contrast with AtAurora1 and 2 that were evenly distributed in the nuclueus, GFP-AtAurora3 concentrated in nuclear dots arranged around the nucleolus and at the nuclear periphery in early prophase cells (Figure 4U). The diffuse perinuclear fluorescence and later, at the position where the poles of the spindle are formed, is presumably artifactual accumulation of the fusion product in an unknown compartment because we have seen this recurrently for other unrelated GFP-fusion constructs in BY-2 lines (Figures 4V to 4X). The strongest concentration of GFP-AtAurora3 occurred at or near the centromeres that became aligned during metaphase (Figures 4V and 4W, arrows). GFP-AtAurora-3 remained at the centromeres until a few minutes after the start of anaphase when the signals divided with chromosomes (Figure 4X, arrows). Then it became distributed to a granular component of the cytoplasm (Figure 4Y).
Recombinant AtAurora1 Phosphorylates Histone H3 in Vitro
We tested the ability of recombinant AtAurora1 to phosphorylate histone H3 in vitro. AtAurora1 was expressed in insect cells using the baculovirus sf9 expression system. The activity of the expressed AtAurora1 was analyzed using a kinase assay with two different substrates. Histone H3 of Arabidopsis was expressed in E. coli to ensure that posttranslational modifications were absent before the kinase assay (Figure 5A). Total histone extracts, including H3 isolated from calf thymus, were used to test histone H3-type specificity of AtAurora1 (Figure 5B). The possible influence of other potential posttranslational modifications of H3 was not studied in detail, but it is known from the literature that H3 methylation and acetylation are competitive processes (Cheung et al., 2000; Rea et al., 2000). The in vitro kinase assay demonstrates that, like human Aurora A and B proteins (Crosio et al., 2002), the immunoprecipitated AtAurora1 showed protein kinase activity toward recombinant histone H3 (Figure 5A, lane 6, arrow). Histone H3 was not phosphorylated when incubated with proteins of uninfected insect cells (Figure 5A, lane 5). The phosphorylation signal was also absent when AtAurora1 antibody was added and when immunoprecipitated AtAurora1 was omitted from the reaction mixtures (lane 7) or contained substrate only (lane 8). The additional signals (Figure 5A, asterisk), which correspond with the size of the antibody light chain, are probably caused by nonspecific autophosphorylation of the antibody-containing fraction. The preferential phosphorylation of histone H3 was demonstrated by the kinase assay using total histones as AtAurora1 substrate. The kinase assay with precipitated AtAurora1 from infected insect cells with antirecombinant AtAurora1 (Figure 5B, lane 3) or with anti-6xHisTag antibodies (lane 4) indicates a preferential H3 phosphorylation by AtAurora1. In addition, a weaker kinase activity toward histone H4 (Figure 5B, lanes 3 and 4, arrowhead) and casein (data not shown) was observed.
Figure 5.
Phosphorylation of Histone H3 at Ser 10 by Recombinant AtAurora1.
(A) and (B) Radioactive in vitro kinase assay performed with immunoprecipitated His-tagged AtAurora1 using recombinant Arabidopsis-type histone H3 (A) or a total histone mix isolated from calf thymus as substrate (B). The amounts of histones in each reaction were monitored by Coomassie staining (lanes 1 to 4 of [A] and lanes 1 and 2 of [B]).
(A) With anti-AtAurora antibodies, immunoprecipitated recombinant AtAurora1 shows kinase activity toward recombinant histone H3 (lane 6). Negative control experiments were performed with proteins of noninfected baculovirus insect cells and substrate (lane 5), with AtAurora antibody, and substrate (lane 7) or substrate only (lane 8).
(B) The kinase assay with recombinant AtAurora1 precipitated with anti-AtAurora (lane 3) or anti-6xHisTag antibodies (lane 4) indicates a preferential phosphorylation of H3. The arrows indicate the bands corresponding to the position of phosphorylated H3. A weak kinase activity was observed toward histone H4 (arrowhead).
(C) Nonradioactive in vitro kinase assay performed with protein extracts from recombinant AtAurora1-infected baculovirus insect cells (inf) and uninfected baculovirus insect cells (con; negative control) using recombinant histone H3 as substrate. Specific phosphorylation at Ser 10 of histone H3 is indicated with an asterisk. Samples were transferred onto polyvinylidene difluoride membranes, and then the membranes were incubated with antibodies against histone H3 (anti-H3, as control), phosphorylated histone H3 at Thr 3 [p(T3)H3], Ser 10 [p(S10)H3], Thr 11 [p(T11)H3], Ser 28 [p(S28)H3], or at Thr 32 [p(T32)H3]. The arrow indicates the band corresponding to the position of histone H3.
We next determined the amino acid residue of histone H3 that is phosphorylated by AtAurora1 (Figure 5C). Nonradioactive in vitro kinase assays (see Figure 5C) performed with protein extracts from recombinant AtAurora1-infected insect cells using recombinant histone H3 as substrate and subsequent immunoblot analyses demonstrate a specific phosphorylation at Ser 10 of histone H3. No significant phosphorylation was detected at the histone H3 sites Ser 28, Thr 3, Thr 11, or Thr 32.
AtAurora Colocalizes with Phosphorylated Histone H3 during Mitosis
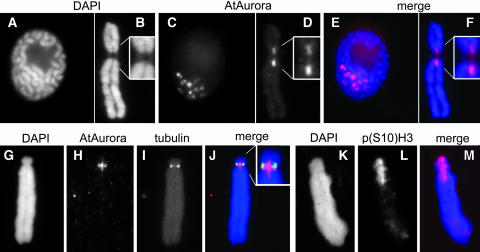
To investigate whether AtAurora1 is colocated on mitotic chromosomes with phosphorylated histone H3, the subchromosomal location of AtAurora was determined in mitotic chromosomes of field bean (Vicia faba) (Figures 6A to 6J). This plant species was used because its large chromosomes allow a precise identification of subchromosomal domains. Immunostaining with the anti-AtAurora antibody yielded mainly specific signals at the internal centromeric region (Figures 6D and 6H). In the majority of the prophase nuclei, the immunosignals appeared as globular domains, which were further characterized by a distinct central signal gap (Figure 6C). Double labeling with anti-AtAurora and anti-α tubulin antibodies revealed a partial overlap between kinetochore-localized tubulin and the AtAurora-specific signals (Figures 6G to 6J). However, AtAurora signal was restricted to centromeric regions where sister chromatid cohesion persists longest (Figures 6D and 6H) and where the highest level of histone H3 phosphorylation at Ser 10 occurs (Figures 6K to 6M).
Figure 6.
Detailed Analysis of AtAurora in Prophase Nuclei and Mitotic Metaphase Chromosomes of Field Bean.
(A) to (F) At prophase and metaphase, immunosignals of the anti-AtAurora antibody are restricted to the pericentromeric regions where sister chromatids cohere chromosomes. DAPI, 4′,6-diamidino-2-phenylindole.
(G) to (J) Metaphase chromosome after simultaneous immunostaining with anti-AtAurora and with anti-α tubulin antibodies.
(K) to (M) Immunodetection of phosphorylated histone H3 at Ser 10 [p(S10)H3] yields centromeric signals, coinciding with the chromosomal position of AtAurora.
Further enlarged pericentromeric domains are shown in insets in (B), (D), and (J).
DISCUSSION
The conservation of these general mechanisms and regulatory pathways emphasize the ancient nature and importance of the processes controlling cell division throughout the eukaryotes. This very conservation allowed a sequence-based identification of mitosis-regulated Aurora kinases from diverse eukaryotes. The data presented here indicate that plants encode Aurora-like kinases, analogous to the yeast Aurora/Ipl1 founding member and the Aurora-related kinases of other organisms. All plant and nonplant Aurora-like kinases share similar structures, with their catalytic domains flanked by N-terminal domains of variable lengths. The plant C-terminal regions are comparable with the notably short C-regions of other mitotic Auroras. In contrast with CALK of Chlamydomonas (Pan and Snell, 2000), Auroras of Arabidopsis possess no putative microtubule binding domain. The absence of a putative signal sequence or transmembrane domain in the AtAurora sequences is consistent with the idea that, like all the other Aurora/Ipl1p protein kinases studied, AtAuroras are cytoplasmic proteins.
Consistent with the mitotic function of Auroras described in other systems, the transcripts of Arabidopsis Auroras are most abundant in tissues containing actively dividing cells. The transcription behavior of all members of the Arabidopsis Aurora gene family is similar, with AtAurora1 as the most actively transcribed kinase. Cell cycle–associated transcription is further underlined by microarray data of synchronized Arabidopsis cells (Menges et al., 2003). In accordance with the transcriptional expression levels, AtAurora1/2 proteins were most abundant in young roots, flowers, and flower buds. Interestingly, in analogy with the ectopic tumor-specific overexpression of human Auroras (Carmena and Earnshaw, 2003), the AtAuroras were highly expressed in fast-growing nondifferentiated callus cells generated from mature leaves.
Evolutionary analysis shows that Aurora kinases, while consistent in sequence theme throughout eukaryotes, have undergone lineage-specific expansions and functional specialization in metazoans. The Aurora-like kinases of plants are clearly separated from animal and fungi Auroras. The occurrences of two subclades in plants raises the interesting possibility that plants possess two different isoforms of Aurora, which we called plant Aurora α and Aurora β (Figure 2). The gene tree revealed α and β kinases to be sister groups. Although only α kinases have so far been found in lower land plants, the tree topology suggests that the gene duplication predates the origin of this group. Thus, we expect that β kinase genes will be found in nearly all plants when genome sequencing advances. The existence of two Aurora α isoforms in Arabidopsis could be explained by a duplication event in the evolutionary lineage of Arabidopsis. This assumption is supported by the highly conserved intron/exon structure and amino acid sequence and the similar dynamic subcellular distribution of both proteins. Therefore, it is likely that both α-type AtAuroras act in the same molecular process in a functionally redundant manner.
Although plant Auroras display some of the dynamic distribution properties typical of the chromosomal passenger Aurora B-type, including association with the centromeres, relocalization to the spindle midzone, and finally concentration at the center of the cytokinetic apparatus where the cell plate is held together by phragmoplast microtubules, a clear classification of plant Auroras into Aurora A, B, and C classes (Adams et al., 2001a) is difficult because Arabidopsis Auroras exhibit several plant-specific characteristics.
AtAurora 1 and 2 associate with plant-specific cytoskeletal structures (preprophase band, phragmoplast, and nascent cell plate) that are necessary for cytokinesis to ensure appropriate positioning and adequate assembly of a new cell wall between the separating sister nuclei. Furthermore, plants do not possess a centrosome-like structure and therefore must perform functions unrelated to those reported for Aurora A (Adams et al., 2001b). However, AtAurora1 and 2 associate with the microtubule spindle as reported for Aurora-type A. Furthermore, both plant Auroras display the dynamic properties of the chromosomal passenger Aurora B-type protein (Bischoff et al., 1998; Bischoff and Plowman, 1999). They associate with the centromere, then relocalize to the midzone of the central spindle and finally concentrate at the midline of the phragmoplast along with the cell plate. Therefore, AtAurora1 and 2 could potentially represent plant-specific intermediates between Aurora-type A and B.
The localization of AtAurora in the pericentromere, the chromosomal site where most of the histone H3 phosphorylation at Ser 10 occurs during mitosis in plants (Gernand et al., 2003), suggests a role of AtAurora in histone H3 phosphorylation. Notably, histone H3 has been reported as substrate for several nonplant Auroras (Adams et al., 2001a). Further evidence that Arabidopsis AtAurora1 may act as histone H3 kinase is provided by in vitro kinase assays. To increase the likelyhood of producing an active AtAurora1 protein, recombinant AtAurora1 was expressed in the eukaryotic insect cell line sf9. For other eukaryotes, it has been shown that the Thr residue present in the conserved activation domain of Aurora needs to be phosphorylated for kinase activity (Eyers et al., 2003). Histone H3 is a classic Aurora substrate, and it was therefore tested in our kinase assays. To ensure histone H3 was susceptible to phosphorylation, it was purified from recombinant bacterial extracts as a positive control. A mixture of all the different plant histones with native posttranslational modifications was also used, and a strong phosphorylation of H3 was found as reported for nonplant Auroras (Scrittori et al., 2001; Crosio et al., 2002). The specificity of phosphorylation of histone H3 supports the possibility that it is a true target for plant AtAurora1 in vivo.
Notably, recombinant AtAurora1 phosphorylates histone H3 at Ser 10. The same amino acid residue specificity has also been demonstrated for Ipl1p of S. cerevisiae and the B-type Auroras of C. elegans, Drosophila, and mammals (Hsu et al., 2000; Giet and Glover, 2001; Crosio et al., 2002; Goto et al., 2002). However, in contrast with the mammalian B-type Aurora (Goto et al., 2002), AtAurora1 of Arabidopsis reveals no kinase activity toward H3 at position Ser 28. This observation suggests the existence of at least a second cell cycle–regulated histone H3 kinase because, during mitosis, the spatial and temporal dynamics of histone H3 phosphorylation at positions Ser 10 and 28 are similar in plants and animals (Gernand et al., 2003). Whether AtAurora2 and 3 or other kinases are involved in the phosphorylation of Ser 28 remains to be determined.
In conclusion, we identified an Aurora-like kinase family in Arabidopsis and other plant species. Phylogenetic analysis suggests that plant Auroras have been separated early in plant evolution into two major subgroups (plant Aurora α and plant Aurora β). Although this split predates the origin of land plants (∼500 million years ago; Sanderson, 2003) they nevertheless have maintained a conserved role in cell cycle–related signal transduction pathways, albeit of some plant-specific specialization of function. The challenge ahead is to define the precise function, physiological regulation, and signaling networks of the plant-specific Aurora kinases.
METHODS
Plant Growth and Harvest
Wild-type (Arabidopsis thaliana var Columbia) plants were cultivated in growth chambers in a 12-h-light/dark cycle at 20 C°. Tissue samples were harvested from 2- to 4-week-old plants and stored at −80 C° until analysis.
RNA Gel Blot Analysis
Total RNA was prepared from seedlings, flower buds, roots, leaves, flowers, and stems. Ten micrograms per sample were separated electrophoretically on 1.5% formaldehyde-agarose gels and transferred on nylon membranes (Hybond N+; Amersham, Buckinghamshire, UK) according to standard procedures. Equal loading of samples after spectrophotometric measurement was monitored by ethidium bromide staining and hybridization of RNA membrane with an actin-specific cDNA probe. cDNA fragments from the genes tested (AtAurora1, AtAurora2, and AtAurora3) were 32P labeled with the Megaprime labeling kit (Amersham Biosciences Europe, Freiburg, Germany) according to the manufacturer's instructions and hybridized to the membranes. Filters were washed at high stringency.
Gene Cloning and RT-PCR Analysis
Oligonucleotide primers corresponding to AtAurora1 (forward, 5′-NNACTAGTGGCGCGCCATGGCGATCCCTACGGAGACACAACACCAG-3′; reverse, 5′-NNGGATCCATTTAAATTTAAACTCTGTAGATTCCAGAAGGATCAGC-3′), AtAurora2 (forward, 5′-NNACTAGTGGAGCGCCATGTTGGTTAAGGAGTCTACGCAACGTTTG-3′; reverse, 5′-NNGGATCCATTTAAATGATTGTTCAAGTGGATCACAAGTTCCCTCC-3′), and AtAurora3 (forward, 5′-NNACTAGTGGCGCGCCATGAGTAAGAAATCGACAGAATCTGACGCTG-3′; reverse, 5′-NNGGATCCATTTAAATTCAAATATCAATTGAGGCACACACCTTTC-3′) full-length cDNAs were used for RT-PCR (RevertAid H Minus First Strand cDNA Synthesis kit; Fermentas, St. Leon-Rot, Germany). At the 5′-end of the primers, a 20-bp-long restriction recognition site specific for SwaI/AscI or BamHI/SpeI was added. Pooled mRNA of seedling, flower bud, root, leave, and, stem RNA was isolated using magnetic beads (Dyanal, Oslo, Norway). The RT-PCR products were cloned in pGEM-T-Easy vector (Promega, Madison, WI). To avoid contamination with genomic DNA, primers overlapping intron/exon junctions (positions indicated in Figure 1C, arrows) were selected for semiquantitative RT-PCR analysis. For RT-PCR on control genes, the following genes/primer pairs were used: cdc6 (At2g29680) (forward, 5′-GGAAATATGTGCAAGAAAAGTATCAG-3′; reverse, 5′-CAGAACAGACTATGATTTGTTGGTG-3′), e2f (At3g48160) (forward, 5′-ATCCCTCCAAAATAGATAACCGAC-3′; reverse, 5′-ATCTAAGGTGTGAGTCTTCTCTATG-3′), actin2 (At3g18780) (forward, 5′-TGGTCGTACAACCGGTATTGTGCTG-3′; reverse, 5′-TTGGAGATCCACATCTGCTGGAATG-3′), eEF1B α2 (At5g19510) (forward, 5′-AAACCTACATCTCCGGGATCAATT-3′; reverse, 5′-ACAGAAGACTTTCCACTCTCTTTAG-3′), and phosphatase TOPP2 (At5g59160) (forward, 5′-NNACTAGTGGCG CGCCATGGCGCAACAAGG-3′; reverse, 5′-NNGGATCCATTTAAATCCTCTTTACTTCAT-3′).
GFP Constructs and Analysis of GFP Signals
Primers were designed to include the putative start and stop codons of the Aurora genes, resulting in the forward 5′ primers 5′-ATGGCGATCCCTACGGAGAC-3′, 5′-ATGTTGTATCAGGCGGCTTCAG-3′, and 5′-ATGAGTAAGAAATCGACAGAATCTGACG-3′ and the reverse 3′ primers 5′-AACTCTGTAGATTCCAGAAGGATCAGC-3′, 5′-TCCTCTGTAAAGGCCTGATGGG-3′, and 5′-AATATCAATTGAGGCACACACACC-3′, to amplify the open reading frames of At4g32830, At2g25880, and At2g45490, respectively. Gateway (Invitrogen, Carlsbad, CA) attB1 and attB2 sequence extensions were added for in-frame cloning into pK7WGF2 to generate N-terminal GFP-aurora fusion products. C-terminal GFP fusion constructs were as described before (Van Damme et al., 2004).
Transgenic BY-2 calli were mixed with 150 μL of 1.5% low melting agarose and spread onto the bottom of a 1-well coverglass chamber (Lab-Tek, Naperville, IL) and covered with fresh 750 μL of fresh BY-2 medium. Chambers were incubated at room temperature for 24 h before imaging with a confocal laser-scanning microscope composed of an inverted Axiovert Zeiss 100M (Jena, Germany) and equipped with a 63× water-corrected objective (numerical aperture of 1.2) and an argon ion laser to generate 488-nm light for GFP excitation. The images were captured with the LSM510 image acquisition software (Zeiss). Images were exported as TIFF files and processed with Adobe Photoshop (Mountain View, CA).
Production of Recombinant Proteins and Antisera
cDNA corresponding to the entire AtAurora1 and histone H3 (At1g09200) genes were cloned into vector pET33 and pET28 (Novagen, Madison, WI), respectively, and expressed in Escherichia coli (strain Rosetta-Tuna) to generate N- and C-6xHis-tag fusion peptides. These fusion peptides were purified on nickel-agarose columns (Ni-NTA Agarose; Qiagen, Valencia, CA) under denaturing conditions with pH-step elution to pH 2.5 and used after dialyses in urea step gradient. Polyclonal AtAurora and histone H3 antibodies were produced in rabbits by repeated injection over a time span of 6 weeks. The reactivity of the histone H3 antiserum was tested with the same mixture of the recombinant histone H3 protein that was used for immunization and by indirect immunolabeling of Arabidopsis nuclei (see Supplemental Figure 2 online).
Extraction of Total Plant Protein
Plant material (200 to 300 mg) was ground under liquid nitrogen and suspended in 1 mL of solubilization buffer (56 mM Na2CO3, 56 mM DTT, 2% SDS, 12% sucrose, and 2 mM EDTA). After 10 min of incubation at 70°C, cell debris were removed by centrifugation. Forty micrograms of protein of each sample were analyzed by protein gel blotting.
PAGE and Protein Gel Blot Analysis
Protein samples were separated by SDS-PAGE in 12% polyacrylamide gels according to Laemmli (1970) and Schagger et al. (1988) and then electrotransferred onto nitrocellulose membranes. After blotting, membranes were reversibly stained with 1% ponceau red. Membranes were incubated for 12 h at 4°C in PBS and 5% low-fat milk or 2× PBS, 5% BSA, 1% PEG 3500, and 1% PVP 10, containing the appropriate antibody. Secondary anti-rabbit (Sigma-Aldrich, St. Louis, MO) antibodies conjugated to horseradish peroxidase were used to reveal immunocomplexes by enhanced chemiluminescence (Pierce, Rockford, IL).
Samples of the nonradioative in vitro kinase assay were transferred onto polyvinylidene difluoride membranes, and then the membranes were incubated with antibodies against histone H3 (anti-H3, as control), phosphorylated histone H3 at Thr 3 (07-424; Upstate Biotechnology, Charlottesville, VA), Ser 10 (06-570; Upstate), Thr 11 (Preuss et al., 2003), Ser 28 (Goto et al., 1999), or at Thr 32 (ab4076; Abcam, Cambridge, UK).
Production of AtAurora1 Protein in Spodoptera frugiperda (Sf9) Insect Cells
The open reading frame of AtAurora1 cDNA was subcloned into the pVL1392-His plasmid (Clontech, Palo Alto, CA) in frame with the N-terminal his-tag. Sf9 cells were then transfected with the pVL1392-His-AtAurora1 plasmid using CellFECTIN (Invitrogen) according to the manufacturer's instructions. Transfected cells were tested by protein gel blot analysis for the presence of 6xHis-At-Aurora protein. The medium from these cells was used as recombinant baculoviruses stock for Sf-9 cell infection (30 mL, serum-free SF-4 medium; PromoCell, Heidelberg, Germany) for 48 h at 28°C for production of AtAurora1 protein.
In Vitro Kinase Assay
For immunoprecipitation, Sf9 cells containing baculovirus recombinant AtAurora1 were lysed in a buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, pH 7.6, 1% Nonidet P-40, and 1 mM phenylmethylsulfonyl fluoride. After sonication, insoluble material was removed by centrifugation at 4°C. Approximately 300 μg of protein were immunoprecipitated by incubation at 4°C overnight with 2 μg of anti-At-Aurora1/2 rabbit antibody. The immunocomplex was precipitated after incubation with protein A-Sepharose (Amersham-Pharmacia Biotech, Uppsala, Sweden) for 1 h. The beads were washed three times with the lysis buffer and twice with PBS containing 4 nM phosphatase inhibitor Calyculin (Sigma-Aldrich).
Immunoprecipitates were resuspended in a kinase assay buffer (50 mM Tris HCl, pH 7.4, 5 mM MnCl2, 10 mM MgCl2, 1 mM DTT, 5 μCi/sample ATP[32P]) and incubated for 60 min at 30°C in the presence of 4 μg of recombinant histone H3 His-tagged or 12 μg of core histone mix from calf thymus (Roche, Indianapolis, IN) together with H1 from calf thymus (Type III-S; Sigma-Aldrich). The kinase reaction was terminated by adding Laemmli sample buffer followed by boiling. Reaction mixtures were resolved by 15% SDS-PAGE, stained with Coomassie Brilliant Blue, dried, and autoradiographed.
Preparation of Isolated Nuclei/Chromosomes and Indirect Immunolabeling
Suspensions of nuclei and mitotic metaphase chromosomes of Vicia faba were prepared as described by Schubert et al. (1993). Immunostaining was performed according to Houben et al. (1999a). Briefly, to avoid nonspecific antibody binding, slides were blocked for 30 min in 4% (w/v) BSA and 0.1% Tween 20 in PBS at room temperature before two washes in PBS for 5 min each, and incubated with the primary antirecombinant AtAurora1, anti-peptide AtAurora1 antibodies, or polyclonal rabbit antibody against histone H3 phosphorylated at Ser position 10 (Upstate Biotechnology). After 12 h incubation at 4°C and washing for 15 min in PBS, the slides were incubated with corresponding secondary antibody-fluorescence conjugates. For simultaneous immunostaining with anti-α-tubulin antibody, a microtubule stabilizing buffer (50 mM Pipes, 5 mM MgSO4, and 5 mM EGTA, pH 6.9) was used instead of PBS. The anti-α-tubulin antibody (N356; Amersham) diluted 1:50 was detected after incubation with fluorescein isothiocyanate–conjugated anti-mouse antibody (Dianova, Hamburg, Germany) diluted 1:100. Fluorescence signals were observed using a microscope equipped with epifluorescence optics. The images, recorded with a cooled CCD camera, were pseudocolored and merged using the program Adobe Photoshop.
Phylogenetic Analyses of Aurora Kinase Domains
Fourty-one amino acid sequences of Aurora kinase-like proteins were inferred from ESTs or genomic DNA sequences from the EMBL nucleotide database. The sequences were aligned in ClustalX, and the alignment was manually justified where necessary. The analyses were restricted to the conserved catalytic domain, as particularly the sequences obtained from Chlamydomonas and Leishmania were much longer and rather diverse toward the C-terminal end compared with other eukaryotic sequences. The final alignment length of this part was 279 amino acids. The sequences were analyzed with neighbor-joining and maximum parsimony algorithms in PAUP* 4.0b10 (Swofford, 2002). Statistical support of the branches was tested with 500 bootstrap resamples (neighbor-joining) and 100,000 resamples with the fast stepwise addition algorithm (maximum parsimony). In all analyses, Leishmania was defined as the outgroup.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers CAH69532, CAH69533, and CAH69534.
Supplementary Material
Acknowledgments
We thank Annegret Tewes for providing tissue cultures, Renate Manteuffel for generation of antibody, Uwe Scholz for sequence search (all Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany), Jeremy Timmis (University of Adelaide, Australia) and Ingo Schubert (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) for critical reading of the manuscript, and anonymous reviewers for helpful comments. The antibodies against H3 phosphorylated at Ser 28 and Thr 11 were kindly provided by M. Inagaki (Aichi Cancer Center Research Institute, Nagoya, Japan) and K.H. Scheidtmann (University of Bonn, Germany), respectively. We are grateful to Margit Hantschmann, Katrin Kumke, and Jana Thurow for excellent technical assistance. D.D. and A.H. were supported by grants of the Deutsche Forschungsgemeinschaft (Ho 1779/2-3). D.V.D. is a predoctoral fellow and D.G. is a postdoctoral fellow supported by the Fund for Scientific Research–Flanders (Belgium).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andreas Houben (houben@ipk-gatersleben.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.029710.
References
- Adams, R.R., Carmena, M., and Earnshaw, W.C. (2001. a). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11, 49–54. [DOI] [PubMed] [Google Scholar]
- Adams, R.R., Carmena, M., and Earnshaw, W.C. (2001. b). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11, 49–54. [DOI] [PubMed] [Google Scholar]
- Adams, R.R., Maiato, H., Earnshaw, W.C., and Carmena, M. (2001. c). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P.D., Knatko, E., Moore, W.J., and Swedlow, J.R. (2003). Mitotic mechanics: The auroras come into view. Curr. Opin. Cell Biol. 15, 672–683. [DOI] [PubMed] [Google Scholar]
- Arlot-Bonnemains, Y., Klotzbucher, A., Giet, R., Uzbekov, R., Bihan, R., and Prigent, C. (2001). Identification of a functional destruction box in the Xenopus laevis aurora-A kinase pEg2. FEBS Lett. 508, 149–152. [DOI] [PubMed] [Google Scholar]
- Becher, M., Talke, I.N., Krall, L., and Kramer, U. (2004). Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 37, 251–268. [DOI] [PubMed] [Google Scholar]
- Berdnik, D., and Knoblich, J.A. (2002). Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 12, 640–647. [DOI] [PubMed] [Google Scholar]
- Biggins, S., and Murray, A.W. (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15, 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J.R., et al. (1998). A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J.R., and Plowman, G.D. (1999). The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9, 454–459. [DOI] [PubMed] [Google Scholar]
- Burssens, S., Montagu, V.M., and Inzé, D. (1998). The cell cycle in Arabidopsis. Plant Physiol. Biochem. 36, 9–19. [Google Scholar]
- Carmena, M., and Earnshaw, W.C. (2003). The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854. [DOI] [PubMed] [Google Scholar]
- Castro, A., Vigneron, S., Bernis, C., Labbe, J.C., Prigent, C., and Lorca, T. (2002). The D-Box-activating domain (DAD) is a new proteolysis signal that stimulates the silent D-Box sequence of Aurora-A. EMBO Rep. 3, 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, P., Tanner, K.G., Cheung, W.L., Sassone-Corsi, P., Denu, J.M., and Allis, C.D. (2000). Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915. [DOI] [PubMed] [Google Scholar]
- Crosio, C., Fimia, G.M., Loury, R., Kimura, M., Okano, Y., Zhou, H., Sen, S., Allis, C.D., and Sassone-Corsi, P. (2002). Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers, P.A., Erikson, E., Chen, L.G., and Maller, J.L. (2003). A novel mechanism for activation of the protein kinase aurora A. Curr. Biol. 13, 691–697. [DOI] [PubMed] [Google Scholar]
- Eyers, P.A., and Maller, J.L. (2004). Regulation of Xenopus aurora A activation by TPX2. J. Biol. Chem. 279, 9008–9015. [DOI] [PubMed] [Google Scholar]
- Fass, E., Shahar, S., Zhao, J., Zemach, A., Avivi, Y., and Grafi, G. (2002). Phosphorylation of histone h3 at serine 10 cannot account directly for the detachment of human heterochromatin protein 1gamma from mitotic chromosomes in plant cells. J. Biol. Chem. 277, 30921–30927. [DOI] [PubMed] [Google Scholar]
- Gernand, D., Demidov, D., and Houben, A. (2003). The temporal and spatial pattern of histone H3 phosphorylation at serine 28 and serine 10 is similar in plants but differs between mono- and polycentric chromosomes. Cytogenet. Genome Res. 101, 172–176. [DOI] [PubMed] [Google Scholar]
- Giet, R., and Glover, D.M. (2001). Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., McLean, D., Descamps, S., Lee, M.J., Raff, J.W., Prigent, C., and Glover, D.M. (2002). Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156, 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., and Prigent, C. (1999). Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci. 112, 3591–3601. [DOI] [PubMed] [Google Scholar]
- Giet, R., Uzbekov, R., Cubizolles, F., Le Guellec, K., and Prigent, C. (1999. a). The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 274, 15005–15013. [DOI] [PubMed] [Google Scholar]
- Giet, R., Uzbekov, R., Kireev, I., and Prigent, C. (1999. b). The Xenopus laevis centrosome aurora/Ipl1-related kinase. Biol. Cell. 91, 461–470. [PubMed] [Google Scholar]
- Glover, D.M., Leibowitz, M.H., McLean, D.A., and Parry, H. (1995). Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81, 95–105. [DOI] [PubMed] [Google Scholar]
- Goto, H., Tomono, Y., Ajiro, K., Kosako, H., Fujita, M., Sakurai, M., Okawa, K., Iwamatsu, A., Okigaki, T., Takahashi, T., and Inagaki, M. (1999). Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 274, 25543–25549. [DOI] [PubMed] [Google Scholar]
- Goto, H., Yasui, Y., Nigg, E.A., and Inagaki, M. (2002). Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 7, 11–17. [DOI] [PubMed] [Google Scholar]
- Honda, K., Mihara, H., Kato, Y., Yamaguchi, A., Tanaka, H., Yasuda, H., Furukawa, K., and Urano, T. (2000). Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene 19, 2812–2819. [DOI] [PubMed] [Google Scholar]
- Houben, A., Wako, T., Furushima-Shimogawara, R., Presting, G., Kunzel, G., Schubert, I., and Fukui, K. (1999. a). The cell cycle dependent phosphorylation of histone H3 is correlated with the condensation of plant mitotic chromosomes. Plant J. 18, 675–679. [DOI] [PubMed] [Google Scholar]
- Houben, A., Wako, T., Furushima-Shimogawara, R., Presting, G., Kunzel, G., Schubert, I.I., and Fukui, K. (1999. b). Short communication: The cell cycle dependent phosphorylation of histone H3 is correlated with the condensation of plant mitotic chromosomes. Plant J. 18, 675–679. [DOI] [PubMed] [Google Scholar]
- Hsu, J.Y., et al. (2000). Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291. [DOI] [PubMed] [Google Scholar]
- Ito, M., Iwase, M., Kodama, H., Lavisse, P., Komamine, A., Nishihama, R., Machida, Y., and Watanabe, A. (1998). A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna, S., Pasierbek, P., Jantsch, M., Loidl, J., and Glotzer, M. (2002). The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12, 798–812. [DOI] [PubMed] [Google Scholar]
- Kallio, M.J., McCleland, M.L., Stukenberg, P.T., and Gorbsky, G.J. (2002). Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12, 900–905. [DOI] [PubMed] [Google Scholar]
- Kaszas, E., and Cande, W.Z. (2000). Phosphorylation of histone H3 is correlated with changes in the maintenance of sister chromatid cohesion during meiosis in maize, rather than the condensation of the chromatin. J. Cell Sci. 113, 3217–3226. [DOI] [PubMed] [Google Scholar]
- Katayama, H., Brinkley, W.R., and Sen, S. (2003). The Aurora kinases: Role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 22, 451–464. [DOI] [PubMed] [Google Scholar]
- Kimura, M., Matsuda, Y., Yoshioka, T., and Okano, Y. (1999). Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J. Biol. Chem. 274, 7334–7340. [DOI] [PubMed] [Google Scholar]
- Kufer, T.A., Nigg, E.A., and Sillje, H.H. (2003). Regulation of Aurora-A kinase on the mitotic spindle. Chromosoma 112, 159–163. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lange, B.M., Rebollo, E., Herold, A., and Gonzalez, C. (2002). Cdc37 is essential for chromosome segregation and cytokinesis in higher eukaryotes. EMBO J. 21, 5364–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanero, S., Arana, P., Puertas, M.J., and Houben, A. (2000). The chromosomal distribution of phosphorylated histone H3 differs between plants and animals at meiosis. Chromosoma 109, 308–317. [DOI] [PubMed] [Google Scholar]
- Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., and Albani, D. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277, 9911–9919. [DOI] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murray, J.A. (2002). Cell cycle-regulated gene expression in Arabidopsis. J. Biol. Chem. 277, 41987–42002. [DOI] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murray, J.A. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53, 423–442. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., Honda, R., and Nigg, E.A. (2002). Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., Honda, R., and Nigg, E.A. (2004). Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr. Opin. Genet. Dev. 14, 29–36. [DOI] [PubMed] [Google Scholar]
- Murata-Hori, M., and Wang, Y.L. (2002). The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12, 894–899. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. (2001). Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32. [DOI] [PubMed] [Google Scholar]
- Pan, J., and Snell, W.J. (2000). Regulated targeting of a protein kinase into an intact flagellum. An aurora/Ipl1p-like protein kinase translocates from the cell body into the flagella during gamete activation in Chlamydomonas. J. Biol. Chem. 275, 24106–24114. [DOI] [PubMed] [Google Scholar]
- Pan, J., Wang, Q., and Snell, W.J. (2004). An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev. Cell 6, 445–451. [DOI] [PubMed] [Google Scholar]
- Preuss, U., Landsberg, G., and Scheidtmann, K.H. (2003). Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 31, 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, G.B.A., Engler, J.D., Ferreira, P.C.G., and Hemerly, A.S. (2001). DNA replication in plants: Characterization of a cdc6 homologue from Arabidopsis thaliana. J. Exp. Bot. 52, 2239–2240. [DOI] [PubMed] [Google Scholar]
- Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B.D., Sun, Z.W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C.P., Allis, C.D., and Jenuwein, T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Rogers, E., Bishop, J.D., Waddle, J.A., Schumacher, J.M., and Lin, R. (2002). The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson, M.J. (2003). Molecular data from 27 proteins do not support a precambrian origin of land plants. Am. J. Bot. 90, 954–956. [DOI] [PubMed] [Google Scholar]
- Schagger, H., Aquila, H., and Jagow, V.G. (1988). Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 173, 201–205. [DOI] [PubMed] [Google Scholar]
- Schubert, I., Dolezel, J., Houben, A., Scherthan, H., and Wanner, G. (1993). Refined examination of plant metaphase chromosome structure at different levels made feasible by new isolation methods. Chromosoma 102, 96–101. [Google Scholar]
- Scrittori, L., Hans, F., Angelov, D., Charra, M., Prigent, C., and Dimitrov, S. (2001). pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopus egg extract. J. Biol. Chem. 276, 30002–30010. [DOI] [PubMed] [Google Scholar]
- Sen, S., et al. (2002). Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J. Natl. Cancer Inst. 94, 1320–1329. [DOI] [PubMed] [Google Scholar]
- Shannon, K.B., and Salmon, E.D. (2002). Chromosome dynamics: New light on Aurora B kinase function. Curr. Biol. 12, R458–R460. [DOI] [PubMed] [Google Scholar]
- Shaul, O., Mironov, V., Burssens, S., Van Montagu, M., and Inze, D. (1996). Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 93, 4868–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. (Sunderland, MA: Sinauer Associates).
- Taguchi, S., Honda, K., Sugiura, K., Yamaguchi, A., Furukawa, K., and Urano, T. (2002). Degradation of human Aurora-A protein kinase is mediated by hCdh1. FEBS Lett. 519, 59–65. [DOI] [PubMed] [Google Scholar]
- Tatsuka, M., Katayama, H., Ota, T., Tanaka, T., Odashima, S., Suzuki, F., and Terada, Y. (1998). Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 58, 4811–4816. [PubMed] [Google Scholar]
- Tseng, T.C., Chen, S.H., Hsu, Y.P., and Tang, T.K. (1998). Protein kinase profile of sperm and eggs: Cloning and characterization of two novel testis-specific protein kinases (AIE1, AIE2) related to yeast and fly chromosome segregation regulators. DNA Cell Biol. 17, 823–833. [DOI] [PubMed] [Google Scholar]
- Tung, H.Y., Wang, W., and Chan, C.S. (1995). Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol. Cell. Biol. 15, 6064–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme, D., Bouget, F.Y., Van Poucke, K., Inze, D., and Geelen, D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 40, 386–398. [DOI] [PubMed] [Google Scholar]
- Walter, A.O., Seghezzi, W., Korver, W., Sheung, J., and Lees, E. (2000). The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 19, 4906–4916. [DOI] [PubMed] [Google Scholar]
- Zeitlin, S.G., Shelby, R.D., and Sullivan, K.F. (2001). CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Kuang, J., Zhong, L., Kuo, W.L., Gray, J.W., Sahin, A., Brinkley, B.R., and Sen, S. (1998). Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20, 189–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.