Abstract
Detailed quantification of neural dynamics across the entire brain will be the key to genuinely understanding perception and behavior. With the recent developments in microscopy and biosensor engineering, the zebrafish has made a grand entrance in neuroscience as its small size and optical transparency enable imaging access to its entire brain at cellular and even subcellular resolution. However, until recently many neurobiological insights were largely correlational or provided little mechanistical insight into the brain-wide population dynamics generated by diverse types of neurons. Now with increasingly sophisticated behavioral, imaging, and causal intervention paradigms, zebrafish are revealing how entire vertebrate brains function. Here we review recent research that fulfills promises made by the early wave of technical advances. These studies reveal new features of brain-wide neural processing and the importance of integrative investigation and computational modelling. Moreover, we outline the future tools necessary for solving broader brain-scale circuit problems.
Keywords: Neuroscience, brain-scale neural circuits, microscopy, behavior, computational tools, zebrafish, optogenetics, calcium imaging, neural dynamics, cell types
Introduction
While the vertebrate brain is large, containing many functionally distinct regions, diverse cell types, and billions of synapses, until recently neuroscience was restricted to studying only small fractions of the brain at a time. Advances in imaging of neural activity [1] and microelectronics [2] now provide activity data from thousands of neurons and suggest that brain function is powered by activity distributed over large numbers of spatially dispersed neurons [3] that encode various information related to perception and behavior. However, these methods still fall short of delivering a complete dynamic view of the brain, as studies in mammals struggle to locate, define, and manipulate cell types or compare distant brain regions. This is why the translucent zebrafish Danio rerio has steadily gained traction for neuroscience [4]. For similar reasons, the fruit fly Drosophila melanogaster [5] and the nematode Caenorhabditis elegans [6] represent excellent systems to study brain-scale dynamics, but the zebrafish remains the only vertebrate permitting brain wide experimental access during behavior. Fast volumetric microscopy techniques [7,8] enable observations of dynamic neural processes across the entire zebrafish brain at micrometer resolution, impossible in other vertebrates [9]. Concurrently, new light-based biosensors have evolved rapidly, allowing for control and visualization of neural activity using high-performance calcium [10], GABA [11], dopamine [12], and bright, stable voltage sensors [•13]. By harnessing these optical advances in combination with detailed characterization of behavior [14], perturbations [15], and modelling [16], the field is beginning to dissect information processing in the zebrafish’s manageable population of ~100,000 neurons. Next to reviewing neurobiological advances in zebrafish, we will also discuss whether inspecting the neural activity of (almost) all neurons is merely convenient for homing in on relevant microcircuitry or whether there are fundamental principles to be learned by querying the entire brain.
The value of a holistic approach to neural recordings
First, what really counts as “whole-brain” recording? Recently, phosphorylated extracellular signal-regulated kinase (pERK) [17] has been used to report snapshot activity across the entire zebrafish brain, yet the neural dynamics are completely lost. On the other hand, traditional two-photon laser scanning microscopy typically record fast neural calcium dynamics on single optical planes, so whole-brain recordings must be assembled computationally [18,19] from sequential acquisitions that are often minutes or hours apart. In other words, while such approaches span the entire brain, they do not capture simultaneous whole-brain activity (with some notable exceptions [20]). Alternatively, volumetric light-sheet microscopy [21] microscopy permits imaging at multiple brain volumes per second, and light-field microscopy offers outstanding temporal resolution but is currently limited in resolution ••[22]. Yet, assuming real-time whole-brain imaging will be routine in zebrafish, what will be gained beyond reaching the abstract goal of completeness? Most brain-wide imaging studies emphasize its utility for agnostic searches for task-activated brain areas (Table 1), although simultaneous whole-brain access is not strictly necessary. While these searches are often used to generate correlational maps, the field is developing new analyses that reveal more about local circuit mechanisms ••[23], and there is a growing appreciation for more holistic views of network-wide activity patterns related to brain state transitions ••[22,24,25]. Beyond completeness, one of the major advantages simultaneous whole-brain analysis is that it may help unravel mysteries of redundancy and state-dependent encoding that are ubiquitous across species [3–6,24]. These redundancies may act as alterative pathways, broadcasters for motor commands downstream of sensory processing, error signals, or other state-dependent information necessary for parallel, multi-scale behavioral coordination, and zebrafish are uniquely positioned to adjudicate between these roles. Yet, while we report here that comprehensive sampling to generate correlation maps have become standard and insights into the encoding of brain states start to emerge, addressing these redundancies remains largely unexplored. Below we review recent progress using various forms of whole-brain analysis to discover both detailed microcircuits and insights about brain network functions underlying a range of behaviors, placing zebrafish at the center of efforts to solve models of whole-brain collaboration.
Table 1:
Overview of recent studies investigating whole brain interactions
| Behavior/phenomenon | Focus Brain areas | Imaging Method | Causal Manipul. | Cell specificity | Model | Remarks | Ref. |
|---|---|---|---|---|---|---|---|
| Nociception | *** | Convolutional Neural Networks | [16] | ||||

|
*** | Heat navigation | [64] | ||||
| Ht |

|

|

|
pERK | [17] | ||
| Ht |

|

|
MultiMAP | •[46] | |||
| Auditory & Vestibular |

|
* | Rotating light-sheet | [8] | |||

|
* | Optical otolith trapping | [37] | ||||

|
* | Sound processing | [39] | ||||
| Visuomotor Behaviors | Hb |

|

|

|
** | Glia cells | ••[23] |
| Th |

|

|
* | Looming detection | [30] | ||

|

|
** | Graph theory | [38] | |||
| Cb |

|

|
* | Electrophysiology | [42] | ||

|
* | Various visual stim. | [4] | ||||
| Ot, Pt |

|
** | Receptive field mapping | [27] | |||

|
* | Vis. stimulus design | [28] | ||||

|
*** | Higher order motion | [29] | ||||
| Pt |

|

|
* | FuGIMA, Atlas | [32] | ||
| Hb |

|
*** | Evidence Accum. | •[18] | |||
| IPN |

|
*** | Evidence Accum. | •[33] | |||
| Hunting Behaviors/Appetite | Ot, Pt |

|

|

|
* | Electroporation | [34] |
| Ht |

|

|

|
* | pERK | [41] | |
| Ha, DRN |

|

|

|
*** | Light-Field Imaging | ••[22] | |
| DRN |

|

|
** | Tracking microscope | ••[24] | ||

|

|

|
Morphology | [35] | |||
| Sleep |

|
* | Polysomnography | •[43] | |||
| - |

|
* | Galanin | [44] | |||
| - |

|

|
Serotonin | [45] | |||
| Miscellaneous | Cb |

|

|
** | Operant conditioning | ••[25] | |

|

|
Holographic photostim. | [67] | ||||
| Sc |

|

|

|
Cell tracking | [7] | ||

|

|
pERK, gene screen | [52] | ||||

|

|
Cellular atlas | •[31] | ||||
| Pt |

|

|
Voltage imaging | •[13] | |||

|

|
Online algorithms | [56] |
Many recent studies aiming to reveal neural mechanisms underlying behavior or perception in zebrafish used cellular resolution functional in vivo imaging approaches such as light-sheet ( ), selective plane illumination microscopy (
), selective plane illumination microscopy ( ), two-photon laser scanning microscopy (
), two-photon laser scanning microscopy ( ), or confocal microscopy
), or confocal microscopy  . Non-live imaging (-) to reveal neural activity using phosphorylated extracellular signal-regulated kinase (pERK), anatomical or molecular features have been used as powerful complementary tools. Most studies, after a brain-wide activity screen, focused on select brain regions to elucidate specific circuit mechanisms in one or more brain area: Ht, hypothalamus; Hb, hindbrain; Th, thalamus; Cb, cerebellum; Pt, Pretectum; Ot, Optic tectum; IPN, interpeduncular nucleus; Ha, habenula; DRN, dorsal raphe nucleus; Sc, spinal cord. To establish casual relationships various specific circuit perturbations were used:
. Non-live imaging (-) to reveal neural activity using phosphorylated extracellular signal-regulated kinase (pERK), anatomical or molecular features have been used as powerful complementary tools. Most studies, after a brain-wide activity screen, focused on select brain regions to elucidate specific circuit mechanisms in one or more brain area: Ht, hypothalamus; Hb, hindbrain; Th, thalamus; Cb, cerebellum; Pt, Pretectum; Ot, Optic tectum; IPN, interpeduncular nucleus; Ha, habenula; DRN, dorsal raphe nucleus; Sc, spinal cord. To establish casual relationships various specific circuit perturbations were used:  laser ablations,
laser ablations,  photoactivation,
photoactivation,  photoinhibition,
photoinhibition,  pharmacological approaches. Increasingly, studies combine functional profiling with cell type specificity (
pharmacological approaches. Increasingly, studies combine functional profiling with cell type specificity ( ), using specific neurotransmitter or Gal4driver lines (
), using specific neurotransmitter or Gal4driver lines ( ), and even genetically unique cell types or tracing of single cells (
), and even genetically unique cell types or tracing of single cells ( ). Finally, analysis and modeling approaches in these investigations vary in their level of complexity from regression mapping and qualitative circuit models (*), highly quantitative descriptions (**), to quantitative circuit models (***).
). Finally, analysis and modeling approaches in these investigations vary in their level of complexity from regression mapping and qualitative circuit models (*), highly quantitative descriptions (**), to quantitative circuit models (***).
Agnostic search strategies to identify behaviorally relevant brain regions
With the advent of large-scale imaging of neural activity, it is now possible to conduct a broad search of the entire zebrafish brain to conduct an unbiased sample of the regions that might contribute to the behavior in question. Such agnostic search strategies allow to determine the neuronal circuitry underlying behavior (Fig. 1a). As the zebrafish visual system is highly conserved [26] and well developed at day five post fertilization, many early studies used this common strategy to present behaviorally relevant visual stimuli during brain-wide imaging, followed by a careful characterization of circuit components [27], often recapitulated with modeling approaches. Exemplifying this strategy, two recent studies paired careful stimulus design with regression analysis to reveal the complex information processing across the brain supporting how local, light-dark transitions [28] and naturalistic higher-order motion detection induce optomotor responses [29]. Using reverse correlations to agnostically search for specific neural activity, these studies deconstructed visual circuits centered around the central retinal targets: the optic tectum, pretectum and parallel thalamic pathways [30]. Still others have used correlational mapping to reveal how various visual stimuli drive convergent activity in neurons in the anterior hindbrain [4], producing intricate topologies of sensory and motor brain areas. Following these functional characterizations, many studies use local circuit analysis to refine connectivity hypotheses after identifying relevant brain areas. For instance, Kramer et al. used precise photoactivation of functionally characterized motion-sensitive pretectal neurons and a single cell brain atlas •[31] to quantify morphological differences and projection patterns [32]. Finally, theoretical modelling approaches are often used to distill neural activity patterns, refine brain-scale mapping, and subsequent local circuit analysis. For instance, publications by Bahl et al. •[18] and Dragomir et al. •[33] demonstrate how distributed neural activity driven by classic random dot motion paradigms can be further screened for neurons whose complex temporal dynamics match specific theoretical aspects of evidence accumulation, such as leaky integrator time constants.
Figure 1. Brain states encoded in network activity and local circuitry.
(a) The common strategy to use whole-brain regression analysis under various conditions and behavioral recordings often reveals widespread sensory (teal) and motor (purple) related signals. After task relevant neurons are located, most studies progress with local circuit analysis to refine response types, connectivity and anatomy, using molecular identity, and through the use of a combination of single cell electroporation, transgenic lines, and causal perturbations, including laser ablations of single neurons or optogenetic activation in concert with behavioral testing. These results are often synthesized into models that range from simple activity maps, suggesting circuit diagrams, to realistic quantitative network simulations using recorded activity dynamics in recurrent neural network replicating biological effective connectivity. Other approaches include probabilistic models linking sensory input to behavioral output and comparison to artificial neural networks.
(b) In a recent study ••[24], focus shifted towards dynamic brain state transitions rather than correlational maps. Left, example behavior trajectories of zebrafish in exploitation (red) and exploration (blue) states. Right, locations of all neurons encoding these brain states, projected across 17 registered fish recorded with a tracking microscope, revealing brain state switches triggered by serotonergic neurons (dashed box). Ro, rostral; C, caudal; L, left; R, right.
(c) PCA trajectory of whole-brain activity (104,142 neurons) from a representative animal, color-coded by the activity of exploitation-state-encoding neurons.
(d) Average activity (mean ± s.d.) of exploitation-state-encoding (red) and exploration-state-encoding (blue) neurons across the transition from exploration to exploitation (top). These dynamics are likely key to understand brain wide states that modulating other processes.
(e) Lin et al. ••[25]. used an operant conditioning assay combined with whole-brain calcium imaging to investigate how decision brain states develop over time. Head-fixed larval zebrafish receive a mildly aversive heat stimulus by an infrared laser (red trapezoid) at the beginning of a trial. The laser is turned off if the fish makes a tail movement in the reward direction and remains on otherwise. In the second training block the reward direction is switched, with each block consisting of 20–25 trials
(f) The learning progress of an example learner fish. Black traces indicate tail positions over time, magenta and green rectangles indicate the duration of the heat stimulus for each incorrect and correct trial, respectively.
(g) Temporal evolution of brain states reveals distributed brain activity and bihemispheric preparatory activity. ARTR, anterior rhombencephalic turning region; Cb, cerebellum; HB, habenula; Te, telencephalon.
(h) Performance-dependent bifurcation of brain states before turn initiation. Left, after heat onset, brain states exhibit similarity along the “Heat ON” dimension (vertical axis), followed by a pre-turn bifurcation toward the correct or incorrect state. Similarity is measured by partial correlation between a given brain state and the average correct, incorrect, or “Heat ON” state. Right, representation of single-trial bifurcation process. Individual correct and incorrect trials are highlighted during the pre-motor period (black diamonds: heat onset, green and magenta dots: turn initiations for correct and incorrect turns, respectively).
(i) 2D representation of single-trial bifurcation process. Individual correct and incorrect trials are highlighted during the pre-motor period (black diamonds: heat onset, green and magenta dots: turn initiations for correct and incorrect turns, respectively).
Whole-brain search has also been used to study zebrafish prey capture behavior, which involves more complex sensorimotor processing than the visual reflexes above. Bianco and colleagues used whole-brain imaging during a virtual prey capture assay to reveal specific retinorecipient neurons serving as command neurons for hunting initiation [34]. This study is a showcase for how the agnostic, systematic search for neural correlates leads to identification and characterization of genetically targetable neurons that link sensory processing to specific motor execution. Follow-up studies show that neural activity in distant cholinergic neurons of the nucleus isthmi [35] appear to sustain binocular, intertectal processing during hunting behaviors [36].
Vestibular processing poses interesting challenges for imaging, but recent use of miniaturized [8] or tilting light-sheet microscopy has permitted brain-scale imaging during physiological or fictive body rotations [37]. These experiments reveal vestibular processing to be a widespread phenomenon, with multiple, distributed, functionally distinct clusters, in addition to evidence supporting bilateral asymmetries. Additionally, presenting water flow with microfluidic devices [38] and auditory stimuli [39] revealed diverse flow and sound responsive neurons distributed across the brain. Ultimately, it will be interesting to learn how the zebrafish brain integrates information across the different modalities activated during behavior.
Towards integrative brain views on brain state transitions
Increasingly, investigations now extend zebrafish brain mapping by focusing on brain state transitions [40]. Recently, innovative tracking microscopy for monitoring brain-wide neural activity while zebrafish hunt for live prey ••[24] revealed opposing oscillatory neural network states during foraging (Fig. 1b–d). These network states correspond to minutes-long exploratory and exploitatory phases in which fish either increase overall locomotion or engage in prey capture behaviors, respectively. Using linear regression to identify neurons with activity profiles correlated with various movement types, eye position, and prey detection and ingestion, Marques et al. show that these internal states are encoded in brain-wide networks. Furthermore, they demonstrate that the anticorrelated, widespread neural activity patterns associated with these internal states is controlled by specific serotonergic neurons coupled with the required sensorimotor processing associated with prey capture. These findings suggest that these serotonergic neurons of the dorsal raphe nucleus (DRN) act as brain state ‘switching’ signal broadcasted to the rest of the brain. Only by recording from the entire brain, they can show that specific clusters of neurons in specific brain regions become activated in response to this broadcast signal, but that other neurons in other brain regions are suppressed by the same signal. This study exemplifies the importance of investigating brain-wide neural activity while monitoring behavior, as the observed neural dynamics related to both stimulus processing and internal brain state changes might only be understood in the context of nuanced behavior over time. Emphasizing the careful analysis of behavior, more static methods (pERK) have recently localized a different set of bidirectional, anticorrelated networks of neurons in the caudal and lateral periventricular zone of the hypothalamus associated with longer term hunger and feeding states [41]. Another study (Fig. 1e–i) discovered the global recurring dynamics preceding a decision by combining fast volumetric light-sheet imaging during a novel operant conditioning task ••[25] in which fish have to flick their tail in the ‘correct’ direction to ‘switch off’ a painful heat stimulus. The authors discovered that the neural dynamics can predict both the time of decision as well as the decision (motor output) itself on a trial-by-trial basis. These dynamics occur in a distributed interhemispheric network, primarily between habenula and cerebellum. The study also highlighted a more cognitive role for the cerebellum, with predictive pre-motor activity more than 10 s before movement initiation being largely localized to granule cells for individual trials. Finally, the authors propose a circuit model in which the ‘timing of movement’ is predicted by ramping bilateral cerebellum neural activity and the ‘tail flick direction’ is computed by ipsi and contralateral cerebellar competition. Together with the recent characterization of visually evoked spatiotemporal dynamics across the entire Purkinje cell population [42], these investigations further underscore the importance of analyzing neural activity in the context of behavior.
Recent holistic neural recordings of sleep deprived zebrafish revealed the neural signatures of sleep, which had been elusive in species lacking traditional cortical structures. Leung et al., using non-invasive polysomnography, show conservation of slow-wave and rapid eye movement sleep, as well as a role for the conserved melanin-concentrating hormone, suggesting that common neural signatures of sleep emerged over 450 million years ago [••35]. Further, brain-wide pERK mapping identified the neuropeptide Galanin in the preoptic hypothalamus as controlling rebound sleep, a state caused by sleep deprivation [44]. While the serotonergic neurons in the raphe appear to be direct modulators of sleep levels in both zebrafish and mice [45], this research suggests that sleep might be a brain-, if not body-, scale neurobiological phenomenon.
Furthermore, recent brain-scale recordings provide insights of how zebrafish process aversive information. By utilizing light-field microscopy during shock-induced passive coping behaviors, Andalman et al. revealed that transitions to coping behaviors are orchestrated by an increased recruitment of habenular neurons ••[22]. Wee et al. used pERK mapping to identify brain regions processing noxious information, revealing convergent nociceptive pathways in zebrafish. Localized to the hypothalamic preoptic area, oxytocin neurons reflect broad classes of noxious stimuli and their activation is sufficient to trigger defensive behavior recruiting brain stem premotor regions [17]. Similarly, whole-brain analysis uncovered diverse peptidergic neurons in the hypothalamus driving defensive behaviors to homeostatic threats, such as high salinity •[46]. These studies are important steps towards integrative understanding of how specific cell types with distinct molecular expression profiles collaborate to regulate brain state transitions.
Experimental access to defined cell types across the brain
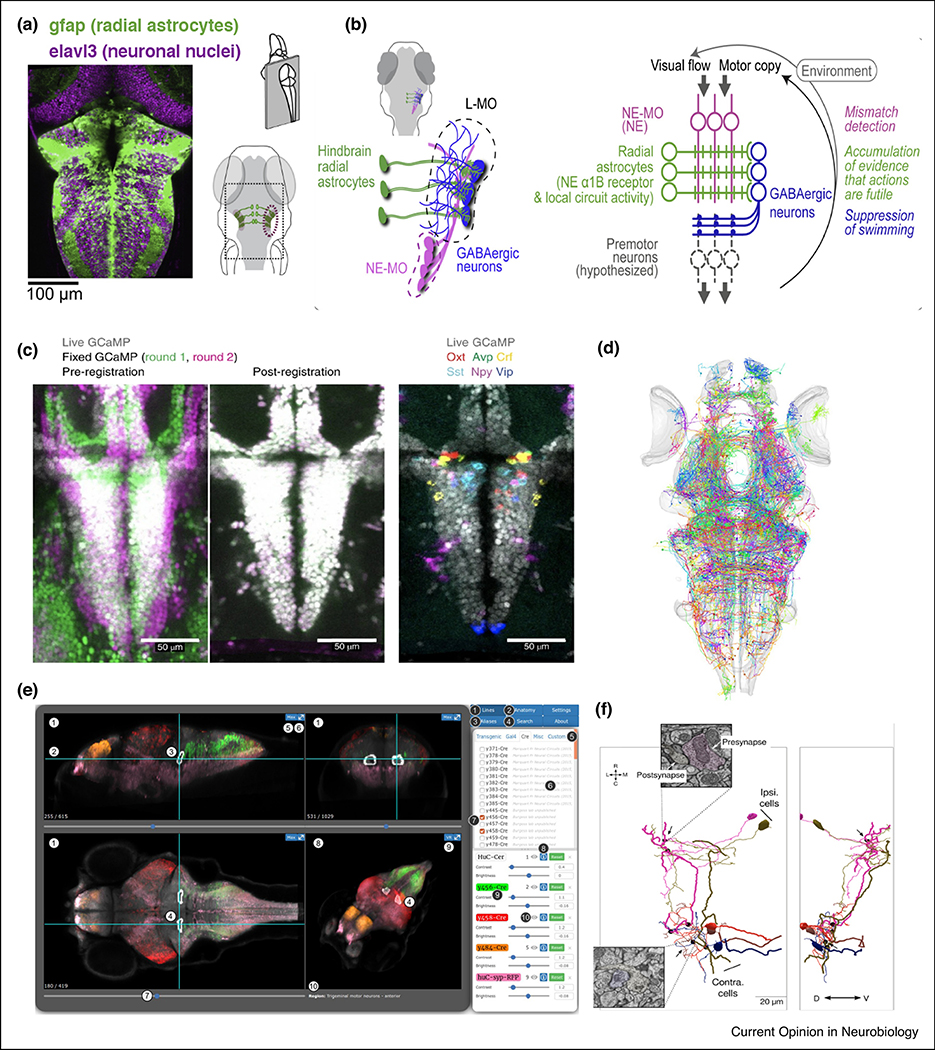
The previous sections described how whole-brain imaging can be used to delineate networks and circuits based on their functional properties. But detailed circuit dissection benefits from genetic methods that target specific cells in the brain. Intersectional genetic methods, e.g. Gal4 driver lines, allow restriction of sensor or actuator expression in only neurons that co-express multiple drivers, granting unique access to genetically defied neural populations [47]. Comprehensive single cell RNA-Seq maps can also be used to develop cell-specific genetic access [48]. Illustrating the power of cell-specific access in the whole-brain zebrafish paradigm, Mu et al. used dual color agnostic activity search for both glia and neurons (Fig. 2a), examining regions involved in futility-induced passivity behavior ••[23]. Specifically, they presented visual motion stimuli in either closed-loop or open-loop configuration. The latter configuration in which the visual stimulus speed is not modulated by the behavior caused the fish to enter a phase of “giving up”, ceasing all swim activity. They demonstrate that these glia cells called radial astrocytes serve as key evidence accumulators before zebrafish “give up” swimming when visual feedback is decoupled from behavior. Suggesting an unexpected computational role, these radial astrocytes integrate signals encoding the lack of visual feedback and are both necessary and sufficient to drive this ‘induced passivity’ behavior through a GABA-mediated mechanism (Fig. 2b). This study exemplifies the power of brain-wide imaging followed by causal manipulations in defined cell types. The authors used targeted optogenetic photoactivation and photoinhibition, specific laser ablations, and pharmacology, showcasing how whole-brain imaging with cell type specificity yields novel insights into global and local circuit function. An alternative strategy for establishing such cell-specific associations is to register brain-scale activity maps with multiplexed gene expression profiling (Fig. 2c). Furthermore, the field is prioritizing online brain atlas repositories [31,47,49,50] to dynamically compare, assess and evaluate incoming findings of anatomy, expression profile, and activity (Fig. 2d–e). The remarkable acceleration of sequencing and molecular techniques have also enabled reconstruction of the entire developmental trajectory of every single cell in the zebrafish [51] and evaluation of neuropsychiatric disease related risk genes [52] using CRISPR/Cas9 [53]. Together with electron microscopy reconstructions revealing connectivity [54,55], the ability to target specific molecularly defined cell types represent important tools to dissect the logic of neural circuits underlying behavior.
Figure 2. The importance of diverse cell types across the brain.
(a) Mu et al. ••[23] studied brain wide responses of radial astrocytes (green) and neurons (magenta) while either providing or withholding visual feedback. Dual color whole-brain neuronal and astrocyte Ca2+ imaging (GCaMP6f and jRGECO1b), concurrent with fictive behavior monitoring and visual stimulus delivery, showed dynamic increases in specific cell populations during epochs of futility-induced passivity. While neuronal brain-wide activity was higher during active behavior than passivity, brain-wide glial Ca2+ signals increased before passivity onset and remained elevated during passivity.
(b) In an experimental tour de force combining optogenetics, specific ablations and pharmacology, radial astrocytes are demonstrated to be necessary and sufficient for induction of passivity. Mismatch signals from norepinephrinergic neurons are integrated over time and using an unknown signaling mechanism behaviors are shut down through local GABAergic inhibition.
(c) Barron-Lovett et al. •[46] investigated how fish respond to aversive stimuli, such as high salinity. By using their potent MultiMAP [75] approach, live neural activity volumes are registered with post-fixation and staining volumes. Here, this approach aligns neuropeptidergic identities with functional properties in the preoptic hypothalamus.
(d) Schematic illustrating the remarkable searchable online repository of over 2000 single neurons registered and aligned by Kunst et al. [31]. This represents a strategy to leverage the single-neuron atlas (also used by Kramer et al.) to determine long range projections that are difficult to trace with current photoactivatable fluorescent proteins.
(e) Graphical user interface of Zbrowser [47] with multiview options of Cre and Gal4 transgenic lines. Similar atlases such as the Z-brain, maintained by Engert and colleagues, and important cellular atlases [31] are invaluable tools elevating zebrafish scientific community by enabling fast comparisons of task relevant brain regions and visual exploration of expression profiles.
(f) Serial electron microscopy to reconstruct neurons identified via functional calcium imaging [55], will allow to determine circuit specific connectivity. Here, three views of synaptically connected integrator neurons encoding eye positions. Synapses from the ipsi-only group onto another neuron from the ipsi-only group (top left) (black circles) and from the ipsi-only group onto contra-only neurons (right). Black arrows indicate the location of the synapses, with insets showing the electron micrographs at two representative locations; colors in the insets are representative of the cells to which the synapses belong.
Towards comprehensive behavior quantification underlying whole-brain dynamics
Ultimately, the brain serves to control behavior, and one of the strengths of the zebrafish is the relative ease to quantify their behavior. Ultimately, unscrambling complicated whole-brain networks will require intricate quantitative descriptions of circuit computations transforming sensory input into behavior. To gain precise control of the sensory environment, various virtual reality paradigms are being used [56,57], e.g. to model responses to light gradients [58]. Importantly, high-quality, open-source methods for tracking [59,60] and unsupervised clustering (Fig. 3a–e) of swim kinematics [14] are setting standards for complex behavioral analysis. Comprehensive, large-scale recordings of thousands of swim events allowed Mearns et al. [61] and Johnsen et al. [62] to visualize entire kinematic ethograms and to generate probabilistic models of behavior, revealing structure of behavioral state transitions on short and long timescales. Both studies highlight the importance of past actions and sensory experience, linking behaviors in statistically predictable ways to generate hypotheses for underlying neural dynamics. In another study, using careful behavioral analysis to generate testable predictions of dynamic computation, 3D tracking and modeling of hunting zebrafish and their live prey suggest that zebrafish brain utilize implicit predictive models the environment [63]. With the technical progress to record from large populations of neurons in freely swimming fish connecting quantitative models of behavior and investigating how internal state variables guiding naturalistic behavior emerge over time appears within reach [24]. Recently, Haesemeyer et al. showed that artificial neural networks (ANN) navigating heat gradients can be a powerful tool for understanding neural dynamics [16], where they reported emergence of artificial neurons encoding stimulus derivative over time in the same way as the zebrafish brain [64]. Furthermore, recurrent neural networks fit to neural responses have been used to predict and explain effective connectivity changes within and between brain regions during behavioral state changes ••[22]. As in other animal models [65], convergent ANNs should enable mechanistic studies impossible in biological tissue. Finally, we note the value of comprehensive neural recordings to investigate how cascading spontaneous activity propagates throughout the entire brain volume, phenomena termed ‘neuronal avalanches’ [66]. Such studies inform models predicting how the entire system responds to external or internal state changes.
Figure 3. Large scale behavioral analysis brings the entire ethogram into view.
(a) Zebrafish swim in discrete swim bouts. These elementary movements can be clustered into 13 distinct swim types [14], as shown here as angle of tail segment (°) versus time (ms), with one hundred randomly picked bouts of each type (black). Cyan lines are the average of all bouts in each category. Colored dots next to bout abbreviation correspond to (b). Such careful assessment of behavior is critical to understand how the brain transforms any signals into distinct types of behavioral outputs. These 13 distinct swim types include: approach swims (AS), two distinct slow swims (Slow 1, Slow 2), capture swims (Short CS, Long CS), burst swims (BS), large turns for prey capture (J-turns), high angle turns (HAT), routine turns (RT), spot avoidance turns (SAT), abrupt high angle turns (O-bend), and two remaining escape responses (c-starts) of long-latency (LLC) or short-latency (SLC).
(b) Bout density versus the first two principal components of the movement space used to categorize all bouts. Colors represent bout categories from (a).
(c) Similar bout-type analysis [61] measures the kinematic difference between every pair of bouts with dynamic time warping (DTW) to conclude that behavior structure is largely continuous. Non-linear isomap embedding shows bouts in single behavioral space, with some local densities representing more stereotyped types of forward swimming. Inset: Dimensionality reduction of the first three ‘Eigenfish’.
(d) Bolton et al. [63] used a novel 3D assay for continuous tracking of eye angle, yaw, pitch, and tail angles mapping prey trajectories to fish movement choices. Cartoon shows a fish hunting live prey, paramecia. Prey features are mapped to a spherical coordinate system originating at the fish’s mouth. Angular prey position and velocity construct a future estimate of prey position to guide hunting behavior.
(e) Johnson et al. [62] used large scale behavioral tracking that allowed them to use probabilistic modeling to characterize the likelihood of visual stimuli preceding for each rightward hunting bout type.
(f) Larsch et al. [72] recently showed that motion cues induce social, shoaling-like behavior in juvenile fish (21 days post fertilization), though imaging of associated brain function remains under way. Schematic of a virtual social assay used in the study, whereby each animal sees black dots underneath at the location of the other three animal(s), virtually pairing them.
(g) Huang et al. [57] recently demonstrated how virtual reality rigs can be used to present visual stimulation to zebrafish in realistic 3-D environments, enabling adult virtual reality.
(h) Dragomir et al. [33] (and Bahl et al) have recently employed classic random dot motion paradigms to flexibly investigate evidence integration. Zebrafish offer a strong behavioral model to investigate sensory integration tasks traditionally reserved for primate research.
(i) Schematic of the rotating light-sheet microscope demonstrated by Migault et al. [8], used to monitor the activity of the vestibular system. This microscope includes separate illumination (LU, light-sheet unit) and detection (DU, detection unit) pathways that rotate with the fish.
(j) Delivery of acoustic stimuli delivered via waterproof speakers [39] allows brain-wide imaging of auditory responses.
Future breakthroughs
Overall, we predict that while high-quality, fast functional imaging will become easier, we must continue to develop other necessary tools. There is a critical need to streamline and integrate data collection, storage, processing, annotation, analysis, visualization, and interpretation. Additionally, the field requires user-friendly computational tools for online imaging analysis that would permit interactive experimental design [56], e.g. by acting on information generated during the experiment. Such methods for adaptive experimental design will be necessary for comprehensive functional connectivity mapping using precise optogenetic manipulation [67]. Routine electron microscopy reconstructions [54,55] and effective transsynaptic tracing methods, like cell type specific viruses [68], will be crucial for confirming hypothesized connectivity. Fortunately, efforts to combine functional imaging and complete synaptic EM reconstructions are underway [69]. Extensions of imaging paradigms to older fish [57], as well as other fish species potentially more suitable for studying complex behaviors and brain function as adults [70,71], may be crucial to model circuits not yet developed in the larval zebrafish. These complex behaviors include important new work on shoaling, which opens up research into how socially relevant stimuli are processed in the brain [72], possibly under genetic control [73]. Ultimately, given the size and complexity of whole-brain zebrafish datasets, the field may benefit most proximally from a common interactive framework to share and search multiscale information, from molecular to morphological to functional neural activity data. Initiatives such as neurodata.io, which supports large-scale storage of anatomical and activity data on the cloud, will serve as strong foundations for collaboration and provide useful datasets for developing new analytic tools and machine learning algorithms [74].
Conclusions
The zebrafish is still relatively young as a systems neuroscience model but has practical advantages that have already generated new insights into brain-wide sensorimotor transformations. Investigating brain-scale neural dynamics in zebrafish has become a routine tool for uncovering regional microcircuit level computations and also for discovering how networks collaborate holistically. Achieving realistic, multiscale dynamic models will require fast imaging, precise perturbation technologies, sensitive biosensors and actuators delivered to specific cell types, advanced computational tools for acquisition, analysis, and annotation. Recent whole-brain studies integrate multiple levels of analysis spanning behavior, neuronal dynamics, genetic identity, and connectivity, and these studies establish new principles of interaction between neurons and brain regions across a wide range of behaviors.
Highlights:
Zebrafish brain-scale imaging is routinely used to identify relevant regions.
Many neurons in multiple brain areas cooperate to generate behaviors.
Specific circuit mechanisms underlying various behaviors are being elucidated.
Causal interrogation, dynamic monitoring, & modelling will be necessary.
Computational tools should integrate acquisition, analysis, and data sharing.
Acknowledgements
We are grateful for careful reading and discussions by Timothy W. Dunn and Misha Ahrens. This work is supported by grants from N.I.H. (BRAIN initiative R34-EB026951-01A1, SBIR R44-OD24879-02), and by the Whitehall Foundation.
Footnotes
Conflict of interest statement
Nothing to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Stirman JN, Smith IT, Kudenov MW, Smith SL: Wide field-of-view, multi-region two-photon imaging of neuronal activity in the mammalian brain. Nat Biotechnol 2016, 34:857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, Barbarits B, Lee AK, Anastassiou CA, Andrei A, Aydın Ç, et al. : Fully integrated silicon probes for high-density recording of neural activity. Nature 2017, 551:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stringer C, Pachitariu M, Steinmetz N, Reddy CB, Carandini M, Harris KD: Spontaneous behaviors drive multidimensional, brainwide activity. Science 2019, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Mu Y, Hu Y, Kuan AT, Nikitchenko M, Randlett O, Chen AB, Gavornik JP, Sompolinsky H, Engert F, et al. : Brain-wide Organization of Neuronal Activity and Convergent Sensorimotor Transformations in Larval Zebrafish. Neuron 2018, 100:876–890.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aimon S, Katsuki T, Jia T, Grosenick L, Broxton M, Deisseroth K, Sejnowski TJ, Greenspan RJ: Fast near-whole–brain imaging in adult Drosophila during responses to stimuli and behavior. PLOS Biol 2019, 17:e2006732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato S, Kaplan HS, Schrödel T, Skora S, Lindsay TH, Yemini E, Lockery S, Zimmer M: Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell 2015, 163:656–669. [DOI] [PubMed] [Google Scholar]
- 7.Wan Y, Wei Z, Looger LL, Koyama M, Druckmann S, Keller PJ: Single-Cell Reconstruction of Emerging Population Activity in an Entire Developing Circuit. Cell 2019, 179:355–372.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migault G, van der Plas TL, Trentesaux H, Panier T, Candelier R, Proville R, Englitz B, Debrégeas G, Bormuth V: Whole-Brain Calcium Imaging during Physiological Vestibular Stimulation in Larval Zebrafish. Curr Biol 2018, 28:3723–3735.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ: Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 2013, 10:413–420. [DOI] [PubMed] [Google Scholar]
- 10.Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. : High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 2019, 16:649–657. [DOI] [PubMed] [Google Scholar]
- 11.Marvin JS, Shimoda Y, Magloire V, Leite M, Kawashima T, Jensen TP, Kolb I, Knott EL, Novak O, Podgorski K, et al. : A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat Methods 2019, 16:763–770. [DOI] [PubMed] [Google Scholar]
- 12.Sun F, Zeng J, Jing M, Zhou J, Feng J, Owen SF, Luo Y, Li F, Wang H, Yamaguchi T, et al. : A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 2018, 174:481–496.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, Shuai Y, Huang Y-C, Campagnola L, Seeman SC, Yu J, et al. : Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 2019, 365:699–704. • Genetically encoded voltage indicators hold the promise to faithfully read out action potentials with extreme temporal precision but have been plagued by poor signal-to-noise and short imaging durations. Abdelfattah et al. generated Voltron, a new chemogenetic voltage indicator with effective subthreshold voltage signals and bright and stable enough to record fluorescence changes at the millisecond-timescale resolving single action potentials deep in the zebrafish brain during visually evoked swimming behavior.
- 14.Marques JC, Lackner S, Félix R, Orger MB: Structure of the Zebrafish Locomotor Repertoire Revealed with Unsupervised Behavioral Clustering. Curr Biol CB 2018, 28:181–195.e5. [DOI] [PubMed] [Google Scholar]
- 15.Antinucci P, Dumitrescu A, Deleuze C, Morley HJ, Leung K, Hagley T, Kubo F, Baier H, Bianco IH, Wyart C: A calibrated optogenetic toolbox of stable zebrafish opsin lines. eLife 2020, 9:e54937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haesemeyer M, Schier AF, Engert F: Convergent Temperature Representations in Artificial and Biological Neural Networks. Neuron 2019, 103:1123–1134.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wee CL, Nikitchenko M, Wang W-C, Luks-Morgan SJ, Song E, Gagnon JA, Randlett O, Bianco IH, Lacoste AMB, Glushenkova E, et al. : Zebrafish oxytocin neurons drive nocifensive behavior via brainstem premotor targets. Nat Neurosci 2019, 22:1477–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bahl A, Engert F: Neural circuits for evidence accumulation and decision making in larval zebrafish. Nat Neurosci 2020, 23:94–102. • Published back-to-back with [33], Bahl and Engert analyzed optomotor behavior and brain-scale two-photon calcium fluorescence changes in neurons responding to random dot motion with varied coherence. They find the behavior is closely modeled by a bounded leaky integrator model represented in subpopulations of hindbrain neurons. Demonstrating the power of agnostic searches to home in on functional cell-types that match computational elements in a theoretical model, creating a plausible local circuit model for evidence accumulation.
- 19.Naumann EA, Fitzgerald JE, Dunn TW, Rihel J, Sompolinsky H, Engert F: From Whole-Brain Data to Functional Circuit Models: The Zebrafish Optomotor Response. Cell 2016, 167:947–960.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazemipour A, Novak O, Flickinger D, Marvin JS, Abdelfattah AS, King J, Borden PM, Kim JJ, Al-Abdullatif SH, Deal PE, et al. : Kilohertz frame-rate two-photon tomography. Nat Methods 2019, 16:778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller PJ, Ahrens MB: Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 2015, 85:462–483. [DOI] [PubMed] [Google Scholar]
- 22. Andalman AS, Burns VM, Lovett-Barron M, Broxton M, Poole B, Yang SJ, Grosenick L, Lerner TN, Chen R, Benster T, et al. : Neuronal Dynamics Regulating Brain and Behavioral State Transitions. Cell 2019, 177:970–985.e20. •• Andalman et al. use brain-wide calcium imaging via simultaneous light-field microscopy to identify habenula and raphe nuclei as important brain regions for passive coping mechanisms by zebrafish when shocked repeatedly. To reveal the neural mechanisms underlying adversity evasion they use region-specific optogenetics and powerful, scalable multi-region recurrent neural network modeling to describe the connectivity within and between brain regions.
- 23. Mu Y, Bennett DV, Rubinov M, Narayan S, Yang C-T, Tanimoto M, Mensh BD, Looger LL, Ahrens MB: Glia Accumulate Evidence that Actions Are Futile and Suppress Unsuccessful Behavior. Cell 2019, 178:27–43.e19. •• Mu et al. used dual color light-sheet imaging to survey single-cell dynamics of brain-wide neurons and glia cells involved in state switching in a model of futility-induced passivity. They not only demonstrate the brain-scale dynamical interactions between neurons and glia, they also reveal hindbrain localized noradrenergic neurons that detect swim failures and radial astrocytes as key evidence accumulators. Using a comprehensive sequence of perturbations including optogenetics, laser ablations, and pharmacology, they show that radial astrocytes are both necessary and sufficient to induce this behavioral state change. This study represents compelling evidence that astrocytes participate in information processing in brain circuits.
- 24. Marques JC, Li M, Schaak D, Robson DN, Li JM: Internal state dynamics shape brainwide activity and foraging behaviour. Nature 2019, doi: 10.1038/s41586-019-1858-z. •• This study is exemplary in using novel tracking microscopy to agnostically examine brain-wide activity of zebrafish while hunting for live prey. The authors show that zebrafish spontaneously alternate between two persistent internal states during foraging, revealing opposing oscillatory neural networks encode the exploitation and exploration states, highlighting a serotonergic subpopulation driving the exploitation state.
- 25. Lin Q, Manley J, Helmreich M, Schlumm F, Li JM, Robson DN, Engert F, Schier A, Nöbauer T, Vaziri A: Cerebellar Neurodynamics Predict Decision Timing and Outcome on the Single-Trial Level. Cell 2020, doi: 10.1016/j.cell.2019.12.018. •• This study investigated explicitly how neural activity in different brain regions collaborates to drive behavior. Combining whole-brain volumetric calcium light-sheet imaging with a novel operant-conditioning task in larval zebrafish, they find global, recurring dynamics of brain states emerging from a distributed network with trial-to-trial functional connectivity changes, which correlate with behavior. As part of these network dynamics, cerebellar (especially granule cell activity) is predictive seconds before moving and decisions about locomotion direction is computed bi-hemispherically.
- 26.Zimmermann MJY, Nevala NE, Yoshimatsu T, Osorio D, Nilsson D-E, Berens P, Baden T: Zebrafish Differentially Process Color across Visual Space to Match Natural Scenes. Curr Biol 2018, 28:2018–2032.e5. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, Hinz J, Zhang Y, Thiele TR, Arrenberg AB: Parallel Channels for Motion Feature Extraction in the Pretectum and Tectum of Larval Zebrafish. Cell Rep 2020, 30:442–453.e6. [DOI] [PubMed] [Google Scholar]
- 28.Kist AM, Portugues R: Optomotor Swimming in Larval Zebrafish Is Driven by Global Whole-Field Visual Motion and Local Light-Dark Transitions. Cell Rep 2019, 29:659–670.e3. [DOI] [PubMed] [Google Scholar]
- 29.Yildizoglu T, Riegler C, Fitzgerald JE, Portugues R: A Neural Representation of Naturalistic Motion-Guided Behavior in the Zebrafish Brain. Curr Biol CB 2020, doi: 10.1016/j.cub.2020.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heap LAL, Vanwalleghem G, Thompson AW, Favre-Bulle IA, Scott EK: Luminance Changes Drive Directional Startle through a Thalamic Pathway. Neuron 2018, 99:293–301.e4. [DOI] [PubMed] [Google Scholar]
- 31. Kunst M, Laurell E, Mokayes N, Kramer A, Kubo F, Fernandes AM, Förster D, Dal Maschio M, Baier H: A Cellular-Resolution Atlas of the Larval Zebrafish Brain. Neuron 2019, doi: 10.1016/j.neuron.2019.04.034. • This study is significant as it presents the first single cell, brain-scale inter-areal wiring diagrams with subcellular resolution in the zebrafish. This digital catalogue serves as a computational resource for the zebrafish neuroscience community with high hopes for a future, common, standardized platform combining all atlases and electron microscope reconstructions as resource.
- 32.Kramer A, Wu Y, Baier H, Kubo F: Neuronal Architecture of a Visual Center that Processes Optic Flow. Neuron 2019, doi: 10.1016/j.neuron.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 33. Dragomir EI, Štih V, Portugues R: Evidence accumulation during a sensorimotor decision task revealed by whole-brain imaging. Nat Neurosci 2020, 23:85–93. • Published back-to-back with [18], Dragomir et al. to reveal graded behavioral sensitivity to coherence in the context of classical dot motion paradigms, with stronger optomotor responses for stronger coherence. Integration of visual motion coherence is shown to be a momentary, highly time-dependent, brain-wide process, with the hindbrain caudal interpeduncular nucleus effectively encoding direction-specific turning rates.
- 34.Antinucci P, Folgueira M, Bianco IH: Pretectal neurons control hunting behaviour. eLife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henriques PM, Rahman N, Jackson SE, Bianco IH: Nucleus Isthmi Is Required to Sustain Target Pursuit during Visually Guided Prey-Catching. Curr Biol CB 2019, 29:1771–1786.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebhardt C, Auer TO, Henriques PM, Rajan G, Duroure K, Bianco IH, Del Bene F: An interhemispheric neural circuit allowing binocular integration in the optic tectum. Nat Commun 2019, 10:5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Favre-Bulle IA, Vanwalleghem G, Taylor MA, Rubinsztein-Dunlop H, Scott EK: Cellular-Resolution Imaging of Vestibular Processing across the Larval Zebrafish Brain. Curr Biol 2018, 28:3711–3722.e3. [DOI] [PubMed] [Google Scholar]
- 38.Vanwalleghem G, Schuster K, Taylor MA, Favre-Bulle IA, Scott EK: Brain-Wide Mapping of Water Flow Perception in Zebrafish. J Neurosci Off J Soc Neurosci 2020, 40:4130–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Privat M, Romano SA, Pietri T, Jouary A, Boulanger-Weill J, Elbaz N, Duchemin A, Soares D, Sumbre G: Sensorimotor Transformations in the Zebrafish Auditory System. Curr Biol 2019, 29:4010–4023.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunn TW, Mu Y, Narayan S, Randlett O, Naumann EA, Yang C-T, Schier AF, Freeman J, Engert F, Ahrens MB: Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. eLife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wee CL, Song EY, Johnson RE, Ailani D, Randlett O, Kim J-Y, Nikitchenko M, Bahl A, Yang C-T, Ahrens MB, et al. : A bidirectional network for appetite control in larval zebrafish. eLife 2019, 8:e43775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knogler LD, Kist AM, Portugues R: Motor context dominates output from purkinje cell functional regions during reflexive visuomotor behaviours. eLife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leung LC, Wang GX, Madelaine R, Skariah G, Kawakami K, Deisseroth K, Urban AE, Mourrain P: Neural signatures of sleep in zebrafish. Nature 2019, 571:198. • This study uses brain-wide imaging in zebrafish to identify slow-wave sleep and propagating wave sleep patterns whose neural signatures resemble slow bursting sleep and rapid eye movement sleep in mammals, respectively. The fluorescence-based polysomnography permits the unbiased cellular level examination of many brain regions and identified centrally located ependymal cells which are activated by melanin-concentrating hormone, implicating that it has a sleep-promoting role in fishes that is similar to its role in mammals.
- 44.Reichert S, Pavón Arocas O, Rihel J: The Neuropeptide Galanin Is Required for Homeostatic Rebound Sleep following Increased Neuronal Activity. Neuron 2019, 104:370–384.e5. [DOI] [PubMed] [Google Scholar]
- 45.Oikonomou G, Altermatt M, Zhang R-W, Coughlin GM, Montz C, Gradinaru V, Prober DA: The Serotonergic Raphe Promote Sleep in Zebrafish and Mice. Neuron 2019, 103:686–701.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lovett-Barron M, Chen R, Bradbury S, Andalman AS, Wagle M, Guo S, Deisseroth K: Multiple convergent hypothalamus-brainstem circuits drive defensive behavior. Nat Neurosci 2020, doi: 10.1038/s41593-020-0655-1. • Lovett-Barron et al. use whole-brain imaging to identify brain regions responsive to homeostatic threats, such as high salinity and employ their powerful Multi-MAP technology, capable of aligning molecular and functional cellular profiles to assign neuropeptidergic identities to behaviorally defined neural classes in the hypothalamus. Recruitment of overlapping sets of peptidergic neurons suggest inherent flexibility of genetically defined cell types to form robust response to aversive stimuli.
- 47.Tabor KM, Marquart GD, Hurt C, Smith TS, Geoca AK, Bhandiwad AA, Subedi A, Sinclair JL, Rose HM, Polys NF, et al. : Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. eLife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey S, Shekhar K, Regev A, Schier AF: Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-Cell RNA-Seq. Curr Biol CB 2018, 28:1052–1065.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Randlett O, Wee CL, Naumann EA, Nnaemeka O, Schoppik D, Fitzgerald JE, Portugues R, Lacoste AMB, Riegler C, Engert F, et al. : Whole-brain activity mapping onto a zebrafish brain atlas. Nat Methods 2015, advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hildebrand DGC, Cicconet M, Torres RM, Choi W, Quan TM, Moon J, Wetzel AW, Scott Champion A, Graham BJ, Randlett O, et al. : Whole-brain serial-section electron microscopy in larval zebrafish. Nature 2017, 545:345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF: Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 2018, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thyme SB, Pieper LM, Li EH, Pandey S, Wang Y, Morris NS, Sha C, Choi JW, Herrera KJ, Soucy ER, et al. : Phenotypic Landscape of Schizophrenia-Associated Genes Defines Candidates and Their Shared Functions. Cell 2019, 177:478–491.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albadri S, De Santis F, Di Donato V, Del Bene F: CRISPR/Cas9-Mediated Knockin and Knockout in Zebrafish. In Genome Editing in Neurosciences. Edited by Jaenisch R, Zhang F, Gage F. Springer; 2017. [PubMed] [Google Scholar]
- 54.Svara FN, Kornfeld J, Denk W, Bollmann JH: Volume EM Reconstruction of Spinal Cord Reveals Wiring Specificity in Speed-Related Motor Circuits. Cell Rep 2018, 23:2942–2954. [DOI] [PubMed] [Google Scholar]
- 55.Vishwanathan A, Daie K, Ramirez AD, Lichtman JW, Aksay ERF, Seung HS: Electron Microscopic Reconstruction of Functionally Identified Cells in a Neural Integrator. Curr Biol CB 2017, 27:2137–2147.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vladimirov N, Wang C, Höckendorf B, Pujala A, Tanimoto M, Mu Y, Yang C-T, Wittenbach JD, Freeman J, Preibisch S, et al. : Brain-wide circuit interrogation at the cellular level guided by online analysis of neuronal function. Nat Methods 2018, 15:1117–1125. [DOI] [PubMed] [Google Scholar]
- 57.Huang K-H, Rupprecht P, Frank T, Kawakami K, Bouwmeester T, Friedrich RW: A virtual reality system to analyze neural activity and behavior in adult zebrafish. Nat Methods 2020, 17:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karpenko S, Wolf S, Lafaye J, Le Goc G, Panier T, Bormuth V, Candelier R, Debrégeas G: From behavior to circuit modeling of light-seeking navigation in zebrafish larvae. eLife 2020, 9:e52882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Štih V, Petrucco L, Kist AM, Portugues R: Stytra: An open-source, integrated system for stimulation, tracking and closed-loop behavioral experiments. PLOS Comput Biol 2019, 15:e1006699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romero-Ferrero F, Bergomi MG, Hinz RC, Heras FJH, de Polavieja GG: idtracker.ai: tracking all individuals in small or large collectives of unmarked animals. Nat Methods 2019, 16:179–182. [DOI] [PubMed] [Google Scholar]
- 61.Mearns DS, Donovan JC, Fernandes AM, Semmelhack JL, Baier H: Deconstructing Hunting Behavior Reveals a Tightly Coupled Stimulus-Response Loop. Curr Biol CB 2020, 30:54–69.e9. [DOI] [PubMed] [Google Scholar]
- 62.Johnson RE, Linderman S, Panier T, Wee CL, Song E, Herrera KJ, Miller A, Engert F: Probabilistic Models of Larval Zebrafish Behavior Reveal Structure on Many Scales. Curr Biol 2020, 30:70–82.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolton AD, Haesemeyer M, Jordi J, Schaechtle U, Saad FA, Mansinghka VK, Tenenbaum JB, Engert F: Elements of a stochastic 3D prediction engine in larval zebrafish prey capture. eLife 2019, 8:e51975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haesemeyer M, Robson DN, Li JM, Schier AF, Engert F: A Brain-wide Circuit Model of Heat-Evoked Swimming Behavior in Larval Zebrafish. Neuron 2018, 98:817–831.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kell AJE, Yamins DLK, Shook EN, Norman-Haignere SV, McDermott JH: A Task-Optimized Neural Network Replicates Human Auditory Behavior, Predicts Brain Responses, and Reveals a Cortical Processing Hierarchy. Neuron 2018, 98:630–644.e16. [DOI] [PubMed] [Google Scholar]
- 66.Ponce-Alvarez A, Jouary A, Privat M, Deco G, Sumbre G: Whole-Brain Neuronal Activity Displays Crackling Noise Dynamics. Neuron 2018, 100:1446–1459.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.dal Maschio M, Donovan JC, Helmbrecht TO, Baier H: Linking Neurons to Network Function and Behavior by Two-Photon Holographic Optogenetics and Volumetric Imaging. Neuron 2017, 94:774–789.e5. [DOI] [PubMed] [Google Scholar]
- 68.Ma M, Kler S, Pan YA: Structural Neural Connectivity Analysis in Zebrafish With Restricted Anterograde Transneuronal Viral Labeling and Quantitative Brain Mapping. Front Neural Circuits 2020, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wanner AA, Vishwanathan A: Methods for Mapping Neuronal Activity to Synaptic Connectivity: Lessons From Larval Zebrafish. Front Neural Circuits 2018, 12:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schulze L, Henninger J, Kadobianskyi M, Chaigne T, Faustino AI, Hakiy N, Albadri S, Schuelke M, Maler L, Bene FD, et al. : Transparent Danionella translucida as a genetically tractable vertebrate brain model. Nat Methods 2018, 15:977–983. [DOI] [PubMed] [Google Scholar]
- 71.Chow DM, Sinefeld D, Kolkman KE, Ouzounov DG, Akbari N, Tatarsky R, Bass A, Xu C, Fetcho JR: Deep three-photon imaging of the brain in intact adult zebrafish. Nat Methods 2020, 17:605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larsch J, Baier H: Biological Motion as an Innate Perceptual Mechanism Driving Social Affiliation. Curr Biol CB 2018, 28:3523–3532.e4. [DOI] [PubMed] [Google Scholar]
- 73.Tang W, Davidson JD, Zhang G, Conen KE, Fang J, Serluca F, Li J, Xiong X, Coble M, Tsai T, et al. : Genetic Control of Collective Behavior in Zebrafish. iScience 2020, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vu M-AT, Adalı T, Ba D, Buzsáki G, Carlson D, Heller K, Liston C, Rudin C, Sohal VS, Widge AS, et al. : A Shared Vision for Machine Learning in Neuroscience. J Neurosci 2018, 38:1601–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lovett-Barron M, Andalman AS, Allen WE, Vesuna S, Kauvar I, Burns VM, Deisseroth K: Ancestral Circuits for the Coordinated Modulation of Brain State. Cell 2017, 171:1411–1423.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]