Abstract
The Cauliflower mosaic virus (CaMV) open reading frame VI product (P6) is essential for the viral infection cycle. It controls translation reinitiation of the viral polycistronic RNAs and forms cytoplasmic inclusion bodies (viroplasms) where virus replication and assembly occur. In this study, the mechanism involved in viroplasm formation was investigated by in vitro and in vivo experiments. Far protein gel blot assays using a collection of P6 deletion mutants demonstrated that the N-terminal α-helix of P6 mediates interaction between P6 molecules. Transient expression in tobacco (Nicotiana tabacum) BY-2 cells of full-length P6 and P6 mutants fused to enhanced green fluorescent protein revealed that viroplasms are formed at the periphery of the nucleus and that the N-terminal domain of P6 is an important determinant in this process. Finally, this study led to the unexpected finding that P6 is a nucleocytoplasmic shuttle protein and that its nuclear export is mediated by a Leu-rich sequence that is part of the α-helix domain implicated in viroplasm formation. The discovery that P6 can localize to the nucleus opens new prospects for understanding yet unknown roles of this viral protein in the course of the CaMV infection cycle.
INTRODUCTION
Cauliflower mosaic virus (CaMV) is the type member of the Caulimovirus genus of the Caulimoviridae family. Its double-stranded circular DNA (∼8 kb) is replicated via an RNA intermediate by a virus-encoded reverse transcriptase, and the virus is classified in the pararetrovirus supergroup, to which animal viruses of the Hepadnaviridae family also belong (for a review, see Haas et al., 2002). The CaMV genome possesses six major open reading frames (ORFs I to VI), which are all located on the same DNA strand. Many functions of the corresponding gene products P1 to P6 have been elucidated, but the mechanisms by which they operate during viral infection are not yet fully understood.
The viral DNA is transcribed by the cellular RNA polymerase II into two major capped and polyadenylated RNAs, a monocistronic 19S mRNA and a pregenomic 35S RNA that serves as a template both for reverse transcription and for translation into P1 to P5. The 35S RNA undergoes alternative splicing events leading to four mRNAs in which ORF I and part of ORF II are deleted (Kiss-Laszlo et al., 1995). Currently, the mechanism regulating the nuclear export of 35S RNA and its spliced forms is unknown. These RNAs are translated by the cellular machinery following two unconventional strategies, ribosomal shunt and termination–reinitiation (for a review, see Ryabova et al., 2002).
The CaMV P6 protein (62 kD), which is expressed specifically from the 19S RNA, is a multifunctional protein that represents a key component in the CaMV infectious cycle. P6 is the major determinant of host specificity and influences symptom severity (Daubert et al., 1984; Daubert and Routh, 1990; Agama et al., 2002). Inoculation of cruciferous and solanaceous plant species with chimeric CaMV genomes bearing ORF VI derived from different CaMV isolates showed that the N-terminal region of P6 is responsible for host specificity (Daubert et al., 1984; Schoelz et al., 1986). Transgenic Arabidopsis thaliana plants expressing P6 display disease symptoms whose severity is related to the expression level of the transgene (Zijlstra et al., 1996). Comparison of the cellular mRNA content of ORF VI-transgenic and control Arabidopsis plants revealed that ORF VI expression downregulates or upregulates several host genes (Geri et al., 1999). Whether P6 plays a direct role in regulating expression of these cellular genes (i.e., by regulating their transcription) has not been determined. Finally, P6 from certain CaMV isolates can also act as an avirulence gene product to promote a hypersensitive response in some Nicotiana species (Palanichelvam et al., 2000; Cole et al., 2001).
P6 trans-activates translation of the viral polycistronic 35S RNA and its spliced versions and hence allows synthesis of a complete set of viral proteins (for a review, see Ryabova et al., 2002). Park et al. (2001) demonstrated that P6 is a translational reinitiation factor that associates with the host translational machinery and thus permits translation of downstream ORFs. This function is mediated by physical interactions between an internal region, including the minimal sequence of P6 required for trans-activation (the mini-TAV; De Tapia et al., 1993), the initiation factor eIF3 (Park et al., 2001), and the ribosomal proteins L13 (Bureau et al., 2004), L18 (Leh et al., 2000), and L24 (Park et al., 2001). The observed interaction between P6 and the CaMV capsid protein (P4) also suggests a role for P6 as a scaffolding protein in the assembly of CaMV particles (Himmelbach et al., 1996).
P6 is an abundantly synthesized CaMV protein, and in the cytoplasm of infected cells it forms amorphous, non-membrane-limited, electron-dense inclusion bodies, also referred to as viroplasms. These electron-dense viroplasms are distinct from the electron-lucent inclusion bodies that are mainly composed of CaMV P2 protein (Espinoza et al., 1991; Drucker et al., 2002). The number and the size (2 to 10 μm in diameter) of the electron-dense viroplasms depend on the stage of the viral cycle but also on the CaMV isolate and the host plant (Shalla et al., 1980). Electron-dense viroplasms are a hallmark of infection of plant cells by caulimoviruses and soymoviruses. They probably play an important role in the infectious cycle because they are the sites of protein synthesis, viral genome replication, morphogenesis, and storage of the newly formed virions (Mazzolini et al., 1989; Rothnie et al., 1994). Other CaMV proteins have been detected in the electron-dense viroplasms (Drucker et al., 2002), but none of them seems to be required for their formation because transgenic Arabidopsis plants expressing P6 alone contain inclusion body-like structures (Cecchini et al., 1997). Previous data from several studies suggested that P6 self-associates (Leh, 1999; Haas et al., 2000), and Li and Leisner (2002) showed that multiple domains within P6 can interact with the full-length protein; they proposed that these interactions might be involved in the formation of viroplasms.
In this article, we demonstrate for the first time that P6 localizes both in the cytoplasm and in the nucleus of plant cells and that the N-terminal region of P6 has a dual function. It is a major determinant for in vitro interaction between P6 molecules and mediates the formation of viroplasms in vivo. It also contains a Leu-rich nuclear export signal that prevents accumulation of P6 molecules within the nucleus.
RESULTS
The N-Terminal Region Mediates P6–P6 Interactions in Vitro
In previously described far protein gel blot experiments (Leh, 1999), proteins from healthy and CaMV-infected turnip (Brassica rapa) or Arabidopsis plants were fractionated by SDS-PAGE and transferred to nitrocellulose. Using P6 expressed in Escherichia coli and 32P-radiolabeled in vitro as overlay, a radioactive signal was detected with proteins from infected plants at the level of a polypeptide of 62 kD that also reacted with anti-P6 antibodies, strongly suggesting that P6 interacts with itself. However, because CaMV-P6 protein downregulates or upregulates the expression of several host protein genes (Geri et al., 1999), it could not be totally excluded that the blot-immobilized species interacting with 32P-P6 in the above experiment was a host protein of similar mobility to P6 that was expressed upon viral infection. To rule out this possibility, we have performed an identical far protein gel blot experiment except that the immobilized proteins on the blot were obtained from an extract of E. coli expressing P6. The 32P-P6 in the overlay again reacted with a 62-kD species (Figure 1C, lane P6), which was also recognized by anti-P6 antibodies (Figure 1B, lane P6), providing independent confirmation that P6 can interact with itself. A similar result was obtained using a pull-down assay (data not shown).
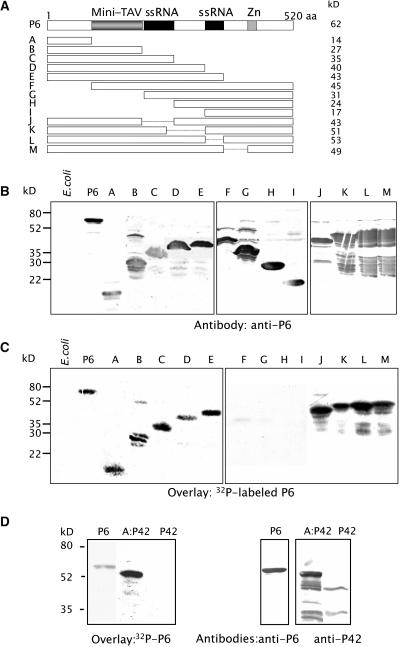
Figure 1.
Mapping of the P6 Domain Involved in P6–P6 Interactions.
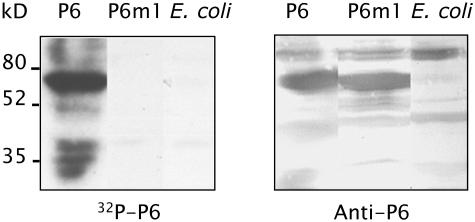
(A) Schematic representation of full-length P6 (520 amino acids [aa]) and P6 deletion mutants used in the far protein gel blot assays. The minimal sequence required for translational transactivation (mini-TAV), the single-stranded RNA binding domains (ssRNA), and the zinc finger motif (Zn) are indicated by dark gray, black, and gray boxes, respectively. The molecular masses of P6 and its deleted versions are indicated to the right.
(B) and (C) Bacterial extracts containing full-length P6 (lane P6), truncated versions of P6 (lanes A to M) or not (lane E. coli) were separated by SDS-PAGE (15%) and transferred onto a nitrocellulose membrane. The membranes were either incubated with antibodies raised against P6 (B) or submitted to a far protein gel blot assay using in vitro 32P-labeled P6 (C).
(D) The 112 N-terminal amino acids of P6 corresponding to mutant A were fused to an unrelated protein, P42 of Beet necrotic yellow vein virus. The fusion protein was expressed in E. coli, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. The membranes were submitted to a far protein gel blot assay using 32P-labeled P6 as a probe (left panel) or to protein gel blotting using specific polyclonal antibodies (right panels). Molecular masses of marker proteins used in the experiments are indicated at the left.
Because P6 contains several domains that can bind single- and/or double-stranded RNA and RNA-DNA heteroduplexes (De Tapia et al., 1993; Cerritelli et al., 1998), far protein gel blot assays were also performed after treatment of both the overlay and the membrane-bound proteins with RNase and DNase. These treatments did not impair formation of the P6-P6 complex, demonstrating that neither RNA nor DNA mediates the P6–P6 interaction and consequently that one or more domains of P6 are directly involved.
To characterize the region(s) required for self-association of P6, we tested the capacity of a series of P6 deletion mutants (Figure 1A) to bind full-length P6. The mutants corresponded to N- and C-terminal recurrent deletions and to P6 bearing internal deletions of previously identified functional domains (i.e., the mini-TAV and RNA binding sites) (De Tapia et al., 1993). The deleted proteins were expressed in E. coli from pET3a recombinant plasmids, separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and incubated in the presence of 32P-labeled P6 in the overlay solution. As shown in Figure 1C, P6 interacted with the C-terminal deletion mutants A, B, C, D, and E and with the internally deleted mutants J, K, L, and M, but not with the N-terminal deletion mutants F, G, H, and I. Because no radioactive signals could be detected with the latter mutants even though they were present in relatively high amounts on the membrane (Figure 1B, lanes F to I), we can rule out the possibility that the signals observed with the other P6 mutants correspond to artifactual binding as a result of gross overloading of the membrane with proteins. The minor signals observed in lanes J to M (Figure 1C) correspond to interactions between the overlay and P6 degradation products rather than to nonspecific interactions with bacteria proteins as demonstrated by the control experiment performed with bacteria transformed with empty vector (Figure 1C, lane E. coli).
The smallest mutant able to interact with P6 was mutant A, which corresponds to the 112 N-terminal amino acids of P6, a region we will refer to as domain A. The latter was fused to the N terminus of a protein from an unrelated virus, P42 of Beet necrotic yellow vein virus, or to the C terminus of glutathione S-transferase (GST) to analyze the ability of domain A to interact with P6 when placed in an unrelated sequence context. P6 bound to A:P42 but not to P42 alone (Figure 1D) and to GST:A but not to GST (see below), thus further demonstrating that domain A can act independently of the rest of the amino acid sequence in mediating P6 self-association in vitro. Taken together, the results of the above far protein gel blot experiments provide evidence that the N-terminal region encompasses the domain required for P6 self-interaction in vitro.
The N Terminus of P6 Is an Essential Determinant for Both the Formation of Viroplasms and Their Localization in the Cytoplasm
To obtain further information about the importance of the N-terminal region of P6 in the formation of inclusion bodies, tobacco (Nicotiana tabacum) BY-2 cells were transfected with recombinant pCK-EGFP plasmids coding for full-length P6 and two deleted versions (A and P6ΔA) fused to the C terminus of the enhanced green fluorescent protein (EGFP). The results described below are representative of at least four independent transfection experiments and observation by confocal laser scanning microscopy (CLSM) at 20 h post-transfection.
After bombardment of BY-2 cells with plasmids expressing the EGFP:P6 fusion protein, ∼80% of transfected cells contained large cytoplasmic inclusion bodies (3 to 5 μm in diameter) with pitted surfaces, generally in the proximity of the nucleus (Figure 2A, panels 1 and 2). The inclusion bodies were formed by numerous smaller aggregates, most of which appeared as hollow donut-like structures (Figure 2A, panel 3). To exclude the possibility that their formation was artifactual (i.e., as a result of the EGFP moiety fused to the P6), protoplasts were prepared from CaMV-infected turnip plants as described by Kobayashi et al. (1998), fixed and immunolabeled with anti-P6 and secondary antibodies coupled to the fluorochrome Alexa 568. Observations by CLSM revealed that the viroplasms thus produced in the context of an authentic viral infection had a similar structure (Figure 2B), demonstrating that the EGFP moiety has no pronounced effect on the self-assembly of EGFP:P6 in tobacco cells. Moreover these results also illustrate that P6 molecules assemble properly and independently of the cellular context because similarly shaped aggregates were formed in cells from host (turnip) and non-host (tobacco) plants.
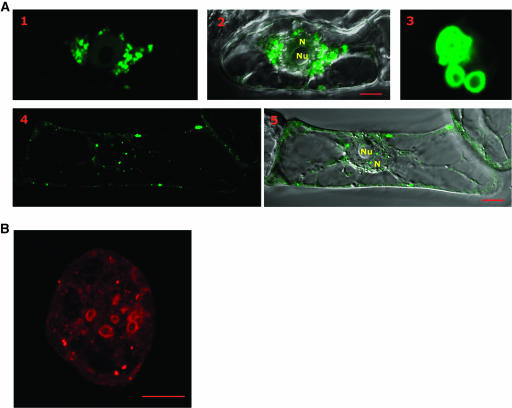
Figure 2.
Subcellular Localization Analysis of P6 Fused to EGFP in Tobacco BY-2 Cells and of P6 in Protoplasts from CaMV-Infected Turnip Plants by CLSM.
(A) Green fluorescent images of EGFP:P6 (images 1 and 4) were taken 20 h after transfection of tobacco cells by bombardment with pCK-EGFP:P6 plasmid. A 0.45-μm-thick optical section was sampled using a single track confocal microscope and appropriate filters. Image 3 corresponds to an enlargement of aggregates similar to those observed in image 1. Images 2 and 5 correspond to the superposition of the fluorescent image and the corresponding differential interference contrast image. Bars = 10 μm. N, nucleus; Nu, nucleolus.
(B) Protoplasts prepared from CaMV-infected turnip leaves were fixed and immunolabeled with rabbit anti-P6 antibodies and mouse anti-rabbit IgG coupled to Alexa 568 as secondary antibody. Shown is the red fluorescent image of a typical protoplast. The confocal images were collected with a focal depth of 0.45 μm. Bar = 10 μm.
Approximately 20% of the tobacco cells expressing the EGFP:P6 fusion protein contained aggregates of variable sizes but <2 μm in diameter (Figure 2A, panels 4 and 5), which probably correspond to early stages of viroplasm formation. The smaller aggregates generally were scattered in the cytoplasm, although they were also sometimes found within the nucleus when the cells were analyzed by CLSM. The presence of such aggregates within the nucleus may indicate that EGFP:P6 molecules were transported to the nucleus (see below) and were then unable to exit after their self-assembly because of the large size of the resulting aggregates.
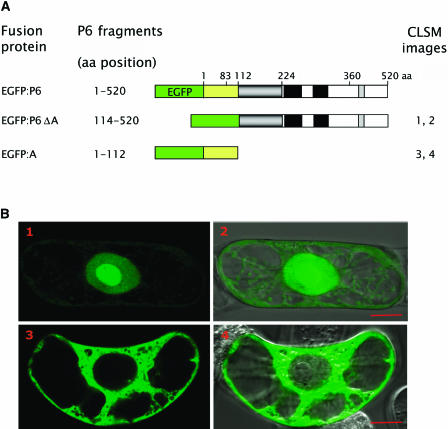
In contrast with EGFP:P6, EGFP:P6ΔA (Figure 3A) did not form aggregates in tobacco cells (Figure 3B, panels 1 and 2) but was mainly found in the nucleus and in particular within the nucleolus. This result strongly suggests that the N-terminal region of P6 is a determinant necessary for the formation of aggregates and that it also contains signal(s) involved in the targeting and/or retention of P6 within the cytoplasm. Similarly to EGFP:P6ΔA, expression of EGFP:A never gave rise to aggregates of any size, but the protein was instead distributed uniformly in the cytoplasm (Figure 3B, panels 3 and 4). The failure of EGFP:A to accumulate in the nucleus to a significant extent was somewhat surprising because it is sufficiently small that it would be expected to be able to traverse nuclear pores by passive diffusion.
Figure 3.
Subcellular Localization of Truncated Versions of EGFP:P6 in Tobacco BY-2 Cells.
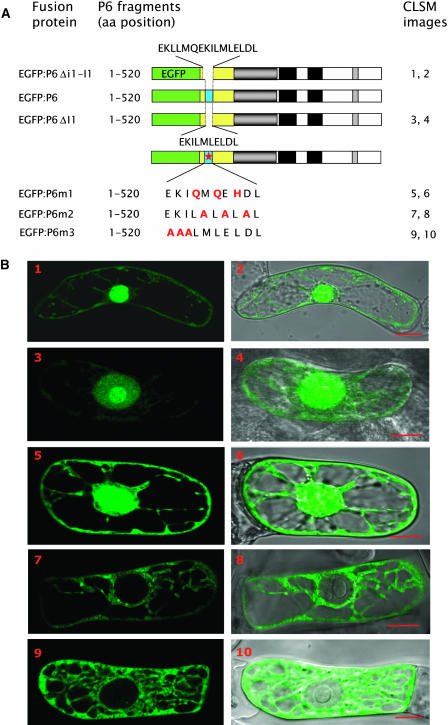
(A) Schematic representation of full-length P6 and of truncated versions fused to the C terminus of EGFP. P6 fragments: the numbers correspond to amino acid positions within the cloned P6 sequence. EGFP is represented by a green box and deleted versions of P6 by an empty box, except domain A, which is yellow (not to scale). The other domains of P6 are represented as in Figure 1. CLSM images: the numbers refer to (B). aa, amino acids.
(B) Subcellular localization of EGFP:P6 mutants (1 to 4) in tobacco BY-2 cells 20 h post-transfection by bombardment. Fluorescence images were collected by CLSM as described in Figure 2. Images 2 and 4 correspond to the superposition of the fluorescent image and the corresponding differential interference contrast image. Bars = 10 μm.
Taken together, these data show that the N-terminal region of P6 is necessary but apparently not sufficient for the formation of viroplasms and, thus, that other region(s) of P6 contribute to this process. Our failure to detect EGFP:A in the nucleus strongly supports the idea that the cytosolic localization of P6 is governed by domain A.
The N-Terminal P6–P6 Interaction Domain Is Conserved among CaMV Strains
Comparison of the N-terminal sequence of P6 from CaMV strain Cabb-B JI with its counterparts from other CaMV strains (Cabb S, Cabb S-Japan, NY8153, CM1841, W260, D/H, D4, B29, Xin/Jin, and Bari) showed identity ranging from 83 to 97%. The region can be divided into two subdomains that we have designated A1 (amino acids 1 to 83) and A2 (amino acids 84 to 112), respectively (Figure 4A). The former is the most conserved (87 to 99% of identity) with two notable invariant sequences: I1 (amino acids 11 to 20) and I2 (amino acids 63 to 83). Note that I1 also contains a pentapeptide motif EKILM (residues 11 to 15) that is identical at four of five positions to the upsteam sequence EKLLM (motif i1; Figure 4B, top). The sequence I1 forms part of a predicted α-helix located near the N terminus (positions 4 to 31) that contains a series of four successive heptad sequences (abcdefg), where residues in positions a and d are hydrophobic as in Leu zipper motifs (Figure 4B). This sequence is predicted by computer analysis (Berger, 1995) to dimerize via a coiled-coil structure with >98% probability. Subdomain A2 is more variable (62 to 93% identity) and does not possess a conserved motif among CaMV strains; it has a predicted β sheet configuration.
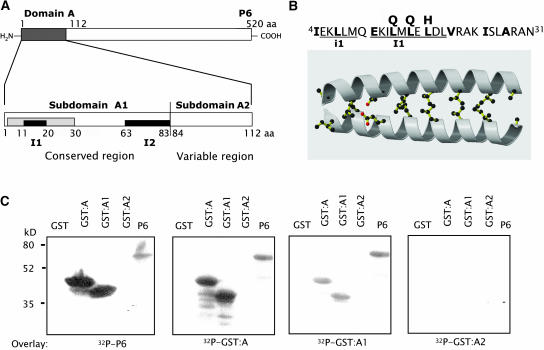
Figure 4.
Fine-Scale Mapping of the P6–P6 Interaction Domain.
(A) Schematic representation of domain A (amino acids 1 to 112). The conserved region can be divided into two subdomains designated A1 (amino acids 1 to 83) and A2 (amino acids 84 to 112). The two invariant sequences (I1 and I2) in domain A are indicated by solid lines and the α-helix by a gray box. aa, amino acids.
(B) Top: the sequence of amino acids 4 to 31 of P6 contains four typical Leu zipper heptad motifs. Hydrophobic residues at heptad positions a and d are in bold. The invariant sequence I1 and the near duplicate sequence i1 are indicated. Leu residues substituted in EGFP:P6m by a Gln (at positions 14 and 16) and by a His (at position 18) are specified. Bottom: a computer-generated model of a parallel coiled-coil structure formed between the N termini of two P6 molecules. The side chains of the residues in positions a and d are shown.
(C) GST:A, GST:A1, and GST:A2 as well as GST and full-length P6 were expressed in E. coli and submitted to far protein gel blot analysis using 32P-labeled P6, GST:A, GST:A1, or GST:A2 as probe in the overlay. The radioactive complexes were detected by autoradiography after an exposure of 24 h. Molecular masses of marker proteins are indicated at the left.
As a first step toward further defining the P6–P6 interaction domain, the sequences coding for subdomains A1 and A2 were amplified by PCR and cloned into the pGEX-2TK vector to produce GST-tagged proteins. The capacity of these fusion proteins to self-interact and to interact with P6 or GST:A were tested by far protein gel blot experiments. Radiolabeled P6 interacted with GST:A and GST:A1 but not with GST:A2 (Figure 4C, left panel), showing that subdomain A1 is responsible for the interaction between P6 molecules. This result was confirmed by the finding that, when either GST:A or GST:A1 was used as overlay, they interacted with P6 and with themselves but not with GST:A2, whereas the latter was unable to bind either to P6 or to any of the fusion proteins (Figure 4C, right panels). Note that none of the fusion proteins interacted with GST, excluding the possibility that the observed interactions were due to dimerization of the tag. Taken together, these results provide evidence that the 83 N-terminal amino acids encompass the domain required for P6 self-interaction in vitro and, hence, probably for the formation of viroplasms.
Mutations in the N-Terminal α-Helix of P6 Affect the Formation of Viroplasms
In view of the fact that subdomain A1 is totally conserved among CaMV strains and is part of the putative Leu zipper–containing α-helix, additional experiments were performed to further investigate its role in viroplasm formation. We removed both the i1 and I1 sequences (amino acids from positions 5 to 20) from P6 (Figure 5A) to see if the reduction of the size of the α-helix impairs the formation of viroplasms. The fluorescence of the corresponding EGFP:P6Δi1-I1 mutant was very abundant and diffuse in the nucleus, whereas only low levels were found in the cytoplasm (Figure 5B, panels 1 and 2). Similar behavior was observed with mutant EGFP:P6ΔI1, in which we deleted only the I1 sequence (Figure 5B, panels 3 and 4). These P6 mutants did not form aggregates, thus reinforcing the hypothesis that the N-terminal α-helix is required for P6 self-assembly. Moreover, these results strongly suggest that the α-helix and/or specific residues of I1 are also implicated in the cytoplasmic localization of P6 because both constructions, EGFP:P6Δi1-I1 and EGFP:P6ΔI1, localized almost totally in the nucleus, in contrast with EGFP:P6 (Figure 2A).
Figure 5.
The N-Terminal α-Helix of P6 Is an Essential Determinant for the Formation of Viroplasms.
(A) Schematic representation of EGFP:P6 and its deleted versions. P6 fragments: the numbers correspond to the amino acid positions within the cloned P6 sequence. EGFP and domain A are represented by green and yellow boxes, respectively (not to scale). The Leu-enriched sequences (i1-I1 and I1) are indicated above EGFP:P6Δi1-I1 and below EGFP:P6ΔI1, respectively. I1 in domain A is represented by a blue box. The empty space and the red star in the blue box indicate the deletion and the point mutations, respectively. The other domains of P6 are represented as in Figure 1. Amino acids mutated in motif I1 of the three EGFP:P6m versions are depicted in red. The other motifs are defined in Figure 1. Legend CLSM images: numbering refers to (B). aa, amino acids.
(B) Subcellular localization of point mutated EGFP:P6 and deleted versions (images 1 to 10). The fluorescent proteins were expressed in BY-2 cells and the fluorescent images (1, 3, 5, 7, and 9) collected by CLSM were superposed on the corresponding differential interference contrast images (2, 4, 6, 8, and 10). Bars = 10 μm.
In summary, the foregoing results strongly suggest that the N-terminal α-helix has structural features important for both the aggregation of P6 and its localization in the cytoplasm. To demonstrate its role in the formation of viroplasm, we mutated three Leu residues of the I1 sequence of P6 (see also below). The Leu residues at positions 14 and 16 were substituted by Gln residues and the Leu at position 18 by a His (Figure 4B, top). Amino acid residues 14 and 18 correspond to position “d” of the second heptad and to position “a” in the third heptad of the Leu zipper. They are predicted to lie on the surface of the α-helix and to be involved in hydrophobic interactions between P6 molecules in the coiled-coil structure (Figure 4B). Leu 16, on the other hand, is not predicted to be surface-located.
First, we tested the capacity of this mutant, named P6m1, to interact with the wild-type P6 by far protein gel blot assay. The protein was expressed in E. coli, fractionated by SDS-PAGE, transferred onto a nitrocellulose membrane, and then incubated with 32P-labeled P6. As shown in Figure 6, P6m1 no longer interacted with P6, suggesting that the putative hydrophobic bonds involving Leu 14 and 18 are crucial for formation of the P6-P6 complex (cf. to lane P6 corresponding to the positive control). We also fused P6m1 to the C terminus of EGFP and expressed it in BY-2 cells. Visualization of EGFP:P6m1 by CLSM revealed that it never produced aggregates but was instead evenly distributed throughout the cytoplasm and the nucleus (Figure 5B, panels 5 and 6) in contrast with EGFP:P6 deleted versions that mainly accumulated within the nucleus (Figure 5B, panels 1 to 4). We also determined by Ala scanning whether other residues of the α-helix are involved in the aggregation of EGFP:P6. In the mutant EGFP:P6m2, the Met, Glu, and Asp residues located between Leu residues of the I1 sequence at positions 15, 17, and 19, respectively, were replaced by Ala residues, and in mutant EGFP:P6m3, the residues of the EKI motif at positions 11 to 13 was substituted by an Ala triplet. In contrast with EGFP:P6m1, when expressed in BY-2 cells both constructions were exclusively found in the cytoplasm; no fluorescence could be detected in the nucleus even after 48 to 72 h incubation of the transfected tobacco cells (Figure 5B, panels 7 to 10). However, EGFP:P6m2 also generated numerous small fluorescent foci that were superimposed on the diffuse fluorescence (Figure 5B, panel 7; see also Figure 8C, panel 5), whereas EGFP:P6m3 was unable to form aggregates.
Figure 6.
Point Mutations in the Invariant Sequence I1 Impair P6–P6 Interactions
Protein extracts of bacteria expressing either wild-type P6 (lane P6), point-mutated P6 (lane P6m1), or an extract of bacteria transformed with empty vector (lane E. coli) were fractionated by SDS-PAGE (15%) and transferred onto a nitrocellulose membrane. The membranes were incubated either with antibodies raised against P6 (right panel) or submitted to a far protein gel blot assay using in vitro 32P-labeled P6 (left panel). Molecular masses of marker proteins are indicated to the left.
Figure 8.
The Leu-Enriched Sequence of Domain A Is Responsible for the Nuclear Export of P6.
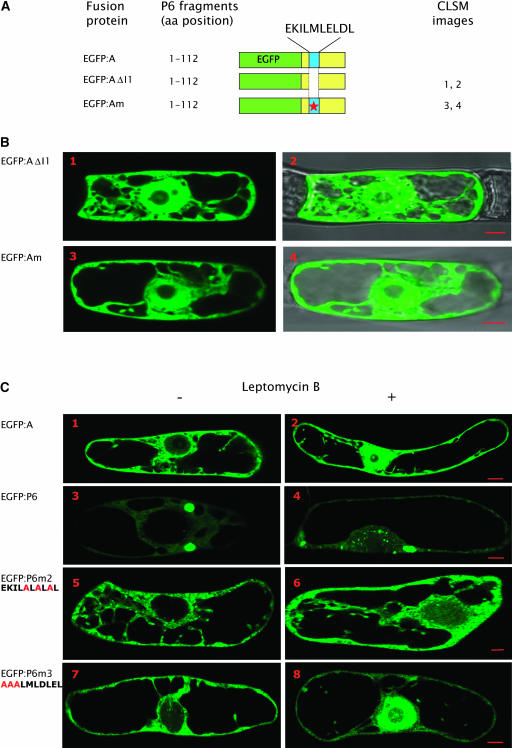
(A) Schematic representation of EGFP:A and mutated versions. EGFP, domain A, and motif I1 of P6 are represented as in Figure 5. The empty space and the red star indicate the deletion and the point mutated motif I1, respectively.
(B) Subcellular localization in BY-2 cells of EGFP:AI1 and EGFP:Am (images 1 to 4). The fluorescent images (1 and 3) observed by CLSM are superposed on the corresponding differential interference contrast images (2 and 4).
(C) Effect of leptomycin B on the subcellular localization of EGFP:A fusion proteins and of wild-type and point mutated forms of EGFP:P6. Fluorescent cells were selected under an HBO lamp, collected in BY-2 cell culture medium containing 100 nM leptomycin B (images to the right) or no leptomycin (images to the left), and incubated 6 h at 24°C with gentle shaking. The point-mutated amino acids within the I1 sequence are indicated. Bars = 10 μm.
Taken together, these results demonstrate that the N-terminal α-helix of P6 is essential for the formation of viroplasms and, furthermore, that its Leu zipper mediates the P6–P6 interaction. Various amino acids of the α-helix, not all of which are located at the interface of the predicted coiled-coil structure (Figure 4B), are important for the aggregation process. If we refer to our model (Figure 4B), Glu from the EKI motif and Leu at positions 14 and 18 are directly involved in the interaction between P6 molecules, thus explaining the inability of EGFP:P6m1 and EGFP:P6m3 to form aggregates. Whether the other residues, in particular those localized between the Leu residues, affect the secondary structure of the N terminus of P6 or the complete protein remains an open question. Indeed, EGFP:P6m2 could still give small aggregates but lost the capacity to assemble into large perinuclear viroplasms.
P6 Is a Nucleocytoplasmic Shuttling Protein
As mentioned at the beginning, small aggregates of EGFP:P6 were found in the nucleus of BY-2 cells (Figure 2, panels 4 and 5) and EGFP:P6ΔA was present mainly in the nucleus (Figure 3B, panels 1 and 2), although the corresponding fusion proteins have approximate molecular masses of 85 and 75 kD, respectively, which are higher than the reported limit for passive diffusion across nuclear pores (Görlich and Kutay, 1999). We first suspected that EGFP:P6 and EGFP:P6ΔA might be cleaved by a cellular protease to produce a species small enough to freely diffuse through the nuclear pores. Such a hypothesis was consistent with the observation that a P6-specific degradation product of 42 kD is frequently found in CaMV-infected plants (Maule et al., 1989) and in heterologous expression systems (Leh et al., 2000). However, protein gel blotting assays performed with proteins from transfected BY-2 cells expressing EGFP:P6 or EGFP:P6ΔA, using antibodies raised against GFP, revealed only polypeptides of 85 and 70 kD, respectively (see supplemental data online), indicating that no significant degradation of the fusion proteins occurred. Consequently, EGFP:P6 and EGFP:P6ΔA are probably transported actively into the nucleus of tobacco cells.
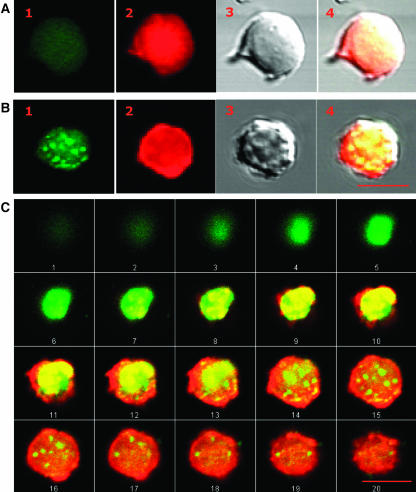
The aforesaid observations raised the question whether full-length P6 might likewise enter the nucleus of host plants during CaMV infection. To answer this question, we prepared protoplasts from systemically CaMV-infected and healthy turnip plants and performed immunodetection of P6 using anti-P6 and secondary antibodies coupled to Alexa 488. Observation of the cytosol and in particular the nucleus by fluorescent microscopy under standard conditions was often hindered by the presence of numerous chloroplasts. Nevertheless, diffuse fluorescence could be visualized within the nucleus but not the nucleolus. Generally, the protoplasts contained numerous P6 aggregates in proximity to the nuclear membrane so that it was difficult to determine whether significant amounts of the immunolabeled P6 were indeed within the interior of the nucleus. Therefore, immunolabeling with anti-P6 antibodies was performed on purified nuclei isolated from healthy and CaMV-infected turnip protoplasts; the nuclei were also stained with propidium iodide (Figures 7A and 7B, panel 2). Nuclei prepared from healthy plants never reacted with anti-P6 antibodies (Figure 7A), whereas ∼50% of those from infected plants were immunolabeled (Figure 7B). P6-Alexa 488 fluorescent foci were observed in the nucleoplasm and/or the nucleolus from infected turnip cells, thus demonstrating that P6 molecules do indeed enter the nucleus during the CaMV infection cycle. The nuclei were often contaminated with P6-containing viroplasms because the latter remained firmly attached to the outer surface even when they were further purified through a composite sucrose/Percoll gradient. Therefore, we realized a series of photographs obtained by CLSM analysis of 0.5-μm-thick sections across purified and P6-immunolabeled nuclei. As shown in Figure 7C, large contaminating viroplasms progressively disappeared from view in successive sections (panels 5 to 14), whereas small P6-labeled aggregates progressively appeared within the organelle (panels 13 to 18), thus providing additional evidence for the presence of P6 within the nucleus. The fluorescent foci might correspond to P6 aggregates and/or to interactions between P6 and specialized nuclear compartments such as speckles (for a review, see Lamond and Spector, 2003) or Cajal bodies (Ochs et al., 1994). The localization of P6 in the nucleolus, observed in some cases, might be related to its capacity to interact with the ribosomal subunits as shown by Park et al. (2001).
Figure 7.
Nuclear Localization of P6.
Nuclei from healthy (A) and CaMV-infected turnip leaves (B) were fixed and immunolabeled with rabbit anti-P6 antibodies and mouse anti-rabbit IgG coupled to Alexa 488 as secondary antibodies (image 1) and stained with propidium iodide (image 2). Panel 3 corresponds to differential interference contrast images and the right-hand images (panel 4) to their superposition with the fluorescent and propidium-stained images. The confocal images were collected with a focal depth of 0.45 μm (C). A series of such optical sections through a nucleus isolated from an infected plant, anti-P6 immunolabeled, and stained with propidium iodide. Bar = 5 μm.
To exclude the possibility that P6 entered the nuclei during their purification (i.e., by diffusion through an altered nuclear envelope), nuclei from healthy protoplasts were incubated during 30 min at 4°C with ∼100 μg of soluble P6 protein expressed in E. coli. The preparation was immunolabeled as describe above. No fluorescence was detectable within the organelles after this treatment, thus reinforcing our conclusion that the P6 protein found in nuclei from infected cells is actively transported there in the course of the CaMV replication cycle.
In conclusion, our data clearly support the idea that P6 can enter the nucleus during viral infection and also indicate that P6 is a nucleocytoplasmic shuttling protein. Furthermore, our findings also strongly suggest that the sequence downstream of domain A is implicated in the nuclear localization of P6, whereas the N-terminal region of P6 might contain a nuclear export signal (NES).
The N Terminus of P6 Contains an NES That Is Recognized by the CRM-1 Nuclear Export Pathway
The latter hypothesis is reinforced by the fact that deletion of the conserved hydrophobic sequence I1 located in subdomain A1 or mutation of the Leu residues at positions 14, 16, and 18 partially abolished nuclear export of P6 (Figure 5B, panels 3 to 6). Moreover, this Leu-rich sequence bears some resemblance to the NES (EKDTLLIDL) found in the BR1 protein of the Squash leaf curl virus, a geminivirus (Ward and Lazarowitz, 1999), and to the NES sequence of several known rapidly shuttling nuclear proteins, such as HIV Rev protein (for a review, see Pollard and Malim, 1998).
To provide further evidence that the aforesaid sequence is an NES, we deleted the sequence I1 or mutated Leu residues 14, 16, and 18 in the EGFP:A fusion protein (Figure 8A) as described above, and the behavior in tobacco BY-2 cells of the mutants was compared with that of the nonmutated protein. These experiments were performed with EGFP:A rather than with the full-length P6 because nuclear accumulation of EGFP:A mutants would be directly relevant to the impairment of the export process, whereas the accumulation of EGFP:P6 mutants in the nucleus does not permit discrimination between an export defect and an active import of the protein. Indeed, the wild-type fusion protein EGFP:A was never found in the nucleus, although it is of a size that should permit it to diffuse freely through the nuclear pore (Figure 3B, panels 3 and 4).
When sequence I1 was removed from domain A, EGFP:AΔI1 was equally distributed in the cytoplasm and the nucleus except for the nucleolus (Figure 8B, panels 1 and 2), whereas EGFP:A localized exclusively to the cytoplasmic compartment (Figure 3B, panels 3 and 4). The subcellular localization in BY-2 cells of mutant EGFP:Am (Figure 8B, panels 3 and 4), in which the three Leu residues of I1 were replaced by polar residues, was similar to that observed with EGFP:AΔI1 (Figure 8B, panels 1 and 2); EGFP:Am was found in both the cytoplasm and in the nucleus but not in the nucleolus. The absence of both EGFP:A mutants in the nucleolus, in contrast with the situation with EGFP:P6ΔI1 and EGFP:P6 point mutated versions (Figure 5), might be due to the fact that the N-terminal region of P6 is unable to interact with ribosomal proteins, whereas the EGFP:P6 mutants still contain the corresponding interaction domains (i.e., mini-TAV and RNA binding domain A) (Leh et al., 2000; Park et al., 2001; Bureau et al., 2004).
All these results support a model in which (1) both EGFP:A and EGFP:A mutants can enter and exit the nucleus by diffusion, (2) EGFP:A molecules but neither EGFP:AΔI1 nor EGFP:Am are rapidly exported to the cytoplasm, and (3) that the sequence I1 functions as an NES because point mutations of Leu residues in I1 impair the export of EGFP:A. Thus, these Leu residues appear to be essential residues for the nuclear export of P6 as already described for nucleocytoplasmic shuttling proteins possessing a Leu-rich NES and in particular for the BR1 protein of Squash leaf curl virus (Ward and Lazarowitz, 1999). However, we cannot totally exclude that the invariant sequence I1 also has properties involved in the retention of fusion protein in the cytosol.
Therefore, the nuclear export of P6 was further investigated, treating BY-2 cells transfected with the aforementioned recombinant plasmids with leptomycin B, which specifically inhibits the CRM-1 pathway involved in the nuclear export of many proteins (Fornerod et al., 1997; Kudo et al., 1998). When BY-2 cells expressing EGFP:A were incubated with 100 nM leptomycin B, 6 h after bombardment, the fluorescent protein accumulated abundantly in the nucleus (Figure 8C, panel 2), whereas no fluorescence was present in this compartment in untreated control cells (Figure 8C, panel 1), thus confirming that EGFP:A molecules are actively exported from the nucleus. The fusion protein EGFP:P6 formed large aggregates in the cytoplasm and was undetectable in the nucleus of transfected tobacco cells (Figure 8C, panel 3), but when the latter were treated with leptomycin B, it was found in both the cytoplasm and in the nucleus (Figure 8C, panel 4). The nuclear compartment contained diffuse fluorescence, accompanied as expected by many small aggregates because P6 was able to self-interact. This result proves that P6 is actively transported between the nucleocytoplasmic compartments. Similar experiments were also performed with point mutated EGFP:P6 versions to determine the functional importance of different residues in the nuclear export of P6. As expected, the mutant EGFP:P6m1 had the same subcellular localization in BY-2 cells treated with leptomycin B (data not shown) or not (Figure 5B, panels 5 and 6) because mutations of the Leu residues are sufficient per se to impair the export of the fusion protein. The fluorescence in the cytoplasm even after leptomycin B treatment suggests that not all EGFP:P6m1 molecules entered the nucleus because they were retained in the cytoplasm or, alternatively, that other residues of the I1 sequence might contribute to the export. In contrast with EGFP:P6m1, leptomycin B had only little effect on the nuclear export of EGFP:P6m2 because almost no fluorescence could be detected within the nucleus (Figure 8C, panels 5 and 6) in most BY-2 cells observed by CLSM; only few cells exhibited a highly fluorescent nucleus. The difference observed in the response to leptomycin B treatment might be due to the fact that (1) the residues mutated in EGFP:P6m2 are indeed essential for export as hypothesized previously or (2) that their mutations modify the conformation of EGFP:P6 in a manner that interferes somehow with the activity of the inhibitor. The EGFP:P6m3 mutant mainly accumulated in the nucleoplasm when the cells were incubated in the presence of leptomycin B, whereas it did not in untreated cells; only low levels of EGFP:P6m3 were found in the cytoplasm of treated cells (Figure 8C, panels 7 and 8). This result suggests that the EKI motif is not part of the NES, or if it is, it has a minor influence on the export of P6 by the CRM-1 pathway.
Together, these data lead to the conclusion that P6 contains at its N terminus an NES and that its Leu residues involved in the nuclear export of P6 are recognized by the CRM-1 pathway. The activity of this export pathway limits the accumulation of P6 in the nucleus that could be deleterious for the CaMV infectious cycle.
DISCUSSION
During the course of a CaMV infection, P6 forms electron-dense viroplasms that are thought to be virion factories that serve as a scaffold for virus replication and assembly. In this study, we have investigated by in vitro and in vivo experimental approaches the mechanism leading to the formation of these viroplasms. Our results show that P6 self-interacts, as already suggested (Haas et al., 2000), and further demonstrate that, at least in vitro, the P6–P6 interactions are exclusively mediated by the N-terminal region encompassing residues 1 to 83 (subdomain A1). No other P6 sequence was able to interact in vitro with full-length P6. The fact that fusion of domain A to unrelated proteins drove this interaction in vitro demonstrates that the N terminus of P6 is sufficient per se to promote self-interaction independently from neighboring sequences. Our results only partially agree with those obtained by Li and Leisner (2002), who used the yeast double hybrid system to show that, in addition to the N terminus, three other domains of P6 are able to bind full-length P6, namely the mini-TAV domain, the downstream adjacent sequence, and the C-terminal region except for a 90-amino-acid-long sequence. Possibly the additional interacting domains characterized in the double hybrid system bind to P6 with an affinity that is too low for detection in far protein gel blot experiments. Incomplete renaturation of the domains in question could also interfere with activity in the in vitro assays. On the other hand, we cannot totally exclude the possibilities that some of the interactions detected in yeast by Li and Leisner (2002) might be mediated by yeast RNA or proteins. Indeed, domains of P6 identified by these authors to be involved in P6–P6 interactions have nucleic acid binding properties and/or interact with cellular proteins, in particular nuclear-localized proteins (Park et al., 2001; Bureau et al., 2004).
The role of subdomain A1 in the formation of viroplasms was investigated by transient expression in tobacco BY-2 cells of full-length P6 and deleted versions of P6 fused to EGFP. Expression of full-length P6 fusion protein gave rise to viroplasms located in the proximity of the nucleus, which are structurally similar to those found in CaMV-infected plants. Indeed, in both cases, the viroplasms appeared by confocal microscopy analysis as aggregates of multiple hollow macromolecular structures, and it is suggested that they might be produced stepwise by the assembly of the donut-like structures that could be visualized in some transfected tobacco cells. Similar hollow structures were also observed when NSP2 and NSP5 proteins, the two major viral components of dense viroplasms induced by rotaviruses, were coexpressed in cultured host cells in the absence of other rotaviral proteins and of rotavirus replication (Fabbretti et al., 1999). We assume that the hollow center of the P6 aggregates corresponds to the electron-lucent holes of viroplasms observed by electron microscopy in CaMV-infected cells (Xiong et al., 1982).
The results obtained by transient expression of truncated versions of P6 clearly demonstrate that, although the 83 N-terminal amino acids of P6 are necessary, they are not sufficient for the formation of viroplasms and that consequently other region(s) of P6 are implicated in this process. The domain between amino acids 289 and 379, referred to as D3 by Li and Leisner (2002), might be one of these sequences because these authors have shown that it plays an important role in P6 self-association when tested in vivo using the yeast two hybrid system. This region of P6 is also engaged in interactions needed for translational transactivation (De Tapia et al., 1993; Park et al., 2001). In any event, our findings suggest that the N-terminally mediated P6–P6 interaction is a prerequisite for further interactions between other P6 sequences and/or for stabilization of macromolecular structures because formation of viroplasms was totally impaired when an N-terminally truncated P6 (P6ΔA) was expressed in tobacco cells.
Computer analysis of subdomain A1 indicated that it forms an amphipathic α-helix at its N terminus (residues 4 to 31). The latter contains a Leu zipper motif that could form a parallel coiled-coil structure (Lupas, 1997), strongly suggesting that such a conformation is implicated in the interaction between P6 molecules. This hypothesis is confirmed by the results of far protein gel blot assays showing that the intermolecular interaction is lost if key hydrophobic amino acids of the Leu zipper motifs are substituted by polar residues. Involvement of such an interaction in the formation of viroplasms is evidenced by the failure of P6 carrying these point mutations in the α-helix to form aggregates in transfected tobacco cells. Currently, we hypothesize that the coiled-coil formation between the N termini of interacting P6 molecules induces conformational changes that allow other regions of P6 to participate in the aggregation process. Indeed, mutations of amino acids that are not located at the coiled-coil interacting surface did not prevent the formation of small aggregates; however, they impaired assembly of these aggregates into viroplasms. It is evident that a further understanding of the mechanism of viroplasm formation will also require characterization of the other domains of P6 implicated in this process as well as possible cellular structures and/or factors that might be involved, such as endomembranes and/or the microtubular network. Nevertheless, no host-specific factors seem to be needed because viroplasms that formed in tobacco cells, a non-host for CaMV, are similar to those found in host cells. As noted earlier, membrane components may function in the early steps of P6 self-assembly, characterized by formation of small aggregates as the latter disappeared upon treatment of tobacco cells with nonionic detergent, whereas the viroplasms located at the nuclear periphery were unaffected (data not shown). Further investigations will be required to determine whether binding to endomembranes is essential for the self-assembly of P6 and whether its hydrophobic N terminus, which presents features of a peptide signal (Blobel and Dobberstein, 1975), plays a role. Because viroplasm formation does not visually perturb the microtubule or the actin filament networks in BY-2 cells, it will be of particular interest to determine whether viroplasm formation involves the cytoskeleton, as described in animal cells for aggresomes and for the viral factories induced by large cytoplasmic DNA viruses such as African swine fever virus (Kopito, 2000; Heath et al., 2001). Johnston et al. (1998) proposed a model in which small aggregates are delivered by retrograde transport along microtubules to the periphery of the nucleus where they are assembled into large structures.
Transient expression of EGFP-tagged P6 led to the unexpected discovery that P6, considered until now as a cytoplasmic protein, is actually a nucleocytoplasmic shuttling protein. The presence of this viral protein within the nucleus was confirmed by its immunodetection in nuclei prepared from CaMV-infected turnip leaves and CLSM-generated optical serial sections through the organelles. Our results, obtained with transfected tobacco cells, indicate that only a small fraction of P6, probably molecules that are not engaged in the aggregation process, enters the nucleus. Indeed, EGFP:P6 mainly formed inclusion bodies in the cytoplasm and was almost undetectable in the nucleus of tobacco cells. Only inhibition of the export process by leptomycin B systematically permitted observation of diffuse EGFP:P6 and small aggregates within the nucleus. Our results also indicate that the export of P6 probably occurs very rapidly in infected cells, so that only low amounts are present in the nucleus at any time. The strong P6 nuclear export activity probably prevents accumulation of P6 within the nucleus, which could be deleterious for CaMV infectivity. In addition, it is more than likely that the nucleocytoplasmic transport is finely regulated during the viral cycle (in other words, that it occurs only at specific stages). These would explain the difficulty we encountered to detect P6 in nuclei from infected plants.
The study of the behavior of P6 mutants in BY-2 cells revealed that the nuclear export activity is associated with the Leu-rich sequence (residues 11 to 20) at the N terminus of P6. Its involvement in nuclear export was demonstrated by the incapacity of P6 to exit the nucleus when Leu residues of the sequence were point mutated and by the fact that mutated domain A of P6 accumulates in the nucleus, in contrast with the wild-type form. The sequence EKI is not implicated in the export of P6 as evidenced by the results of experiments performed with leptomycin B, but it is an important determinant for the formation of viroplasms. Concerning the involvement of the other residues of the I1 invariant sequence, further investigations are necessary to definitively answer this question. Interestingly, the NES is part of the α-helix that is involved in P6 self-assembly and this fact might explain why deletion of the first 90 nucleotides of the CaMV ORF VI abolishes systemic infection and significantly reduces the replication of the genome in single cells (Kobayashi and Hohn, 2003). The overlap between domains involved in P6 export and self-assembly also raises the question of how these two activities are regulated during the viral cycle. We hypothesize that P6 protein shuttling between the nuclear and cytoplasmic compartments primarily involves a population of P6 monomers (or dimers) that have escaped the aggregation process. Recent studies have demonstrated that importins fulfill a dual function as a nuclear import receptor and cytoplasmic chaperone for nuclear imported proteins (Jäkel et al., 2002). Such an antiaggregation mechanism might also be involved for P6 molecules. This does not, however, exclude the possibility that P6 could be incorporated into viroplasms after their export from the nucleus.
The discovery that P6 is a nucleocytoplasmic shuttle protein opens new prospects for understanding the mechanisms by which this viral protein regulates the CaMV infectious cycle. The function(s) of P6 in the nucleus can only be a matter for speculation at present. P6 might have a role similar to the Rev protein of HIV-1 (Pollard and Malim, 1998) in controlling export of CaMV 35S RNA and its spliced versions because it also has the capacity to bind single- and double-stranded RNA (De Tapia et al., 1993; Cerritelli et al., 1998). The presence of P6 in the nucleolus, where assembly of ribosomal subunits occurs, raises the possibility that P6 might interact directly with ribosomes before their export to render them competent for translation of the CaMV polycistronic mRNA. The ribosomal proteins L18 and L24, which interact with the mini-TAV (Leh et al., 2000) and RNA binding domains (Park et al., 2001) of P6, respectively, could be targets for P6 because they participate in the formation of the 60S subunit in the nucleolus (Andersen et al., 2002). Other functions might also be associated with the nucleocytoplasmic localization of P6 (i.e., inhibition of nonsense-mediated mRNA decay to prevent degradation of the 35S RNA and its spliced versions) (for a review, see Maquat and Carmichael, 2001). These hypotheses are supported by the finding that P6 nuclear export is mediated by the CRM-1 pathway (Kudo et al., 1998), which is known to be specifically used for export of the ribosomal subunits and of some cellular mRNAs (for a review, see Weis, 2002).
METHODS
Construction of Recombinant Plasmids
Recombinant plasmids were constructed by insertion of viral sequences into the pET3a derivatives pETKaKS (Leh et al., 2000), pGEX-2TK (Amersham-Pharmacia Biotech, Uppsala, Sweden), and pCK-EGFP (Clontech, Palo Alto, CA). DNA fragments flanked by appropriate restriction cloning sites were generated using PCR; the oligonucleotides used for PCR are listed in Table 1.
Table 1.
Oligonucleotides Used as Primers to Generate the PCR Products Cloned into pGEX-2TK, pCK-EGFP, pETP42, and pETKaKS.6 Vectors
| Name | Sequence | Restriction Site |
|---|---|---|
| A | Cloning of sequences corresponding to CaMV P6 truncated versions | |
| ABam(+) | gccggatccATGGAGAACGAAAAACTC | BamHI |
| AEco(−) | gatgaattcTCATGGAATTCCCTGATGAGG | EcoRI |
| A1Eco(−) | cacgaatccCTAAGCCATCAACGGATTTG | EcoRI |
| A2Bam(+) | ggcggatccTCCAATATCTTGTCAAAAGAT | BamHI |
| A2Eco(−) | cacgaattcCTATTCTGCTCTGAGAGGAGC | EcoRI |
| B | Cloning of sequences corresponding to CaMV P6 protein and its truncated versions | |
| Bsr mut(+) | GACTGGGGTTGTACTAAGG | |
| Bsr mut(−) | CCGCCTTAGTACAACC | |
| P6Bsr(+) | catgtacaagATGGAGAACATAGAAAAACTCATAGTACAACC | BsrGI |
| PBsr(+) | gcatgtacaagCTCAAGATCAGAAGTACTATTC | BsrGI |
| P6ΔABsr(+) | gcatgtacaagATCCCACAAAAATCTGAGCTTAA | BsrGI |
| QBsr(+) | gcatgtacaagCCAATCCCACAAAAATCTGAG | BsrGI |
| P6Xba(−) | gattctagaTCAATCCACTTGCTTTGAAGAC | XbaI |
| AXba(−) | gattctagaTCATGGAATTCCCTGATGAGG | XbaI |
| A1Xba(−) | gattctagaTCAAGCCATCAACGGATTTGT | XbaI |
| BXba(−) | gattctagaTCAGGAGATCTCTTTTGGGGC | XbaI |
| QXba(−) | gattctagaTCAAAATATGTCTTTCTCTGTGTTCTTG | XbaI |
| C | Cloning of the sequence corresponding to the P6 N-terminal domain (amino acids 1 to 112) | |
| ANco(+) | gatccatggATGGAGAACATAGAAAAACTC | NcoI |
| ANco(−) | ctaccatggtAATTCCCTGATGAGGACG | NcoI |
| D | Site-directed mutagenesis | |
| NESmut(+) | AAAATACAAATGCAAGAACACGATCTA | |
| ΔNES(−) | CTCTTGCATGAGGAGTTTTT | |
| NES(+) | GTAAGAGCAAAAATAAGCTTA | |
| Δi1(−) | TATGTTCTCCATCTTGTACAGC |
Forward primer (+) and reverse primer (−). Restriction sites in the primer sequences are represented by bold-faced lower-case letters. PCR products were cloned into pGEX-2TK (A), pCK-EGFP (B and D), pETP42 (C), and pETKaKS.6 (D) vectors.
CaMV ORF VI and its derivatives were cloned either into the KpnI and SacI sites or into the SacI site of the pETKaKS plasmid. Viral DNA sequences were amplified from plasmid pMD324 containing the CaMV Cabb-JI genome (Delseny and Hull, 1983) using two primers bearing at their 5′ termini KpnI and SacI sites, respectively, or SacI sites. The DNA fragments were digested with the appropriate restriction enzymes and introduced into pETKaKS cleaved with KpnI and SacI or with SacI. All constructs were confirmed to be error free by sequencing. Expression of the recombinant plasmids in Escherichia coli generates fusion proteins containing at their N terminus the decapeptide Met-Arg-Arg-Ala-Ser-Val-Gly-Ser-Gly-Thr, which can be phosphorylated in vitro by a protein kinase from bovine heart muscle (the phosphorylation site is in bold-faced type).
The DNA sequence encoding the N terminus of the CaMV P6 protein (nucleotides 1 to 336 of ORF VI) was amplified from the pETKaKS.6 recombinant plasmid encompassing the complete ORF VI using primers carrying an NcoI restriction site at their 5′ end. The PCR fragment was digested with NcoI and cloned into pETP42 (Lauber et al., 1998), which had been cleaved with the same enzyme to produce pET-A:P42. Plasmids pGST:A (ORF VI nucleotides 1 to 336), pGST:A1 (ORF VI nucleotides 1 to 249), and pGST:A2 (ORF VI nucleotides 250 to 336), coding for different regions of the P6 N terminus fused to GST, were obtained by PCR amplification of different ORF VI sequences with primers containing 5′ terminal BamHI and EcoRI sites (Table 1). The amplified DNA fragments were digested with the appropriate restriction endonucleases and cloned into linearized pGEX-2TK.
The pCK-EGFP vector was used to construct the recombinant plasmids coding for fusion proteins between EGFP and wild-type CaMV P6 or P6 mutants. The corresponding ORF VI sequences were amplified by PCR from pMD324 using two primers carrying at their 5′ ends BsrGI and XbaI sites, respectively (Table 1).
Deletions and point mutations were introduced in EGFP:P6 and EGFP:A by site-directed mutagenesis (Stratagene, La Jolla, CA). The recombinant plasmids were amplified by PCR using pfu Turbo polymerase according to the manufacturer's instructions and designed internal oligonucleotides as primers (Table 1). The mixture was then incubated with two units of DpnI for 2 h at 37°C to destroy the template. The 5′ ends of PCR products were phosphorylated by T4 polynucleotide kinase in the presence of 1 mM ATP for subsequent ligation. Error-free recombinant plasmids were identified by DNA sequencing.
Production and Phosphorylation of Recombinant Proteins
E. coli BL21/DE3(pLysS) strain was transformed by electroporation with pETKaKS and pGEX-2TK recombinant plasmids coding for full-length P6 or P6 mutants. Expression of the heterologous proteins was induced with 1 mM isopropyl β-thiogalactoside for 2 h once the bacterial culture had reached the exponential phase. Bacteria were collected by centrifugation at 5000g for 5 min, resuspended in HMK buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 12 mM MgCl2), and lysed by sonication (three pulses for 20 s at 50 W). After centrifugation at 12,000g for 10 min, the supernatant was discarded and the inclusion bodies resuspended in 500 μL of HMK buffer.
Full-length P6 and the P6 fragments A, A1, and A2 expressed from pETKaKS and pGEX-2TK vectors, respectively, were labeled in the presence of [γ-32P]ATP (3000 Ci/mmole) and bovine heart muscle protein kinase (10 units) for 2 h at room temperature, according to the instructions of the manufacturer (Sigma-Aldrich, St. Louis, MO). Excess radioactive ATP was eliminated by filtration through a Sephadex G50 or G25 column (Amersham-Pharmacia Biotech) depending on the molecular mass of the labeled fusion proteins.
Protein Gel Blot Analysis
Proteins from recombinant bacteria were separated by SDS-PAGE and electrophoretically transferred onto a nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany). The membranes were blocked overnight in 5% nonfat dried milk in PBS buffer (140 mM NaCl, 2.7 mM KCl, and 8.1 mM Na2HPO4, pH 7.3) containing 0.1% Tween 20 and then incubated for 4 h at room temperature with specific rabbit or sheep polyclonal antibodies raised against P6 (1:10,000 dilution) or GST (1:5,000 dilution), respectively. The membranes were washed with PBS buffer and treated with goat anti-rabbit IgG antibodies, respectively, conjugated either to alkaline phosphatase or peroxidase, at the dilution recommended by the manufacturer.
Far Protein Gel Blot Assays
A protein blotting overlay technique was used to detect interactions between proteins. Proteins were resolved by SDS-PAGE and transferred onto a nitrocellulose membrane. Membranes were washed several times at 4°C in HM buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 25 mM MgCl2) containing 5% nonfat dried milk and incubated for 12 h at 4°C with gentle shaking in the same buffer containing the [32P]-labeled protein in the overlay. After three washes in HM buffer, the membranes were dried and radioactive complexes were detected by autoradiography.
Transient Expression in Tobacco BY-2 Cells
The CaMV P6 protein and its deleted versions fused to EGFP were transiently expressed in BY-2 tobacco suspension cells (Nicotiana tabacum cv Bright Yellow 2) maintained as described by Banjoko and Trelease (1995). Cells were subcultured each 7 d and harvested 3 d after medium renewal for biolistic transfection. Cells were filtered onto Whatman disks and placed for 2 to 4 h on 0.8% agar MS media plates supplemented with 0.1 M mannitol and 0.1 M sorbitol. Particle preparation and bombardment assays were performed as described by Hunold et al. (1995) with modifications: 2 mg of 1.1 μm tungsten particles (Bio-Rad, Hercules, CA) were immersed in 1 mL of absolute alcohol for 20 min. Dried particles were then successively mixed with 10 μg of recombinant plasmid DNA (pCK-EGFP vector) supplemented with 18% glycerol, 0.75 M CaCl2, and 90 mM spermidine in a final volume of 90 μL. The firing distance was 11 cm and the helium pressure was 7 bars. After bombardment, cells were transferred to 0.8% agar MS media plates and incubated in the dark at 28°C. BY-2 transfected cells were collected under HBO binoculars (excitation/emission wavelength 488/505 to 545 nm) 20 h after bombardment and cultured in MS liquid medium before further treatment and/or CLSM observations.
Virus and Host Plant
Turnips (Brassica rapa cv Just Right F1 hybrid, provided by Takii and Co., Kyoto, Japan) were mechanically inoculated at the four leaf stage (Jacquot et al., 1998) with CaMV Cabb-JI and grown in a greenhouse at 22°C for 5 weeks before preparation of protoplasts from infected leaves.
Isolation of Turnip Nuclei
Protoplasts prepared from CaMV-infected and noninfected turnip leaves (Kobayashi et al., 1998) were used to isolate nuclei. Approximately 8 × 106 protoplasts were washed twice in nuclei buffer (250 mM sucrose, 25 mM Mes, 0.5 mM EDTA, 1 mM MgCl2, 1 mM EGTA, pH 5.5, and a complete cocktail of protease inhibitors [Roche, Indianapolis, IN]). After centrifugation for 5 min at 100g, the protoplasts were resuspended in 50 mL of cold nuclei buffer containing 1 mM DTT, 0.025% Nonidet P-40, and 1 mM phenylmethylsulfonyl fluoride and shaken slowly for 20 min at 4°C. Nuclei were isolated by filtering the suspension through 50-μm mesh nylon and collected at 4°C by centrifugation for 5 min at 550g. They were resuspended in the nuclei buffer and centrifuged at 4°C through a discontinuous gradient composed of 18% Ficoll and 85% Percoll for 15 min at 8000g. The band containing the nuclei, located between the Ficoll and Percoll layers, was diluted threefold with 10 mM Pipes-KOH, pH 7.0, and centrifuged at 600g during 10 min.
Fluorescence Analysis
Fluorescent BY-2 tobacco cells, transfected with EGFP or a protein fused to EGFP, were observed between a slide and cover slip with a Zeiss LSM510 confocal microscope (Jena, Germany). EGFP was viewed by excitation at 488 nm with an argon laser using an appropriate emission filter to collect the green signal from the optical section. Fluorescent cells were also observed under the same conditions after incubation for 8 h at 24°C with gentle shaking in the BY-2 cell culture medium containing 100 nM leptomycin B.
For immunofluorescence studies, protoplasts or nuclei prepared as described above were harvested and fixed for 15 min with gentle shaking in protoplast or nuclei-specific medium, respectively, containing 4% glutaraldehyde. Thereafter, they were washed three times with the appropriate medium, once with the medium diluted volume to volume with PBS, then again with PBS and finally resuspended in PBS buffer. A sample of protoplasts or nuclei was mounted on a poly-l-Lys–coated cover slip, allowed to settle for 1 h at room temperature, and then treated overnight at 4°C in a 0.1% sodium borohydride solution. Protoplasts and nuclei were incubated for 1 h in a blocking solution (5% acetylated BSA [Aurion, Wageningen, The Netherlands], 5% normal goat serum, and 0.1% cold water fish skin gelatin prepared in PBS) and then overnight with the polyclonal anti-P6 antibodies. After six washes with 0.1% BSAc in PBS, protoplasts or nuclei were treated with goat anti-rabbit antibodies coupled to Alexa 488 (Molecular Probes, Eugene, OR), respectively, for 12 h. After removal of excess secondary antibodies by six washes in 0.1% BSAc in PBS, the protoplasts and nuclei were subsequently examined with a Zeiss LSM510 confocal microscope.
Acknowledgments
We thank Marc Bergdoll for the three-dimensional modeling of P6 and John Stanley for providing us with the CaMV Cabb-JI genome sequence. We are most grateful to Christiane Garaud and Jérôme Mutterer for advice on CLSM and to Ken Richards for critical reading of the manuscript. The Inter-Institute Confocal Microscopy Platform was cofinanced by the Région Alsace, Centre National de la Recherche Scientifique, the Université Louis Pasteur, and the Association de la Recherche pour le Cancer. This work was supported by the Centre National de la Recherche Scientifique and by the Université Louis Pasteur of Strasbourg.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mario Keller (mario.keller@ibmp-ulp.u-strasbg.fr).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.029017.
References
- Agama, K., Beach, S., Schoelz, J., and Leisner, S.M. (2002). The 5′ third of Cauliflower mosaic virus Gene VI conditions resistance breakage in Arabidopsis ecotype Tsu-0. Virology 92, 190–196. [DOI] [PubMed] [Google Scholar]
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K.L., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Banjoko, A., and Trelease, R.N. (1995). Development and application of an in vivo plant peroxisome import system. Plant Physiol. 107, 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, B. (1995). Algorithms for protein structural motif recognition. J. Comput. Biol. 2, 125–138. [DOI] [PubMed] [Google Scholar]
- Blobel, G., and Dobberstein, B. (1975). Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67, 835–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, M., Leh, V., Haas, M., Geldreich, A., Ryabova, L., Yot, P., and Keller, M. (2004). P6 protein of Cauliflower mosaic virus, a translational reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. J. Gen. Virol. 85, 3765–3775. [DOI] [PubMed] [Google Scholar]
- Cecchini, E., Gong, Z., Geri, C., Covey, S.N., and Milner, J.J. (1997). Transgenic Arabidopsis lines expressing gene VI from Cauliflower mosaic virus variants exhibit a range of symptom-like phenotypes and accumulate inclusion bodies. Mol. Plant Microbe Interact. 10, 1094–1120. [DOI] [PubMed] [Google Scholar]
- Cerritelli, S., Fedoroff, O., Reid, B., and Crouch, R. (1998). A common 40 amino acid motif in eukaryotic RNases H1 and caulimovirus ORF VI proteins binds to duplex RNAs. Nucleic Acids Res. 26, 1834–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, A.B., Kiraly, L., Ross, K., and Schoelz, J.E. (2001). Uncoupling resistance from cell death in the hypersensitive response of Nicotiana species to Cauliflower mosaic virus infection. Mol. Plant Microbe Interact. 14, 31–41. [DOI] [PubMed] [Google Scholar]
- Daubert, S.D., and Routh, G. (1990). Point mutations in Cauliflower mosaic virus gene VI confer host-specific symptom changes. Mol. Plant Microbe Interact. 3, 341–345. [DOI] [PubMed] [Google Scholar]
- Daubert, S.D., Schoelz, J., Debao, L., and Shepherd, R.J. (1984). Expression of disease symptoms in Cauliflower mosaic virus genomic hybrids. J. Mol. Appl. Genet. 2, 537–547. [PubMed] [Google Scholar]
- Delseny, M., and Hull, R. (1983). Isolation and characterization of faithful altered clones of the genomes of Cauliflower mosaic virus isolates Cabb B-JI, CM4-184 and Bari. Plasmid 9, 31–41. [DOI] [PubMed] [Google Scholar]
- De Tapia, M., Himmelbach, A., and Hohn, T. (1993). Molecular dissection of the Cauliflower mosaic virus translation transactivator. EMBO J. 12, 3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker, M., Froissart, R., Hébrard, E., Uzest, M., Ravallec, M., Espérandieu, P., Mani, J.-L., Pugnière, M., Roquet, F., Fereres, A., and Blanc, S. (2002). Intracellular distribution of viral gene products regulates a complex mechanism of Cauliflower mosaic virus acquisition by its aphid vector. Proc. Natl. Acad. Sci. USA 99, 2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza, A.M., Medina, V., Hull, R., and Markham, P.G. (1991). Cauliflower mosaic virus gene II products forms distinct inclusion bodies in infected plant cells. Virology 185, 337–344. [DOI] [PubMed] [Google Scholar]
- Fabbretti, E., Afrikanova, I., Vascotto, F., and Burrone, O.R. (1999). Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80, 333–339. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I.W. (1997). CRM1 is an export receptor for leucine-rich nuclear exports signals. Cell 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Geri, C., Cecchini, E., Giannakou, M., Covey, S., and Milner, J. (1999). Altered patterns of gene expression in Arabidopsis elicited by Cauliflower mosaic virus (CaMV) infection and by a CaMV gene VI transgene. Mol. Plant Microbe Interact. 12, 377–384. [DOI] [PubMed] [Google Scholar]
- Görlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Haas, M., Bureau, M., Geldreich, A., Yot, P., and Keller, M. (2002). Cauliflower mosaic virus: Still in the news. Mol. Plant Pathol. 3, 419–429. [DOI] [PubMed] [Google Scholar]
- Haas, M., Leh, V., Yot, P., and Keller, M. (2000). Caractérisation du domaine de la protéine P6 du Virus de la mosaïque du chou-fleur impliqué dans la formation des viroplasmes. Virologie 4, 165. [Google Scholar]
- Heath, C.M., Windsor, M., and Wileman, T. (2001). Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A., Chapdelaine, Y., and Hohn, T. (1996). Interaction between Cauliflower mosaic virus inclusion body protein and capsid protein: Implication for viral assembly. Virology 217, 147–157. [DOI] [PubMed] [Google Scholar]
- Hunold, R., Burrus, M., Bronner, R., Duret, J.P., and Hahne, G. (1995). Transient gene expression in sunflower (Helianthus annuus L.) following microprojectile bombardment. Plant Sci. 105, 95–109. [Google Scholar]
- Jacquot, E., Geldreich, A., Keller, M., and Yot, P. (1998). Mapping regions of the Cauliflower mosaic virus ORF III product required for infectivity. Virology 242, 395–402. [DOI] [PubMed] [Google Scholar]
- Jäkel, S., Mingot, J.-M., Schwarzmaier, P., Hartmann, E., and Görlich, D. (2002). Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, J.A., Ward, C.L., and Kopito, R.R. (1998). Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Blanc, S., and Hohn, T. (1995). Splicing of Cauliflower mosaic virus 35S RNA is essential for viral infectivity. EMBO J. 14, 3552–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, K., and Hohn, T. (2003). Dissection of cauliflower mosaic virus transactivator/viroplasmin reveals distinct essential functions in basic virus replication. J. Virol. 77, 8577–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, K., Nakayashiki, H., Tsuge, S., Mise, K., and Furusawa, I. (1998). Accumulation kinetics of viral gene products in Cauliflower mosaic virus-infected turnip protoplast. Microbiol. Immunol. 42, 65–69. [DOI] [PubMed] [Google Scholar]
- Kopito, R.R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E.P., Yoneda, Y., Yanagida, M., Horinouchi, S., and Yoshida, M. (1998). Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 1, 540–547. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and Spector, D.L. (2003). Nuclear speckles: A model for nuclear organelles. Mol. Cell. Biol. 4, 605–611. [DOI] [PubMed] [Google Scholar]
- Lauber, E., Bleykasten-Grosshans, C., Erhardt, M., Bouzoubaa, S., Jonard, G., Richards, K., and Guilley, H. (1998). Cell-to-cell movement of Beet necrotic yellow vein virus: I. Heterologous complementation experiments provide evidence for specific interactions among the triple gene block proteins. Mol. Plant Microbe Interact. 11, 618–625. [DOI] [PubMed] [Google Scholar]
- Leh, V. (1999). Etude des Interactions entre Protéines Engageant les Produits des ORFIII et ORFVI du Virus de la Mosaïque du Chou-Fleur. PhD dissertation (Strasbourg, France: Université Louis Pasteur).
- Leh, V., Yot, P., and Keller, M. (2000). The Cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology 266, 1–7. [DOI] [PubMed] [Google Scholar]
- Li, Y., and Leisner, S.C. (2002). Multiple domains within the Cauliflower mosaic virus gene VI product interact with the full-length protein. Mol. Plant Microbe Interact. 15, 1050–1057. [DOI] [PubMed] [Google Scholar]
- Lupas, A. (1997). Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 7, 388–393. [DOI] [PubMed] [Google Scholar]
- Maquat, L., and Carmichael, G. (2001). Quality control of mRNA function. Cell 104, 173–176. [DOI] [PubMed] [Google Scholar]
- Maule, A.J., Harker, C.L., and Wilson, I.G. (1989). The pattern of accumulation of Cauliflower mosaic virus specific products in infected turnips. Virology 169, 436–446. [DOI] [PubMed] [Google Scholar]
- Mazzolini, L., Dabos, P., Constantin, S., and Yot, P. (1989). Further evidence that viroplasms are the site of Cauliflower mosaic virus genome replication by reverse transcription during viral infection. J. Gen. Virol. 70, 3439–3449. [Google Scholar]
- Ochs, R.L., Stein, T.W., and Tan, E.M.M. (1994). Coiled bodies in the nucleolus of breast cancer cells. J. Cell Sci. 107, 385–399. [DOI] [PubMed] [Google Scholar]
- Palanichelvam, K., Cole, A.B., Shababi, M., and Schoelz, J.E. (2000). Agroinfiltration of Cauliflower mosaic virus gene VI elicits hypersensitive response in Nicotiana species. Mol. Plant Microbe Interact. 13, 1275–1279. [DOI] [PubMed] [Google Scholar]
- Park, H.-S., Himmelbach, A., Browning, K.S., Hohn, T., and Ryabova, L.A. (2001). A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106, 723–733. [DOI] [PubMed] [Google Scholar]
- Pollard, V.W., and Malim, M.H. (1998). The HIV-1 Rev protein. Annu. Rev. Microbiol. 52, 491–532. [DOI] [PubMed] [Google Scholar]
- Rothnie, H., Chapdelaine, Y., and Hohn, T. (1994). Pararetroviruses and retroviruses: A comparative review of viral structure and gene expression strategies. Adv. Virus Res. 44, 1–67. [DOI] [PubMed] [Google Scholar]
- Ryabova, L.A., Pooggin, M.M., and Hohn, T. (2002). Viral strategies of translation initiation: Ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol 272, 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoelz, J.E., Shepherd, R.J., and Daubert, S.D. (1986). Gene VI of CaMV encodes a host range determinant. Mol. Cell. Biol. 6, 2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalla, T.A., Shepherd, R.J., and Petersen, L.J. (1980). Comparative cytology of nine isolates of Cauliflower mosaic virus. Virology 102, 381–388. [DOI] [PubMed] [Google Scholar]
- Ward, B.M., and Lazarowitz, S.G. (1999). Nuclear export in plants. Use of geminivirus movement proteins for a cell-based export assay. Plant Cell 11, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, K. (2002). Nucleocytoplasmic transport: Cargo trafficking across the border. Curr. Opin. Cell Biol. 14, 328–335. [DOI] [PubMed] [Google Scholar]
- Xiong, C., Balàzs, E., Lebeurier, G., Hindenlang, C., Stoeckel, M.E., and Porte, A. (1982). Comparative cytology of two isolates of Cauliflower mosaic virus. J. Gen. Virol. 61, 75–81. [Google Scholar]
- Zijlstra, C., Schärer-Hernandez, N., Gal, S., and Hohn, T. (1996). Arabidopsis thaliana expressing the Cauliflower mosaic virus ORF VI transgene has a late flowering phenotype. Virus Genes 13, 5–17. [DOI] [PubMed] [Google Scholar]