Abstract
A 38-kDa major outer membrane protein (OMP) was isolated from the nitrogen-fixing enterobacterium Rahnella aquatilis CF3. This protein exists as a stable trimer in the presence of 2% sodium dodecyl sulfate at temperatures below 60°C. Single channel experiments showed that this major OMP of R. aquatilis CF3 is able to form pores in the planar lipid membrane. Two oligonucleotides encoding the N-terminal portion of the 38-kDa OMP and C-terminal portion of OmpC were used to amplify the 38-kDa gene by PCR. The deduced amino acid sequence showed a strong homology with Escherichia coli, Klebsiella pneumoniae, Salmonella typhi, and Serratia marcescens OmpC sequences, except loops L6 and L7, which are postulated to be cell surface exposed. On the basis of the OmpF-PhoE three-dimensional structure, it seems likely that this 38-kDa organizes three 16-strand β-barrel subunits. The relationship between the structure and the double functionality of this protein as porin and as a root adhesin is discussed.
Rahnella aquatilis is a gram-negative enteric bacterium. It was isolated first from drinking and river water (15) and subsequently from human clinical specimens (27) and from the rhizospheres of different plants (5).
R. aquatilis CF3 appears to lack fimbriae which could mediate the adhesive mechanism of other bacteria such as Klebsiella sp. (18). Since the R. aquatilis major outer membrane protein (OMP), which has an apparent molecular mass of 38 kDa, was shown to be involved in the adhesion of this organism to wheat roots (1), we consider OMPs to be important in the interaction between this R. aquatilis strain and roots of its host plant. The previously determined N-terminal amino acid sequence (1) indicates that this protein could be related to the enterobacterial porin family. These major OMPs are organized in a trimeric structure and are usually found in gram-negative bacteria (25). They form three water-filled channels that allow diffusion of small nutrients through the outer membrane (24). Porins might also be involved in other functions, such as those described during the invasion of epithelial cells by Salmonella typhimurium (9) and Shigella flexneri (6).
We had previously reported that the N-terminal sequence of the major OMP (38 kDa) of R. aquatilis CF3 showed strong homology with enterobacterial porins (1). As this protein seems to be involved in the adhesion of R. aquatilis to plant roots (1), we sought to characterize it.
MATERIALS AND METHODS
Bacterial strains.
Rahnella aquatilis CF3, a nitrogen-fixing enterobacterium isolated from the rhizosphere of wheat (5), and Escherichia coli HB101 (7) were used in this study.
Isolation of OMPs.
Bacteria were grown at 28°C in Luria-Bertani medium and harvested during the stationary phase. OMPs were isolated as described by Hurlbert and Gross (13). Bacterial cells were harvested by centrifugation, resuspended in distilled water (5 ml g of cells−1), and then sonicated three times for 30 s (100 W) at 4°C. Cell debris and unbroken cells were removed by low-speed centrifugation (5,000 × g) for 10 min. Subsequently, cell envelopes were pelleted at 48,000 × g for 60 min at 4°C and extracted with N-lauroyl sarcosinate (final concentration, 1%) in 10 mM Tris-HCl (pH 7.5) for 30 min at 28°C. After centrifugation at 48,000 × g for 60 min at 4°C, the final membrane pellet was suspended in water (1 ml g−1 [fresh weight]) and stored at −80°C. Protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide, 0.26% bisacrylamide) in the presence of 4 M urea.
Solubilization of the major OMP.
The OMP preparation was suspended in 0.5% Zwittergent 3-16 (Zw 3-16; Calbiochem) solution in 10 mM Tris-HCl buffer, pH 8.0, and incubated for 1 h at 37°C. The nonsolubilized material was removed by centrifugation at 50,000 × g for 1 h at 4°C. The supernatant containing the major OMP was collected and stored at −80°C.
The denaturing temperature effect on protein solubilization was analyzed by SDS-PAGE. Gels were stained as described by Neuhoff et al. (23).
Reconstitution in planar lipid bilayers.
The phospholipids 1-palmitoyl-2-oleoylphosphocholine (POPC) and dioleoyl-phosphatidyl ethanolamine (DOPE) were from Avanti Polar Lipids (Birmingham, Ala.). From a POPC-DOPE mixture (7:3) in hexane (0.5%), virtually solvent-free planar lipid bilayers were formed by the apposition of two monolayers (22) on a 150-μm-diameter hole in a thin piece of Teflon film (thickness, 10 μm) sandwiched between two half glass and pretreated with hexadecane-hexane (1:40, vol/vol). The electrolyte solution was 1 or 0.1 M NaCl buffered with 10 mM Tris, pH 7.4. The 0.1-to-1-M gradient was used for measuring ion selectivity. Bilayer formation was monitored by the capacitance response prior to protein addition. The bulk concentration of reincorporated protein was about 500 μg ml−1. The current fluctuations were recorded through a bilayer amplifier (Model 1200; Biologic) and stored on a DTR 2100 apparatus (Biologic). The stored signals were transferred to a computer for analysis (current traces and amplitude histograms) by using software from Intracell (Royston, United Kingdom).
Synthesis of oligonucleotides and PCR.
The following oligonucleotide primers corresponding to the 38-kDa N terminus (DGNKLD) (1) and to the enterobacterial OmpC C terminus (GLVYQF) were synthesized (Eurogentec): forward, 5′-GACGGTAATAAACTCGAT-3′, and reverse, 5′-GAACTGRTANACCAGACC-3′. PCR amplification was done with an automated PCR thermoblock (Hybaid).
PCR product direct sequencing.
The 1-kbp PCR product was purified on a 1% low-melting-point agarose gel. It was directly sequenced by using a protocol described by Anderson et al. (4). Four primers were used in the sequencing reaction mixture (sequences obtained with two primers described below allowed us to determine two others). Sequence analysis was done with the Genomyx system (Beckman). Sequence data were assembled and analyzed by using the PC Gene package (IntelliGenetics, Mountain View, Calif.). For data searches, FASTA (26) and BLASTP (3) programs were used.
Nucleotide sequence accession number.
The nucleotide sequence of the gene encoding the 38-kDa R. aquatilis OMP has been deposited in the EMBL database under accession no. AJ002879.
RESULTS
Solubilization of the major OMP of R. aquatilis.
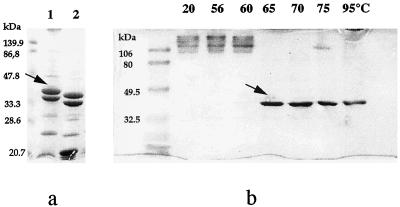
SDS-PAGE analysis of R. aquatilis OMPs showed a pattern quite similar to that of E. coli, with a slight difference in the migration of two major OMPs (apparent molecular weights of approximately 38,000 and 35,000 for R. aquatilis CF3, and 36,000 and 33,000 for E. coli HB101) (Fig. 1a).
FIG. 1.
Coomassie blue-stained gels, resolved by SDS-PAGE, of OMPs of R. aquatilis CF3 (lane 1) and E. coli HB101 (lane 2) (a) and Zw 3-16 partially purified 38-kDa OMP solubilized at various temperatures (b). Solubilization temperatures, from left to right, were 20, 37, 55, 65, 70, 80, and 96°C. For both panels, arrows show the position of the 38-kDa monomer and molecular mass position markers are given on the left.
The 38-kDa protein was solubilized in the presence of 0.5% Zw 3-16, after a 1-h incubation at 37°C. The Zw 3-16-solubilized 38-kDa protein of R. aquatilis CF3 contained only negligible amounts of contaminants. The native 38-kDa protein forms a stable trimer (∼100 kDa), even in the presence of 2% SDS, at temperatures below 65°C (Fig. 1b). Above 65°C, only the monomeric form was detected. This suggests that the R. aquatilis porin is more thermally unstable than the E. coli OmpF porin, which requires temperatures of >75°C to dissociate (11).
Channel-forming properties.
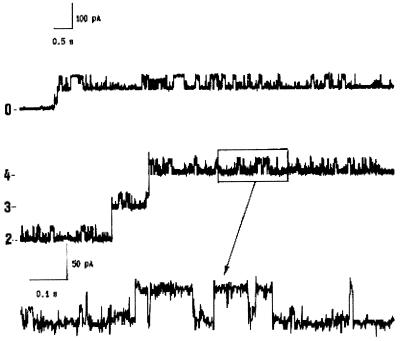
The reincorporation of purified 38-kDa protein in POPC-DOPE (7:3) bilayers (1 M NaCl) induced long-duration steps (conductance, 2,400 pS) after application of a 55-mV potential (Fig. 2), and on the application of these steps, smaller and faster fluctuations were observed. Upon scale enlargement, the quickly fluctuating levels appeared well defined and were estimated to have a conductance of 800 pS.
FIG. 2.
Single-channel current records of POPC-DOPE bilayers formed in 1 M NaCl at 25°C in the presence of 0.5 μg of the 38-kDa protein of R. aquatilis per ml. Data were recorded at 2,000 Hz with a 300-Hz filter. The applied membrane potential was 55 mV. The lower trace shows an expanded recording of the box marked on the plot above it.
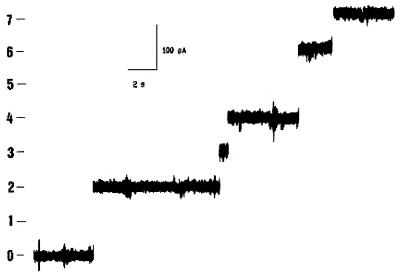
When the protein concentration was increased, the 800-pS conductance states disappeared and the current increased in a stepwise fashion (Fig. 3), with conductance values of 2,400 and 4,800 pS for a 30-mV potential. Transmembrane voltages higher than 130 mV did not cause channel closing.
FIG. 3.
Single-channel records of porin under same conditions as those described for Fig. 2, but with a protein concentration of 1 μg/ml. The trace was recorded at 500 Hz and nonfiltered after 30 min of standby. The applied potential was 30 mV.
In asymmetrical NaCl media (0.1-to-1-M gradient) the reversal potential was 20 mV after the junction potential correction due to the gradient. This observed shift allows us to estimate, from the application of the Hodgkin-Goldman-Katz equation, the PNa/PCl ratio to be about 2.6 (12).
Identification of an ompC-like gene in R. aquatilis.
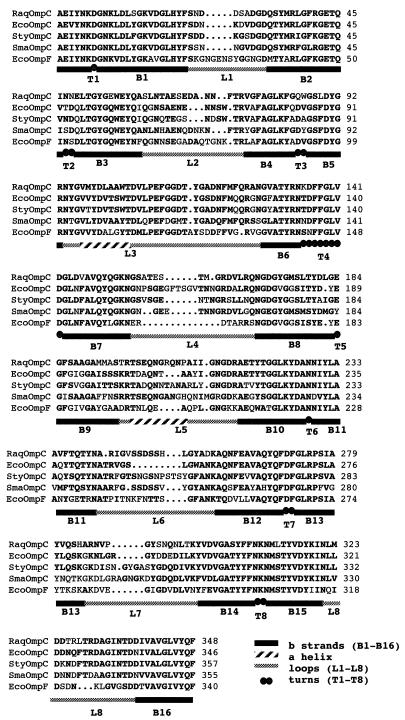
Using primers corresponding to the 38-kDa protein N terminus (DGNKLD) and to the enterobacterial OmpC C terminus (GLVYQF), we amplified a 1-kbp fragment from total R. aquatilis DNA and directly sequenced it. The deduced protein sequence was used to conduct a comparison with sequences available in the GenBank database. The sequences were aligned by using the Clustal program and then manipulated manually on SeqApp to provide the best alignments. When aligned with the amino acid sequence of other enterobacterial porins, the sequence of the 38-kDa protein most closely resembles OmpC. The highest homologies, 66.8, 66.5, and 65.4% were obtained with E. coli, Salmonella typhi, and Serratia marcescens OmpC porin sequences, respectively (Fig. 4), and fitted in with the predicted β-strands. Homology was only 54% with E. coli OmpF. The loop alignment also showed that there is conservation between loop 6 of the R. aquatilis 38-kDa protein and the corresponding loop in S. marcescens (14). Postulated loop 7 is characterized by the lowest homology (∼40%) compared with the predicted external loop 3 (79% homology) (Table 1). Loop 3, which contains the PEFGG motif that forms a turn, is important because it is responsible for the size constriction of channels by bending them into the lumen (8).
FIG. 4.
Multiple alignment of the amino acid sequence deduced from the 1-kbp PCR product encoding the 38-kDa OMP from R. aquatilis (Raq) with the corresponding sequences of mature porins of E. coli (Eco), S. typhi (Sty), and S. marcescens (Sma) OmpC. b strands, β-strands; a helix, α-helix.
TABLE 1.
Comparison of the percentages of homology within each of the eight external loops of the R. aquatilis 38-kDa protein, E. coli OmpC, and S. marcescens OmpC
| Loop | % Similarity to OmpC from:
|
|
|---|---|---|
| E. coli | S. marcescens | |
| 1 | 56 | 56 |
| 2 | 53 | 65 |
| 3 | 79 | 71 |
| 4 | 41 | 60 |
| 5 | 58 | 58 |
| 6 | 41 | 65 |
| 7 | 38 | 32 |
| 8 | 78 | 61 |
| Entire protein | 66.8 | 65.4 |
DISCUSSION
The isolated R. aquatilis 38-kDa OMP forms a homotrimer which is stable in the presence of 2% SDS, even at 60°C, whereas E. coli porins become unstable at temperatures over 70°C. Thus, the energy necessary to dissociate the 38-kDa trimer is weaker than that for E. coli porins. This might be a consequence of an adaptation of R. aquatilis to the relatively low temperature in soil and of E. coli to the higher temperature of the human body. Fourel et al. (11) showed that a G-to-D substitution at position 119 in the PEFGG motif of E. coli OmpF affects its thermal stability. This possibility can be excluded in our case, because this motif is well conserved in the 38-kDa protein. It has been reported that the interaction among loops 2, 3, and 4 plays a strategic role in the stability of the trimeric organization (8, 10). Interestingly, loop 4 (Table 1) shows a great divergence, suggesting a different evolution. This variability in loop 4 could explain the reduced thermostability of the 38-kDa trimer.
The results of the single-channel experiments show that the major OMP from R. aquatilis is able to form pores in planar lipid membranes. The conductance values derived (shown in Fig. 3) indicate that the faster superimposed channel of 800 pS more or less than the large 2,400-pS level, i.e., one-third of the value, could be the active monomeric form. This behavior has been observed for other porins, like OmpF of E. coli (19). Like OmpC and OmpF from E. coli, which are cationically selective (10 and 3.8, respectively), the 38-kDa OMP exhibits a moderate cation selectivity (2.6).
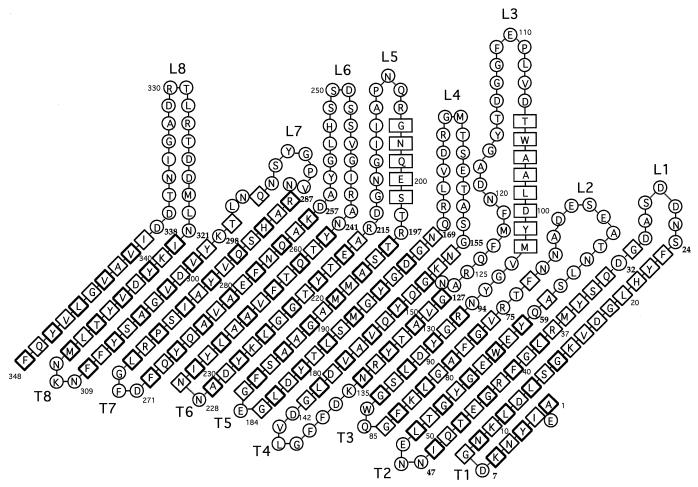
When the amino acid sequence deduced from gene sequencing of the R. aquatilis 38-kDa OMP was compared to those of the OmpC from E. coli, S. typhi, and S. marcescens, a high degree of homology was observed (∼65%). In such enterobacterial porins, the predicted β-strands are highly conserved. Thus, it seems likely that sequence insertions and deletions, which evolved over time and resulted in different lengths of porin sequences, occurred in corresponding predicted loop regions (Table 1). On the basis of the high homology between E. coli OmpC, OmpF, and the 38-kDa protein investigated in the present study, we propose the topological model presented in Fig. 5.
FIG. 5.
Theoretical folding pattern of the 38-kDa OMP. Rectangles indicate residues forming α-helices, diamonds indicate those making up β-strands (in boldface print if their side chains are external), and circles indicate those constituting hydrogen-bonded reverse turns and loops.
Interestingly, the pentapeptide motif PEFGG, mentioned by Jeanteur et al. (16), is located in the predicted loop L3. This important internal loop, which defined the constriction zone of the pore lumen in crystallographic studies (8), is almost completely conserved between the four porins. In addition, three strategic Arg residues are present in the 38-kDa product, i.e., in positions 37 (37 in E. coli OmpC), 75 (74 in E. coli OmpC), and 125 (124 in E. coli OmpC). These three Arg residues have a strategic role in the organization of the NH3+ row in the constriction area of the channel (8). Substitutions of these residues alter the pore properties, leading to an increase in pore size (20), clearly evidencing the structure-function relationship of this region (17, 20, 28). The high conservation of these strategic domains supports the hypothesis that the 38-kDa product exerts a pore function.
Consequently, we conclude that the R. aquatilis 38-kDa protein is a porin. With respect to the high homology of the primary structures observed for OmpC of E. coli, S. typhi, and S. marcescens and the 38-kDa protein of R. aquatilis, we propose that this R. aquatilis 38-kDa protein should be named OmpC.
It has been reported that outer membrane porins, such as OmpD from S. typhimurium (9), OmpK36 from Klebsiella pneumoniae (2), and OmpC from S. flexneri (6), could act as virulence factors during the invasive process of these bacteria. Recent studies indicate that the 38-kDa OMP of R. aquatilis is involved in the adhesion of this bacterium to wheat roots (1). As loops are exhibited at the bacterial surface, bacteria might interact with other surfaces through them. Loop 6 showed high homology with the corresponding loop in S. marcescens OmpC (65%), while it showed only 41% homology with the corresponding loop in E. coli OmpC. R. aquatilis and S. marcescens share the motif SSDSS in loop 6, which is not present in the other enterobacterial porins. Unlike the other enterobacteria whose porins have been characterized (human and or animal pathogens), R. aquatilis and S. marcescens were isolated from soil and water. The loop 6 structure might hence be responsible for the interaction of these bacteria with environmental compounds. Loop 7, showing the lowest conservation level, is the most prominent external loop in the three-dimensional structure (2): it could be an attractive candidate for the binding function. Alberti et al. (2) recently proposed that loop 7 might be involved in the cell surface-exposed receptor site for C1q. In addition, Fourel et al. (10) reported that the substitution in the loop 7 of OmpF eliminates the colicin N receptor site. It would be now interesting to investigate the interaction of R. aquatilis with wheat roots by introducing deletions or performing site-directed mutagenesis in this region.
ACKNOWLEDGMENT
We thank Michael Lebuhn for critically reading the manuscript.
REFERENCES
- 1.Achouak W, De Mot R, Heulin T. Purification and partial characterization of an outer membrane protein involved in the adhesion of Rahnella aquatilis to wheat roots. FEMS Microbiol Ecol. 1994;16:19–24. [Google Scholar]
- 2.Alberti S, Rodríguez-Quiñones F, Schirmer T, Rummel G, Tomás J M, Rosenbusch J P, Benedí V J. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect Immun. 1995;63:903–910. doi: 10.1128/iai.63.3.903-910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Stephen F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Anderson R D, Bao C-Y, Minnick D T, Veigel M, Sedwick W D. Optimization of double-stranded DNA sequencing for polymerase chain reaction products. USB Editorial Comments. 1992;19:39–40. , 57–58. [Google Scholar]
- 5.Berge O, Heulin T, Achouak W, Richard C, Bally R, Balandreau J. Rahnella aquatilis, a nitrogen-fixing enteric bacterium associated with the rhizosphere of wheat and maize. Can J Microbiol. 1991;37:195–203. [Google Scholar]
- 6.Bernardini M L, Sanna M G, Fontaine A, Sansonetti P. OmpC is involved in invasion of epithelial cells by Shigella flexneri. Infect Immun. 1993;61:3625–3635. doi: 10.1128/iai.61.9.3625-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 8.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 9.Dorman C J, Chatfield S, Higgins C F, Hayward C, Dougan G. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect Immun. 1989;57:2136–2140. doi: 10.1128/iai.57.7.2136-2140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fourel D, Bernadac A, Pagès J M. Involvement of exposed polypeptide loops in trimeric stability and membrane insertion of Escherichia coli OmpF porin. Eur J Biochem. 1994;222:625–630. doi: 10.1111/j.1432-1033.1994.tb18905.x. [DOI] [PubMed] [Google Scholar]
- 11.Fourel D, Mizushima S, Bernadac A, Pagès J-M. Specific regions of Escherichia coli OmpF protein involved in antigenic and colicin receptor sites and in stable trimerization. J Bacteriol. 1993;175:2754–2757. doi: 10.1128/jb.175.9.2754-2757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hille B. Ionic channels of excitable membranes. Sunderland, Mass: Sinauer Associates; 1984. pp. 226–246. [Google Scholar]
- 13.Hurlbert R E, Gross D C. Isolation and partial characterization of the cell wall of Pseudomonas syringae HS 191: comparison of outer membrane proteins of HS191 with those of two plasmidless derivatives. J Gen Microbiol. 1983;129:2241. [Google Scholar]
- 14.Hustul J-A, Worobec E. Molecular characterization of a 40 kDa OmpC-like porin from Serratia marcescens. Microbiology. 1994;140:379–387. doi: 10.1099/13500872-140-2-379. [DOI] [PubMed] [Google Scholar]
- 15.Izard D, Gavini F, Trinel P A, Leclerc H. Rahnella aquatilis, nouveau membre de la famille des Enterobacteriaceae. Ann Microbiol (Inst Pasteur) 1979;0A:163–177. [PubMed] [Google Scholar]
- 16.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: Sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 17.Karshikoff A, Cowan S W, Spassov V, Ladenstein R, Schirmer T. Electrostatic properties of two porin channels from E. coli. J Mol Biol. 1994;240:372–384. doi: 10.1006/jmbi.1994.1451. [DOI] [PubMed] [Google Scholar]
- 18.Korhonen T K, Tarkka E, Ranta H, Haahtela K. Type 3 fimbriae of Klebsiella sp.: molecular characterization and role in bacterial adhesion to plant roots. J Bacteriol. 1983;155:860–865. doi: 10.1128/jb.155.2.860-865.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakey J H, Pattus F. The voltage-dependent ability of Escherichia coli porins in different planar bilayers reconstitution. Eur J Biochem. 1989;186:303–308. doi: 10.1111/j.1432-1033.1989.tb15209.x. [DOI] [PubMed] [Google Scholar]
- 20.Lou K L, Saint N, Prilipov A, Rummel G, Benson S A, Rosenbusch J P, Schirmer T. Structural and functional characterization of OmpF porin mutants selected for larger pore size. Crystallographic analysis. J Biol Chem. 1996;271:20669–20675. [PubMed] [Google Scholar]
- 21.Misra R, Benson S A. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J Bacteriol. 1988;170:3611–3617. doi: 10.1128/jb.170.8.3611-3617.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montal M, Mueller P. Formation of bimolecular membrane from lipid monolayers and a study of their electrical properties. Proc Natl Acad Sci USA. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 25.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson W. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 27.Richard C. Nouvelles Enterobacteriaceae rencontrées en bactériologie médicale: Moellerella wisconsensis, Koserella trabulsii, Lecleria adecarboxylata, Escherichia fergusonii, Enterobacter asburiae, Rahnella aquatilis. Ann Biol Clin. 1989;47:231–236. [PubMed] [Google Scholar]
- 28.Saint N, Lou K-L, Widmer C, Luckey M, Schirmer T, Rosenbusch J P. Structural and functional characterization of OmpF porin mutants selected for larger pore size. Functional characterization. J Biol Chem. 1996;271:20676–20680. [PubMed] [Google Scholar]