Abstract
Jasmonic acid (JA) is a lipid-derived signal that regulates plant defense responses to biotic stress. Here, we report the characterization of a JA-deficient mutant of tomato (Lycopersicon esculentum) that lacks local and systemic expression of defensive proteinase inhibitors (PIs) in response to wounding. Map-based cloning studies demonstrated that this phenotype results from loss of function of an acyl-CoA oxidase (ACX1A) that catalyzes the first step in the peroxisomal β-oxidation stage of JA biosynthesis. Recombinant ACX1A exhibited a preference for C12 and C14 straight-chain acyl-CoAs and also was active in the metabolism of C18 cyclopentanoid-CoA precursors of JA. The overall growth, development, and reproduction of acx1 plants were similar to wild-type plants. However, the mutant was compromised in its defense against tobacco hornworm (Manduca sexta) attack. Grafting experiments showed that loss of ACX1A function disrupts the production of the transmissible signal for wound-induced PI expression but does not affect the recognition of this signal in undamaged responding leaves. We conclude that ACX1A is essential for the β-oxidation stage of JA biosynthesis and that JA or its derivatives is required both for antiherbivore resistance and the production of the systemic wound signal. These findings support a role for peroxisomes in the production of lipid-based signaling molecules that promote systemic defense responses.
INTRODUCTION
Jasmonic acid (JA) and its cyclic precursors and derivatives, collectively referred to as jasmonates (JAs), constitute a family of bioactive oxylipins that regulate plant responses to environmental and developmental cues. JAs are perhaps best known for their role in orchestrating plant defense responses to herbivores and certain microbial pathogens (Liechti and Farmer, 2002; Turner et al., 2002). Activation of jasmonate-mediated defenses is typically preceded by accumulation of JAs in response to biotic stress (Wasternack and Hause, 2002). JAs are synthesized by the so-called octadecanoid pathway that involves enzymes located in two different subcellular compartments (Figure 1; Vick and Zimmerman, 1984; Schaller, 2001; Wasternack and Hause, 2002). The first part of the pathway directs the conversion of linolenic acid to 12-oxo-phytodienoic acid (OPDA) by the sequential action of the plastid enzymes lipoxygenase (LOX), allene oxide synthase, and allene oxide cyclase. The second part of the pathway takes place in peroxisomes, where OPDA is reduced by OPDA reductase (OPR3) to give 3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid (OPC8). Removal of six carbons from the octanoate side chain of OPC8 yields JA.
Figure 1.
Octadecanoid Pathway for JA Biosynthesis.
OPDA produced in the chloroplast is transported to the peroxisome for subsequent conversion to JA via the action of OPR3 and β-oxidation enzymes. The relative order in which OPR3 and ACS act in the pathway is not known. See text for details. LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPDA, 12-oxo-phytodienoic acid; OPR3, OPDA reductase; ACS, acyl-CoA synthetase; MFP, multifunctional protein containing 2-trans-enoyl-CoA hydratase and l-3-hydroxyacyl-CoA dehydrogenase activities; KAT, 3-ketoacyl-CoA thiolase; OPC8, 3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid; JA, (+)-7-iso-jasmonic acid.
Studies on β-oxidation in plants have mainly focused on its role in the breakdown of storage lipids in germinating seeds (Graham and Eastmond, 2002). Increasing evidence, however, indicates that nonfatty plant tissues also depend on β-oxidation for several other processes, including the synthesis of indole-acetic acid and JA (Graham and Eastmond, 2002; Zolman and Bartel, 2004). Metabolic labeling experiments with 18O-OPDA provided the first evidence that the conversion of OPC8 to JA involves three cycles of β-oxidation (Vick and Zimmerman, 1984). These early studies are supported by recent work showing that OPR3 is located in peroxisomes (Stintzi and Browse, 2000; Strassner et al., 2002), which are thought to be the exclusive site of fatty acid β-oxidation in higher plants (Graham and Eastmond, 2002; Tilton et al., 2004). Thus, it is generally assumed that the final steps of JA synthesis are catalyzed by the three core enzymes of the β-oxidation cycle, namely acyl-CoA oxidase (ACX), the multifunctional protein (containing 2-trans-enoyl-CoA hydratase and l-3-hydroxyacyl-CoA dehydrogenase activities), and 3-ketoacyl-CoA thiolase (KAT). An additional thioesterase activity is also presumably involved in the release of JA from JA-CoA, the product of the final round of β-oxidation (Figure 1). Identification of specific gene products that participate in the conversion of OPC8 to JA has been hampered by the fact that ACX, multifunctional protein, KAT, and thioesterases are encoded by small gene families (Graham and Eastmond, 2002; Shockey et al., 2003; Reumann et al., 2004; Tilton et al., 2004). Furthermore, isoforms within a particular enzyme family often exhibit overlapping substrate specificity (Graham and Eastmond, 2002). Recent antisense experiments conducted in Arabidopsis thaliana have provided evidence that specific isoforms of ACX (ACX1) and KAT (KAT2) play a role in wound-induced JA synthesis (Cruz Castillo et al., 2004). However, the precise contribution of these family members to JA production in healthy and damaged tissues remains to be determined.
A role for peroxisomal β-oxidation in the production of bioactive plant oxylipins contrasts the well-established function of this catabolic pathway in the inactivation of arachidonic acid–derived prostanoids in animal cells. Only in rare exceptions has a β-oxidation metabolite of a prostanoid been shown to be bioactive (Hou et al., 2001). Although JA is often regarded as the physiological signal for jasmonate-mediated responses, increasing evidence indicates that C18 precursors of JA exert bioactivity in the absence of their conversion to JA. For example, structure-activity studies have shown that exogenous OPDA is more potent than JA in its ability to activate the tendril coiling response of Bryonia dioica to mechanical stimulation (Weiler et al., 1993; Blechert et al., 1999). OPDA also is more effective than JA in eliciting the synthesis of certain diterpenoid volatiles in lima bean (Phaseolus lunatus) (Koch et al., 1999) as well as the accumulation of glyceollin phytoalexins in soybean (Glycine max) (Fliegmann et al., 2003). These observations have led to the proposal that plant cells contain receptors that are selective for OPDA and JA (Blechert et al., 1999; Koch et al., 1999). The notion that JA and OPDA function as discrete signaling entities is supported by the observation that some JA-mediated processes, such as male fertility in Arabidopsis, are not promoted by OPDA (McConn and Browse, 1996; Stintzi and Browse, 2000). Structure-activity studies also indicate that the biosynthesis of alkaloids in Eschscholtzia californica cell cultures, and the expression of defensive proteinase inhibitor (PI) genes in tomato (Lycopersicon esculentum), is mediated by JA rather than OPDA (Haider et al., 2000; Miersch and Wasternack, 2000). Because this collective body of work implies that conversion of OPDA to JA is important for some but not all jasmonate-mediated processes, a clear understanding of the role of β-oxidation in jasmonate signaling has not yet emerged.
The interpretation of structure-activity studies aimed at defining the in vivo signaling activity of specific JAs is complicated by several factors. First, the concentration of exogenous JAs used for structure-activity studies may not accurately reflect in vivo concentration of the compound. Furthermore, different concentrations of the same compound may evoke different responses, as was shown to be the case for OPDA-induced volatile emission in lima bean (Koch et al., 1999). Second, exogenous JAs are subjected to a wide variety of metabolic transformations that profoundly affect their biological activity (Wasternack and Hause, 2002; Swiatek et al., 2004; Staswick and Tiryaki, 2004). Experiments involving exogenous OPDA are further complicated by its conversion to JA in planta. Finally, increasing evidence indicates that JA biosynthesis is restricted to specific cell types within the vascular system and that JA produced at this location exerts its effects in different cell types (Li et al., 2002b; Hause et al., 2003; Howe, 2005). Thus, exogenous compounds may activate the jasmonate signaling pathway by a mechanism that bypasses intracellular or intercellular transport steps that are normally required for the action of endogenous JAs. A fascinating natural example of this phenomenon is the induction of JA responses by the phytotoxin coronatine, which is thought to be a structural mimic of OPDA or JA-Ile (Weiler et al., 1994; Beale and Ward, 1998; Staswick and Tiryaki, 2004). After its introduction into plant cells by virulent strains of Pseudomonas syringae, coronatine exerts its virulence effects by activating the host JA signal transduction pathway in a manner that is independent of the biosynthesis of endogenous JAs (Weiler et al., 1994; Zhao et al., 2003).
Genetic manipulation of the octadecanoid pathway offers an alternative and complementary approach to elucidate the biological activity of specific pathway intermediates. Genetic studies with the Arabidopsis opr3 mutant have provided some of the best evidence to date that OPDA per se can function as a signal for plant defensive responses. Although opr3 plants fail to accumulate biologically active JA, Stintzi et al. (2001) discovered that this mutant retains jasmonate-mediated resistance to insect and fungal attack. This finding demonstrates that resistance in opr3 plants is mediated by an octadecanoid pathway-derived compound other than JA. The ability of opr3 plants to express defense-related genes in response to exogenous OPDA suggested that OPDA itself may fulfill this role. It was thus proposed that JA and OPDA work together in wild-type plants to coordinate the expression of appropriate sets of target genes (Stintzi et al., 2001; Weber, 2002). At present, the relative contribution of cyclopentenone (e.g., OPDA) and cyclopentanone (JA) JAs to host resistance is unclear. Identification and characterization of mutants that are defective in the conversion of OPC8 to JA is needed to rigorously address this question. Cruz Castillo et al. (2004) recently reported that antisense suppression of ACX1 and KAT2 in Arabidopsis reduces wound-induced JA production and activation of wound-responsive genes. However, defense-related or other physiological phenotypes of these lines were not reported.
Here, we describe the characterization of a wound-response mutant of tomato that is deficient in JA biosynthesis. Positional cloning studies demonstrated that this defect results from loss of function of a member (ACX1A) of the ACX family of enzymes that participate in peroxisomal β-oxidation. Results obtained from characterization of acx1 mutant plants advance our knowledge of the jasmonate pathway in several ways. First, we show that ACX1A uses OPC8-CoA as a substrate and that this isoform participates in the synthesis of the vast majority of wound-induced JA in tomato leaves. Second, we demonstrate that JA or a derivative of JA, rather than C18 precursors of JA, is the physiological signal for induced defense responses to insect attack. Finally, grafting experiments indicate that the β-oxidation stage of JA biosynthesis is required for the production of the transmissible wound signal. These findings support the notion that JA is an essential component of the long-distance signal for induced resistance to herbivores.
RESULTS
Wound-Response Phenotype of JL1 Plants
The JL1 line of tomato harbors an ethyl methanesulfonate–induced recessive mutation that impairs the accumulation of two Ser PIs (PI-I and PI-II) in response to mechanical wounding (Lightner et al., 1993). Our further characterization of JL1 was performed with a homozygous line obtained from three successive backcrosses of the original mutant to the wild-type parental cultivar. Wound-response assays with this line showed that damaged JL1 leaves (local response) accumulated low but detectable levels of PI-II protein, whereas no PI-II protein was detected in undamaged leaves (systemic response) of wounded JL1 plants (Figure 2A). Treatment of JL1 plants with exogenous JA, however, was sufficient to induce the accumulation of wild-type levels of PI-II (Figure 2A). These findings are consistent with the results of Lightner et al. (1993) and suggest that the JL1 plants are defective in the synthesis rather than the action of JA.
Figure 2.
JL1 Plants Are Defective in Wound-Induced Gene Expression.
(A) PI-II accumulation in tomato leaves in response to mechanical wounding or treatment with JA. All leaflets on the lower leaf of 18-d-old wild-type (closed bar) and JL1 (open bar, labeled acx1) plants were wounded with a hemostat. This wounding regimen was repeated 3 h later. Twenty-four hours after the second wounding, PI-II levels were measured in the wounded leaf (Loc) and the upper unwounded leaf (Sys). As controls, PI-II levels also were determined in the pooled upper and lower leaves of unwounded (UW) plants. A separate set of 16-d-old plants was excised at the base of the stem and supplied with 15 mM phosphate buffer (Buf) or JA (10 nmol/plant, dissolved in 15 mM phosphate buffer). Twenty-four hours after elicitation, PI-II levels in leaves were determined. Values represent means ± sd of six plants per genotype.
(B) Time course of gene expression in response to mechanical wounding. Wild-type and JL1 plants (16 d old) containing two fully expanded leaves were wounded with a hemostat on both leaves. At various times (hours) thereafter, wounded leaves were harvested for RNA extraction. RNA gel blots were hybridized to 32P-labeled cDNA probes for PI-II, cathepsin D inhibitor (CDI), LoxD, or OPR3. As a loading control, a duplicate blot was hybridized to a labeled cDNA for translation initiation factor eIF4A.
RNA gel blot analysis was used to determine whether the expression of wound-responsive genes is affected in JL1 plants. Two general classes of genes that differ in their timing of wound-induced expression have been described in tomato. So-called late genes include those encoding PIs and other defensive proteins, whereas early genes encode various signaling-related proteins including JA biosynthetic enzymes (Ryan, 2000; Lee and Howe, 2003; Howe, 2005). The level of wound-induced PI-II (a late gene) transcripts in severely wounded JL1 leaves was <10% of that observed in the wild type (Figure 2B). Thus, the deficiency in PI-II protein accumulation in JL1 can be attributed to a defect in the activation of PI-II gene expression. That the time course of the residual PI-II expression in damaged JL1 leaves was similar to that of wound-induced PI-II expression in wild-type plants indicates that JL1 is affected in the amplitude rather than the timing of wound-induced signaling. Wound-induced expression of another PI gene (cathepsin D inhibitor) as well as the early wound response genes LoxD and OPR3 was also abrogated in JL1 (Figure 2B). These results demonstrate that JL1 plants are impaired in the signal transduction pathway for wound-induced local and systemic expression of defense-related genes.
The Wound-Response Phenotype of JL1 Results from a Defect in ACX
The deficiency in wound-induced PI-II protein accumulation in JL1 provided a facile assay for map-based cloning studies aimed at determining the molecular basis of this defect. A combination of amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP) markers was used to position the target gene to a region on the long arm of chromosome 8 between RFLP markers CT111 and cLED14K7 (Figure 3A). Linkage analysis of additional RFLP markers in this interval showed that the target locus cosegregated with marker TG510 in a mapping population of 1200 backcrossed (BC1) plants. Screening of a Lycopersicon cheesmanii BAC library with TG510 as a probe resulted in the identification of two positive clones (166B24 and 232L13). BAC-end mapping experiments showed that these clones were contiguous and that the target gene was physically located on 232L13 (Figure 3A). A shotgun DNA sequencing strategy was used to obtain approximately sixfold sequencing coverage of BAC 232L13. The resulting sequence information was compiled and analyzed with gene prediction software and BLAST searches (Altschul et al., 1990) against the tomato EST database (http://www.tigr.org). The DNA sequence of the cosegregating TG510 marker was found to reside within the open reading frame of a hypothetical gene represented by a single tomato EST (EST475746). This gene was located adjacent to two putative ACX-encoding genes, which we designated LcACX1A and LcACX1B (Figure 3B). Given the likely role of ACX in JA biosynthesis and wound signaling (Vick and Zimmerman, 1984; Cruz Castillo et al., 2004), subsequent experiments were focused on testing the hypothesis that the wound-response phenotype of JL1 results from a defect in ACX1A or ACX1B.
Figure 3.
Map-Based Cloning of the ACX1A Gene.
(A) Fine genetic and physical mapping of ACX1A. The target gene was mapped to a genetic interval between RFLP marker GP40 and cLED14K7 on tomato chromosome 8. Analysis of a BC1 mapping population of 1200 plants delimited the gene to a region covered by two overlapping BAC clones 166B24 and 232L13. Numbers in parentheses indicate the number of recombination events identified between markers and the target gene.
(B) Structures of three L. cheesmanii genes (indicated by arrows) identified on an ∼20-kb region of BAC232L13. Boxes indicate exons, and lines between boxes indicate introns or intergenic regions. A hypothetical gene consisting of two exons was identified upstream of LcACX1A. The TG510 marker that cosegregated with the target locus (ACX1A) is located in the second intron of this hypothetical gene. The asterisk denotes the location of the missense mutation (T138I) identified in the ACX1A gene from L. esculentum.
(C) Phylogenetic relationship of LeACX1A and LeACX1B to ACXs from other plants. Shown is an unrooted phylogenetic tree constructed from the predicted amino acid sequences of representative plant ACXs. Gray boxes indicate the substrate preference of ACX subgroups for acyl-CoA substrates of various chain lengths. Tomato sequences are underlined. U220503, U223228, and U216994 are additional tomato ACX sequences compiled from the EST sequence database at the Solanaceae Genomics Network (http://www.sgn.cornell.edu). Another sequence (U229242) in the tomato EST database appears to be homologous to the long-chain ACXs (e.g., AtACX2) but, because of its partial length, was not included in the phylogeny. Species names and accession numbers are listed in the Methods.
Plant ACXs comprise a family of flavoenzymes that catalyze the initial step of peroxisomal β-oxidation of a variety of acyl-CoA substrates (Graham and Eastmond, 2002). As a first step toward characterizing the tomato ACX1A and ACX1B genes, we used RT-PCR to obtain full-length cDNAs (designated LeACX1A and LeACX1B) from wild-type (cv Castlemart) L. esculentum. The predicted amino acid sequence of the two tomato ACXs are 81.7% identical. ACX1A and ACX1B are most similar to ACXs characterized from Arabidopsis (80.1 and 73.3% identical to AtACX1, respectively) and soybean (81.8 and 75.7% identical to GmACX1-1, respectively) that have broad specificity for medium- to long-chain acyl-CoAs (Figure 3C; Hooks et al., 1999; Agarwal et al., 2001). Both LeACX1A and LeACX1B possess a C-terminal peroxisomal targeting signal type 1 (PTS1) motif (see Supplemental Figure 1 online; Reumann et al., 2004). The C-terminal region of ACX1B lacks a 15–amino acid sequence that is highly conserved in other medium- to long-chain plant ACXs (see Supplemental Figure 1 online). Comparison of the ACX1B genomic and cDNA sequences suggested that this polymorphism results from alternative splicing at cryptic donor and acceptor splice sites.
Comparison of the sequence of wild-type– and JL1-derived ACX cDNAs revealed a single C-to-T nucleotide substitution in LeACX1A. No sequence differences were found in LeACX1B. The single-base change in LeACX1A, which was confirmed by sequencing of PCR-amplified genomic DNA from JL1, is predicted to replace a highly conserved (i.e., invariant among all plant and animal ACXs) Thr residue at position 138 with an Ile (see Supplemental Figure 1 online). The three-dimensional structure of mammalian ACX (Nakajima et al., 2002) and its conserved mitochondrial counterpart, acyl-CoA dehydrogenase (Kim and Miura, 2004), has shown that this Thr binds the flavin ring of FAD that defines the active site of the enzyme.
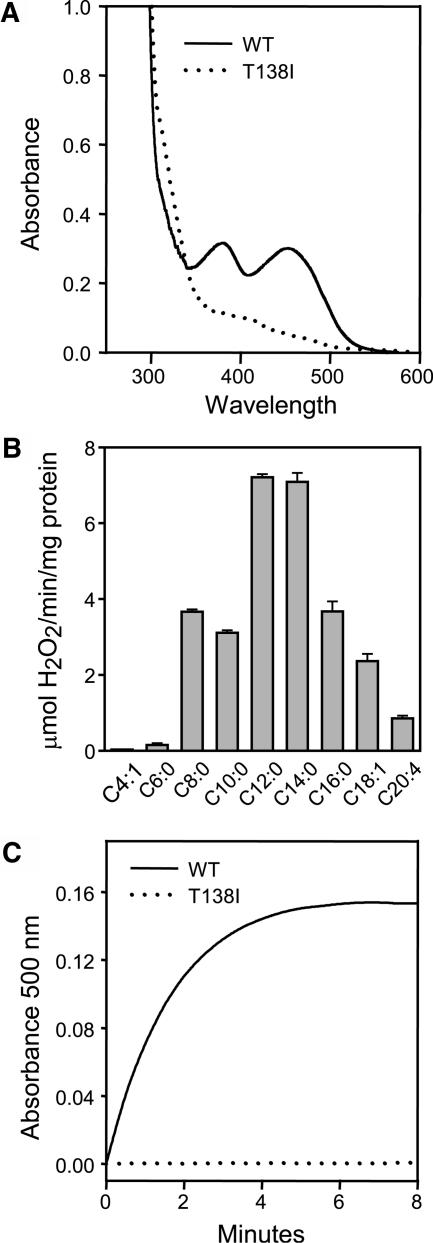
To determine whether this mutation (T138I) affects the biochemical activity of ACX1A, we expressed His-tagged derivatives of the wild-type (ACX1wt) and mutant (ACX1T138I) proteins in Escherichia coli. Affinity-purified ACX1wt displayed 381- and 452-nm absorption peaks that are indicative of flavoenzymes (Figure 4A). An equivalent amount of affinity-purified ACX1T138I lacked this spectral signature. In vitro activity assays with straight-chain acyl-CoAs showed that recombinant ACX1wt has a preference for C12 and C14 substrates (Figure 4B). The enzyme was less active with C18 and C20 substrates and showed little or no activity with short-chain (≤C6) acyl-CoAs. ACX1T138I showed no detectable activity against 14:0-CoA, which was a preferred substrate of the wild-type enzyme (Figure 4C). These results indicate that the T138I mutation in JL1 renders ACX1A inactive, most likely by disrupting FAD binding to the apoprotein.
Figure 4.
Biochemical Characterization of Wild-Type and Mutant Forms of ACX1A.
(A) UV-visible spectra of recombinant ACX1wt (solid line) and ACX1T138I (dotted line) expressed in E. coli. Absorption spectra were recorded with affinity-purified protein at a concentration of 2 mg/mL. The 381- and 452-nm absorption peaks exhibited by ACX1wt are indicative of FAD binding.
(B) Substrate specificity of ACX1wt. The activity of recombinant ACX1wt against various fatty acyl-CoA esters was measured with a spectrophotometric assay linked to H2O2 production. Data show the mean ± sd of triplicate activity measurements with the same extract and are representative of independent experiments performed with different enzyme preparations.
(C) ACX activity of recombinant ACX1wt (solid line) and ACX1T138I (dotted line) against 14:0-CoA. The change in absorbance at 500 nm results from a chromagen whose formation is linked to ACX-catalyzed H2O2 production.
Genetic complementation experiments were performed to determine whether the missense mutation in LeACX1A was responsible for the deficiency in wound-induced PI-II expression in JL1 plants. The wild-type LeACX1A cDNA was placed under the control of the 35S promoter of Cauliflower mosaic virus in T-DNA vector pBI121. This construct was introduced into the JL1 genetic background by Agrobacterium tumefaciens–mediated transformation. From 33 independent 35S-ACX1A transgenic plants confirmed by genomic PCR, 20 primary lines (T0) showed normal levels of wound-induced PI-II expression both in the wounded leaf and in the systemic unwounded leaves (Figure 5). Further characterization of two of these lines showed that the complemented phenotype was inherited in the T1 generation (data not shown). These results demonstrate that the wound-response phenotype of JL1 results from loss of function of ACX1A. We henceforth refer to the JL1 as acx1, which more accurately describes the biochemical defect in the mutant.
Figure 5.
Genetic Complementation of the Wound-Response Phenotype of JL1.
Wild-type, JL1 mutant (labeled acx1), and 35S-ACX1A–expressing mutant plants (at the five- to six-leaf stage) were wounded once across the midvein of each leaflet on the two lower leaves. The same leaflets were wounded again 3 h later. Twenty-four hours after the initial wounding, PI-II levels in the lower wounded leaves (open bars) and the upper unwounded leaves (closed bars) were measured. Data show the mean ± sd (n = 7 for the wild type and acx1; n = 20 independent transgenic lines confirmed by PCR to contain the 35S-ACX1A transgene). PI-II levels in unwounded control plants were below the detection limit of the assay (∼5 μg PI-II/mL leaf juice).
ACX1A Is Required for JA Biosynthesis
Current knowledge of the JA biosynthetic pathway predicts that OPC8-CoA is the substrate for entry into the β-oxidation cycle via the action of ACX (Figure 1). To determine whether ACX1A is capable of metabolizing OPC8-CoA, we used an in vitro assay that couples ACX1A to a yeast acyl-CoA synthetase (ACS) that activates the fatty acid substrate to the corresponding CoA ester. Addition of 50 μM OPC8 to the assay resulted in ACX1A activity that was significantly greater than that obtained with the same concentration of 14:0 (Figure 6A). Control experiments showed that ACX1A activity in these experiments was dependent on ACS and CoA and was proportional to the amount of substrate added (data not shown). Interestingly, OPDA was comparable to OPC8 in its ability to promote ACX1A activity. These results provide evidence that ACX1A can metabolize OPC8-CoA and that reduction of the double bond in the cyclopentenone ring of OPDA is not required for the enzyme's activity.
Figure 6.
ACX1A Catalyzes an Essential Step in JA Biosynthesis.
(A) In vitro metabolism of OPC8 and OPDA in a coupled ACS-ACX reaction. The indicated substrates (5 μL in 0.25% Triton X-100) or mock control were added to a reaction mixture containing yeast ACS, CoA, and ATP. This reaction proceeded for 0.5 h at 25°C, at which time the ACX reaction was initiated by addition of 1 μg of purified LeACX1A. ACX1A-catalyzed production of H2O2 was monitored spectrophotometrically as described in Methods.
(B) JA levels in response to wounding. Two-leaf-stage wild-type (closed circles) and acx1 (open circles) plants were mechanically wounded with a hemostat at the distal end of each leaflet. Leaf tissue was harvested for quantification of JA levels at various times after wounding. Unwounded plants were used for the 0 h time point. Data show the mean ± sd of three independent sample preparations and are representative of three independent experiments. FW, fresh weight.
(C) OPDA levels in response to wounding. Two-leaf-stage wild-type (closed bars) and acx1 (open bars) plants were mechanically wounded as described above, and OPDA levels were measured 1 h after wounding (wounded). OPDA was also extracted from leaves of unwounded plants (control). Data show the mean ± sd of three independent samples and are indicative of three independent experiments.
To determine the contribution of ACX1A to JA biosynthesis in tomato leaves, we used gas chromatography–mass spectrometry to measure JA levels in wild-type and mutant plants (Figure 6B). Whereas wild-type leaves exhibited a rapid and transient increase in JA accumulation in response to mechanical wounding, damaged acx1 plants produced very little JA; the peak level of JA in wounded acx1 leaves at 1 h was ∼5% of that in wild-type plants. We consistently observed that the JA content in unwounded acx1 leaves was significantly lower than that in the wild type. In three independent experiments, the mean JA level in unwounded wild-type and acx1 leaves was 18 ± 7 and 5 ± 2, respectively (P = 0.0013, Student's t test). These results indicate that ACX1A is required for the majority of wound-induced JA accumulation in tomato leaves and that this ACX isoform also contributes to the maintenance of basal JA levels in leaf tissue. The accumulated level of OPDA in control and wounded acx1 plants was slightly reduced in comparison with the wild type, but this difference was not statistically significant (Figure 6C).
acx1 Plants Are Compromised in Resistance to Insect Attack
The inability of acx1 plants to express significant levels of defensive PIs in response to mechanical wounding suggested that the mutant might be compromised in resistance to herbivorous insects. To test this possibility, 6-week-old (experiment 1) or 4-week-old (experiment 2) wild-type and acx1 plants were challenged with Manduca sexta (tobacco hornworm) larvae. After termination of the feeding trial, PI-II protein accumulation in undamaged and damaged leaf tissue from challenged plants was measured. In contrast with high levels of PI-II accumulation in herbivore-damaged wild-type leaves, very little or no PI-II protein accumulation was detected in hornworm-attacked acx1 plants (Table 1). RNA gel blot analysis showed that the deficiency in PI-II protein accumulation resulted from a block in hornworm-induced local and systemic expression of the PI-II gene, which was accompanied by reduced expression of several other defense-related genes (Figure 7A). Failure of the mutant to activate defense gene expression was correlated with severely reduced levels of JA in herbivore-attacked plants (Figure 7B). It was readily apparent that larvae consumed much more foliage from acx1 than from wild-type plants (Figures 7C and 7D). Consistent with this observation, the average weight of larvae reared on the mutant was 3.5-fold (experiment 1) and 1.9-fold (experiment 2) greater than that of larvae reared on wild-type plants (P < 0.001 for both experiments) (Table 1; Figure 7E). These results demonstrate that ACX1A is required for JA-mediated resistance of tomato to attacking hornworm larvae.
Table 1.
Tobacco Hornworm Feeding Assay on Wild-Type and acx1 Plants
| Experimenta | Genotype | Damaged Leaf PI-II (μg/mL) | Undamaged Leaf PI-II (μg/mL) | Larval Weight (g)b |
|---|---|---|---|---|
| 1 | Wild type | 225 ± 47 | 199 ± 66 | 0.99 ± 0.41 (n = 38) |
| acx1 | Not detectable | Not detectable | 3.45 ± 1.68 (n = 42) | |
| 2 | Wild type | 141 ± 73 | 151 ± 26 | 0.10 ± 0.04 (n = 38) |
| acx1 | 15 ± 24 | Not detectable | 0.19 ± 0.05 (n = 41) |
In experiment 1, 11 newly hatched larvae were placed on leaves of each of five separately potted 6-week-old plants of each genotype. In experiment 2, 60 newly hatched larvae were placed randomly on leaves of 20 4-week-old plants of each genotype. Larvae were allowed to move freely between plants of the same genotype. Experiments 1 and 2 were terminated 10 and 3 d after the start of the feeding trial, respectively. At that time, larvae were recovered from plants and their weight measured. Also, PI-II levels in damaged and undamaged leaf tissue were measured. PI-II data represent the mean ± sd of at least six independent measurements. The detection limit of the PI-II assay was ∼5 μg PI-II/mL leaf juice.
In both experiments, the weight of larvae grown on wild-type and mutant plants was significantly different at P < 0.001 (Student's t test).
Figure 7.
acx1 Plants Are Compromised in JA-Mediated Resistance to Tobacco Hornworm Larvae.
(A) Newly hatched hornworm larvae were allowed to feed on 4-week-old wild-type and acx1 plants for 3 d. Total RNA was isolated separately from hornworm-damaged (D) and undamaged (U) leaves of the same genotype. RNA was also isolated from unchallenged plants as a control (C). RNA gel blots were hybridized to cDNA probes for the genes indicated on the left.
(B) Three-week-old wild-type (closed bar) and acx1 (open bar) plants were challenged for ∼4 h with third-instar tobacco hornworm larvae. JA was extracted from damaged leaf tissue (+THW) and from leaves of unchallenged plants (control) and quantified by GC-MS. Data show the mean ± sd of three independent JA extractions.
(C) Representative wild-type plants at the end of experiment 1 feeding trial.
(D) Representative acx1 plants at the end of experiment 1 feeding trial.
(E) Hornworm larvae recovered from representative wild-type (left column) and acx1 (right column) plants at the end of experiment 1 feeding trial.
acx1 Plants Fail to Produce the Systemic Wound Signal for Defense Gene Expression
The deficiency of wound-induced systemic gene expression in acx1 plants could result either from a defect in production of a long-distance wound signal or a defect in the recognition of that signal in distal undamaged leaves. To address this question, wound-induced PI-II expression was analyzed in reciprocal grafts between wild-type and acx1 plants (Figure 8). Four-week-old plants were grafted such that both the rootstock (stock) and the scion contained at least two healthy leaves. After the graft junction healed, stock leaves were wounded and PI-II mRNA levels were measured 11 h later in both the damaged stock leaves (local response) and the undamaged scion leaves (systemic response). Wounding of wild-type stock leaves resulted in local and systemic accumulation of PI-II transcripts to levels well above that observed in unwounded control plants that had also been grafted (Figure 8, lanes 1 and 2). This finding indicates that wounded wild-type leaves produce a graft-transmissible signal that is recognized in undamaged scion leaves. By contrast, wounding of acx1 stock leaves resulted in a weak local response and an undetectable systemic response (Figure 8, lanes 3 and 4). Analysis of acx1/wild-type hybrid grafts showed that upon wounding of acx1 stock leaves, wild-type scions failed to activate PI-II expression (Figure 8, lanes 5 and 6). The accumulation of PI-II mRNA in wild-type scions from this graft combination results from wounds inflicted during the grafting procedure. Analysis of the reciprocal graft combination showed that acx1 scions responded to a signal emanating from wounded leaves of wild-type rootstock (Figure 8, lanes 7 and 8). These experiments demonstrate that acx1 disrupts wound-induced systemic PI expression by blocking the production of the transmissible signal in damaged leaves, rather than by blocking the recognition of the signal in undamaged scion leaves.
Figure 8.
Systemic Wound Signaling in Grafts between Wild-Type and acx1 Plants.
Wild-type and acx1 plants were grafted in the four combinations indicated using the method of Li et al. (2002b). The genotypes listed above and below the horizontal line correspond to the scion and rootstock, respectively. For each graft combination, plants were divided into a control (−) and experimental (+) group consisting of four grafted plants per group. For the experimental group, each leaflet on the rootstock was mechanically injured with a hemostat. Eleven hours after wounding, leaf tissue was harvested separately from wounded rootstock leaves (stock) and undamaged scion leaves (scion) for RNA extraction. The control set of plants received no wounding, other than that inflicted by the grafting procedure itself. Levels of PI-II mRNA were analyzed by RNA gel blot analysis using an eIF4A cDNA probe as a loading control. The results shown are representative of three independent experiments.
DISCUSSION
Role of ACX1A in JA Biosynthesis
Here, we provide several lines of evidence demonstrating that the wound-signaling defect in the JL1 mutant of tomato (renamed here acx1) results from a block in the β-oxidation stage of the octadecanoid pathway. First, acx1 plants harbor a defective member (ACX1A) of the ACX family of enzymes that catalyze the first step in the β-oxidation spiral. Second, recombinant ACX1A metabolizes OPC8-CoA, which is the presumptive substrate for the first β-oxidation cycle in the JA biosynthetic pathway. Finally, acx1 leaves accumulate very little (∼5% wild-type levels) JA in response to wounding. This deficiency in JA accounts for the lack of wound-induced PI expression in acx1 plants and is consistent with the ability of the mutant to express PIs in response to exogenous JA (Figure 2). We thus conclude that ACX1A is responsible for the majority of wound-induced JA in tomato leaves. The reduced level of JA in undamaged acx1 leaves further indicates that this ACX isoform participates in the maintenance of basal levels of the phytohormone.
The deduced amino acid sequence and substrate specificity of LeACX1A is most similar to that of ACX1 and ACX5 of Arabidopsis and two ACXs from soybean (Hooks et al., 1999; Eastmond et al., 2000a; Agarwal et al., 2001). These ACXs have relatively broad substrate specificity and, as yet, no definitive physiological function. Several observations lead us to propose that LeACX1A and other members of the so-called medium- to long-chain subfamily of ACXs (Figure 3C) play a prominent role in JA biosynthesis. First and foremost is the JA-deficient phenotype of acx1 plants. Second, results from the coupled ACS-ACX assay indicate that ACX1A exhibits a preference for C18 cyclopentanoid-CoAs over 14:0-CoA (Figure 6A). This observation suggests that ACX1A and related enzymes possess structural features that facilitate the metabolism of C18 cyclopentanoid fatty acids. Third, recent studies have implicated Arabidopsis ACX1 in wound-induced JA synthesis and expression of wound-responsive genes (Cruz Castillo et al., 2004). Unlike the severe JA deficiency in tomato acx1 plants, Arabidopsis acx1 antisense lines retained 50 to 60% of wild-type levels of wound-induced JA. This relatively weak phenotype may reflect incomplete suppression of AtACX1 or the capacity of other ACXs (e.g., ACX5) to participate in JA biosynthesis in wounded Arabidopsis leaves. Hooks et al. (1999) reported that anti-ACX1 lines of Arabidopsis exhibit a modest reduction in root growth, but JA levels were not assessed in that study.
Identification of LeACX1A as an essential component of the octadecanoid pathway supports the original proposal by Vick and Zimmerman (1984) that JA biosynthesis involves β-oxidation enzymes that remove six carbons from the octanoate side chain of OPC8. The presence of a PTS1 sequence at the C terminus of LeACX1A, together with the fact that fatty acid β-oxidation in plants occurs in peroxisomes, are consistent with the notion that ACX1A is a peroxisomal protein. Subcellular localization experiments are needed to confirm this. Based on the ability of ACX1A to use OPC8-CoA and the location of OPR3 in peroxisomes (Strassner et al., 2002), our results indicate that the likely in vivo substrate for ACX1A is a peroxisomal pool of OPC8-CoA (Figure 1). At present, the temporal sequence of metabolic events involved in the conversion of plastid-localized OPDA to OPC8-CoA remains to be established. One possibility is that OPDA is transported from the plastid as a CoA ester and is then converted by OPR3 to OPC8-CoA in the peroxisome. Because OPR3 readily accepts free OPDA as a substrate (Schaller, 2001; Strassner et al., 2002), an alternative scheme is that OPR3 first reduces OPDA to OPC8, which is then converted to OPC8-CoA. Identification of enzymes involved in the synthesis and transport of C18 cyclopentanoid-CoAs may help to distinguish these possibilities.
Our results do not exclude the possibility that conversion of OPC8-CoA to JA-CoA involves multiple ACXs that specifically act on OPC8-CoA, OPC6-CoA, or OPC4-CoA. However, the ability of LeACX1A to metabolize a broad range of fatty acid chain lengths (Figure 4B) argues against this hypothesis. Determination of the relative specificity of ACX1A for different OPC-CoA derivatives would be helpful to further address this issue, as would the identification of OPC-CoA intermediates that accumulate in acx1 tissues. In considering the role of ACX1A in JA biosynthesis, it is noteworthy that the enzyme accepts both OPC8- and OPDA-CoAs as substrates in the ACS-ACX coupled assay (Figure 6A). Because we did not determine the extent to which each substrate was activated to the CoA ester after the ACS reaction, it is not possible to draw firm conclusions about the relative preference of ACX1A for different substrates. However, assuming that OPC8 and OPDA were equally converted to the CoA ester, it would appear that ACX1A does not discriminate between the cyclopentenone (i.e., OPDA) and the cyclopentanone (i.e., OPC8) derivatives. This observation raises the possibility that OPR3 activity is not strictly required for entry of OPDA into the β-oxidation cycle. Complete β-oxidation of OPDA is expected to yield 4,5-didehydro-JA. Such a pathway may explain the production of this JA derivative in fungi (Miersch et al., 1989) and plants (Dathe et al., 1989).
The acx1 mutation affects a Thr residue (Thr138) that plays a critical role in positioning the FAD cofactor at the active site of the enzyme (Kim and Miura, 2004). That recombinant ACX1T138I lacked detectable activity against 14:0-CoA indicates that acx1 is most likely a null mutation. If this is the case, the residual JA production in acx1 plants can be attributed to one or more other ACX family members. The sequence of BAC232L13 revealed that LcACX1A is located adjacent to a closely related family member, designated LcACX1B (Figure 3B). Although RT-PCR experiments confirmed that ACX1B is expressed in L. esculentum leaves, the presence of a 15–amino acid deletion in the deduced sequence of ACX1B (see Supplemental Figure 1 online) raises the question of whether this gene encodes an active enzyme. Attempts to express ACX1B in E. coli resulted in failure to detect accumulation of the fusion protein (L. Katsir, A.L. Schilmiller, and G.A. Howe, unpublished results). Inspection of the tomato EST database (http://www.sgn.cornell.edu) provided evidence for additional ACX genes that are homologous to the short-chain (AtACX4) and medium-chain (AtACX3) ACXs of Arabidopsis (Figure 3C). An additional partial-length sequence (SGN-U229242) appears to be homologous to the Arabidopsis long-chain ACX (AtACX2). Given the distinct but overlapping substrate specificity of different ACX family members (Eastmond et al., 2000a; Graham and Eastmond, 2002), it is reasonable to propose that one or more of these tomato ACXs is responsible for the residual JA production in acx1 leaves.
In higher plants, β-oxidation has been extensively studied for its role in converting storage lipids to sucrose during early stages of seedling development. The importance of this process in the plant life cycle is highlighted by the fact that mutants defective in peroxisomal β-oxidation often exhibit developmental arrest at the seedling stage (Hayashi et al., 1998; Zolman et al., 2001; Footitt et al., 2002). It is possible that ACX1A, in addition to its role in JA biosynthesis, also is involved in fatty acid catabolism during seed germination and seedling growth. This idea is supported by the ability of ACX1A to use straight-chain fatty acyl-CoAs (Figure 4B) as well as the accumulation of ACX1A transcripts in germinating tomato seedlings (A.L. Schilmiller and G.A. Howe, unpublished results). On the other hand, the ability of acx1 seedlings to establish photoautotrophic growth in the absence of sucrose (see Supplemental Figure 2 online) indicates that if ACX1A does participate in seed lipid mobilization, this role is not required for seedling establishment. A nonessential role for ACX1A in seed lipid mobilization is consistent with detailed biochemical analysis of the ACX family in Arabidopsis (Hooks et al., 1999; Eastmond et al., 2000a, 2000b; Froman et al., 2000). These collective studies indicate that the substrate specificities of AtACX2 and AtACX3 are likely sufficient to metabolize, in the absence of AtACX1, the full range of medium- and long-chain acyl-CoAs that exist in vivo.
β-Oxidation in nonfatty plant tissues has been implicated in diverse physiological processes, including floral development, synthesis of indole-acetic acid and JA, and production of acetyl-CoA substrate for primary and secondary metabolism (Graham and Eastmond, 2002; Zolman and Bartel, 2004). Aside from defects in JA biosynthesis and JA-mediated defense responses, adult acx1 plants did not exhibit overt morphological or reproductive phenotypes. Based on these observations, we suggest that a primary function of ACX1A in tomato leaves is catalysis of JA synthesis. As mentioned above, we cannot rule out the possibility that the enzyme also participates in β-oxidation of other substrates or that it is involved in JA biosynthesis in nonleaf tissues.
We previously showed that the F-box protein CORONATINE-INSENSITIVE1, which is an essential component of the JA signaling pathway, is required for female fertility in tomato (Li et al., 2004). This finding implies that normal female reproductive development in tomato requires the action of endogenous JAs, which are known to accumulate to high levels in reproductive organs (Hause et al., 2000). Although acx1 homozygotes produce normal amounts of seed, we have observed that the rate of acx1 seed germination varies considerably (from <15 to >90%) between different seed batches. This variability may be related to environmental growth conditions (see Methods). The ability of acx1 plants to produce viable seed is similar to that of the spr2 tomato mutant that is defective in the production of linolenic acid, the major precursor of JAs (Li et al., 2003). We thus suggest that reproductive tissues of acx1 plants, like those of spr2, are capable of producing JAs at a level that is sufficient to promote seed production. Assuming that acx1 is a null mutation, this explanation implies that other ACX isoforms can participate in JA biosynthesis in reproductive tissues.
JA Is Required for Induced Defense Responses in Tomato
The original model for wound signaling in tomato indicated that the terminal product of the octadecanoid pathway, JA, is the active signal for expression of defensive PIs (Farmer and Ryan, 1992). Subsequent studies showing that OPDA exhibits biological activity in some plants (Weiler et al., 1993; Blechert et al., 1999; Koch et al., 1999) raised the possibility that metabolism of OPDA to JA may not be required for wound-induced PI expression. Miersch and Wasternack (2000) addressed this question by testing the PI-inducing activity of various OPC8 derivatives that differ in their ability to undergo β-oxidation. They showed that treatment of tomato leaves with even chain-length derivatives (OPC6 and OPC4) of OPC8 that are β-oxidized to JA resulted in PI-II expression, whereas odd chain-length derivatives (OPC7, OPC5, and OPC3) that are not converted to JA were inactive. These results indicate that activation of PI expression by exogenous OPDA and OPC8 involves β-oxidation of these compounds (Miersch and Wasternack, 2000). Here, we extend this finding by showing that a genetic block in β-oxidation abrogates PI expression in response to mechanical wounding and insect attack. Because acx1 plants are not affected in the biosynthesis of OPDA, we conclude that JA (or a JA derivative) is the biological signal for wound-induced PI expression in tomato leaves.
The increased susceptibility of acx1 plants to tobacco hornworm attack further demonstrates that induced resistance of tomato to this herbivore is mediated by JA rather than by C18 precursors of JA. This conclusion contrasts with previous work suggesting that OPDA is the signal for opr3-mediated resistance of Arabidopsis to insect (Bradysia impatiens) and fungal (Alternaria brassicicola) pests (Stintzi et al., 2001). This disparity may reflect the use of different biotic agents (e.g., chewing versus saprophagous insects) or differences in the mechanism by which octadecanoid pathway-derived signals confer resistance in different plants. The contrasting defense phenotypes of tomato acx1 and Arabidopsis opr3 mutants may also reflect the fact that these mutations affect different steps in the pathway. In tomato acx1 plants, OPDA is presumably converted to OPC8-CoA, which is unable to enter the β-oxidation cycle. Disruption of OPR3, on the other hand, prevents the conversion of OPDA to OPC8. It is possible that OPDA or a related bioactive signal produced as a result of OPR3 disruption (but not ACX1 disruption) activates defense responses that are normally promoted by JA in wild-type plants. This interpretation is consistent with the ability of exogenous OPDA to activate defense gene expression in opr3 plants (Stintzi et al., 2001). The ability of LeACX1A to metabolize OPDA-CoA in vitro also raises the possibility that OPDA may enter the β-oxidation cycle in opr3 plants. Future research aimed at determining the resistance phenotype of JA-deficient β-oxidation mutants of Arabidopsis should be useful to clarify the extent to which OPDA functions as a defense signal in the presence of OPR3. Characterization of tomato mutants defective in LeOPR3 (Strassner et al., 2002) should also provide insight into this question.
Role of β-Oxidation in Systemic Wound Signaling
Grafting experiments with wound-signaling mutants of tomato have provided evidence that JAs act non-cell autonomously to promote systemic expression of defensive PIs and that the peptide signal systemin functions at or near the site of wounding to amplify the octadecanoid signaling pathway (Li et al., 2002b; Ryan and Moura, 2002; Lee and Howe, 2003; Stratmann, 2003; Howe, 2005). Experiments with the spr2 mutant that is defective in a plastidic ω-3 fatty acid desaturase showed that the octadecanoid pathway is required for production of the systemic signal (Li et al., 2002b, 2003). However, because spr2 plants are deficient in all oxylipins derived from trienoic fatty acids (16:3 and 18:3), the precise nature of the signal could not be determined. Results reported herein show that acx1 plants, like the spr2 mutant, lack the ability to produce the graft-transmissible signal for PI expression. Thus, the active compound involved in the production of (or acting as) the systemic signal must be a product of β-oxidation rather than a C18 substrate for β-oxidation. Consistent with previous results (Li et al., 2002b), the ability of acx1 scions to respond to the long-distance signal indicates that de novo synthesis of JA is not required in cells that perceive and respond to the systemic signal. Grafting experiments with a JA-insensitive mutant (jai1) have shown, however, that undamaged scion leaves require a functional JA signaling pathway for recognition of the systemic signal (Li et al., 2002b, 2004). These results add to a growing body of evidence indicating that JA is an essential component of the transmissible wound signal (Howe, 2005).
Current evidence does not exclude the possibility that JA is further metabolized to a compound that is systemically active or that JA induces the production of another signal that is mobilized to distal tissues. In this context, it is interesting to note that the ACX-catalyzed step of β-oxidation produces H2O2 (Figure 1), which was shown to be a second messenger for wound-induced PI expression (Orozco-Cardenas et al., 2001; Sagi et al., 2004). The obligatory coupling of JA biosynthesis to H2O2 production (in a ratio of one JA to three H2O2 molecules) would appear to provide an efficient mechanism to ensure that both JA and H2O2 are present in a cell at the same time. Additional work is needed to determine whether ACX-catalyzed production of H2O2 in response to wounding or systemin plays a role in systemic signaling.
The involvement of β-oxidation in JA biosynthesis implies an important role for peroxisomes in orchestrating plant responses to this stress hormone. A remarkable feature of plant peroxisomes is their capacity to alter their enzymatic content in response to environmental or developmental cues (Olsen, 1998; Corpas et al., 2001). The induced expression of genes involved in β-oxidation and peroxisome biogenesis in response to wounding and pathogen infection further suggests that biotic stress may be a trigger for peroxisome proliferation in certain plant tissues (Lopez-Huertas et al., 2000; Mysore et al., 2002; Schenk et al., 2003; Cruz Castillo et al., 2004). As noted by Strassner et al. (2002), this degree of metabolic plasticity raises the possibility that the terminal steps in JA biosynthesis occur in a specialized type of peroxisome. Our results are consistent with this idea and further suggest that JA-producing peroxisomes play an important role in the generation of signals that mediate systemic defense responses to herbivore attack. Recent studies have shown that plastid enzymes of the octadecanoid pathway are spatially restricted to the companion cell-sieve tube element complex of the vascular bundle (Ryan, 2000; Hause et al., 2003; Howe, 2005). That wound-induced JA accumulation is also enriched in vascular tissues (Stenzel et al., 2003) would suggest that ACX1 and other peroxisomal enzymes of the pathway are located in these cell types as well. Support for this idea comes from studies showing that peroxisomes exist in sieve tube elements of root phloem (Jedd and Chua, 2002) and that soybean ACX1 is localized in the phloem cells of hypocotyl tissue (Agarwal et al., 2001). Future studies aimed at determining the location of JA-producing peroxisomes may provide additional insight into the role of oxylipins in long-distance wound signaling.
METHODS
Plant Materials and Growth Conditions
Tomato (Lycopersicon esculentum) cv Castlemart was used as the wild-type parent for all experiments. Seed for the acx1 mutant was collected from an acx1/acx1 homozygous line derived from successive backcrosses of the original mutant (previously called JL1; Lightner et al., 1993) to the wild type. Seedlings were grown in Jiffy peat pots (Hummert International, Earth City, MO) in a growth chamber maintained under 17 h of light (200 μE/m2/s) at 28°C and 7 h of dark at 18°C. Adult acx1 plants were morphologically indistinguishable from wild-type plants and exhibited normal fruit and seed production. The germination rate of acx1/acx1 seeds ranged between 10 and 92%, depending on the seed batch. We reproducibly observed that the germination rate of acx1/acx1 seed collected from field-grown plants was much higher than that of seed obtained from potted plants grown in the greenhouse (A.L. Schilmiller, C. Li, and G.A. Howe, unpublished results). The overall growth rate of acx1 seedlings grown on MS medium either in the presence or absence of light was comparable to that of wild-type seedlings (see Supplemental Figure 2 online). Seeds of L. pennellii (LA716) and the introgression lines (Eshed and Zamir, 1994) were obtained from the C.M. Rick Tomato Genetic Resource Center (University of California at Davis).
PI-II Assays
Wound-induced PI-II levels in tomato leaves were determined by a radial immunodiffusion assay as previously described (Li et al., 2003; Zhao et al., 2003). The wound-response phenotype of plants within segregating populations was typically performed with 16-d-old plants containing two expanded leaves and a third emerging leaf. A hemostat was employed to produce a crushing-type wound across the midrib of all leaflets of the lower (oldest) leaf. After 3 h, a second wound was inflicted on the same leaflet, proximal to the petiole. Wounded plants were incubated for 24 h under standard growth conditions, after which the wounded leaf (local response) and an upper unwounded leaf (systemic response) were harvested separately for determination of PI-II protein levels. JA feeding experiments were performed as previously described (Lee and Howe, 2003; Li et al., 2003).
Map-Based Cloning
Map-based cloning procedures similar to those described (Li et al., 2002a, 2003) were used to identify the LeACX1A gene. A homozygous acx1 plant (L. esculentum) was crossed to the wild tomato species L. pennellii (LA716), and the resulting F1 plant was backcrossed to the acx1 parental line to generate a BC1 mapping population. The wound-response phenotype of individual BC1 plants was scored by measuring PI-II protein levels in response to mechanical wounding, as described above. Bulked segregant analysis was used in combination with AFLP to identify molecular markers linked to the target locus. Equal amounts of genomic DNA from 10 randomly selected wound-responsive (i.e., wild-type) and 10 nonresponsive (i.e., mutant) BC1 plants were pooled to construct a wild-type DNA bulk (B+) and a mutant DNA bulk (B−), respectively. AFLP markers that distinguished the B+ DNA pool from the B− DNA pool were identified as previously described (Li et al., 2003). One such marker of ∼260 bp in size was identified with the AFLP primer combination E-AAC/M-CTT. This DNA fragment was cloned into a plasmid vector pGEM-T Easy (Promega, Madison, WI) and converted to a RFLP marker as described previously (Li et al., 2002a). Cosegregation of this marker with the wound-response phenotype of the 20 BC1 individuals used to construct the bulks confirmed the tight linkage to the target locus (data not shown). Tomato introgression lines (ILs) harboring defined L. pennellii chromosome segments in the L. esculentum background (Eshed and Zamir, 1994) were used to map the marker to the IL8-2 region on chromosome 8 (data not shown). Subsequent mapping experiments with RFLP markers in this region (Tanksley et al., 1992) further refined the target locus to the region between RFLP marker CT111 and a second marker constructed from tomato EST clone cLED14K7 (C. Li and G. Howe, unpublished results) (Figure 3A).
For fine mapping of the target gene, the size of the mapping population was expanded to 1200 BC1 plants that were scored for wound-induced PI-II expression and subsequently genotyped. Sixty plants showing recombination between CT111 and cLED14K7 were identified. These recombinants were then genotyped with two additional markers (GP40 and TG510) located within the target interval. Results from these experiments showed that the target gene was flanked by RFLP marker cLED14K7 at a distance of 1.6 centimorgans (20 recombinants) and at a distance of 1 centimorgan (12 recombinants) by GP40. TG510 cosegregated with the target gene in all 1200 BC1 plants (Figure 3A). TG510 was therefore used to screen a BAC library constructed from L. esculentum genomic DNA (Budiman et al., 2000). For unknown reasons, positive clones were not identified from this BAC library. Screening of an L. cheesmanii BAC library with TG510 resulted in identification of two overlapping BAC clones, 166B24 and 232L13. DNA clones corresponding to the BAC ends were isolated by thermal asymmetric interlaced–PCR as previously described (Li et al., 2003). These clones were hybridized to BAC DNAs to determine the relative orientation of the two BACs to each other. BAC ends were also converted to RFLP markers and mapped relative to the target locus, using the 32 recombinants identified in the GP40-TG510-cLED14k7 interval. The left end of BAC166B24 (166B24L) mapped one recombination event to the left of the target gene. Both the right end of BAC166B24 (166B24R) and the left end of BAC232L13 (232L13L) cosegregated with the target gene. A cloned fragment corresponding to the right end of BAC232L13 (232L13R1) was not mapped because a suitable marker polymorphism could not be identified between L. pennellii and L. esculentum. A marker from the right end of BAC232L13 that shows recombination with the target gene was developed as follows. BAC232L13 DNA was digested to completion with HindIII, and the resulting fragments were shotgun cloned into pGEM-7Zf(+). One of these subclones, designated 232L13R2, was mapped two recombinants to the right of the target gene (Figure 3A). These results provided strong evidence that the target gene is physically located on BAC232L13.
Isolation of LeACX cDNAs and DNA Sequence Analysis
Shotgun DNA sequencing of L. cheesmanii BAC232L13 and subsequent assembly of the sequence was performed by Lark Technologies (Houston, TX). The shotgun library of fragmented BAC DNA was constructed in cloning vector pUC19. In total, 1536 sequencing reactions were performed and analyzed on ABI3730xl DNA analyzers (Applied Biosystems, Foster City, CA). The reads were screened for quality and vector sequence with the Phred-Phrap software package (Ewing et al., 1998). A total of 1228 reads, having an average read length of >600 bp, passed the screen. The raw data from the 1228 reads provided approximately sixfold coverage of the BAC clone. The sequence of a 30-kb contig that contains the L. cheesmanii ACX1A and ACX1B genes was deposited in GenBank with accession number AY817111.
RT-PCR of RNA isolated from 5-d-old wild-type and acx1 seedlings was employed to obtain full-length LeACX1A and LeACX1B cDNAs. The forward and reverse oligonucleotide primers for amplification of the LeACX1A cDNA were 5′-CTGAGAGTAAGAGAGATGGAG-3′ and 5′-CTGGGAGGAAAAGAAGCCAAA-3′, respectively. The RNA template for RT-PCR amplification of LeACX1B was prepared from leaves of 4-week-old plants. Forward and reverse primers for these reactions were 5′-TTGTATTCTATTTCATCCCTGAGAG-3′ and 5′-CTCCCATTTTCTGCAGTTCTTC-3′, respectively, which yielded a 2005-bp product.
For construction of the ACX phylogenetic tree (Figure 3C), full-length amino acid sequences were aligned using ClustalX v.1.81 for MAC (Thompson et al., 1997). Mean character distances were used to construct an unrooted neighbor-joining phylogeny using PAUP 4.0*, version 4.0b10 for MAC PowerPC (Swofford, 2000).
Construction of the L. cheesmanii BAC Library
L. cheesmannii seedlings (accession LA483; C. Rick Tomato Genetic Resource Center, Davis, CA) were greenhouse grown for 1 month after sewing and then dark grown for 3 d before harvest to minimize starch content. Young expanding leaves were collected for isolation of nuclei. Extraction of high molecular weight genomic DNA from isolated nuclei was performed as described by Zhang et al. (1996). Resulting high molecular weight tomato genomic DNA was partially digested with HindIII, followed by two rounds of size selection from agarose gels. In the first size selection, pulsed field gel electrophoresis was performed with a 70-s pulse at 5.5 V/cm for a total run time of 18 h at 11°C. DNA gel slices, including 150- to 500-kb fragments, were excised from the gel based on λ-DNA molecular weight markers (New England Biolabs, Beverly, MA) and used in the next size selection. The second size selection gel was run using a 4-s pulse at 5 V/cm for 11 h at 11°C, and the compressed band representing DNA fragments >100 kb was excised. Before ligation, the final size-selected DNA was released from agarose by electroelution using 1-cm dialysis tubing (Gibco BRL, Rockville, MD). Electroelution was performed at 11°C for 2 h at 5 V/cm followed by a 90-s reversed-current pulse. Eluted DNA was quantified by inspection on an agarose gel. HindIII-digested and dephosphorylated pBeloBAC II vector (Shizuya et al., 1992) was used for library construction. A molar ratio of ∼3:1 (vector:insert) was used for ligation. Bacterial transformation was performed by electroporation using Gene Hogs electrocompetent Escherichia coli cells (DH10B) with procedures provided by the supplier (Invitrogen, Carlsbad, CA). Transformed cells were plated onto LB agar (Difco, Sparks, MD) supplemented with 12.5 μg/mL of chloramphenicol and 0.25% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and 104,832 white colonies were picked and stored at −80°C in 273 384-well microtiter plates as glycerol stocks. High-density nylon hybridization filters were created with a Q-bot robotic workstation (Genentix, New Milton, UK).
Biochemical Characterization of LeACX1A
A full-length LeACX1A EST clone (cLES14H13) obtained from Clemson University Genomics Institute was used as the template for a PCR-based approach to construct a vector for expression of ACX1A with an N-terminal His6 tag. The forward primer (5′-GAGCTCGTAAGAGAGATGGAGGGTGTA-3′) was designed with a SacI site and the reverse primer (5′-CCGCTCGAGCGGAACAGTTTGCTGCAGCTC-3′) spanned a PstI site located in the 3′-untranslated region of LeACX1A. PCR amplification yielded a 2047-bp product that was subcloned into pGEM-T easy (Promega). After digestion with SacI and PstI, the LeACX1A cDNA insert in this construct was cloned into the same sites of expression vector pQE30 (Qiagen, Valencia, CA). The resulting construct, which added 19 amino acids (MRGSHHHHHHGSACELVRE) to the N terminus of ACX1A, was transformed into the Rosetta strain of E. coli (Novagen, Madison, WI). The expression construct for ACXT138I was prepared by the same procedure, except the template for PCR was the LeACX1A cDNA derived from the acx1 mutant.
Expression of recombinant ACX1A proteins (ACX1wt and ACX1T138I) was initiated by inoculating 1 mL of an overnight culture into 200 mL of Terrific Broth medium supplemented with 100 μg/mL of ampicillin and 12.5 μg/mL of chloramphenicol. Bacteria were grown at 37°C in a shaker (250 rpm) to an OD600 of 0.5. Cultures were then cooled to 30°C, and isopropyl-thio-β-d-galactopyranoside (Roche, Indianapolis, IN) was added to a final concentration of 0.1 mM. The induced cultures were incubated at 30°C for 18 h with gentle shaking (120 rpm). Cells were harvested by centrifugation and either stored at −20°C or used immediately for ACX purification. The cell paste was resuspended in 50 mM potassium phosphate, pH 7.6, 150 mM NaCl, and 10% (v/v) glycerol and then lysed by sonication. His-tagged ACX proteins were purified from the cleared lysate by nickel affinity chromatography (nickel-nitrilotriacetic acid agarose resin; Qiagen) according to manufacturer's directions. During the purification procedure, FAD (Sigma-Aldrich, St. Louis, MO) was added to all buffers at a final concentration of 10 μM. Protein measurements were performed by the BCA assay (Pierce, Rockford, IL), with BSA as a standard. ACX activity with commercially available acyl-CoA substrates (Sigma-Aldrich) was measured with an H2O2-coupled spectrophotometric assay as described previously (Hryb and Hogg, 1979). Typical enzyme assays contained 0.5 μg of affinity-purified ACX and 50 μM of acyl-CoA substrate.
ACX1A-catalyzed metabolism of OPDA-CoA and OPC8-CoA was measured with an enzymatic colorimetric assay (Roche) that is typically used to measure free fatty acid levels in serum or plasma (Shimizu et al., 1980). The method is based on the activation of nonesterified fatty acids by ACS to the corresponding CoA ester, followed by detection of ACX-catalyzed H2O2 production (Shimizu et al., 1980). The test kit provided by the manufacture uses a yeast ACS and an unspecified ACX for the coupled reaction. For our studies, the ACX provided by the manufacturer was replaced with affinity-purified LeACX1A. Fatty acid substrate (dissolved in 0.25% Triton X-100) was added to 100 μL of the manufacturer's reaction mix A (containing ATP, CoA, ACS, and sodium phosphate, pH 7.8). Concentrated stocks of OPDA and OPC8 in ethanol were diluted directly in 0.25% Triton X-100 (v/v). The sodium salt of myristic acid (14:0) was prepared according to Shimizu et al. (1980). Reactions were allowed to proceed for 30 min at 25°C, at which time excess CoA was trapped by alkylation via the addition of 5 μL of N-ethyl-maleinimide. The ACX reaction was then initiated by addition of 1 μg of purified LeACX1A. H2O2 produced in this reaction was used by a peroxidase to convert 2,4,6-tribromo-3-hydroxy-benzoic acid and 4-aminoantipyrine to a red dye that was measured at 546 nm with a Beckman DU530 spectrophotometer (Fullerton, CA). OPC8 was chemically synthesized according to the procedure of Ainai et al. (2003). NMR spectroscopy showed that the OPC8 was >97% pure and that the ratio of the cis and trans isomer was >20:1. OPDA was obtained from Larodan Fine Chemicals (Malmö, Sweden).
Agrobacterium tumefaciens–Mediated Transformation
A full-length LeACX1A EST clone (cLES14H13) was used as the template for a PCR-based approach to construct a binary vector for expression of LeACX1A in the acx1 genetic background. The forward and reverse primer pair for this reaction was 5′-GCTCTAGAGCGTAAGAGAGATGGAGGGT-3′ and 5′-CGAGCTCGAACAGTTTGCTGCAGCTCTCG-3′, respectively. The resulting 2047-bp PCR product was cloned into the XbaI and SacI sites of binary vector pBI121 (Clontech, Palo Alto, CA) under the control of the 35S promoter of Cauliflower mosaic virus. This construct was transformed into Agrobacterium strain AGLO, which was used to perform Agrobacterium-mediated transformation of acx1 cotyledon explants as previously described (Li et al., 2003). Kanamycin-resistant regenerated transformants in which the presence of the 35S-ACX1A transgene was confirmed by PCR were potted into standard soil mix and grown in a growth chamber under standard conditions. Approximately 3 weeks after transfer to soil, these T0 plants were assayed for PI-II accumulation in response to mechanical wounding.
JA and OPDA Measurements
Leaves of 3-week-old tomato plants were wounded with a hemostat as described above or were subject to damage by Manduca sexta larvae. Wounded leaf tissue (500 mg fresh weight) from three different plants was pooled and immediately frozen in liquid nitrogen. Leaf tissue was also harvested from unwounded plants as a control. JA was extracted and quantified as described by Schmelz et al. (2004). Briefly, 50 ng of dihydrojasmonic acid was added to each sample as an internal standard. Frozen tissue was ground to a fine powder with a mortar and pestle and then transferred to a tube containing 0.6 mL of 1-propanol (67% v/v). After brief vortexing of this mixture, 1 mL of methylene chloride was added and the pH was adjusted to 4.0 with HCl. This mixture was vortexed and then centrifuged at 4000g for 5 min. After transfer to a new tube, the organic layer was derivatized with 2 μL of trimethylsilyldiazomethane (Sigma-Aldrich) for 30 min at room temperature. Methylated compounds in this mixture were volatilized by heating and collected on a volatile collection trap (Analytical Research Systems, Gainesville, FL) as described previously (Schmelz et al., 2004). Compounds were eluted from the volatile collection trap resin with methylene chloride and subsequently analyzed by gas chromatography–mass spectrometry (GC-MS).
GC-MS analysis was performed in the selected ion monitoring mode with isobutane chemical ionization (Schmelz et al., 2004). The GC-MS system consisted of a 6890 Network GC connected to a 5973 inert Mass Selective Detector (Agilent, Palo Alto, CA). Compounds were separated on an HP5MS column (30 m × 0.25 mm × 0.25 μm). The temperature regime for GC was 40°C for 1 min after injection, followed by sequential temperature ramps of 25°C/min to 150°C, 5°C/min to 200°C, and a 10°C/min ramp to 240°C. The 240°C temperature was maintained for 10 min. The identity of methyl-JA (MeJA) in plant samples was confirmed by comparison of the elution time and mass spectra to an authentic MeJA standard (Sigma-Aldrich). The retention time of the cis and trans isomers of MeJA was 12.65 and 13.18 min, respectively. Both isomers generated a [M + H]+ parent ion at a mass-to-charge ratio of 225. The retention time of the cis and trans isomers of the methyl-dihydrojasmonic acid internal standard was 12.72 and 13.20 min, respectively, both of which produced a [M + H]+ parent ion at a mass-to-charge ratio of 227. The combined peak area of the cis and trans isomers of each compound was used for quantification of total JA. OPDA was extracted and analyzed as described previously (Li et al., 2002b). Tetrahydro-OPDA was used as an internal standard for quantification. Standard curves were constructed with known quantities of MeJA and methyl-OPDA, together with their corresponding internal standards.
Nucleic Acid Analysis
DNA and RNA extraction and gel blot analyses were conducted as described previously (Li et al., 2003, 2004). cDNA clones for CDI, LoxD, and OPR3 were as described by Li et al. (2004). Duplicate RNA gels were stained with ethidium bromide to ensure equal loading of samples and quality of the RNA. A cDNA for tomato translation initiation factor eIF4a (cLED1D24) was hybridized to duplicate RNA gel blots as a loading control.
Insect Feeding Trials
Tobacco hornworm (M. sexta) eggs and artificial diet for hornworm larvae were obtained from Carolina Biological Supply Company (Burlington, NC). Eggs were hatched at 26°C as recommended by the supplier. Hatched larvae were reared on the artificial diet for 3 d before transfer to tomato plants. The average weight of larvae at the beginning of the feeding trial was ∼5 mg.
Sequence data for the full-length cDNA for LeACX1A and LeACX1B have been deposited with the EMBL/GenBank data libraries under accession numbers AY817109 and AY817110, respectively. Accession numbers for the sequences shown in Figure 3C are as follows: AtACX1, T52121; AtACX2, T52120; AtACX3, AAM20431; AtACX4, AAM19813; AtACX5, NP_181112; AtACX6, NP_172120; GmACX1-1, AAL01887; GmACX1-2, AAL01888; HvACX, CAA04688; LeACX1A, AY817109; LeACX1B, AY817110; PcACX, AAF14635; Phalaenopsis ACX, AAB67883; pumpkin ACX, T07901; ZmACX, AY105897 and AY104290. U220503, U223228, and U216994 are L. esculentum unigene sequences compiled from the EST sequence database at the Solanaceae Genomics Network (http://www.sgn.cornell.edu).
Supplementary Material
Acknowledgments
We acknowledge Wayne Riekhof and Brian Barnett for assistance with the preliminary characterization of acx1 plants and Bonnie McCaig for assistance with the ACX phylogeny. We thank Hans Weber (University of Lausanne, Switzerland), Eric Schmeltz (USDA–Agricultural Research Service, Gainesville, FL), and the MSU Mass Spectrometry Facility (East Lansing, MI) for helpful advice with JA and OPDA measurements. We also acknowledge Steve Tanksley (Cornell University, Ithaca, NY) for providing the tomato RFLP markers used in this study. Tomato EST clones were obtained from the Clemson University Genomics Institute (Clemson, SC). This research was supported by the National Institutes of Health (Grant GM57795 to G.A.H.) and by the U.S. Department of Energy (Grant DE-FG02-91ER20021 to G.A.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gregg A. Howe (howeg@msu.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.029108.
References
- Agarwal, A.K., Qi, Y., Bhat, D.G., Woerner, B.M., and Brown, S.M. (2001). Gene isolation and characterization of two acyl CoA oxidases from soybean with broad substrate specificities and enhanced expression in the growing seedling axis. Plant Mol. Biol. 47, 519–531. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ainai, T., Matsuumi, M., and Kobayashi, Y. (2003). Efficient total synthesis of 12-oxo-PDA and OPC-8:0. J. Org. Chem. 68, 7825–7832. [DOI] [PubMed] [Google Scholar]
- Beale, M.H., and Ward, J.L. (1998). Jasmonates: Key players in the plant defence. Nat. Prod. Rep. 15, 533–548. [DOI] [PubMed] [Google Scholar]
- Blechert, S., Bockelmann, C., Füsslein, M., Schrader, T.V., Stelmach, B., Niesel, U., and Weiler, E.W. (1999). Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207, 470–479. [Google Scholar]
- Budiman, M.A., Mao, L., Wood, T.C., and Wing, R.A. (2000). A deep-coverage tomato BAC library and prospects towards development of an STC framework for genome sequencing. Genome Res. 10, 129–136. [PMC free article] [PubMed] [Google Scholar]
- Corpas, F.J., Barroso, J.B., and del Rio, L.A. (2001). Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 6, 145–150. [DOI] [PubMed] [Google Scholar]
- Cruz Castillo, M., Martinez, C., Buchala, A., Métraux, J.P., and León, J. (2004). Gene-specific involvement of beta-oxidation in wound-activated responses in Arabidopsis. Plant Physiol. 135, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe, W., Miersch, O., and Schmidt, J. (1989). Occurance of jasmonic acid, related compounds and abscisic acid in fertile and sterile fronds of three Equisetum species. Biochem. Physiol. Pflanzen 185, 83–92. [Google Scholar]
- Eastmond, P.J., Eastmond, P.J., Hooks, M.A., Williams, D., Lange, P., Bechtold, N., Sarrobert, C., Nussaume, L., and Graham, I.A. (2000. b). Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J. Biol. Chem. 275, 34375–34381. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., Hooks, M.A., and Graham, I.A. (2000. a). The Arabidopsis acyl-CoA oxidase gene family. Biochem. Soc. Trans. 28, 755–757. [PubMed] [Google Scholar]
- Eshed, Y., and Zamir, D. (1994). A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 79, 175–179. [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E., and Ryan, C.A. (1992). Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann, J., Schuler, G., Boland, W., Ebel, J., and Mithofer, A. (2003). The role of octadecanoids and functional mimics in soybean defense responses. Biol. Chem. 384, 437–446. [DOI] [PubMed] [Google Scholar]
- Footitt, S., Slocombe, S.P., Larner, V., Kurup, S., Wu, Y., Larson, T., Graham, I., Baker, A., and Holdsworth, M. (2002). Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 21, 2912–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froman, B.E., Edwards, P.C., Bursch, A.G., and Dehesh, K. (2000). ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol. 123, 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, I.A., and Eastmond, P.J. (2002). Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog. Lipid Res. 41, 156–181. [DOI] [PubMed] [Google Scholar]
- Haider, G., von Schrader, T., Fusslein, M., Blechert, S., and Kutchan, T.M. (2000). Structure-activity relationships of synthetic analogs of jasmonic acid and coronatine on induction of benzo[c]phenanthridine alkaloid accumulation in Eschscholzia californica cell cultures. Biol. Chem. 381, 741–748. [DOI] [PubMed] [Google Scholar]
- Hause, B., Hause, G., Kutter, C., Miersch, O., and Wasternack, C. (2003). Enzymes of jasmonate biosynthesis occur in tomato sieve elements. Plant Cell Physiol. 44, 643–648. [DOI] [PubMed] [Google Scholar]
- Hause, B., Stenzel, I., Miersch, O., Maucher, H., Kramell, R., Ziegler, J., and Wasternack, C. (2000). Tissue-specific oxylipin signature of tomato flowers: Allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 24, 113–126. [DOI] [PubMed] [Google Scholar]
- Hayashi, M., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks, M.A., Kellas, F., and Graham, I.A. (1999). Long-chain acyl-CoA oxidases of Arabidopsis. Plant J. 20, 1–13. [DOI] [PubMed] [Google Scholar]
- Hou, X., Roberts, L.J., Taber, D.F., Morrow, J.D., Kanai, K., Gobeil, F., Beauchamp, M.H., Bernier, S.G., Lepage, G., Varma, D.R., and Chemtob, S. (2001). 2,3-Dinor-5,6-dihydro-15-F(2t)-isoprostane: A bioactive prostanoid metabolite. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R391–R400. [DOI] [PubMed] [Google Scholar]
- Howe, G.A. (2005). Jasmonates as signals in the wound response. J. Plant Growth Regul., in press.
- Hryb, D.J., and Hogg, J.F. (1979). Chain length specificities of peroxisomal and mitochondrial β-oxidation in rat liver. Biochem. Biophys. Res. Commun. 87, 1200–1206. [DOI] [PubMed] [Google Scholar]
- Jedd, G., and Chua, N.H. (2002). Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 43, 384–392. [DOI] [PubMed] [Google Scholar]
- Kim, J.J., and Miura, R. (2004). Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences. Eur. J. Biochem. 271, 483–493. [DOI] [PubMed] [Google Scholar]
- Koch, T., Krumm, T., Jung, V., Engelberth, J., and Boland, W. (1999). Differential induction of plant biosynthesis in the lima bean by early and late intermediates of the octadecanoid-signaling pathway. Plant Physiol. 121, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G.I., and Howe, G.A. (2003). The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 33, 567–576. [DOI] [PubMed] [Google Scholar]
- Li, C., Liu, G., Xu, C., Lee, G.I., Bauer, P., Ling, H.Q., Ganal, M., and Howe, G.A. (2003). The tomato Suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15, 1646–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Williams, M.M., Loh, Y.T., Lee, G.I., and Howe, G.A. (2002. a). Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 130, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Li, C., Lee, G.I., and Howe, G.A. (2002. b). Distinct roles for jasmonic acid synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 99, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zhao, Y., McCaig, B.C., Wingerd, B.A., Wang, J., Whalon, M.E., Pichersky, E., and Howe, G.A. (2004). The tomato homolog of COI1 is required for maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti, R., and Farmer, E.E. (2002). The jasmonate pathway. Science 296, 1649–1650. [DOI] [PubMed] [Google Scholar]
- Lightner, J., Pearce, G., Ryan, C.A., and Browse, J. (1993). Isolation of signaling mutants of tomato (Lycopersicon esculentum). Mol. Gen. Genet. 241, 595–601. [DOI] [PubMed] [Google Scholar]
- Lopez-Huertas, E., Charlton, W.L., Johnson, B., Graham, I.A., and Baker, A. (2000). Stress induces peroxisome biogenesis genes. EMBO J. 19, 6770–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch, O., Schmidt, J., Sembdner, G., and Schreiber, K. (1989). Jasmonic acid-like substances from the culture filtrate of Botryodiplodia theobromae. Phytochemistry 28, 1303–1305. [Google Scholar]
- Miersch, O., and Wasternack, C. (2000). Octadecanoid and jasmonate signaling in tomato (Lycopersicon esculentum Mill.) leaves: Endogenous jasmonates do not induce jasmonate biosynthesis. Biol. Chem. 381, 715–722. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Crasta, O.R., Tuori, R.P., Folkerts, O., Swirsky, P.B., and Martin, G.B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32, 299–315. [DOI] [PubMed] [Google Scholar]
- Nakajima, Y., Miyahara, I., Hirotsu, K., Nishina, Y., Shiga, K., Setoyama, C., Tamaoki, H., and Miura, R. (2002). Three-dimensional structure of the flavoenzyme acyl-CoA oxidase-II from rat liver, the peroxisomal counterpart of mitochondrial acyl-CoA dehydrogenase. J. Biochem. 131, 365–374. [DOI] [PubMed] [Google Scholar]
- Olsen, L.J. (1998). The surprising complexity of peroxisome biogenesis. Plant Mol. Biol. 38, 163–189. [PubMed] [Google Scholar]
- Orozco-Cardenas, M.L., Narváez-Vásquez, J., and Ryan, C.A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13, 179–191. [PMC free article] [PubMed] [Google Scholar]
- Reumann, S., Ma, C., Lemke, S., and Babujee, L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136, 2587–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C.A. (2000). The systemin signaling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 1477, 112–121. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A., and Moura, D.S. (2002). Systemic wound signaling in plants: A new perception. Proc. Natl. Acad. Sci. USA 99, 6519–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, M., Davydov, O., Orazova, S., Yesbergenova, Z., Ophir, R., Stratmann, J.W., and Fluhr, R. (2004). Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16, 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, F. (2001). Enzymes of the biosynthesis of octadecanoid-derived signaling molecules. J. Exp. Bot. 52, 11–23. [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Manners, J.M., Anderson, J.P., Simpson, R.S., Wilson, I.W., Somerville, S.C., and Maclean, D.J. (2003). Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 132, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz, E.A., Engelberth, J., Tumlinson, J.H., Block, A., and Alborn, T.H. (2004). The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39, 790–808. [DOI] [PubMed] [Google Scholar]
- Shimizu, S., Tani, Y., Yamada, H., Tabata, M., and Murachi, T. (1980). Enzymatic determination of serum-free fatty acids: A colorimetric method. Anal. Biochem. 107, 193–198. [DOI] [PubMed] [Google Scholar]
- Shizuya, A., Birren, B., Kim, U., Mancino, V., Slepak, T., Tachiiri, Y., and Simon, M. (1992). Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in E. coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89, 8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey, J.M., Fulda, M.S., and Browse, J. (2003). Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol. 132, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel, I., Hause, B., Maucher, H., Pitzschke, A., Miersch, O., Ziegler, J., Ryan, C.A., and Wasternack, C. (2003). Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato—Amplification in wound signalling. Plant J. 33, 577–589. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner, J., Schaller, F., Frick, U., Howe, G.A., Weiler, E.W., Amrhein, N., Macheroux, P., and Schaller, A. (2002). Characterization and cDNA microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in local versus systemic wound responses. Plant J. 32, 585–601. [DOI] [PubMed] [Google Scholar]
- Stratmann, J.W. (2003). Long distance run in the wound response—Jasmonic acid is pulling ahead. Trends Plant Sci. 8, 247–250. [DOI] [PubMed] [Google Scholar]
- Swiatek, A., Van Dongen, W., Esmans, E.L., and Van Onckelen, H. (2004). Metabolic fate of jasmonates in tobacco bright yellow-2 cells. Plant Physiol. 135, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (2000). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates).
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilton, G.B., Shockey, J.M., and Browse, J. (2004). Biochemical and molecular characterization of ACH2, an acyl-CoA thioesterase from Arabidopsis thaliana. J. Biol. Chem. 279, 7487–7494. [DOI] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.), S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick, B., and Zimmerman, D.C. (1984). Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 75, 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C., and Hause, B. (2002). Jasmonates and octadecanoids: Signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 72, 165–221. [DOI] [PubMed] [Google Scholar]
- Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. [DOI] [PubMed] [Google Scholar]
- Weiler, E.W., Albrecht, T., Groth, B., Xia, Z.-Q., Luxem, M., Liss, H., Andert, L., and Spengler, P. (1993). Evidence for the involvement of jasmonates and their octadecanoid precursors in the tendril coiling response of Bryonia dioica. Phytochemistry 32, 591–600. [Google Scholar]
- Weiler, E.W., Kutchan, T.M., Gorba, T., Brodschelm, W., Niesel, U., and Bublitz, F. (1994). The Pseudomonas phytotoxin coronatine mimics octadecanoid signaling molecules of higher plants. FEBS Lett. 345, 9–13. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Woo, S., and Wing, R. (1996). BAC, YAC and cosmid library construction. In Plant Gene Isolation: Principles and Practice, G. Foster and D. Twell, eds (Chichester, UK: John Wiley & Sons), pp. 75–99.
- Zhao, Y., Thilmony, R., Bender, C., He, S.Y., and Howe, G.A. (2003). The Hrp type III secretion system and coronatine of Pseudomonas syringae pv. tomato coordinately modify host defense by targeting the jasmonate signaling pathway in tomato. Plant J. 36, 485–499. [DOI] [PubMed] [Google Scholar]
- Zolman, B.K., and Bartel, B. (2004). An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc. Natl. Acad. Sci. USA 101, 1786–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman, B.K., Silva, I.D., and Bartel, B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127, 1266–1278. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.