Abstract
Plants treated with the nonprotein amino acid β-aminobutyric acid (BABA) develop an enhanced capacity to resist biotic and abiotic stresses. This BABA-induced resistance (BABA-IR) is associated with an augmented capacity to express basal defense responses, a phenomenon known as priming. Based on the observation that high amounts of BABA induce sterility in Arabidopsis thaliana, a mutagenesis screen was performed to select mutants impaired in BABA-induced sterility (ibs). Here, we report the isolation and subsequent characterization of three T-DNA–tagged ibs mutants. Mutant ibs1 is affected in a cyclin-dependent kinase–like protein, and ibs2 is defective in AtSAC1b encoding a polyphosphoinositide phosphatase. Mutant ibs3 is affected in the regulation of the ABA1 gene encoding the abscisic acid (ABA) biosynthetic enzyme zeaxanthin epoxidase. To elucidate the function of the three IBS genes in plant resistance, the mutants were tested for BABA-IR against the bacterium Pseudomonas syringae pv tomato, the oomycete Hyaloperonospora parasitica, and BABA-induced tolerance to salt. All three ibs mutants were compromised in BABA-IR against H. parasitica, although to a different extent. Whereas ibs1 was reduced in priming for salicylate (SA)-dependent trailing necrosis, mutants ibs2 and ibs3 were affected in the priming for callose deposition. Only ibs1 failed to express BABA-IR against P. syringae, which coincided with a defect in priming for SA-inducible PR-1 gene expression. By contrast, ibs2 and ibs3 showed reduced BABA-induced tolerance to salt, which correlated with an affected priming for ABA-inducible gene expression. For all three ibs alleles, the defects in BABA-induced sterility and BABA-induced protection against P. syringae, H. parasitica, and salt could be confirmed in independent mutants. The data presented here introduce three novel regulatory genes involved in priming for different defense responses.
INTRODUCTION
Plants have evolved sophisticated mechanisms to defend themselves against pathogens. Besides constitutively expressed barriers, plants can recognize the presence of a pathogen and respond by activating defense reactions. The success of this defense response depends on the speed by which the plant recognizes the attacking pathogen and the intensity by which the appropriate defense mechanism is activated. If the plant fails to respond in time, the appropriate defenses are activated too late, and the pathogen can colonize the plant tissue. In this situation, the plant activates defense mechanisms around the sites of pathogen invasion. These pathogen-inducible defenses are part of the basal resistance response and contribute to slowing down the colonization by the pathogen. The effectiveness of this basal resistance can be enhanced by specific biotic or abiotic stimuli experienced by the plant before contact with the pathogen (Sticher et al., 1997; Mauch-Mani and Métraux, 1998; Pieterse et al., 1998; Zimmerli et al., 2000; Ton et al., 2002). This phenomenon is generally known as induced resistance.
The classical type of induced resistance is often referred to as systemic acquired resistance (SAR) and occurs both in adjacent and distal plant parts upon localized infection by a necrotizing pathogen (Sticher et al., 1997). The signaling pathway controlling SAR requires endogenous accumulation of the stress hormone salicylic acid (SA; Gaffney et al., 1993; Nawrath and Métraux, 1999) and an intact defense regulatory protein No PR-1/No Immunity 1/Salicylic Acid Insensitive 1 (NPR1/NIM1/SAI1) (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). The expression of SAR, triggered by either pathogen infection or treatment with SA or its functional analogs 2,6-dichloroisonicotinic acid or benzothiadiazole (BTH), is tightly associated with the transcriptional activation of genes encoding pathogenesis-related proteins (PRs; Van Loon, 1997).
The nonprotein amino acid β-aminobutyric acid (BABA) activates an induced resistance response as well. Although the protective effect of BABA against numerous plant diseases is well documented (Jakab et al., 2001; Cohen, 2002), the molecular mechanisms behind this type of induced resistance response are less well studied than those of SAR. Only recently, it became clear that the signaling pathway controlling BABA-induced resistance (BABA-IR) is partially different from that of SAR. BABA-IR against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 and the fungal pathogen Botrytis cinerea resembles SAR in that it requires endogenous accumulation of SA and an intact NPR1/NIM1/SAI1 protein (Zimmerli et al., 2000, 2001). However, BABA-IR against the oomycetous pathogen Hyaloperonospora parasitica, as well as BABA-IR against the necrotrophic fungus Plectosphaerella cucumerina, is unaffected in SA-nonaccumulating NahG plants and SA-insensitive npr1-1 plants (Zimmerli et al., 2000; Ton and Mauch-Mani, 2004). By contrast, mutants impaired in the production or sensitivity to the stress hormone abscisic acid (ABA) are blocked in BABA-IR against P. cucumerina (Ton and Mauch-Mani, 2004). This points to the existence of an additional ABA-dependent defense signaling pathway that functions independently of SA and NPR1/NIM1/SAI1. Hence, BABA-IR involves both SA-dependent and ABA-dependent defense mechanisms, and the importance of these defenses varies according to the nature of the challenging pathogen.
An intriguing aspect of BABA-IR is that it confers protection against an extraordinarily wide range of biotic and abiotic stresses (Cohen, 2002). In Arabidopsis thaliana, BABA-IR has been shown to be effective against both necrotrophic and biotrophic pathogens, as well as certain types of abiotic stress, such as drought and salinity (Jakab et al., 2001; G. Jakab and J. Ton, unpublished results). This poses the question: How can BABA induce resistance against so many stresses without causing major trade-off effects on plant growth because of energy investments in costly defense mechanisms? A plausible explanation is that BABA does not directly activate the plant's defense arsenal, but rather conditions the plant for a faster and stronger activation of stress-specific defense mechanisms upon exposure to stress (Zimmerli et al., 2000; Jakab et al., 2001; Ton and Mauch-Mani, 2004). This mode of protection confers enhanced resistance without direct activation of defense mechanisms upon induction treatment. By analogy with a phenotypically similar phenomenon in animals and humans (Hayes et al., 1991; Wyatt et al., 1996), this enhanced capacity to express basal defense mechanisms is called sensitization, potentiation, or priming (Conrath et al., 2002).
The physiological and molecular mechanisms underlying priming are widely unknown. Historically, induced resistance research has mostly been concentrating on direct activation of defense mechanisms upon treatment with the resistance-inducing agent. The possibility of priming as a mechanism of protection has often been overseen because it only becomes apparent in stressed plants. First reports suggesting the occurrence of priming in relation to induced resistance were purely descriptive. In several systems, the timing of the onset of defense was found to be altered: induced plants responded more efficiently to pathogen attack by a faster and stronger accumulation of callose, a higher level of cell wall lignification, or a faster activation of defense-related gene expression. In all these cases, the augmented defense reaction correlated with an enhanced resistance (Skipp and Deverall, 1973; Dean and Kuc, 1987; Kovats et al., 1991a, 1991b; Silverman et al., 1995). The first systematic investigation of priming was initiated by Kauss and colleagues (Kauss et al., 1992a, 1992b). Using the model system of parsley (Petroselinum crispum) cell suspension cultures, they showed that pretreatment with SA, or its functional analogs 2,6-dichloroisonicotinic acid and BTH, primed the cells to secrete elevated amounts of derivatives of the phytoalexin coumarin after treatment with the pmg elicitor from Phytophthora megasperma (Kauss et al., 1992a; Katz et al., 1998; Kohler et al., 2002). Additionally, Mur et al. (1996) showed that tobacco (Nicotiana tabacum) plants pretreated with SA displayed augmented expression of defense-related genes that are usually not directly responsive to SA itself. They proposed a dual role for SA in induced resistance: a direct one leading to the expression of some PR proteins and an indirect one regulating augmented expression of defense-related genes, such as phenylalanine ammonia-lyase, upon elicitation of defense.
In Arabidopsis, treatment with BABA results in a priming for multiple defense mechanisms. In the first place, BABA primes for SA-dependent defense mechanisms, as evidenced by an augmented expression of the SA-inducible marker gene PR-1 upon infection by P. syringae pv tomato or B. cinerea (Zimmerli et al., 2000, 2001). The fact that NahG and npr1-1 plants are unable to express this BABA-IR suggests that the priming for SA-inducible defenses is critical for the protection against these pathogens (Zimmerli et al., 2000, 2001). During the expression of BABA-IR against the necrotrophic fungi P. cucumerina or Alternaria brassicicola, there is a similar augmentation in the expression of SA-inducible defenses (Ton and Mauch-Mani, 2004). However, activation of SA-inducible defenses by treatment with BTH fails to induce resistance against these fungi, and both NahG and npr1-1 plants are unaffected in BABA-IR against P. cucumerina (Ton and Mauch-Mani, 2004). This indicates that the priming for SA-dependent resistance does not contribute to the level of BABA-IR against necrotrophic fungi. Apart from priming for SA-dependent resistance, BABA also primes for a faster and stronger deposition of callose-rich papillae under the appressoria of pathogenic fungi and oomycetes. Recently, we found that the callose-deficient mutant pmr4-1 (Nishimura et al., 2003) is blocked in BABA-IR against P. cucumerina and A. brassicicola (Ton and Mauch-Mani, 2004; J. Ton, unpublished results), suggesting that the augmented callose deposition is important for BABA-IR against these fungi. Hence, BABA primes for different defense mechanisms, and the effectiveness of these augmented defense reactions depends on the challenging pathogen.
To learn more about the molecular mechanisms behind BABA-IR, we performed a mutagenesis screen to select for Arabidopsis mutants that are impaired in the responsiveness to BABA. Based on the observation that repeated treatment with high doses of BABA triggers female sterility (Jakab et al., 2001), we have isolated mutants with an impaired BABA-induced sterility (ibs) response. Evaluation of the defense-related phenotype of three of these ibs mutants revealed distinct mutant phenotypes in the BABA-induced priming for specific defense mechanisms. Here, we introduce three novel regulatory genes that play a role in the priming for defense.
RESULTS
Isolation of Arabidopsis ibs Mutants
The mutagenesis screen for ibs mutants is based on the observation that higher doses of BABA induce sterility in Arabidopsis (Jakab et al., 2001). A T-DNA–mutagenized population of Arabidopsis (accession Wassilewskija-0 [Ws-0]; Feldmann, 1991) was screened for their inability to express female sterility after repeated treatment with 300 μM BABA, applied as a soil-drench. Of the ∼90,000 T3 plants screened, 73 putative ibs mutants were identified showing formation of siliques after repeated treatment with BABA. Rescreening the progeny of the 17 most pronounced mutants confirmed all ibs phenotypes. Subsequently, each of these 17 mutants was tested by DNA gel blot analysis to determine the number of T-DNA insertions. Mutants carrying only one or two T-DNA insertions were further selected for plasmid rescue to identify flanking genomic DNA sequences. Sequence analysis of the rescued plasmids revealed genomic DNA sequences from four mutants, designated ibs1, ibs2, ibs3, and ibs4.
IBS1 Encodes a Cyclin-Dependent Kinase–Like Protein
Mutants ibs1 and ibs4 have a T-DNA insertion at exactly the same locus that maps to the upper arm of chromosome I. The T-DNA is inserted downstream of the open reading frame of a putative kinase gene (At1g18670; Figure 1A). Mutant ibs4 has an additional T-DNA insertion that has not been mapped. Cloning a wild-type cDNA fragment of the 3′-end of the IBS1 gene revealed an unpredicted splicing site at 12 bp upstream of the predicted stop codon. An alternative stop codon was found at 3452 bp downstream of the start codon, which is 718 bp further than the predicted stop codon. The poly(A) tail of the mRNA starts at 344 bp downstream of the stop codon, which is 32 bp upstream of the T-DNA insertion in ibs1 and ibs4. To examine whether the IBS1 gene plays a role in BABA responsiveness, three sequence-indexed T-DNA insertion lines from the Syngenta SAIL collection (accession Columbia-0 [Col-0]) were tested for BABA-induced sterility. The s-634 mutant has a T-DNA insertion in the IBS1 promoter region at 707 bp upstream of the start codon, whereas mutants s-111 and s-765 contain a T-DNA insertion in the second exon of the IBS1 gene (Figure 1A). Upon BABA treatment, mutants s-111 and s-765 produced significantly more seeds than wild-type plants, which was comparable to the amount of seeds produced by BABA-treated ibs1 plants (Figure 1B). This confirms that mutations in the IBS1 gene cause the ibs1 phenotype. By contrast, the promoter-tagged line s-634 displayed a similar level of BABA-induced sterility as wild-type plants, indicating that the T-DNA insertion at 707 bp from the start codon does not cause the ibs1 mutant phenotype. The predicted translation product of IBS1 contains a Ser/Thr protein kinase domain and an N-terminal myristoylation sequence (Figure 2). BLAST analysis of the IBS1 protein revealed 80% sequence identity with a cyclin-dependent kinase (CDK)–like protein (At1G74330) from Arabidopsis (Figure 2). Furthermore, IBS1 has 48% sequence identity with the CDC2 protein from Arabidopsis, a CDK involved in meiotic and mitotic cell divisions (Moran and Walker, 1993), and 44% sequence identity with the CRK1 protein of Beta vulgaris, which belongs to the superfamily of calcium-dependent protein kinases (Hrabak et al., 2003).
Figure 1.
T-DNA Insertions in Different Arabidopsis Mutants, Their Corresponding ibs Phenotypes, and Genetic Complementation of the ibs3 Mutant for BABA-Induced Sterility and BABA-Induced Tolerance to Salt Stress.
(A) Genomic sequences of IBS genes and T-DNA locations in the different ibs mutants. White triangles represent T-DNA insertions in the original Ws-0 mutants ibs1, ibs2, and ibs3; black triangles represent T-DNA insertions in sequence-indexed knockout Col-0 mutants s-111, s-634, s-765, and s-031243. Locations of the ethyl methanesulfonate mutations in the ABA1/IBS3 gene of Col-0 mutants aba1-5 and npq2-1 are unknown.
(B) Quantification of BABA-induced sterility in 6-week-old plants. Four-week-old plants were repeatedly soil-drenched with water or BABA to a final concentration of 300 μM. Values presented are means (± sd) of the number of seeds per silique (n = 20).
(C) Quantification of BABA-induced sterility in Ws-0, ibs3, and T-IBS3g plants upon soil-drench treatment with increasing concentrations of BABA. T-IBS3g plants were generated by transforming ibs3 plants with a construct containing the ABA1/IBS3 genomic clone, including 805-kb of the promoter region and a 2352-bp region downstream of the stop codon of ABA1/IBS3. Values presented are means (± sd) of the number of seeds per silique (n = 20).
(D) Quantification of BABA-induced tolerance to salt stress in Ws-0, ibs3, and T-IBS3g plants. Four-week-old plants were soil-drenched with BABA to a concentration of 250 μM and 2 d later soil-drenched with NaCl to a concentration of 300 mM. Values presented are means (± sd) of the percentage of wilted plants per pot at 3 d after salt treatment. Asterisks indicate statistically significant differences between control and BABA-treated plants (Student's t test; α = 0.05; n = 10).
Figure 2.
Amino Acid Sequence Comparison between IBS1 and Protein Kinases from Arabidopsis (At1G74330 and CDC2C) and B. vulgaris (CRK1).
Identical and similar amino acid sequences to IBS1 are indicated with black and gray shading, respectively. Asterisks indicate the conserved Ser/Thr protein kinase domain, and black circles indicate the N-terminal myristoylation domain.
IBS2 Encodes a Phosphoinositide Phosphatase
Mutant ibs2 contains one truncated version of the T-DNA on the lower arm of chromosome V. The insertion is in the 5′-untranslated region (5′-UTR) of the AtSAC1b gene (At5g66020) at 80 bp from the start codon (Figure 1A). The AtSAC1b gene has recently been characterized as a polyphosphoinositide phosphatase that modulates cellular phosphoinositide levels (Despres et al., 2003). To confirm the involvement of the AtSAC1b/IBS2 gene in BABA responsiveness, we have tested a sequence-indexed T-DNA insertion line from the Salk Institute collection (accession Col-0) for BABA-induced sterility. This s-031243 mutant contains a T-DNA insertion in the 5′-UTR of the AtSAC1b/IBS2 gene at 76 bp upstream of the start codon (Figure 1A). Mutant s-031243 displayed a similar reduction in BABA-induced sterility as ibs2 plants (Figure 1B), confirming that mutations in the 5′-UTR of AtSAC1b/IBS2 cause the ibs2 phenotype.
IBS3 Encodes a Zeaxanthin Epoxidase
The ibs3 mutant contains a T-DNA insertion that maps to the lower arm of chromosome V. The T-DNA is inserted downstream of the 3′-UTR of the ABA1 gene (At5g67030) at 223 bp from of the poly(A) site and 592 bp from the stop codon (Figure 1A). This gene encodes a single-copy zeaxanthin epoxidase gene that functions in the biosynthesis pathway of the abiotic stress hormone ABA (Koornneef et al., 1982; Zhu, 2002). To examine whether independent mutations in the ABA1/IBS3 gene also cause the ibs3 phenotype, we quantified BABA-induced sterility in Col-0 mutants aba1-5 and npq2-1 that both carry ethyl methanesulfonate–induced mutations in this gene (Niyogi et al., 1998). Similarly to ibs3 plants, aba1-5 and npq2-1 produced significantly more seeds than the corresponding wild-type plants upon treatment with BABA (Figure 1B). This indicates that the ABA1/IBS3 gene plays a role in the BABA-induced sterility response.
Because the T-DNA insertion in ibs3 is relatively far from the transcribed region of the ABA/IBS3 gene (233 bp downstream of the poly(A) site), we examined whether this T-DNA insertion is solely responsible for the mutant phenotype. To this end, ibs3 plants were transformed with a genomic DNA fragment from 805 bp upstream of the start codon to 2352 bp downstream of the stop codon of ABA1/IBS3. The resulting complementation line T-IBS3g was tested for BABA-induced sterility and BABA-induced protection against salt stress. Upon treatment with increasing concentrations of BABA, T-IBS3g plants displayed a similar reduction in the number of seeds per silique as wild-type plants, whereas ibs3 plants were significantly reduced in this BABA-induced sterility (Figure 1C). Additionally, both T-IBS3g and wild-type plants showed a statistically significant reduction in the number of wilting plants upon treatment with BABA and subsequent soil-drench treatment with 300 mM NaCl, whereas ibs3 plants completely failed to develop this BABA-induced tolerance (Figure 1D). Hence, transformation with the genomic fragment of ABA1/IBS3 complements for the ibs3 mutation, demonstrating that the T-DNA insertion at 223 bp from the 3′-translated region of the ABA1/IBS3 gene is responsible for the ibs3 mutant phenotype.
IBS1 Regulates Priming for SA-Dependent Defenses against P. syringae
Treatment of Arabidopsis with BABA induces resistance against the bacterial pathogen P. syringae pv tomato DC3000. This BABA-IR depends on the SA- and NPR1/NIM1/SAI1-dependent defense pathway and is marked by an augmented expression of the SA-inducible marker gene PR-1 (Zimmerli et al., 2000). To test the involvement of the three IBS genes in BABA-IR against P. syringae, 5-week-old plants were soil-drenched with 250 μM BABA and subsequently inoculated with the bacteria. Three days after inoculation, the level of disease and bacterial growth in the leaves was quantified. All noninduced mutant plants showed a similar level of susceptibility to P. syringae as noninduced wild-type plants (Figure 3), indicating that the three ibs mutations do not affect basal resistance against the pathogen. In wild-type plants, as well as ibs2 and ibs3 plants, treatment with BABA caused a significant reduction in bacterial proliferation and disease severity (Figures 3A and 3B), indicating that ibs2 and ibs3 are not impaired by BABA-IR against P. syringae. By contrast, ibs1 plants failed to develop this BABA-IR (Figure 3). This demonstrates that the ibs1 mutant is blocked in BABA-IR against P. syringae. Mutants s-111 and s-765, which both contain a T-DNA insertion in the open reading frame of the IBS1 gene, were similarly affected in BABA-IR against P. syringae (see Supplemental Figure 1 online). It can thus be concluded that mutations in the IBS1 gene block the plant's ability to express BABA-IR against P. syringae.
Figure 3.
BABA-IR against P. syringae pv tomato DC3000 in Wild-Type Plants (Ws-0) and ibs Mutants of Arabidopsis.
Six-week-old plants were soil-drenched with BABA to a final concentration of 250 μM and 2 d later challenge-inoculated with a bacterial suspension of P. syringae pv tomato DC3000 at 1.5 × 107 colony-forming units·mL−1. Data are from a representative experiment that was repeated with similar results.
(A) Bacterial growth in the leaves was determined over a 3-d time interval. Values presented are means (± sd) of the log of the proliferation values. Asterisks indicate statistically significant differences compared with noninduced control plants (Student's t test; α = 0.05; n = 5).
(B) Disease symptoms were quantified at 3 d after inoculation and quantified as the proportion of leaves with symptoms. Data presented are means of the average percentage of diseased leaves per plant (± sd). Asterisks indicate statistically significant differences compared with noninduced control plants (Student's t test; α = 0.05; n = 20 to 25).
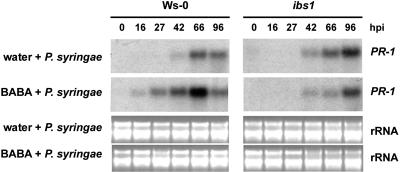
To investigate whether the impaired BABA-IR against P. syringae in ibs1 is related to a defect in the priming for SA-dependent defenses, we monitored the expression of the SA-inducible marker gene PR-1 in wild-type and ibs1 plants upon infection by P. syringae. Noninduced plants showed a transient increase in the level of PR-1 expression that started at 42 h postinoculation (hpi; Figure 4). This pattern of PR-1 gene expression was similar in both wild-type and ibs1 plants, indicating that the ibs1 mutation does not affect the basal activation of the PR-1 gene. Upon treatment with BABA, wild-type plants showed an earlier and stronger expression of the PR-1 gene, which started at 16 hpi and reached a maximum at 66 hpi. By contrast, BABA-treated ibs1 plants failed to show this augmented expression of the PR-1 gene (Figure 4). This indicates that the IBS1 gene regulates priming for SA-dependent resistance.
Figure 4.
RNA Gel Blot Analysis of the SA-Inducible PR-1 Gene Expression in Wild-Type Plants (Ws-0) and ibs1 Plants.
Six-week-old plants were treated with either water or BABA (250 μM). At 2 d after induction treatment, the plants were inoculated with P. syringae pv tomato DC3000. Total RNA was extracted at different time points after inoculation. Each time point represents infected leaves from five different plants. RNA gel blots were hybridized with a P32-labeled probe against PR-1. Ethidium bromide staining of the RNA gel (rRNA) was used to show equal loading. This experiment was repeated twice, yielding similar results.
Mutants ibs1, ibs2, and ibs3 Are Differentially Impaired in BABA-IR against H. parasitica
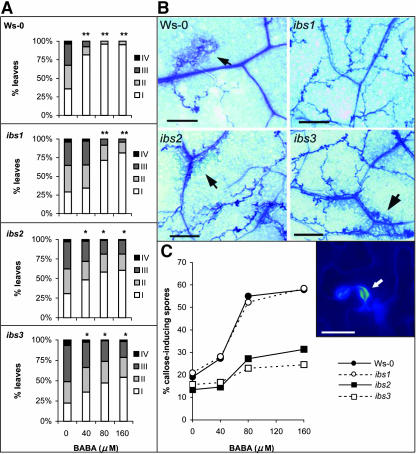
Treatment of Arabidopsis with BABA triggers resistance against the oomycete H. parasitica (Zimmerli et al., 2000). The expression of this BABA-IR correlates with enhanced levels of trailing necrosis around the pathogen hyphae and an augmented deposition of callose under the appressoria of the oomycete (Zimmerli et al., 2000). To determine whether ibs1, ibs2, and ibs3 are impaired in BABA-IR against H. parasitica, 4-week-old plants were soil-drenched with increasing concentrations of BABA and subsequently inoculated with a suspension containing the asexual spores (conidia) of the pathogen. The level of disease was assessed at 8 d after inoculation. Treating wild-type plants with the relatively low dose of 40 μM BABA reduced the number of sporulating leaves by 71% (Figure 5A). Treatment of ibs2 and ibs3 plants with 40 μM BABA resulted in 23 and 20% reduction of sporulating leaves, respectively. Although this level of disease suppression was statistically significant (0.01 < P < 0.05), it was considerably less than that in BABA-treated wild-type plants (Figure 5A). Apparently, ibs2 and ibs3 are partially affected in BABA-IR against H. parasitica upon induction by 40 μM BABA. On the other hand, ibs1 plants showed no statistically significant reduction in the number of sporulating leaves upon treatment with 40 μM BABA, indicating that this mutant is completely blocked in BABA-IR against H. parasitica (Figure 5A). Mutants s-111 and s-765, which carry a T-DNA in the IBS1 gene, displayed a similar nonresponsiveness to treatment with 40 μM BABA as ibs1 plants (see Supplemental Figure 2 online). Hence, IBS1 regulates a component of BABA-IR that is already active upon induction by relatively low amounts of BABA.
Figure 5.
BABA-IR against H. parasitica Strain EMWA in Wild-Type Plants (Ws-0) and ibs Mutants.
Four-week-old plants were soil-drenched with BABA to the indicated concentrations and 2 d later challenge inoculated by spraying a suspension of 5 × 104 conidiospores/mL onto the leaves.
(A) The level of disease severity was quantified at 8 d after inoculation. Disease rating is expressed as the percentages of leaves in disease classes I (no sporulation), II (<50% of the leaf area covered by sporangia), III (>50% of the leaf area covered by sporangia), and IV (heavily covered with sporangia, with additional chlorosis and leaf collapse). Asterisks indicate statistically significant different distributions of the disease severity classes compared with the noninduced control treatments (χ2; single asterisks, P < 0.05; double asterisks, P < 0.001). The data presented are from a representative experiment that was repeated with similar results.
(B) Colonization of BABA-treated leaves by H. parasitica at 4 d after challenge inoculation. Leaves were stained with lactophenol/trypan-blue and analyzed by light microscopy. Arrows indicate trailing necrosis. Bars = 100 μm.
(C) Quantification of callose deposition at 2 d after challenge inoculation. Leaves were stained with calcofluor/aniline-blue and analyzed by epifluorecence microscopy (UV). Callose deposition was quantified by determining the percentage of callose-inducing spores in the epidermal cell layer. Inset shows a representative example of a germinating H. parasitica spore triggering callose deposition in an epidermal cell. Bar = 15 μm.
Because resistance induced by low amounts of BABA is related to enhanced trailing necrosis around the growing hyphae of H. parasitica (Zimmerli et al., 2000), infected leaves were microscopically examined at 4 d after challenge inoculation. Staining the leaves with lactophenol trypan-blue revealed that BABA-treated ibs1 plants failed to show enhanced levels of trailing necrosis, whereas ibs2 and ibs3 were unaffected in this priming response (Figure 5B). In a separate experiment, SA-nonaccumulating NahG plants, SA-insensitive npr1-1 plants, and s-111 and s-765 plants showed a similar defect in the augmented formation of trailing necrosis as ibs1 plants (data not shown). This not only indicates that mutations in the IBS1 gene block the BABA-induced priming for trailing necrosis, but it also shows that the augmented expression of the trailing necrosis depends on the SA-dependent defense pathway.
Upon induction with higher concentrations of BABA (80 and 160 μM), wild-type plants almost entirely suppressed sporulation by H. parasitica (Figure 5A). This relatively high level of resistance correlated with a threefold increase in the number of spores that induce callose depositions in the epidermal cell layer (Figure 5C). Like wild-type plants, ibs1, s-111, and s-765 plants displayed relatively high levels of BABA-IR at 80 and 160 μM BABA (Figure 5A; see Supplemental Figure 2A online). Furthermore, the augmented deposition of callose in BABA-treated ibs1, s-111, and s-765 was comparable to that observed in BABA-treated wild-type plants (Figure 5C; see Supplemental Figure 2B online). This indicates that mutations in the IBS1 gene do not have an effect on the priming for callose and therefore hardly affect the resistance against H. parasitica upon induction by relatively high doses of BABA. By contrast, ibs2 and ibs3 plants were significantly reduced in BABA-IR against H. parasitica at 80 and 160 μM BABA (Figure 5A). This decrease in BABA-IR correlated with reduced levels of callose accumulation (Figure 5C). Mutant s-031243, carrying a T-DNA in the 5′-UTR of the AtSAC1b/IBS2 gene, and mutants npq2-1 and aba1-5, containing ethyl methanesulfonate mutations in the ABA1/IBS3 gene (Niyogi et al., 1998), were similarly affected in BABA-IR and callose deposition (see Supplemental Figure 2 online). This indicates that mutations in AtSAC1b/IBS2 and ABA1/IBS3 affect the augmented callose response, which consequently reduces the level of BABA-IR against H. parasitica.
Mutants ibs2 and ibs3 Are Impaired in Priming for ABA-Dependent Defenses against Salt Stress
BABA protects Arabidopsis against osmotic stress. This BABA response requires an intact ABA-dependent signaling pathway (Jakab et al., 2001; G. Jakab and J. Ton, unpublished results). To investigate the involvement of the IBS genes in this BABA-induced tolerance, 4-week-old Ws-0, ibs1, ibs2, and ibs3 plants were treated with either water or 250 μM BABA and subsequently soil-drenched with 300 mM NaCl. At different time points after salt application, the percentage of wilting plants was determined. Noninduced wild-type plants showed a steady increase in the number of wilting plants up to 70% at 5 d after salt treatment (Figure 6). Pretreatment with BABA resulted in a statistically significant reduction in the number of wilting plants. At 5 d after salt treatment, the number of BABA-treated plants with wilting symptoms was reduced by 60% in comparison with noninduced control plants (Figure 6). The ibs1 mutant expressed wild-type levels of BABA-induced tolerance, indicating that the ibs1 mutation does not affect BABA-induced tolerance to salt. By contrast, the level of BABA-induced tolerance was considerably less pronounced in ibs2 plants: at 1, 2, and 3 d after salt application, there was no statistically significant reduction in the number of wilting plants by BABA. Only at 5 d after salt application, BABA-treated ibs2 plants showed statistically significant reduction in the number of wilted plants (35%; P = 0.004). The allelic ibs2 mutant s-031243 displayed a comparable reduction in BABA-induced tolerance (see Supplemental Figure 3 online). In ibs3 plants, no significant effect by BABA on the number of wilting plants could be observed at all time points, indicating that this mutant is completely blocked in BABA-induced tolerance to salt. Consistent with this, mutants npq2-1 and aba1-5 showed a similar block in the BABA-induced tolerance to salt (see Supplemental Figure 3 online). Apart from the defects in BABA-induced protection, noninduced ibs3, npq2-1, and aba1-5 plants also displayed reduced levels of salt tolerance compared with noninduced wild-type plants (Figure 6; see Supplemental Figure 3 online). Apparently, mutations in the ABA1/IBS3 gene not only block BABA-induced tolerance, but also affect basal tolerance to salt stress.
Figure 6.
BABA-Induced Tolerance to Salt Stress in Wild-Type Plants (Ws-0) and ibs Mutants.
Four-week-old plants were soil-drenched with either water or BABA to a concentration of 250 μM. At 2 d after induction treatment, the plants were soil-drenched with NaCl to a concentration of 300 mM. Values presented are means (± sd) of the percentage of wilted plants per pot. Asterisks indicate statistically significant differences between control and BABA-treated plants (Student's t test; α = 0.05; n = 6 to 8). Data are from a representative experiment that was repeated with similar results.
To examine whether the impaired BABA-induced tolerance in ibs2 and ibs3 is related to a defect in the priming for ABA-dependent defense mechanisms, we quantified expression of the ABA-inducible genes RAB18 and RD29A upon application of increasing amounts of salt. Noninduced wild-type plants showed little induction of the RAB18 and RD29A genes at 100 mM NaCl. Increasing the salt concentrations in the soil from 250 to 500 mM resulted in a dose-dependent activation of RAB18 and RD29A (Figure 7). Wild-type plants pretreated with 250 μM BABA showed enhanced levels of RAB18 and RD29A expression at 100 and 250 mM salt compared with noninduced plants. This augmentation in ABA-induced gene expression was considerably less pronounced in BABA-treated ibs2 plants and absent in ibs3 plants (Figure 7). These results indicate that mutations in AtSAC1b/IBS2 and ABA1/IBS3 affect the priming for ABA-dependent defenses, causing defects in BABA-induced tolerance to salt.
Figure 7.
RNA Gel Blot Analysis of the ABA-Inducible Genes RAB18 and RD29A in Wild-Type (Ws-0), ibs2, and ibs3 Plants.
Four-week-old plants were treated with either water or BABA (250 μM). Two days after induction treatment, the plants were treated with increasing concentrations of NaCl. Total RNA was extracted from leaves of ±20 plants at 1 d after stress treatment with salt. RNA gel blots were hybridized with P32-labeled probes against RAB18 and RD29A. Ethidium bromide staining of the RNA gel (rRNA) was used to show equal loading. This experiment was repeated with similar results.
DISCUSSION
Screening for Plants Impaired in BABA-Induced Sterility Yields Novel Mutants Affected in Priming for Defense
The mutagenesis screen used in this study is based on the effect that relatively high amounts of BABA induce female sterility (Jakab et al., 2001). Out of 90,000 plants screened, 73 putative mutants with an impaired BABA-induced sterility (ibs) phenotype were selected. Further characterization of three of these ibs mutants revealed distinct mutant phenotypes with respect to BABA-IR against biotic and abiotic stresses. Interestingly, none of the three ibs mutants was blocked in BABA-IR against all three stresses tested. This indicates that ibs1, ibs2, and ibs3 are not disturbed in the uptake or perception of BABA, but rather in the priming for specific defense reactions. Additionally, we found evidence that the three IBS genes encode regulatory proteins that are involved in distinct signal transduction pathways. This suggests that BABA-induced sterility is achieved through a combined action of different signaling pathways and that each of these pathways play a specific role in the priming for defense against biotic or abiotic stress. Consistent with this, we found that every ibs mutant still showed a residual reduction in seed production upon treatment with BABA (Figure 1B). Apparently, multiple ibs mutations in different signaling pathways are needed to accomplish a complete block in BABA-induced sterility.
IBS1: A New Regulatory Gene in the Priming for SA-Dependent Resistance
The ibs1 mutant carries a single T-DNA insertion at the 3′-end of a gene encoding a CDK-like protein (Figures 1A and 2). Additionally, two sequence-indexed mutants with a T-DNA insertion in the second exon of this gene displayed similar phenotypes as ibs1 plants regarding BABA-induced sterility and BABA-IR. This demonstrates the involvement of IBS1 in different plant responses to BABA. The predicted translation product of IBS1 shares 80% sequence identity with another CDK-like protein kinase from Arabidopsis and 48% with CDC2C (At5g39420) from Arabidopsis (Figure 2). Most CDKs have been implicated in cell cycle regulation in association to their cyclin partners. However, some CDKs have been reported to control signal transduction pathways regulating gene transcription (Morgan, 1997; Barroco et al., 2003). Additionally, IBS1 shares 44% sequence identity with the CRK1 protein from B. vulgaris (Figure 2). This protein kinase belongs to the family of calcium-dependent protein kinases, which have functions in a wide array of developmental and stress-related responses in plants (Hrabak et al., 2003). The N-terminal part of IBS1 contains an N-terminal myristoylation sequence (Figure 2). N-myristoylation is an irreversible modification that affects the membrane binding properties of many different signal transduction proteins. Interestingly, Boisson et al. (2004) recently found that the N-myristoylated proteome of Arabidopsis contains a remarkably high fraction of cytoplasmic proteins related to defense signaling.
Evaluation of the defense-related phenotype of three allelic ibs1 mutants revealed that IBS1 controls BABA-IR against two biotrophic pathogens. The mutants failed to express BABA-IR against P. syringae and were blocked in BABA-IR against H. parasitica upon treatment with low amounts of BABA. Strikingly, both defects in BABA-IR correlated with a defect in the priming for SA-dependent defenses: Upon infection by P. syringae, BABA-treated ibs1 plants failed to show augmented expression of the SA-inducible PR-1 gene (Figure 4), and during colonization by H. parasitica, BABA-treated ibs1 plants lacked augmented formation of SA-dependent trailing necrosis (Figure 5B). Despite the defects in priming for SA-dependent defense reactions, ibs1 plants did not show a phenotype consistent with a direct blockage of the SA-dependent defense pathway (Cao et al., 1994; Delaney et al., 1995; Nawrath and Métraux, 1999). For instance, we could not observe an enhanced disease susceptibility to P. syringae and H. parasitica in the different ibs1 mutants (Figures 3 and 5; see Supplemental Figures 1 and 2 online). Furthermore, ibs1 plants were not affected or delayed in the activation of the PR-1 gene upon infection by P. syringae (Figure 4). These phenotypes indicate that mutations in IBS1 do not directly affect the SA-dependent defense pathway, but rather block the priming for this pathway. We therefore propose that IBS1 functions as a BABA-induced accelerator of the SA-dependent defense pathway, mediating the augmented expression of resistance against P. syringae and H. parasitica (Figure 8).
Figure 8.
Model for the Priming Mechanisms behind BABA-IR against Different Biotic and Abiotic Stresses.
Infection by P. syringae and H. parasitica leads to activation of the SA-dependent defense pathway. Treatment with BABA primes for SA-dependent defenses, resulting in augmented expression of SA-dependent resistance during infection by P. syringae or H. parasitica. The expression of this BABA-IR is marked by enhanced expression of the PR-1 gene and enhanced formation of trailing necrosis upon infection by P. syringae and H. parasitica, respectively. Infection by H. parasitica also activates a defense pathway that leads to callose deposition. Treatment with BABA generates a PtdIns- and ABA-dependent priming for this defense response, leading to augmented deposition of callose and enhanced resistance against H. parasitica. Salt stress activates an ABA-dependent defense response. Treatment with BABA triggers a PtdIns-dependent priming for ABA-dependent defenses, causing augmented expression of ABA-dependent defenses upon exposure to salt stress. This BABA-induced tolerance to salt is marked by enhanced expression of the ABA-inducible genes RAB18 and RD29A.
Involvement of Phosphoinositide Signaling in BABA-Induced Protection against H. parasitica and Salt Stress
The ibs2 mutant contains a truncated T-DNA in the 5′-UTR of the AtSAC1b gene (Figure 1A). The sequence-indexed mutant s-031243 with a T-DNA insertion at nearly the same locus displayed a similar level of BABA insensitivity, confirming the involvement of AtSAC1b/IBS2 in BABA-induced sterility (Figures 1A and 1B). The predicted translation product of AtSAC1b/IBS2 shows 50% identity with the SAC1 protein of yeast (Despres et al., 2003). This yeast protein functions as a polyphosphoinositide phosphatase, which converts phosphatidylinositol (PtdIns) 3-phosphate, PtdIns 4-phosphate, and PtdIns 3,5-biphosphate into PtdIns (Guo et al., 1999). In yeast, null mutations of the SAC1 gene result in enhanced accumulation of PtdIns(4)P and PtdIns(4,5)P2, affecting protein trafficking and organization of the actin cytoskeleton (Hama et al., 1999; Foti et al., 2001; Schorr et al. 2001). Interestingly, Despres et al. (2003) recently demonstrated that transformation of a yeast sac1 null-mutant with the Arabidopsis AtSAC1b/IBS2 gene results in full complementation of the sac1 mutant phenotype. This demonstrates that the AtSAC1b/IBS2 protein in Arabidopsis fulfills similar cellular functions as the SAC1 protein in yeast. It is therefore plausible to assume that the ibs2 mutations in the 5′-UTR of the AtSAC1b/IBS2 gene cause phenotypes that are related to disruptions in the PtdIns-dependent signaling pathway.
The involvement of PtdIns signaling in plant stress responses to osmotic stress is well documented (Zhu, 2002). In Arabidopsis, osmotic stress leads to enhanced expression of AtPLC1 encoding a PtdIns-specific phospholipase C (Hirayama et al., 1995). Moreover, application of salt has been shown to trigger a rapid accumulation of PtdIns(4,5)P2 and inositol-1,4,5-trisphosphate (De Wald et al., 2001). Hence, osmotic stress stimulates the PtdIns signaling pathway in Arabidopsis. Here, we have shown that two independent mutations in the 5′-UTR of the AtSAC1b/IBS2 gene compromise the plant's ability to express BABA-induced protection against salt stress (Figure 6; see Supplemental Figure 3 online). This defect correlated with a defect in the priming for ABA-inducible gene expression (Figure 7), indicating that AtSAC1b/IBS2 regulates the priming for ABA-dependent defenses. Together, these findings suggest that BABA primes for ABA-inducible defenses via a PtdIns-dependent signal transduction pathway. Subsequent exposure to salt stress results in augmented expression of ABA-dependent defense mechanisms, causing enhanced tolerance to salt (Figure 8).
PtdIns signaling has also been implicated in plant defense against pathogens. A rapid accumulation of inositol-1,4,5-trisphosphate has been observed in citrus infected with the fungus Alternaria alternata (Ortega and Perez, 2001). Furthermore, treatment of rice (Oryza sativa) with SA and jasmonic acid has been reported to rapidly induce the expression of a PtdIns-specific phospholipase C, which correlated with induced resistance against the blast fungus Magnaporthe grisea (Song and Goodman, 2002). Here, we showed that two independent mutations in the 5′-UTR of AtSAC1b/IBS2 cause reduced levels of callose deposition during the expression of BABA-IR against H. parasitica (Figure 5; see Supplemental Figure 2 online). Hence, PtdIns signaling plays an important role in the augmented deposition of callose during expression of BABA-IR against H. parasitica (Figure 8). Because PtdIns signaling regulates the organization of the actin cytoskeleton in yeast (Caroni, 2001; Foti et al., 2001), it is tempting to speculate that PtdIns signaling also regulates cytoskeleton rearrangements in the plant cell during infection by pathogenic fungi or oomycetes. These ultrastructural changes are thought to direct secretory vesicles to the sites of pathogen attack, mediating the delivery of the necessary substrate or synthetic machinery for callose production (Staiger, 2000). The finding that inhibitors of actin microfilament function reduce pathogen-induced callose deposition in barley (Hordeum vulgare) and cowpea (Vigna unguiculata) supports this hypothesis (Skalamera and Heath, 1996; Kobayashi et al., 1997).
ABA Regulates BABA-Induced Protection against Salt Stress and H. parasitica
The third mutant characterized, ibs3, contains a T-DNA insertion at 592 bp downstream of the stop codon of the ABA1/IBS3 gene (Figure 1A). This gene encodes a zeaxanthin epoxidase, which mediates an essential step in the biosynthetic pathway of the stress hormone ABA (Koornneef et al., 1982; Zhu, 2002). Further characterization revealed that this mutant is also affected in BABA-IR against H. parastica and BABA-induced protection against salt. Two other mutants in the ABA1/IBS3 gene displayed similar phenotypes (see Supplemental Figures 2 and 3 online), indicating involvement of ABA1/IBS3 in BABA-induced defense responses. Moreover, transforming the ibs3 mutant with a genomic clone of ABA1/IBS3 complemented for BABA-induced sterility and BABA-induced protection against salt (Figures 1C and 1D). Together, these findings strongly indicate that the ibs3 phenotype is caused by a defect in the functioning of the ABA1/IBS3 gene.
The failure of ibs3 plants to express BABA-induced tolerance to salt correlated with an impaired priming for ABA-inducible gene expression (Figures 6 and 7). This suggests that the augmented expression of ABA-dependent defenses plays an important role in the BABA-induced protection against salt. In addition, noninduced ibs3 plants, like aba1-5 and npq2-1, displayed enhanced levels of susceptibility to salt stress (Figure 6; see Supplemental Figure 3 online). Hence, all three mutations in ABA1/IBS3 affect both basal and BABA-induced tolerance to salt. However, the mutant phenotype of ibs3 was not entirely comparable to that of aba1-5 and npq2-1. For instance, ibs3 plants did not exhibit the enhanced susceptibility to drought that is typical for mutants aba1-5 and npq2-1 (data not shown). Furthermore, control-treated ibs3 plants were not severely affected in the salt-induced activation of the ABA-inducible genes RAB18 and RD29A (Figure 7). This suggests that the T-DNA insertion at the 3′-end of the ABA1/IBS3 gene does not strongly affect the basal responsiveness to osmotic stress, but rather confers a regulatory defect that predominantly affects the augmented expression of ABA-dependent defenses (Figure 8).
Besides the well-established role of ABA in the regulation of plant responses to abiotic stress (Xiong et al., 2002; Zhu, 2002), there is increasing evidence that ABA is also involved in responses to pathogen attack. However, the exact role of ABA in pathogen resistance seems controversial. Audenaert et al. (2002) reported that ABA-treated tomato (Lycopersicon esculentum) showed enhanced levels of susceptibility to B. cinerea, whereas the ABA-deficient tomato mutant sitiens displayed enhanced resistance against this pathogen. In addition, Mohr and Cahill (2003) reported that treatment of Arabidopsis with ABA conferred partial loss of resistance against an avirulent isolate of P. syringae pv tomato. Contrary to these reports, Wiese et al. (2004) recently described a positive role for ABA in pathogen resistance. The authors showed that treatment of barley with ABA primed for papillae-mediated resistance against powdery mildew fungus (Blumeria graminis f sp hordei). In support of this, we reported previously that treatment with ABA triggers callose-mediated resistance in Arabidopsis against the necrotrophic pathogens A. brassicicola and P. cucumerina (Ton and Mauch-Mani, 2004). This BABA-IR was blocked in the Arabidopsis mutants aba1-5 and abi4-1, which correlated with a defect in the priming for callose. These findings strongly indicate that ABA plays an important role in the regulation of rapid callose deposition during the early stages of fungal infection. In this study, we found that mutations in the ABA biosynthetic gene ABA1/IBS3 affect the augmented deposition of callose and the level of BABA-IR against H. parasitica (Figure 5; see Supplemental Figure 2 online). Interestingly, these mutations did not affect the basal level of callose deposition in control-treated plants (Figure 5C; see Supplemental Figure 2 online). We therefore propose that ABA, like PtdIns, contributes to BABA-IR against H. parasitica by predominantly regulating the priming for callose (Figure 8).
BABA Primes for Different Defense Pathways Yielding Broad-Spectrum Protection against Biotic and Abiotic Stresses
It is evident from the data presented in this study that BABA induces resistance against a broad spectrum of biotic and abiotic stresses. We have identified three genes that are differentially involved in the broad-spectrum protection by BABA-IR. Moreover, we showed that these genes regulate BABA-induced priming for different defense responses. Notably, the three ibs mutations did not directly affect defense signaling pathways, but blocked the priming for the corresponding defense response. This strongly suggests that priming for defense is essential for the expression of BABA-IR. In addition, this demonstrates that priming requires specific cellular signaling components.
Physiologically, priming provides an efficient mechanism to obtain broad-spectrum stress resistance. Plants that are primed to effectively resist biotic and abiotic stresses might not suffer from costly energy investments in defense mechanisms because their complete defense arsenal is not activated before stress exposure. Accordingly, plants in the primed state exhibit an enhanced defensive capacity against a wide range of stresses without harmful trade-off effects on commercially important traits such as growth and fruit set. Better knowledge of the priming mechanisms behind BABA-IR could provide valuable insights for generating crop varieties with an enhanced defensive capacity against biotic and abiotic stresses.
METHODS
Plant Lines
The Arabidopsis thaliana T-DNA collection used for the mutagenesis screen is in the background of accession Ws-0 and has been described by Feldmann (1991). Arabidopsis accession Col-0 was obtained from Lehle Seeds (Round Rock, TX), and the Col-0 mutants aba1-5 and npq2-1 were obtained from the Nottingham Arabidopsis Stock Centre (Nottingham, UK). The Col-0 mutant npr1-1 and the Ws-0 transgenic Arabidopsis NahG line were kindly provided by X. Dong (Duke University, Durham, NC) and F. Mauch (University of Fribourg, Switzerland), respectively. The IBS1 T-DNA insertion lines SAIL_634_B09 (s-634), SAIL_111_B09 (s-111), and SAIL_765_A05 (s-765) were obtained from the Arabidopsis T-DNA insertion collection of Syngenta (Research Triangle Park, NC) (Sessions et al., 2002). The IBS2 T-DNA insertion line SALK_031243 (s-031243) was obtained from the Arabidopsis T-DNA insertion collection of the Salk Institute (Alonso et al., 2003). To select plants homozygous for the T-DNA insertion, gene-specific primers (forward and reverse) 5′-TTTAAAATTGGTAGTTGTATC-3′ and 5′-CTAGCGGCGACCAGTGTGAGA-3′, 5′-CAAGAGCGGGAAGAAGAGTAGTAG-3′ and 5′-CCCGAGCAAAAGTTCAAGTGGAC-3′, 5′-GGTAGTGAGTTGGGTAGCGATTTG-3′ and 5′-CAGTTTGTGGGTGACCTCAGTTCT-3′, and 5′-CCACTGAATTGAGTTGTTGGGC-3′ and 5′-CCAAACATGAAGCGGAGGAACC-3′ were used for lines s-634, s-111, s-765, and s-031243, respectively. Plants yielding no PCR product with the gene-specific primers were subsequently tested for the presence of the T-DNA insertion, using the gene-specific forward primer in combination with the T-DNA left border specific primer 5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′.
Plant Growth Conditions and Cultivation of Pathogens
Plants for the mutagenesis screen were cultivated on a mixture of steam-sterilized commercial potting soil and perlite (3:1) at 60% RH under continuous light at 22 ± 2°C to promote flowering. For the induced resistance assays, seedlings were germinated on the same soil/perlite mixture and individually transferred to 33- or 150-mL pots when 2 weeks old. For the duration of the experiment, plants were maintained at 20°C day/18°C night temperature with 8.5 h of light per 24 h and 60% RH. Hyaloperonospora parasitica strains EMWA and WACO9 were maintained on Arabidopsis accessions Ws-0 and Col-0, respectively. Conidiospores were obtained by washing the sporulating leaves in 10 mM MgSO4, collected by centrifugation, and resuspended in 10 mM MgSO4 to a final density of 5 × 104 conidiospores/mL. The virulent strain of Pseudomonas syringae pv tomato DC3000 (Whalen et al., 1991) was grown overnight at 28°C in liquid King's B medium (King et al., 1954).
DNA Gel Blot Analysis and Plasmid Rescue
Genomic DNA was isolated from 5-week-old plants according to Dellaporta et al. (1983). PstI-, BamHI-, and SalI-digested DNA samples (2 μg) were electrophoresed on 1% (w/v) agarose gels, denaturated, and transferred onto Hybond-N+ membranes (Amersham, Buckinghamshire, UK) by capillary transfer. Membranes were hybridized with different probes synthesized from three T-DNA regions (left and right borders and pBR322) by random primer labeling with [α-32P]dCTP according to the supplier's instructions (Promega, Madison, WI) and subsequently exposed to X-Omat or Biomax films (Kodak, Rochester, NY). Based on the data of the DNA gel blots, plasmids from the inserted T-DNA from ibs1, ibs2, ibs3, and ibs4 have been rescued according to Behringer and Medford (1992) and sequenced to identify flanking genomic DNA sequences.
RNA Gel Blot Analysis
Total RNA was extracted by homogenizing frozen leaf tissue in extraction buffer (0.35 M glycine, 0.048 N NaOH, 0.34 M NaCl, 0.04 M EDTA, and 4% [w/v] SDS; 1 mL/g leaf tissue). The homogenates were extracted with phenol and chloroform, and the RNA was precipitated using LiCl as described by Sambrook et al. (1989). For RNA gel blot analysis, 12.5 μg of RNA was denatured using glyoxal and DMSO (Sambrook et al., 1989). Subsequently, samples were electrophoretically separated on 1.5% agarose gels and blotted onto Hybond-N+ membranes by capillary transfer. The electrophoresis buffer and blotting buffer consisted of 10 and 25 mM sodium phosphate, pH 7.0, respectively. RNA gel blots were hybridized and washed as described previously (Pieterse et al., 1994) and exposed to X-Omat or Biomax films. DNA probes were labeled with [α-32P]dCTP by random primer labeling according to the manufacturer's instructions (Promega). Probes to detect RAB18 and RD29A transcripts were prepared by PCR using primers based on GenBank sequences At5g52310 and At5g66400, respectively. Probes to detect PR-1 transcripts were derived from a PR-1 cDNA clone (Uknes et al., 1992). Equal loading was visualized by ethidium bromide staining of the rRNA.
Cloning of IBS1 cDNA
A cDNA library of BABA-treated Arabidopsis (accession Col-0) was screened for IBS1 cDNA, using a probe derived from PCR amplification of the seventh predicted exon of the genomic sequence of IBS1 (Atg18670; forward primer, 5′-TCCCAGAAGTTTCAGAAC-3′; reverse primer, 5′-TGCCTTTTCTTCTCGC-3′). One positive clone was identified and sequenced. Construction of the cDNA library in λUni-ZAP XR, screening, and in vivo excision of the IBS1 clone into Escherichia coli was performed according to the manufacturer's instructions (Stratagene, La Jolla, CA).
Construction of the ibs3 Complementation Line T-IBS3G
The ABA1/IBS3 genomic region (At5g67030) was amplified by high-fidelity PCR with gene-specific primers designed to include the 805-bp promoter region and the 2352-bp region downstream of the stop codon (forward primer, 5′-GTGAGCTCTAGCCTCTAGGCTATGG-3′; reverse primer, 5′-CAGGTACCCCAACCATGTTACAATATC-3′). The resulting 6.5-kb fragment was cloned in the plasmid pCR2.1-TOPO (Invitrogen, Carlsbad, CA), digested by SacI and KpnI, and subcloned into the binary vector pCAMBIA1300. Arabidopsis ibs3 plants were transformed using the Agrobacterium tumefaciens–mediated flower dipping method (Clough and Bent, 1998). T2 and T3 plants were selected on 0.5% (v/w) MS plates containing 30 μg/mL of hygromycin. Line T-IBS3G was selected from the T4 generation and tested for BABA-induced sterility and BABA-induced tolerance to salt stress.
Mutagenesis Screen and BABA-Induced Sterility Assays
T-DNA–mutagenized plants (Feldmann, 1991) were cultivated under continuous light to accelerate flowering. Plants were soil-drenched to 300 μM BABA starting at 4 weeks after sowing. One to two weeks after the onset of flowering, plants were screened for silique formation. Seeds were collected from plants that were still capable of producing full siliques after repeated treatment with BABA. To confirm putative ibs phenotypes, 2-week-old seedlings were transferred to 150-mL pots (four plants per pot) and soil-drenched with BABA to 300 μM starting at 4 weeks after sowing. To quantify BABA-induced sterility, the average number of seeds/silique was determined in 6-week-old plants.
P. syringae Bioassays
Two-week-old seedlings were individually transferred to 33-mL pots. Five-week-old plants were soil-drenched with water (control) or BABA to a final concentration of 250 μM. Two days after chemical treatment, plants were inoculated by dipping the leaves in a suspension of virulent P. syringae pv tomato DC3000, containing 1.5 × 107 colony-forming units·mL−1 in 10 mM MgSO4 and 0.01% (v/v) Silwet L-77. Three days after challenge inoculation, the percentage of leaves with symptoms was determined per plant (n = 20 to 25). Leaves showing necrotic or water-soaked lesions surrounded by chlorosis were scored as diseased. Bacterial growth in the leaves was determined by collecting replicate samples from five plants per genotype. Approximately 30 min after challenge inoculation and 3 d later, leaf samples were collected, weighed, rinsed in water, and homogenized in 10 mM MgSO4. Serial dilutions were plated on selective King's medium B agar supplemented with 100 mg/L cycloheximide and 50 mg/L rifampicin. After incubation at 28°C for 2 d, the number of rifampicin-resistant colony-forming units per gram of infected leaf tissue was determined, and bacterial proliferation over the 3-d time interval was calculated.
H. parasitica Bioassays
Two-week-old seedlings were transferred to 150-mL pots (15 plants per pot) and 1 week later soil-drenched with BABA to the indicated concentrations. Two days after BABA induction, plants were challenge inoculated with H. parasitica strain EMWA or WACO9 by spraying with 10 mM MgSO4 containing 5 × 104 conidiospores per mL to immanent runoff. After inoculation, the plants were kept at 100% RH for 1 d to ensure infection. Five days after infection, plants were placed back to 100% RH for 1 d to induce sporulation. Disease symptoms were scored for ∼200 leaves per treatment at 8 d after inoculation. Disease rating was expressed as intensity of disease symptoms and pathogen sporulation on each leaf: I, no sporulation; II, <50% of the leaf area covered by sporangia; III, >50% of the leaf area covered by sporangia; IV, heavily covered with sporangia, with additional chlorosis and leaf collapse. For visualizing trailing necrosis, infected leaves were stained with lactophenol trypan-blue and examined microscopically at 5 d after inoculation as described by Koch and Slusarenko (1990). For quantification of callose deposition, leaves were collected at 2 d after inoculation and incubated overnight in 95% ethanol. Destained leaves were washed in 0.07 M phosphate buffer, pH 9, incubated for 15 min in 0.07 M phosphate buffer containing 0.005% calcofluor (fluorescent brightener; Sigma-Aldrich, St. Louis, MO) and 0.01% aniline-blue (water blue; Fluka, Buchs, Switzerland), and subsequently washed in 0.07 M phosphate buffer containing only 0.01% aniline-blue to discard excess amounts of the calcofluor. Observations were performed with an epifluorescence microscope with UV filter (band-pass 340 to 380 nm, long-path 425 nm). Callose depositions were quantified by determining the percentage of callose-inducing conidiospores per infected leaf.
Salt Stress Bioassays
Two-week-old seedlings were transferred to 150-mL pots (15 plants per pot) and 2 weeks later soil-drenched with BABA to a final concentration of 250 μM. Two days after BABA induction, plants were soil-drenched with NaCl to 300 mM final concentration. After salt treatment, plants were not watered for the duration of the experiment. Salt stress was quantified by determining the average percentage of wilted plants per pot (n = 8 to 10).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers At1G74330 and CDC2C.
Supplementary Material
Acknowledgments
We thank Felix Mauch for critically reading the manuscript. This work was supported by a grant from the National Center of Competence on Plant Survival in Natural and Agricultural Ecosystems and Swiss National Foundation Grant 31-064024 to B.M.-M., Swiss National Foundation Grant 3100A0-104224-1 to J.-P.M., and Netherlands Organization for Scientific Research Grant 863.04.019 to J.T.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Brigitte Mauch-Mani (brigitte.mauch@unine.ch).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.029728.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Audenaert, K., De Meyer, G.B., and Höfte, M.M. (2002). Abscisic acid determined basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroco, R.M., De Veylder, L., Magyar, Z., Engler, G., Inze, D., and Mironov, V. (2003). Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell. Mol. Life Sci. 60, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer, F.J., and Medford, J.I. (1992). A plasmid rescue technique for the recovery of plant DNA disrupted by T-DNA insertion. Plant Mol. Biol. Rep. 10, 190–194. [Google Scholar]
- Boisson, B., Giglione, C., and Meinnel, T. (2003). Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 44, 43418–43429. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroni, P. (2001). Actin cytoskeleton regulation through modulation of PI (4,5) P2 rafts. EMBO J. 20, 4332–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, Y.R. (2002). β-Amino-butyric acid-induced resistance against plant pathogens. Plant Dis. 86, 448–457. [DOI] [PubMed] [Google Scholar]
- Conrath, U., Pieterse, C.M.J., and Mauch-Mani, B. (2002). Priming in plant-pathogen interactions. Trends Plant Sci. 7, 210–216. [DOI] [PubMed] [Google Scholar]
- Dean, R.A., and Kuc, J. (1987). Rapid lignification in response to wounding and infection as a mechanism for induced systemic protection in cucumber. Physiol. Mol. Plant Pathol. 31, 69–81. [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced systemic disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Despres, B., Bouissonnie, F., Wu, H.J., Gomord, V., Guilleminot, J., Grellet, F., Berger, F., Delseny, M., and Devic, M. (2003). Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J. 34, 293–306. [DOI] [PubMed] [Google Scholar]
- DeWald, D.B., Torabinejad, J., Jones, C.A., Shope, J.C., Cangelosi, A.R., Thompson, J.E., Prestwich, G.D., and Hama, H. (2001). Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 126, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, K.A. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1, 71–82. [Google Scholar]
- Foti, M., Audhya, A., and Emr, S.D. (2001). Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 12, 2396–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T.P., Friedrich, L., Vernooij, B., Negrotto, D., Neye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Guo, S.L., Stolz, L.E., Lemrow, S.M., and York, J.D. (1999). SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274, 12990–12995. [DOI] [PubMed] [Google Scholar]
- Hama, H., Schnieders, E.A., Thorner, J., Takemoto, J.Y., and DeWald, D.B. (1999). Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 34294–34300. [DOI] [PubMed] [Google Scholar]
- Hayes, M.P., Enterline, J.C., Gerrard, T.L., and Zoon, K.C. (1991). Regulation of the interferon-production by human monocytes: Requirements for priming for lipopolysaccharide-induced production. J. Leukoc. Biol. 50, 176–181. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., Ohto, C., Mizoguchi, T., and Shinozaki, K. (1995). A gene encoding a phosphatidylinositol-specific phospholipase-C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 92, 3903–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak, E.M., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 660–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab, G., Cottier, V., Toquin, V., Rigoli, G., Zimmerli, L., Métraux, J.-P., and Mauch-Mani, B. (2001). β-Aminobutyric acid-induced resistance in plants. Eur. J. Plant Pathol. 107, 29–37. [Google Scholar]
- Katz, V.A., Thulke, O.U., and Conrath, U. (1998). A benzothiadiazole primes parsley cells for augmented elicitation of defense responses. Plant Physiol. 117, 1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss, H., Krause, K., and Jeblick, W. (1992. b). Methyl jasmonate conditions parsley suspension cells for increased elicitation of phenylpropanoid defense responses. Biochem. Biophys. Res. Commun. 189, 304–308. [DOI] [PubMed] [Google Scholar]
- Kauss, H., Theisinger-Hinkel, E., Mindermann, R., and Conrath, U. (1992. a). Dichloroisonicotinic and salicylic acid, inducers of systemic acquired resistance, enhance fungal elicitor responses in parsley cells. Plant J. 2, 655–660. [Google Scholar]
- King, E.O., Ward, M.K., and Raney, D.E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Kobayashi, I., Kobayashi, Y., Funaki, Y., Fujimoto, S., Takemoto, T., and Kunoh, H. (1997). Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. Plant J. 11, 525–537. [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, A., Schwindling, S., and Conrath, U. (2002). Benzothiadiazol-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 128, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Derswan, D.L.C.B., and Karssen, C.M. (1982). The isolation of abscisic-acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Kovats, K., Binder, A., and Hohl, H.R. (1991. a). Cytology of systemic induced resistance of cucumber to Colletotrichum lagenarium. Planta 183, 484–490. [DOI] [PubMed] [Google Scholar]
- Kovats, K., Binder, A., and Hohl, H.R. (1991. b). Cytology of systemic induced resistance of tomato to Phytophthora infestans. Planta 183, 491–496. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani, B., and Métraux, J.-P. (1998). Salicylic acid and systemic acquired resistance to pathogen attack. Ann. Bot. 82, 535–540. [Google Scholar]
- Mohr, P.G., and Cahill, D.M. (2003). Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Functional Plant Biol. 30, 461–469. [DOI] [PubMed] [Google Scholar]
- Moran, T.V., and Walker, J.C. (1993). Molecular cloning of 2 novel protein-kinase genes from Arabidopsis thaliana. Biochim. Biophys. Acta 1216, 8–14. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261–291. [DOI] [PubMed] [Google Scholar]
- Mur, L.A., Taylor, G., Warner, S.A.J., Sugars, J.M., White, R.F., and Draper, J. (1996). Salicylic acid potentiates defense gene expression in tissue exhibiting acquired resistance to pathogen attack. Plant J. 9, 559–571. [Google Scholar]
- Nawrath, C., and Métraux, J.-P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Grossman, A.R., and Bjorkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, X., and Perez, L.M. (2001). Participation of the phosphoinositide metabolism in the hypersensitive response of Citrus limon against Alternaria alternata. Biol. Res. 34, 43–50. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., Derksen, A.-M.C.E., Folders, J., and Govers, F. (1994). Expression of the Phytophthora infestans ipiB and ipiO genes in planta and in vitro. Mol. Gen. Genet. 244, 269–277. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., Van Wees, S.C.M., Van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and Van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr, M., Then, A., Tahirovic, S., Hug, N., and Mayinger, P. (2001). The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 11, 1421–1426. [DOI] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana identified in a selective screen utilizing the SA-inducible expression of the TMS2 gene. Mol. Plant Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Métraux, J.-P., and Raskin, I. (1995). Salicylic acid in rice. Biosynthesis, conjugation, and possible role. Plant Physiol. 108, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalamera, D., and Heath, M.C. (1996). Cellular mechanisms of callose deposition in response to fungal infection or chemical damage. Can. J. Bot. 74, 1236–1242. [Google Scholar]
- Skipp, R.A., and Deverall, B.J. (1973). Studies on cross-protection in anthracnose disease of bean. Physiol. Plant Pathol. 2, 357–374. [Google Scholar]
- Song, F.M., and Goodman, R.M. (2002). Molecular cloning and characterization of a rice phosphoinositide-specific phospholipase C gene, OsPI-PLC1, that is activated in systemic acquired resistance. Physiol. Mol. Plant Pathol. 61, 31–40. [Google Scholar]
- Staiger, C.J. (2000). Signalling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 257–288. [DOI] [PubMed] [Google Scholar]
- Sticher, L., Mauch-Mani, B., and Métraux, J.-P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35, 235–270. [DOI] [PubMed] [Google Scholar]
- Ton, J., and Mauch-Mani, B. (2004). β-Amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130. [DOI] [PubMed] [Google Scholar]
- Ton, J., Van Pelt, J.A., Van Loon, L.C., and Pieterse, C.M.J. (2002). Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol. Plant Microbe Interact. 15, 27–34. [DOI] [PubMed] [Google Scholar]
- Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S.C., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L.C. (1997). Induced resistance in plants and the role of pathogenesis-related proteins. Eur. J. Plant Pathol. 103, 753–765. [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, J., Kranz, T., and Schubert, S. (2004). Induction of pathogen resistance in barley by abiotic stress. Plant Biol. 6, 529–536. [DOI] [PubMed] [Google Scholar]
- Wyatt, G.R., Braun, R.P., and Zhang, J. (1996). Priming effect in gene activation by juvenile hormone in locust fat body. Arch. Insect Biochem. Physiol. 32, 633–640. [Google Scholar]
- Xiong, L.M., Schumaker, K.S., and Zhu, J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.), S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli, L., Jakab, G., Métraux, J.-P., and Mauch-Mani, B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proc. Natl. Acad. Sci. USA 97, 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli, L., Métraux, J.-P., and Mauch-Mani, B. (2001). β-Aminobutyric acid-induced protection of Arabidopsis against the necrotrophic fungus Botrytis cinerea. Plant Phys. 126, 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.