Abstract
The development of single cell approaches has facilitated the investigation of cellular heterogeneity and cell type-specific gene expression in complex tissues. Adipose tissue depots contain lipid storing adipocytes as well as a diverse array of cell types that form the adipocyte niche and regulate adipose tissue function. Here, I describe two protocols for the isolation of single cells and nuclei from white and brown adipose tissue. Additionally, I provide a detailed workflow for isolation of cell type- or lineage-specific single nuclei using nuclear tagging and translating ribosome affinity purification (NuTRAP) mouse models.
Keywords: Adipose tissue, Adipocyte progenitors, Transcriptomics, scRNA-seq, snRNA-seq
1. Introduction
Adipose tissue plays a major role in the regulation of energy homeostasis. White adipose tissue (WAT) stores excess calories in the form of lipid droplets, while brown adipose tissue (BAT) is primarily responsible for regulating body temperature through adaptive thermogenesis. The ability of BAT to dissipate chemical energy as heat offers an attractive strategy against obesity and its cardiometabolic sequelae. A critical barrier toward harnessing the potential of adipose thermogenesis to enhance cardiometabolic health in humans is the lack of understanding of the full range of cellular source(s) and pathways involved in the expansion of thermogenic adipocytes.
Adipose depots are ensembles of mature adipocytes, adipocyte progenitors, immune cells, endothelial cells, smooth muscle cells, pericytes, neurons, and Schwann cells [1]. Adipocytes are terminally differentiated cells. Therefore, the renewal and expansion of the adipocyte pool require de novo differentiation of tissue-resident adipocyte progenitors. Adipocyte progenitors are present in the stromal vascular fraction of the tissue and are capable of self-renewal and adipogenic differentiation throughout development and in response to stimulations [2].
The conventional method of studying adipocyte progenitors involves physical dissociation of tissue followed by density-based depletion of floating lipid-laden cells and collection of all the other cell types in the stromal vascular fraction (SVF). However, SVF is a heterogeneous mixture of several distinct cell types, and therefore the conventional bulk approaches do not provide in-depth insights into the contribution of different cell types to adipose function and homeostasis. We and others have recently used single cell transcriptome analysis to dissect the cellular heterogeneity of multiple adipose tissue depots [3-8]. These studies uncovered the presence of several types of adipocyte progenitors that are distinct in lineage [3, 4], cellular state, and adipogenic potential [5-7].
Despite the massive expansion of single cell-based methodologies in the last few years, the application of these approaches to studying adipocytes has remained challenging. The major challenge in transcriptome profiling along the entire adipogenic trajectory at the single cell level is the isolation of intact single adipocytes. The large size, fragile nature, and high buoyancy of mature adipocytes make them notoriously challenging to study at the single cell level. Alternative methods using nuclei isolated from fresh or frozen adipose tissue have been used to fill this gap [9, 10]. However, the nuclear transcripts represent only a fraction of the cellular transcriptome. Furthermore, comparing the single nuclei with the whole cell transcriptome of human adipocytes and preadipocytes revealed inherent transcript enrichment and detection biases in single nucleus RNA-sequencing (snRNA-seq) [11]. Another disadvantage of working with nuclei is that it limits the use of antibody-based positive or negative selection methods for enriching specific cell types and lineages before transcriptome profiling. Thus, single cell and single nucleus transcriptomic methods are complementary approaches with distinct applications in studying adipose tissue.
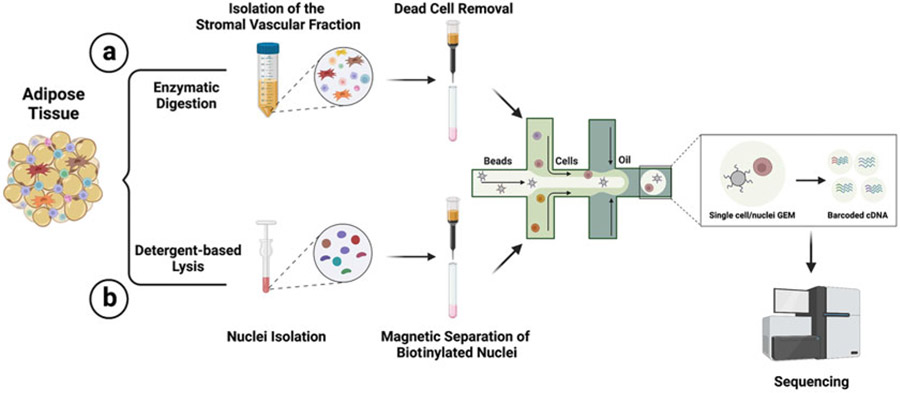
Using single cell transcriptome analysis and genetic lineage tracing, we have recently identified Trpv1-expressing cells as a new source of thermogenic adipocytes in BAT and WAT [3]. Here I describe the methods for isolation of the SVF from murine adipose tissue before single cell RNA-sequencing. Next, I describe an approach for the labeling and isolation of nuclei from specific cell types or lineages in adipose tissue using nuclear tagging and translating ribosome affinity purification (NuTRAP) [12] (Fig. 1).
Fig. 1.
Overview of the experimental steps for (a) single cell transcriptome profiling of adipose tissue and (b) enrichment of cell type- or lineage-specific nuclei using NuTRAP strain
The NuTRAP mouse strain was originally generated for simultaneous transcriptional and epigenomic profiling from specific cell types [12]. NuTRAP mice harbor a single polycistronic element targeted to the Rosa26 locus that encodes (1) the E. coli biotin ligase BirA, (2) the mouse nuclear membrane RanGAP1 protein tagged with a biotin ligase recognition peptide (BLRP) fused to mCherry, and (3) the 60S ribosomal subunit L10a fused to EGFP, each separated by a self-cleaving viral 2A peptide. The presence of a loxP-stop-loxP sequence in front of this polycistronic element stops the expression of these components in the absence of Cre recombinase. Crossing the NuTRAP mice with a cell type-specific Cre line will allow the cell type-specific labeling of the nuclear membrane with biotin and mCherry and subsequent purification using both affinity- and fluorescent-based purification.
2. Materials
2.1. Isolation of the Stromal Vascular Fraction from Mouse BAT or WAT for Single Cell Transcriptomics
Digestion media: Prepare digestion media by dissolving collagenase I (final concentration 1.5 mg/mL) and BSA (final concentration 2%) in HBSS buffer. Mix with Dispase II solution (final concentration 2.5 U/mL). Warm to 37 °C.
Growth media: Prepare growth media by adding FBS (10%) to DMEM, high glucose. Warm to 37 °C.
Ammonium chloride-potassium (ACK) lysis buffer (Lonza).
100 μM cell strainer.
40 μM cell strainer.
Dead Cell Removal Kit (Miltenyi Biotec).
MS columns (Miltenyi Biotec).
MACS separator (Miltenyi Biotec).
Trypan blue.
Automated cell counter or hemocytometer.
2.2. Isolation of Cell Type-/Lineage-Specific Nuclei Using NuTRAP Strain
Lysis buffer: 10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% NP-40 in nuclease-free water.
Nuclei wash and resuspension buffer: 1× PBS with 1% biotin-free bovine serum albumin (BSA) and 0.2 U/μL RNase inhibitor.
Labeling and separation buffer: 1× PBS (pH 7.2) supplemented with 2 mM EDTA, 1% biotin-free BSA, and 0.2 U/μL RNase inhibitor. Keep buffer cold (4 °C). Degas buffer before use, as air bubbles could block the column.
30 μM cell strainer.
Streptavidin MicroBeads (Miltenyi Biotec).
MS columns (Miltenyi Biotec).
MACS separator (Miltenyi Biotec).
Trypan blue.
Automated cell counter or hemocytometer.
3. Method
3.1. Isolation of the Stromal Vascular Fraction from Mouse BAT or WAT for Single Cell Transcriptomics
Sacrifice the mouse and dissect the adipose tissue. If tissues from multiple animals are being dissected, store them in HBSS until all tissues are dissected.
Mince the tissue to very fine pieces in a 50 mL canonical tube. Add 10 mL of the digestion media to each tube.
Place the tubes in a water bath or incubator with a shaker/rotator at 37 °C for 45 minutes.
Remove the tissue from the incubator and vortex for 10 seconds.
Centrifuge at 300 × g at 4 °C for 10 minutes in a swinging bucket centrifuge.
Aspirate the supernatant carefully so as not to disturb the pellet of SVF cells.
Resuspend the pellet in 10 mL of growth media.
Filter through a 100 μM cell strainer into a fresh 50 mL tube. Wash the tube with an additional 10 mL and filter through the cell strainer.
Centrifuge at 300 × g for 7 minutes.
Completely remove the supernatant and resuspend the pellet in 2 mL sterile ACK lysis buffer; place on ice for 5 minutes.
Filter through a 40 μM cell strainer into a fresh 50 mL tube. Wash the tube with 20 mL growth media and filter through the cell strainer.
Centrifuge at 300 × g for 7 minutes.
Resuspend the pellet in 1 mL of 1.5% BSA in PBS.
Use 10 μl of the cell suspension for cell counting and viability assessment. The cell viability of >90% is recommended for obtaining high quality single cell RNA-sequencing data. To achieve high cell viability, an additional step using the Dead Cell Removal Kit (Miltenyi Biotec) can be included to magnetically label and remove cell debris, dead cells, and dying cells. The dead cell removal step is performed according to the instructions for Dead Cell Removal Kit from Miltenyi Biotec described in steps 15–21 (see Notes 1 and 2).
Centrifuge the cell suspension at 300 × g for 5 minutes.
Resuspend the cells in 100 μL of dead cell removal bead solution. Incubate the cell suspension for 15 minutes at room temperature.
Prepare the binding solution by diluting the 20× solution in sterile ddH2O.
Place the MS columns on the MACS separator. Prepare each column by rinsing it with 0.5 mL 1× binding solution. Let the solution pass through the column.
Add 900 μL of 1× binding solution to each sample and apply cell suspension onto the column.
Collect effluent in a 2 mL tube as live cell fraction.
Rinse the column with an additional 1 mL of 1× binding solution.
Use 10 μL of the sample for cell counting and viability assessment.
Centrifuge the cell suspension at 300 × g for 5 minutes.
Resuspend the cells in the appropriate volume of 1.5% BSA in PBS. The recommended loading concentration for most standard single cell RNA-sequencing applications is 700–1200 cells/μL. Lower concentrations in the range of 100–600 cells/μL can be used for low throughput applications (e.g., using Chromium Next GEM Single Cell 3′ LT v3.1 Kit).
Keep the cell suspension on ice and proceed to the single cell isolation workflow. Minimize the time between cell preparation and chip loading.
3.2. Isolation of Cell Type-/Lineage-Specific Nuclei Using NuTRAP Strain
Here, I describe a protocol for isolation of the vascular smooth muscle-derived progenitors and their progenies using Trpv1cre Rosa26nuTRAP mice. Following the nuclei isolation from fresh adipose tissue, magnetic separation of the biotinylated nuclei using Streptavidin MicroBeads enables gentle purification of nuclei from all Trpv1-expressing cells and their progenies, regardless of the cellular state. The MicroBeads used in this protocol are small enough (20–100 nM) that they do not interfere with partitioning or emulsion stability in the single cell gene expression workflow.
3.2.1. Tissue Homogenization and Nuclei Isolation
Sacrifice the mouse and dissect the adipose tissue. If tissues from multiple animals are being dissected, store them in PBS until all tissues are dissected. Place the tissue on ice.
Mince to small <1 mM pieces with scissors.
Transfer into a 1 mL Dounce homogenizer and add 1 mL chilled lysis buffer on ice.
Homogenize with 40 strokes (20 strokes using the loose pestle, 20 strokes using the tight pestle), and transfer back to a 15 mL canonical tube.
Add 4 mL chilled lysis buffer to the tissue and incubate on ice for 7 minutes. Gently swirl to mix and repeat two to three times during the incubation.
Add 10 mL nuclei wash and resuspension buffer to the lysed tissue.
Filter through a 30 μM cell strainer to remove cell debris and large clumps.
Centrifuge the nuclei at 500 g for 5 minutes at 4 °C.
Remove the supernatant without disrupting the nuclei pellet.
Resuspend the pellet with 1 mL nuclei wash and resuspension buffer, and filter it through a 30 μM strainer to a new 15 mL Falcon tube (to avoid the lipid stuck on the wall). Fill the tube with 10 mL nuclei wash and resuspension buffer.
Centrifuge the nuclei at 500 g for 5 minutes at 4 °C. Remove the supernatant without disrupting the nuclei pellet (see Note 3).
Resuspend cell pellet in 90 μL of labeling buffer.
Visually assess and count the nuclei by mixing 10 μL of the nuclei suspension with 10 μL of trypan blue.
If debris is present, wash the nuclei with 1 mL nuclei wash and resuspension buffer, and centrifuge the nuclei at 500 g for 5 minutes at 4 °C. Remove the supernatant without disrupting the nuclei pellet. Resuspend cell pellet in 90 μL of labeling buffer.
Keep the nuclei on ice and proceed immediately with the labeling and purification step.
3.2.2. Labeling and Purification of Biotinylated Nuclei
Add 10 μL of Streptavidin MicroBeads per 107 total nuclei.
Mix well and refrigerate for 15 minutes (4–8 °C). Higher temperatures and/or longer incubation times lead to nonspecific labeling.
Wash nuclei by adding 1–2 mL of buffer per 107 cells and centrifuge at 500 × g for 5 minutes. Aspirate supernatant completely.
Resuspend up to 108 nuclei in 500 μL of separation buffer. For higher cell numbers, scale up the buffer volume accordingly. Proceed to magnetic separation.
3.2.3. Magnetic Separation with MS Columns
Place column in the magnetic field of a suitable MACS separator.
Prepare column by rinsing with 500 μL of separation buffer.
Apply nuclei suspension onto the column.
Collect unlabeled cells that pass through, and wash the column with 500 μL of separation buffer. Perform washing steps by adding separation buffer three times. Only add new buffer when the column reservoir is empty. Collect total effluent. This is the unlabeled cell fraction.
Remove the column from the separator and place it on a new collection tube.
Pipette 1 mL of separation buffer onto the column. Immediately flush out the magnetically labeled cells by firmly pushing the plunger into the column.
Centrifuge the nuclei at 500 × g for 5 minutes at 4 °C. Remove the supernatant without disrupting the nuclei pellet. Resuspend cell pellet in the appropriate amount of labeling buffer to have a concentration of 700–1200 nuclei/μL.
Determine the nuclei concentration using an automated cell counter or hemocytometer.
Keep the nuclei on ice and proceed immediately with the single nuclei isolation workflow. Minimize the time between nuclei preparation and chip loading.
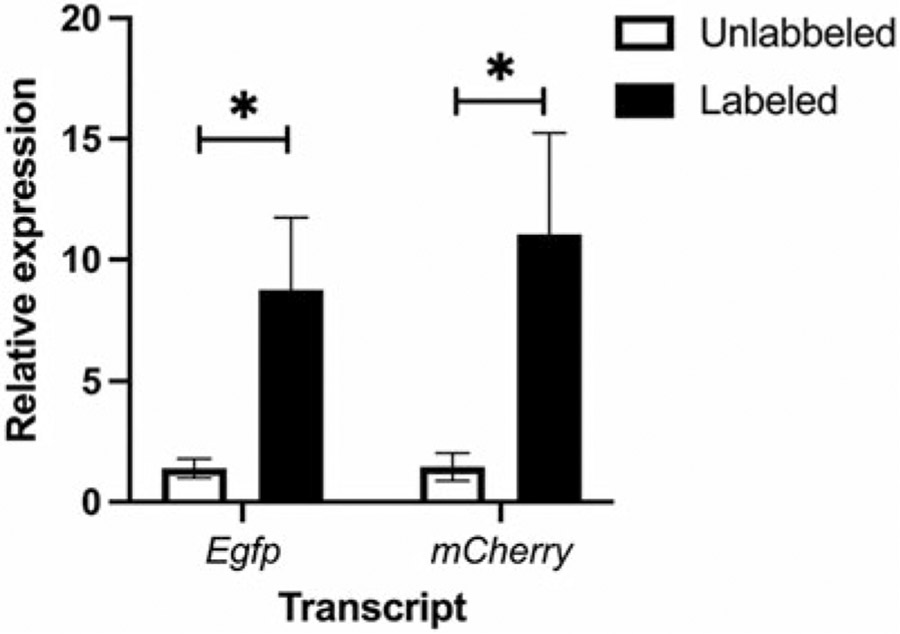
Gene expression analysis of the purified nuclei shows the clear enrichment of mCherry and EGFP transcripts in the labeled cells compared to the cells in the unlabeled fraction (Fig. 2).
Fig. 2.
Enrichment of cell type-specific nuclei using Trpv1cre Rosa26nuTRAP mice. The expression of Egfp and mCherry in the nuclei labeled with Streptavidin MicroBeads and the unlabeled nuclei from BAT of Trpv1cre Rosa26nuTRAP mice
4. Notes
Isolation of a viable cell suspension is the first and most important step in every single cell RNA-sequencing experiment. Optimization of tissue dissociation protocol for each adipose depot, rapid processing, and keeping cell suspensions on ice can improve cell viability.
Accurate counting and quality assessment of cells during the isolation and before loading them onto the microfluidic chip are essential. Cell suspensions should be checked visually. The presence of debris or cell aggregates results in inaccurate cell counts. Filter cell suspensions as needed to remove any debris or cell aggregates. Small volumes of cell suspension can be filtered using a 40 μM Bel-Art Flowmi™ Cell Strainer.
When working with a new adipose depot, it is recommended to optimize lysis time. After step 10, lysis efficacy should be assessed by staining the nuclei with trypan blue, and viability should be assessed through cell counting/microscopy. If a high fraction of viable cells is still present, centrifuge the nuclei at 500 g for 5 minutes at 4 °C, add 5 mL chilled lysis buffer, and incrementally increase the lysis time, monitoring efficacy via microscopy. When optimal lysis has occurred, repeat steps 7–11.
References
- 1.Shamsi F, Wang CH, Tseng YH (2021) The evolving view of thermogenic adipocytes – ontogeny, niche and function. Nat Rev Endocrinol 17(12):726–744. 10.1038/s41574-021-00562-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang QA, Tao C, Gupta RK, Scherer PE (2013) Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med 19(10):1338–1344. 10.1038/nm.3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shamsi F, Piper M, Ho LL, Huang TL, Gupta A, Streets A, Lynes MD, Tseng YH (2021) Vascular smooth muscle-derived Trpv1(+) progenitors are a source of cold-induced thermogenic adipocytes. Nat Metab 3(4):485–495. 10.1038/s42255-021-00373-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angueira AR, Sakers AP, Holman CD, Cheng L, Arbocco MN, Shamsi F, Lynes MD, Shrestha R, Okada C, Batmanov K, Susztak K, Tseng YH, Liaw L, Seale P (2021) Defining the lineage of thermogenic perivascular adipose tissue. Nat Metab 3(4):469–484. 10.1038/s42255-021-00380-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, Seale P (2019) Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364(6438). 10.1126/science.aav2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, Caprara C, Sun W, Schlaudraff KU, Soldati G, Wolfrum C, Deplancke B (2018) A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559(7712):103–108. 10.1038/s41586-018-0226-8 [DOI] [PubMed] [Google Scholar]
- 7.Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, Gupta RK (2018) Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. elife 7. 10.7554/eLife.39636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burl RB, Ramseyer VD, Rondini EA, Pique-Regi R, Lee YH, Granneman JG (2018) Deconstructing adipogenesis induced by beta3-adrenergic receptor activation with single-cell expression profiling. Cell Metab 28(2):300–309 e304. 10.1016/j.cmet.2018.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun W, Dong H, Balaz M, Slyper M, Drokhlyansky E, Colleluori G, Giordano A, Kovanicova Z, Stefanicka P, Balazova L, Ding L, Husted AS, Rudofsky G, Ukropec J, Cinti S, Schwartz TW, Regev A, Wolfrum C (2020) snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587(7832):98–102. 10.1038/s41586-020-2856-x [DOI] [PubMed] [Google Scholar]
- 10.Emont MP, Jacobs C, Essene AL, Pant D, Tenen D, Colleluori G, Di Vincenzo A, Jorgensen AM, Dashti H, Stefek A, McGonagle E, Strobel S, Laber S, Agrawal S, Westcott GP, Kar A, Veregge ML, Gulko A, Srinivasan H, Kramer Z, De Filippis E, Merkel E, Ducie J, Boyd CG, Gourash W, Courcoulas A, Lin SJ, Lee BT, Morris D, Tobias A, Khera AV, Claussnitzer M, Pers TH, Giordano A, Ashenberg O, Regev A, Tsai LT, Rosen ED (2022) A single-cell atlas of human and mouse white adipose tissue. Nature 603(7903):926–933. 10.1038/s41586-022-04518-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta A, Shamsi F, Altemose N, Dorlhiac GF, Cypess AM, White AP, Yosef N, Patti ME, Tseng YH, Streets A (2022) Characterization of transcript enrichment and detection bias in single-nucleus RNA-seq for mapping of distinct human adipocyte lineages. Genome Res 32(2):242–257. 10.1101/gr.275509.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roh HC, Tsai LT, Lyubetskaya A, Tenen D, Kumari M, Rosen ED (2017) Simultaneous transcriptional and epigenomic profiling from specific cell types within heterogeneous tissues in vivo. Cell Rep 18(4):1048–1061. 10.1016/j.celrep.2016.12.087 [DOI] [PMC free article] [PubMed] [Google Scholar]