Abstract
The Tap (taxis toward peptides) receptor and the periplasmic dipeptide-binding protein (DBP) of Escherichia coli together mediate chemotactic responses to dipeptides. Tap is a low-abundance receptor. It is present in 5- to 10-fold-fewer copies than high-abundance receptors like Tar and Tsr. Cells expressing Tap as the sole receptor, even from a multicopy plasmid at 5- to 10-fold-overexpressed levels, do not generate sufficient clockwise (CW) signal to tumble and thus swim exclusively smoothly (run). To study the signaling properties of Tap in detail, we constructed reciprocal hybrids between Tap and Tar fused in the linker region between the periplasmic and cytoplasmic domains. The Tapr hybrid senses dipeptides and is a good CW-signal generator, whereas the Tarp hybrid senses aspartate but is a poor CW-signal generator. Thus, the poor CW signaling of Tap is a property of its cytoplasmic domain. Eighteen residues at the carboxyl terminus of high-abundance receptors, including the NWETF sequence that binds the CheR methylesterase, are missing in Tap. The Tart protein, created by removing these 18 residues from Tar, has diminished CW-signaling ability. The Tapl protein, made by adding the last 18 residues of Tar to the carboxyl terminus of Tap, also does not support CW flagellar rotation. However, Tart and Tapl cross-react well with antibody directed against the conserved cytoplasmic region of Tsr, whereas Tap does not cross-react with this antibody. Tap does cross-react, however, with antibody directed against the low-abundance chemoreceptor Trg. The hybrid, truncated, and extended receptors exhibit various levels of methylation. However, Tar and Tapl, which contain a consensus CheR-binding motif (NWETF) at their carboxyl termini, exhibit the highest basal levels of methylation, as expected. We conclude that no simple correlation exists between the abundance of a receptor, its methylation level, and its CW-signaling ability.
Unstimulated Escherichia coli cells swim smoothly for a period of one to several seconds (a run), during which the flagellar motor rotates counterclockwise (CCW). A reversal to clockwise (CW) flagellar rotation causes a brief episode of uncoordinated thrashing (a tumble) that randomly reorients the subsequent run. Alternating runs and tumbles generate a three-dimensional random walk. In a gradient of an attractant chemical, the random walk is biased so that when a cell swims toward higher concentrations of an attractant, tumbles are suppressed and runs are extended (8).
Chemicals in the environment are sensed via chemoreceptors that span the cell membrane (33, 42, 51). These chemoreceptors modulate the activity of CheA, a cytoplasmic histidine protein kinase that is capable of autophosphorylation. The phosphate is transferred from CheA to the small cytoplasmic protein CheY. The rapid spontaneous decay of phospho-CheY is accelerated by the CheZ protein. Phospho-CheY induces CW rotation of the flagella. Unliganded receptors stimulate CheA activity, whereas attractant-bound receptors suppress CheA autophosphorylation and, in concert with CheZ, reduce the amount of cytoplasmic phospho-CheY.
Chemotactic adaptation is accomplished by reversible methylation of particular glutamate residues in the cytoplasmic domain of the receptors. The signal initiated by attractant binding is canceled by receptor methylation, which increases the mobility of the receptors during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (21, 47, 49). When the level of methylation balances an attractant signal, the receptor returns to its prestimulus signaling state, and the adapted cells return to their prestimulus behavior.
Adaptation reflects a kinetic competition between the activities of CheR and CheB, the latter of which is active in its phosphorylated form and is a substrate for phosphotransfer from phospho-CheA. It may also be influenced by the availability of the glutamate or glutamyl-methyl ester residues as substrates for CheR and CheB. Depending on the concentration and potency of the attractant, the adaptation time can range from a few seconds to many minutes.
Five chemoreceptors have been found in E. coli: Tar, Tsr, Tap, Trg, and the recently discovered receptor for oxygen and redox taxis, Aer (9, 43). Tar and Tsr bind aspartate and serine, respectively (36, 49). Sugars and dipeptides bind to periplasmic substrate-binding proteins, which then interact with the chemoreceptors: maltose-binding protein with Tar (14, 22), dipeptide-binding protein with Tap (1, 34), and galactose- and glucose-binding protein and ribose-binding protein with Trg (2, 3, 23, 26).
Cells lacking Tsr, Tar, Tap, and Trg (ΔT strains) always swim smoothly because they have low basal activities of CheA (13, 15) and are nonchemotactic. Either Tsr or Tar, which are normally present at several thousand copies per cell, can maintain normal run-tumble behavior when present as the sole chemoreceptor. Tap and Trg are present at only a few hundred copies per cell (25, 50), but even when they are overexpressed from a multicopy plasmid, they are less effective at generating CW rotation (see reference 18 and this study).
Tsr, Tar, Tap, and Trg consist of periplasmic, transmembrane, and cytoplasmic domains (12, 29). The amino terminus is in the cytoplasm and connects through the first transmembrane segment (TM1) to the periplasmic receptor domain. The periplasmic domain connects to the cytoplasmic domain by a second transmembrane segment (TM2). The amino-terminal portion of the cytoplasmic domain contains a sequence of unknown function known as the linker region. The cytoplasmic domain also contains the regions required for CW and CCW signaling and for adaptive methylation. Within a sliding window of 29 residues, Tsr, Tar, and Tap have sequence identities that range from 60 to 100% over their cytoplasmic domains. A 48-residue sequence at the core of the signaling region is identical in Tsr and Tar, and it differs in Tap by a single, chemically conservative change.
The periplasmic and cytoplasmic domains of Tsr and Tar can be swapped to create functional chimeras (28). In contrast, two Tar-Tap fusion proteins generated by homologous recombination between these tandem genes did not mediate aspartate or maltose taxis, although both hybrids had an intact Tar receptor domain. The hybrid receptor that contained more of Tap was a dominant CCW signaler, a property that was attributed to “domain incompatibility” (48).
High- and low-abundance chemoreceptors differ at their extreme carboxyl termini. Tap and Trg lack 18 and 28 residues, respectively, present in Tsr and Tar (12, 29). The importance of the extreme C terminus of Tar from Salmonella typhimurium is also highlighted by the finding that its loss leads to severe defects in receptor function and to disruptions in adaptation (44).
The NdeI site in sequences encoding the cytoplasmic linker regions of Tsr and Tar has been used to construct Tsr-Tar and Tar-Tsr hybrids (28). We used the same strategy to engineer similar hybrids between Tar and Tap. (Conceptually similar studies with the Tsr and Trg receptors were reported recently by Baumgartner et al. [18].) To study the function of the extreme carboxyl terminus of Tar, we also truncated Tar by introducing a nonsense codon into tar at the position of the stop codon of tap and lengthened Tap by adding the last 18 residues of Tar to Tap. The behavior of cells producing these various engineered receptors helps elucidate the functions of particular domains and subdomains of high- and low-abundance receptors.
MATERIALS AND METHODS
Bacterial strains.
E. coli VB13 is a thr+ eda+ Δtsr7021 trg::Tn10 Δtar-tap5201 derivative of strain RP437 (41). Strain MM509 is an eda+ Δtar-tap5201 derivative of strain RP437. Strain CJ236 is a dut1 ung1 thi1 relA1 strain, containing plasmid pCJ105 (24), that was used to generate single-stranded plasmids for site-directed mutagenesis.
Plasmids.
Plasmid pVB8 (11) confers Ampr, carries the laclq gene, and expresses the tap gene from the tac promoter. The single-stranded origin from plasmid pZ150 (58) was introduced into pVB8 to create pSW1. An NdeI site was introduced by site-directed mutagenesis at codon 255 of tap in plasmid pSW1 to create plasmid pSW2. Plasmid pMK113 contains the E. coli tar gene and contains the single-stranded origin of phage M13 from plasmid pZ150 (19).
Plasmids pSW2 and pMK113 were digested with NdeI. Each yielded two fragments, since both vectors contain one NdeI site outside of their respective receptor genes. All four fragments were electroeluted from agarose gels. The fragment encoding the carboxyl-terminal end of Tar was ligated with the fragment encoding the amino-terminal end of Tap to create pTapr (Fig. 1). Plasmid pTarp was created by ligating the remaining two fragments. Plasmid pTart was generated by introducing a UAA triplet at codon 536 of tar in plasmid pMK113. Plasmid pTapl was produced by introducing a unique NheI site into pMK113 at codon 530 of tar and into pSW1 at codon 530 of tap. The NheI-PstI fragment from pMK113 carrying the last 18 codons of tar was ligated with a PstI-NheI fragment containing the bulk of the 5′ end of the tap gene to produce an in-frame translational fusion between Tap and the carboxyl-terminal extension of Tar.
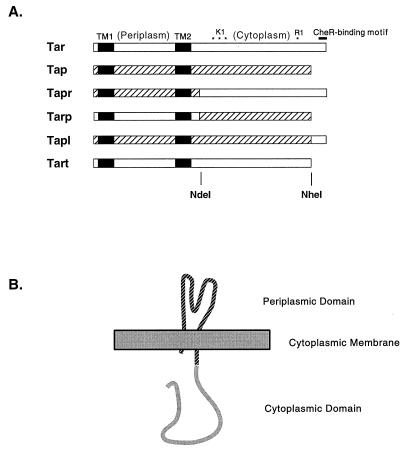
FIG. 1.
Structure of Tar and Tap chimeras. (A) Primary structures. Regions derived from Tar are shown in white, whereas regions derived from Tap are hatched. The TM regions are depicted as solid black rectangles. The positions of glutamate residues subject to methylation are indicated with asterisks above Tar: three methylation sites are in the K1 tryptic peptide, and one methylation site is in the R1 tryptic peptide. The NWETF motif for binding of CheR is depicted as a bold black line above the right-hand (carboxyl-terminal) end of Tar. The periplasmic and cytoplasmic regions of Tar are also labeled. The protein-equivalent positions of the naturally occurring (tar) or introduced (tap) NdeI restriction site and of the introduced NheI restriction site are indicated beneath Tart. (B) Membrane topology of the chimeric Tapr receptor.
Media.
Strains carrying plasmids were selected and maintained on Luria-Bertani agar (37) supplemented with 50 μg of ampicillin and/or 15 μg of tetracycline per ml. Minimal swarm plates contained 0.325% Difco Bacto-Agar and Che salts [10 mM potassium phosphate (pH 7.0); 1 mM (NH4)2SO4, 1 mM MgSO4] supplemented with 20 μg of l-threonine, l-leucine, l-histidine, and l-methionine per ml; 1 μg of thiamine per ml; and 25 μg of ampicillin per ml. The attractants l-aspartate and maltose were added to swarm plates to final concentrations of 100 μM, and the attractant l-prolyl-l-leucine (Pro-Leu) was added to 50 μM. Swarm plates containing aspartate or Pro-Leu were supplemented with 0.5% NaCl and 1 mM glycerol.
Cells for tethering and methylation assays were grown in H1 minimal medium (37) containing 0.5% glycerol; 0.2% Casamino Acids (Difco); 100 μg of l-threonine, l-leucine, l-histidine, and l-methionine per ml; 1 μg of thiamine per ml; and 25 μg of ampicillin per ml. For cultures that were to be tested for their responses to maltose, expression of the proteins of the maltose regulon was induced by supplementing the medium with 0.2% maltose. To induce transcription from the tac promoter, the media contained 1 mM isopropylthiogalactoside (IPTG). All reagents were purchased from Sigma Chemical Co. unless otherwise indicated.
Site-directed mutagenesis.
Site-directed mutagenesis was done according to the method of Kunkel et al. (30), with M13 origin-containing plasmid pMK113 or pSW1. Single-stranded templates for mutagenesis reactions were made by infecting strain CJ236 carrying either of these plasmids with M13 phage derivative KO7 (54), whose DNA competes poorly with plasmid DNA for packaging into virions. Introduction of site-directed mutations was confirmed by DNA sequencing.
Chemotactic swarm assays.
Fresh, overnight colonies from Luria-Bertani–ampicillin agar were inoculated into minimal swarm plates. Plates containing l-aspartate were scored after 8 h of incubation at 30°C, and plates containing maltose or Pro-Leu were scored after 16 h of incubation at 30°C. When expression of Tap or Tap hybrid proteins from the tac promoter was desired, the agar also contained 1 mM IPTG.
Tethered-cell assays.
Cells were inoculated into supplemented minimal-glycerol medium from a fresh, overnight culture grown in the same medium. The optical density at 578 nm (OD578) after inoculation into 10 ml of medium was 0.05, and cells were grown with vigorous swirling at 32°C to an OD578 of 0.5. The flagellar filaments of cells in the culture were then sheared by 5 pulses of 10 s each at the highest power setting of a Waring blender, with a 50-ml stainless-steel blender cup. Cells were washed twice with tethering buffer (10 mM potassium phosphate [pH 7.0], 0.1 M NaCl, 100 μM EDTA, 10 μM l-methionine, 20 mM sodium l-lactate). Tethering buffer also contained chloramphenicol at 20 μg/ml to prevent regrowth of filaments. The washed cells were finally resuspended in tethering buffer at an OD578 of 0.5. Thirty microliters of this suspension was mixed with 40 μl of tethering buffer and 30 μl of antifilament antibody (diluted 1:250 in tethering buffer). Fifty-microliter aliquots of this cell resuspension were placed inside a thin ring of Apiezon L grease on a 12-mm-diameter round glass coverslip that had been cleaned with fuming nitric acid and extensively rinsed with deionized water. Cells were allowed to settle onto the coverslip for at least 30 min at room temperature in a humidity chamber. The coverslip was then affixed to a flow chamber (7) with a seal created by the Apiezon L grease. The cells were viewed at ×400 magnification with an Olympus reverse-phase-contrast microscope. The microscopic images were recorded with a video camera attached to a video recorder, and the behavior of the cells was analyzed from a real-time or one-quarter-speed playback of the videotapes. An average of 30 tethered cells spun per microscope field. Aspartate, Pro-Leu, or maltose was added at 1 mM in tethering buffer during a 15-s flow at a rate of about 1 ml/min. The adaptation times of responding cells were measured as the elapsed time of exclusively CCW flagellar rotation after addition of the attractant until the first reversal to CW flagellar rotation. For each strain and treatment, the mean adaptation time for 40 cells was calculated.
Determination of receptor methylation levels.
Methylation assays were adapted from previously published methods (17, 31, 56). To determine steady-state methylation levels, cells were grown to an OD578 of 0.7 in the same medium as that used to grow cells for the tethering assay. Cells from 1 ml of the culture were pelleted, washed once in cold phosphate buffer (PE; 10 mM potassium phosphate [pH 7.0], 100 μM EDTA), and resuspended in 100 μl of SDS-PAGE sample buffer. Aliquots of 30 μl were loaded per lane for SDS-PAGE. The gels contained either 8 or 10% acrylamide and were run for 4 h at 35 mA. To look at attractant-induced changes in methylation, cells from 1 ml of the culture at an OD578 of 0.7 were pelleted and washed three times in cold PE and resuspended in 1 ml of PE containing 200 μg of chloramphenicol per ml, 10−5 M l-methionine, and 10 mM sodium d-lactate. Samples were incubated for 20 min with shaking at 30°C, and then attractants were added to a final concentration of 10 mM. After another 20 min, the reaction was terminated by adding 100 μl of cold 100% trichloracetic acid. After 15 min on ice, the samples were washed with cold 1% trichloroacetic acid and then acetone and were allowed to air dry. The samples were resuspended in 200 μl of SDS-PAGE sample buffer, and 40 μl of each resuspension was loaded per lane for SDS-PAGE. The 8% acrylamide gel was run at 35 mA for 4 h.
Protein blotting to nitrocellulose filters was done as described by Towbin et al. (52). The filters were incubated with antiserum (obtained from J. S. Parkinson) raised against the conserved signaling domain of Tsr (TsrCSR) or antisera raised against the entire E. coli Tar and Trg proteins (obtained from Robert Bourret and Gerald Hazelbauer, respectively). Antigen-bound rabbit immunoglobulins were visualized with goat anti-rabbit immunoglobulin A–alkaline phosphatase conjugate (Bio-Rad). Methylation states of the receptors were inferred from the number and position of the stained bands. (The mobility of the receptors during SDS-PAGE corresponds to their extent of methylation, with more highly methylated receptors migrating more rapidly.)
DNA sequence analysis.
Double-stranded templates for DNA sequencing were generated from plasmids in strain VB13 by an alkaline lysis procedure (10). The dideoxynucleotide chain termination method (45) was used for double-stranded sequencing (27). Sequenase version 2.0 kits were purchased from U.S. Biochemicals.
RESULTS
Construction of hybrids between Tar and Tap.
The tsr and tar genes have a conserved NdeI site 46 codons downstream of the sequences that encode their respective TM2 segments. The reciprocal fusion proteins encoded by hybrid genes joined at this shared NdeI site have been called Tsar and Tasr, contain the periplasmic and both TM segments of Tsr or Tar, and function in serine and aspartate sensing, respectively (28). The fusion joint is three residues amino terminal to a natural proteolysis-sensitive site in S. typhimurium Tar (39). Cleavage of Tar at this site produces two relatively stable fragments, suggesting that this region of the receptors forms a link between domains that are at least partially structurally independent.
Since the tap gene does not contain this NdeI site, the site was introduced by site-directed mutagenesis. This change converted a Thr residue in Tap into a His residue, which is found at this position in Tar. However, the chemotactic rings formed on Pro-Leu swarm plates were identical for strain MM509 (tsr+ Δtar-tap) expressing the mutant Tap protein from plasmid pSW2 or the wild-type protein from plasmid pSW1, so the His-for-Thr substitution had no obvious phenotype in this context. The NdeI site introduced into tap was then used to construct translationally fused genes encoding the Tarp and Tapr chimeric receptors (Fig. 1).
Chemotactic behavior of cells expressing Tarp or Tapr.
Assays with tethered cells (46) were used to measure the chemotactic responses mediated by the hybrid receptors (Table 1). The basal CW/CCW ratio of flagellar rotation was measured in the absence of chemoeffectors. Upon addition of attractant, responsive receptors normally promote exclusively CCW flagellar rotation until adaptation restores CW rotation.
TABLE 1.
Rotational biases and attractant responses of tethered cells expressing derivatives of Tar and Tap receptorsa
| Plasmid gene-encoded construct | % of time rotating CCW for unstimulated cells | Adaptation time (min) for attractant:
|
||
|---|---|---|---|---|
| Asp | Maltose | Pro-Leu | ||
| Strain VB13 (Δtar-tap Δtsr trg::Tn10) | ||||
| Tar | 85 | 48 | 17 | —b |
| Tap | 100 | NDc | ND | ND |
| Tarp | 100 | ND | ND | ND |
| Tapr | 74 | — | — | 2.3 |
| Tart | 100 | ND | ND | ND |
| Tapl | 100 | ND | ND | ND |
| Strain MM509S (Δtar-tap tsr+trg+) | ||||
| Tap | 62 | — | — | 0.5 |
| Tarp | 49 | 23 | 7.3 | — |
| Tart | 63 | 21 | 6 | — |
| Tapl | 71 | — | — | 0.6 |
Chimeric or otherwise engineered receptors are depicted in Fig. 1. Rotational biases, expressed as percentage of time rotating CCW, were measured manually from videotapes for 50 cells per strain and represent the behavior during a 30-s observation for each cell. Attractants were added to the flow chamber at 1 mM. Adaptation times were measured as the period of exclusively CCW flagellar rotation from 5 s after the onset of the attractant flow until the first reversal to CW flagellar rotation. The values given for adaptation times are the means for 40 cells per strain.
—, no measurable response (because of inappropriate attractant for plasmid gene-encoded receptor).
ND, not detectable (because cells were already turning their flagella exclusively CCW).
Cells of strain VB13 (ΔT) rotated their flagella only CCW when Tap was the sole receptor, even when expression of plasmid-encoded Tap was induced by addition of 1 mM IPTG. However, in strain MM509, in which CW signal is provided by chromosomally encoded Tsr, Tap did mediate a brief (0.5 min) CCW response to 10 mM Pro-Leu. Similarly, cells of strain VB13 expressing plasmid-encoded Tarp as their only receptor also rotated their flagella only CCW, but in strain MM509, Tarp mediated very robust responses to both aspartate and maltose. Plasmid-encoded Tapr supported CW flagellar rotation in strain VB13, and it also mediated good responses to Pro-Leu.
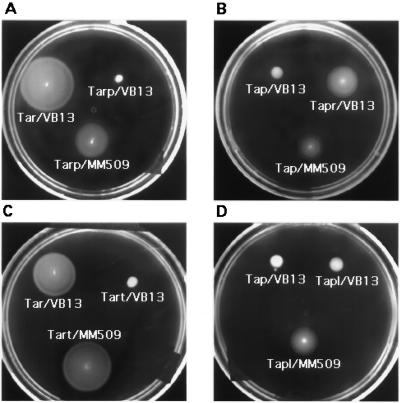
The behavior of strains producing hybrid receptors on swarm plates confirmed the results from tethered-cell assays. Strain VB13 expressing Tarp (Fig. 2A) or Tap (Fig. 2B) was defective in forming a spreading colony in aspartate or Pro-Leu semisolid agar. However, strain MM509 expressing Tarp or Tap produced sizeable chemotactic rings on aspartate (Fig. 2A) or Pro-Leu (Fig. 2B) swarm plates. Strain VB13 expressing Tapr formed distinct chemotactic rings on Pro-Leu swarm plates (Fig. 2B), confirming that Tapr is competent to support CW flagellar rotation and the run-tumble behavior that is essential for the spreading of a colony in semisolid agar (55).
FIG. 2.
Chemotactic responses mediated by wild-type and hybrid receptors. Swarms were formed by colonies of strain VB13 (ΔT) or strain MM509 (Δtar-tap) carrying plasmids containing genes encoding the wild-type or hybrid receptors. (A and C) Aspartate swarm plates. (B and D) Pro-Leu swarm plates. Large, rapidly forming swarms with thick outer rings are characteristic of wild-type aspartate taxis, whereas more slowly developing, thin, sharp rings on the surface of the agar are characteristic of wild-type dipeptide taxis.
The last 18 residues of Tar are necessary but insufficient for generating CW flagellar rotation.
Because Tap lacks the 18 carboxyl-terminal residues present in Tsr or Tar (29), we determined the signaling phenotype of a Tar protein lacking these residues. A UAA stop codon was introduced into the tar gene carried on plasmid pMK113 (19) at the same relative position as the natural termination codon of tap. The truncated Tar (Tart) thus generated ends at residue 535 (Fig. 1). Cells of strain VB13 expressing plasmid-encoded Tart did not rotate their flagella CW (Table 1) and were incapable of spreading on aspartate swarm plates (Fig. 2C). As anticipated, however, strain MM509 expressing Tart responded to aspartate and maltose in the tethered-cell assay (Table 1) and produced normal chemotactic rings on aspartate (Fig. 2C) and maltose (data not shown) swarm plates. Thus, Tart mediated responses to aspartate and maltose if CW signal was provided in trans.
We also added the last 23 residues of Tar onto Tap, including the 18 residues that were deleted from Tar to form Tart (Fig. 1). Strain VB13 cells producing this lengthened edition of Tap (Tapl) neither generated CW flagellar rotation nor spread on Pro-Leu swarm plates. However, Tapl did enable strain MM509 to form chemotactic rings on Pro-Leu swarm plates (Fig. 2D) and to mediate a response to Pro-Leu (0.6 min) about as long as that mediated by wild-type Tap (0.5 min) in this strain (Table 1). Thus, addition of the carboxyl-terminal 18 residues of Tar onto Tap was insufficient to confer the ability to produce CW rotation, but the added residues did not interfere with the ability of Tap to generate CCW signal in response to Pro-Leu.
Methylation properties of the hybrid receptors.
Recent work (57) has shown that the last five residues (NWETF) of the high-abundance receptors constitute a binding site for the CheR methylesterase. Methylation levels affect the signaling state of the receptor in such a way that undermethylation generally leads to a more CCW-biased signal output. Therefore, the inability of Tap to support CW flagellar rotation when it is the sole receptor (except Aer) present in the cell could be due to its decreased basal level of methylation.
Tar expressed in strain VB13 is methylated to some extent even in the absence of aspartate (Fig. 3). Mowbray and Koshland (40) estimated that there are about 0.5 methyl groups per Tar receptor in its unstimulated state. We accordingly infer that the two bands seen on immunoblots in lanes containing Tar from unstimulated cells correspond to the unmethylated protein (upper band) and a singly methylated protein (lower band), each present in approximately equal amounts. Addition of aspartate to the cells leads to a higher level of methylation, seen as additional, faster-migrating bands.
FIG. 3.
Changes in methylation of Tar and hybrid receptors in cells exposed to attractants. Approximately equal amounts of total protein extracted from cells of strain VB13 (ΔT) were loaded in each lane. The samples in the left three lanes were from cells containing plasmid pMK113 (tar+), the samples in the middle three lanes were from cells containing plasmid pTarp, and the samples in the right three lanes were from cells containing plasmid pTapr and induced with 1 mM IPTG. Within each set of three lanes, the sample loaded on the left was from unstimulated cells, the sample loaded in the middle was from cells exposed and adapted to 1 mM Pro-Leu, and the sample loaded on the right was from cells exposed and adapted to 10 mM l-aspartate. The proteins were visualized after incubation with antiserum against the entire E. coli wild-type Tar protein. SDS-PAGE was carried out with an 8% acrylamide gel.
The Tar antibody did not label the Tap protein under any conditions (data not shown), but it did recognize the Tarp and Tapr chimeras (Fig. 3). Tarp expressed in strain VB13 produced a single, faint band, which may represent the unmethylated protein. A slight increase in methylation of Tarp was observed when the protein was obtained from cells exposed to aspartate. Tapr migrated as two bands, with the upper band (unmethylated form of the protein?) being more intense. However, no change in the methylation pattern was seen when Tapr came from cells exposed to saturating concentrations of Pro-Leu.
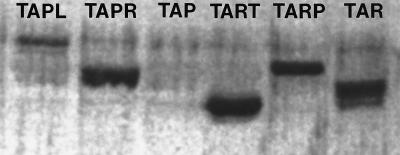
Antibody raised against the TsrCSR also failed to recognize Tap (Fig. 4). The blot was analyzed with a PhosphorImager to increase sensitivity so as to detect poorly cross-reacting or nonabundant proteins. The background level of nonspecific or “spillover” banding was rather high, but no band unequivocally assignable to Tap was present. The TsrCSR antibody did, to various degrees, label the other five receptors. The Tapl band or bands were not very intense, but the other four receptors produced heavy bands. Multiple bands were detected with Tapr, Tart, and Tar, confirming that these proteins experience at least some level of methylation in the absence of attractant. At the level of resolution of the gel, Tarp appeared to form only one band, as it did with Tar antibody (Fig. 3).
FIG. 4.
Immunoblot analysis of hybrid and engineered receptors with TsrCSR antiserum. Samples containing approximately equal amounts of protein were loaded in each lane. Samples came from cells of strain VB13 containing the following plasmids (from left to right): pTapl, pTapr, pSW2 (tap+), pTart, pTarp, and pMK113 (tar+). Cultures of cells containing pTapl, pTapr, and pSW2 contained 1 mM IPTG. The proteins were visualized after incubation with the TsrCSR antiserum. SDS-PAGE was carried out with a 10% acrylamide gel.
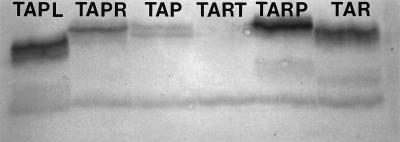
Developing the immunoblots with antibody against Trg produced a very different band pattern (Fig. 5). Tart, which was intensely labeled with TsrCSR antibody, was not labeled at all by Trg antibody. In contrast, Tap, which was essentially invisible on immunoblots made with Tar or TsrCSR antibody, appeared as two distinct bands when the Trg antibody was used. The remaining four proteins (Tapl, Tapr, Tarp, and Tar) were visualized with both antisera (compare Fig. 4 and 5). Tapr and Tar were labeled more strongly with the TsrCSR, whereas Tapl and Tarp were labeled more strongly with the Trg antibody. The relative positions of the bands for a given receptor were also different in the immunoblots visualized with the TsrCSR and Trg antibodies, probably because different acrylamide concentrations were used in the two gels (10% with the TsrCSR blot, 8% with the Trg blot).
FIG. 5.
Immunoblot analysis of hybrid and engineered receptors with Trg antiserum. Samples containing approximately equal amounts of protein were loaded in each lane. Samples came from cells of strain VB13 containing the following plasmids (from left to right): pTapl, pTapr, pSW2 (tap+), pTart, pTarp, and pMK113 (tar+). Cultures of cells containing pTapl, pTapr, and pSW2 contained 1 mM IPTG. The proteins were visualized after incubation with antiserum against the entire E. coli wild-type Trg protein. SDS-PAGE was carried out with an 8% acrylamide gel.
DISCUSSION
The results reported here extend our knowledge of the plasticity of bacterial chemoreceptors. Previous work (28) had shown that the sensing domain of one high-abundance receptor (Tsr or Tar) can be functionally coupled with the cytoplasmic signaling domain of the other. Our work extends this observation to include hybrids of high-abundance (Tar) and low-abundance (Tap) receptors, which differ more in their signaling properties than do Tsr and Tar. For the Tar-Tap (Tarp) and Tap-Tar (Tapr) hybrids, as for the Tsar and Tasr hybrids studied earlier, ligand recognition is determined by the receptor domain, whereas signaling properties are conferred by the cytoplasmic domain. Generally similar conclusions were obtained recently with hybrids constructed from another pair of high- and low-abundance receptors, Tsr and Trg (18).
Functional chimeras have also been made between the receptor and transmembrane domains of Tar (53) or Trg (5) and the signaling domain of the EnvZ osmosensor of E. coli, and even between Tar and the tyrosine kinase domain of the insulin receptor (38). In each case, the activity of the hybrid protein came under control of the appropriate chemoattractant (aspartate for Tar, ribose or galactose for Trg), although no biological function was associated with this control. In contrast, the Tsr-Tar, Tsr-Trg, and Tar-Tap chimeric receptors mediate chemotaxis.
The fusion joints of all of these chimeric receptors lie in the linker region that connects the periplasmic ligand-binding domain and transmembrane domain to the cytoplasmic signaling domain. Construction of the original fusions utilized a naturally occurring, shared NdeI site in the DNA sequences encoding the Tsr and Tar linker regions (29) or an NdeI site introduced into the equivalent position in the other receptor genes. Hybrid receptors with periplasmic ligand-binding domains incorporating elements of Tar and Tsr have also yielded functional chimeras (31). However, Slocum et al. (48) found that Tar-Tap hybrid receptors with fusion joints within the signaling or methylation regions produced nonfunctional or dominant-negative chimeras.
The success of efforts to use the linker region as a site for fusing radically different receptors supports biochemical studies that indicate that the portions of the receptors amino terminal and carboxyl terminal to the linker are structurally autonomous (4, 39). The fact that these domains can be paired in a variety of functional combinations suggests that common mechanisms of signaling exist in a wide range of homodimeric transmembrane receptors (50).
The behavior of the truncated Tar receptor (Tart) and the lengthened Tap receptor (Tapl) provides further clues about the role of the extreme carboxyl-terminal regions of the high-abundance receptors in chemotactic signaling. Tart lacks the last 18 residues of Tar, including the carboxyl-terminal residues NWETF, which constitute a binding motif for CheR. Tart is defective in generating CW signal (Table 1), although it appears to be present in the cell at about the same level as Tar (Fig. 4). Despite the absence of the CheR-binding consensus, it also seems to form at least one methylated species. Tapl, which has the last 18 residues of Tar fused to the carboxyl terminus of Tap, is also defective in generating CW signal. However, the Tapl bands on immunoblots (Fig. 4 and 5) were substantially more intense than those formed by Tap when it was expressed from the same promoter. The multiple-banding pattern shown by Tapl also suggests that it consists of at least two methylated species, even in an unstimulated cell (Fig. 5). These results rule out any simple relationship between methylation state, CW-signaling capacity, and receptor abundance or stability.
Several correlations of receptor structure and chemotactic behavior do emerge from the data, however (Table 1). First, receptors acquiring the bulk of their cytoplasmic domains from Tap (Tap, Tarp, and Tapl) fail to support CW rotation when they are present as the sole receptor species in a cell. Second, receptors with the intact cytoplasmic signaling domain of Tar (Tar and Tapr) can generate CW rotation when they are the only receptor present. Third, the adaptation times of Pro-Leu responses (mediated by dipeptide-binding protein) of cells expressing receptors containing the periplasmic domain of Tap (Tap, Tapr, and Tapl) are always briefer than the maltose responses (mediated by maltose-binding protein) of cells expressing comparable receptors containing the periplasmic domain of Tar (Tar, Tarp, and Tart). Fourth, receptors containing the carboxyl-terminal NWETF sequence that binds CheR methylesterase (57) have increased levels of methylation relative to comparable receptors lacking this motif (see the comparison of Tapl with Tap and Tarp [Fig. 5]).
Tar, Tap, and Trg are all quite dissimilar in the amino acid sequences of their periplasmic domains (12, 29). Thus, we anticipate that the Trg antiserum reacts primarily with epitopes in the more highly conserved cytoplasmic domain of Tar and Tap, in this respect behaving much like antibodies directed against the conserved region of the cytoplasmic domain of Tsr (TsrCSR). The very different labeling patterns achieved with these two antibodies (Fig. 4 and 5) were therefore unexpected. The proteins are presumably denatured during SDS-PAGE, but after transfer to the blotting filters, they must refold into conformations that can be discriminated by these antibodies.
Even though Tap is identical to Tsr and Tar at 47 out of 48 residues in the most highly conserved region of its signaling domain (29), TsrCSR recognizes Tap poorly, if at all (Fig. 4). Trg is identical to Tap at only 37 of the same 48 residues, yet the Trg antibody labels Tap much better than does the TsrCSR antibody (Fig. 5). The reverse is true for Tart, which reacts well with the TsrCSR antibody (Fig. 4) but not at all with the Trg antibody (Fig. 5). Tarp, in which the Tap cytoplasmic domain is fused to the Tar periplasmic domain, is recognized by both antibodies. So is Tapl, which differs from Tap only in having 18 additional residues from Tar at its carboxyl terminus.
These results imply that the structures of the periplasmic and transmembrane domains profoundly affect the conformation of the cytoplasmic domain to which they are coupled. Furthermore, the data suggest that different subregions within the cytoplasmic domain interact in a complex and subtle manner to modulate its structure and, presumably, its function. It is possible that differently methylated species of the same receptor react differently with a single antiserum. We have therefore deferred investigation of adaptive methylation of Tap or Tap-derived receptors until we obtain Tap antisera or until we develop a reliable autoradiographic method, using [methyl-3H]methionine, to determine methylation levels of Tap.
Like Tap, Trg is deficient in CW-signal generation (18). Thus, the cytoplasmic domains of Trg and Tap may be considerably more similar in overall tertiary and quaternary structure than either of them is to the cytoplasmic domain of Tsr or Tar, which are good CW-signal generators. Preliminary characterization of the TsrCSR antiserum has indicated that it primarily recognizes epitopes comprised of residues 290 to 350 of Tsr (42a). This part of the cytoplasmic domain contains the first methylation region, but it does not include the 48 highly conserved residues that have been implicated in interaction with CheA and CheW (4). The TsrCSR antibody may interact with a part of the protein whose conformation regulates the ability of the receptor to stimulate CheA kinase activity. If the Trg antibody has a similar specificity, the TsrCSR and Trg antisera may preferentially cross-react with forms of the receptor that are more and less, respectively, effective in stimulating CheA kinase activity. Therefore, the relative affinities of a receptor for these TsrCSR and Trg antibodies may be general predictors of the ability of each receptor to activate CheA kinase in vitro and generate CW signal in vivo.
With the hybrid receptors described here, we can address several questions whose answers have proven elusive. In vitro studies of chemotactic signaling through Tap and Trg have been problematic, since their cognate binding proteins have a low affinity for the receptors. For example, the Kd for the interaction of maltose-binding protein with Tar in vivo is 250 μM (35). This problem can be overcome with Tarp, which has the signaling characteristics of Tap but recognizes aspartate as a ligand. The Kd of Tarp for aspartate is probably, like that of Tar, in the low-micromolar range. Using membranes containing Tarp, we can determine in vitro whether CheA activity can be stimulated by the cytoplasmic domain of Tap and whether this activity can be down-regulated by aspartate. Also, if the ability to stimulate CheA activity and the affinity for TsrCSR antibody are positively correlated, then membranes containing Tap and Tapl will be less effective in activating CheA than membranes containing an equivalent amount of Tarp. With the in vitro system developed by Ames and Parkinson (4), we can also examine stimulation of CheA activity by the isolated cytoplasmic domain of Tap and compare it with the stimulation of CheA activity by isolated membranes containing intact Tap or Tarp proteins.
Attractants block synthesis of the tumble generator phospho-CheY by inhibiting the stimulation of CheA activity by dimeric chemoreceptors (16). A change in net receptor occupancy by an attractant of <1% generates a physiologically significant signal (6), but the mechanism by which a cis-acting negative signal can be amplified remains an enigma. Gardina and Manson (20) recently suggested a trans-inactivation model for attractant signaling, in which a receptor dimer occupied by an attractant can shut off CW signal generated at unoccupied receptors. By this logic, Tap, Tarp, and Tapl, and perhaps also Tart, may only be capable of signaling via a trans-inactivating mechanism, which could be facilitated by localization of the receptors in a polar patch in the cell (32). We are testing this hypothesis through in vivo and in vitro experiments.
ACKNOWLEDGMENTS
We are grateful to Jerry Hazelbauer for sharing unpublished data prior to publication, for critically reading a draft of the manuscript, and for supplying Trg antibody. Sandy Parkinson and Bob Bourret earned our thanks by providing us with strains and antisera. Paul Gardina was a source of inspiration and enlightenment, just when we most needed it, throughout the course of this work.
This research was support by NIH grant GM39736.
REFERENCES
- 1.Abouhamad W N, Manson M D, Gibson M M, Higgins C F. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol Microbiol. 1991;5:1035–1047. doi: 10.1111/j.1365-2958.1991.tb01876.x. [DOI] [PubMed] [Google Scholar]
- 2.Adler J, Hazelbauer G L, Dahl M M. Chemotaxis towards sugars in Escherichia coli. J Bacteriol. 1973;115:824–847. doi: 10.1128/jb.115.3.824-847.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksamit R R, Koshland D E., Jr Identification of the ribose binding protein as receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry. 1974;13:4473–4478. doi: 10.1021/bi00719a001. [DOI] [PubMed] [Google Scholar]
- 4.Ames P, Parkinson J S. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgartner J W, Kim C, Brissette R E, Inouye M, Park C, Hazelbauer G L. Transmembrane signalling by a hybrid protein: communication from the domain of the chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg H C. Bacterial microprocessing. Cold Spring Harbor Symp Quant Biol. 1990;55:539–545. doi: 10.1101/sqb.1990.055.01.052. [DOI] [PubMed] [Google Scholar]
- 7.Berg H C, Block S M. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J Gen Microbiol. 1984;130:2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- 8.Berg H C, Brown D A. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 9.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blank V. Diploma thesis. Konstanz, Germany: University of Konstanz; 1987. [Google Scholar]
- 12.Bollinger J, Park C, Harayama S, Hazelbauer G L. Structure of the Trg protein: homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borkovich K A, Simon M I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 14.Brass J M, Manson M D. Reconstitution of maltose chemotaxis in Escherichia coli by addition of maltose-binding protein to calcium-treated cells of maltose regulon mutants. J Bacteriol. 1984;157:881–890. doi: 10.1128/jb.157.3.881-890.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clegg D O, Koshland D E., Jr The role of a signaling protein in bacterial sensing: behavioral effects of increased gene expression. Proc Natl Acad Sci USA. 1984;81:5056–5060. doi: 10.1073/pnas.81.16.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochran A G, Kim P S. Imitation of Escherichia coli aspartate receptor signaling in engineered dimers of the cytoplasmic domain. Science. 1996;271:1113–1116. doi: 10.1126/science.271.5252.1113. [DOI] [PubMed] [Google Scholar]
- 17.Engstrom P, Hazelbauer G L. Methyl-accepting chemotaxis proteins are distributed in the membrane independently from basal ends of bacterial flagella. Biochim Biophys Acta. 1982;686:19–26. doi: 10.1016/0005-2736(82)90147-x. [DOI] [PubMed] [Google Scholar]
- 18.Feng, X., J. W. Baumgartner, and G. L. Hazelbauer. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 179:6714–6720. [DOI] [PMC free article] [PubMed]
- 19.Gardina P, Conway C, Kossman M, Manson M. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J Bacteriol. 1992;174:1528–1536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardina P J, Manson M D. Attractant signaling by an aspartate chemoreceptor dimer with a single cytoplasmic domain. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 21.Goy M F, Springer M S, Adler J. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory transduction. Proc Natl Acad Sci USA. 1977;74:4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazelbauer G L. Maltose chemoreceptor of Escherichia coli. J Bacteriol. 1975;122:206–214. doi: 10.1128/jb.122.1.206-214.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazelbauer G L, Adler J. Role of the galactose-binding protein in chemotaxis of E. coli toward galactose. Nature New Biol. 1971;230:101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- 24.Joyce C M, Grindley N D F. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koman A, Harayama S, Hazelbauer G L. Relation of chemotactic response to the amount of receptor: evidence for different efficiencies of signal transduction. J Bacteriol. 1979;138:739–747. doi: 10.1128/jb.138.3.739-747.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondoh H, Ball C B, Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci USA. 1979;76:260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft R, Tardiff J, Krauter K S, Leinwand L A. Using mini-prep plasmid DNA for sequencing double-stranded templates with Sequenase. BioTechniques. 1988;6:544–547. [PubMed] [Google Scholar]
- 28.Krikos A, Conley M P, Boyd A, Berg H C, Simon M I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krikos A, Mutoh N, Boyd A, Simon M I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983;33:615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- 30.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 31.Lee L, Imae Y. Role of threonine residue 154 in ligand recognition of the Tar chemoreceptor in Escherichia coli. J Bacteriol. 1990;172:377–382. doi: 10.1128/jb.172.1.377-382.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:717–723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 33.Manson M D. Bacterial motility and chemotaxis. Adv Microb Physiol. 1992;33:277–346. doi: 10.1016/s0065-2911(08)60219-2. [DOI] [PubMed] [Google Scholar]
- 34.Manson M D, Blank V, Brade G, Higgins C F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature. 1986;321:253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- 35.Manson M D, Boos W, Bassford P J, Jr, Rasmussen B A. Dependence of maltose transport and chemotaxis on the amount of maltose-binding protein. J Biol Chem. 1985;260:9727–9733. [PubMed] [Google Scholar]
- 36.Mesibov R, Adler J. Chemotaxis towards amino acids in Escherichia coli. J Bacteriol. 1972;112:315–326. doi: 10.1128/jb.112.1.315-326.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 38.Moe G R, Koshland D E., Jr . Transmembrane signalling through the aspartate receptor. In: Youvan D C, Dahl F, editors. Microbial energy transduction. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. pp. 163–168. [Google Scholar]
- 39.Mowbray S L, Foster D L, Koshland D E., Jr Proteolytic fragments identified with domains of the aspartate chemoreceptor. J Biol Chem. 1985;260:11711–11718. [PubMed] [Google Scholar]
- 40.Mowbray S L, Koshland D E., Jr Additive and independent responses in a single receptor: aspartate and maltose stimuli on the Tar protein. Cell. 1987;50:171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- 41.Parkinson J S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkinson J S. Signal transduction schemes in bacteria. Cell. 1993;72:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 42a.Parkinson, J. S. Personal communication.
- 43.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. Aer and Tsr transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russo A F, Koshland D E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983;220:1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- 45.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silverman M, Simon M. Flagellar rotation and the nature of bacterial motility. Nature. 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 47.Silverman M, Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci USA. 1977;74:3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slocum M K, Halden N F, Parkinson J S. Hybrid Escherichia coli sensory transducers with altered stimulus detection and signaling properties. J Bacteriol. 1987;169:2938–2944. doi: 10.1128/jb.169.7.2938-2944.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Springer M S, Goy M F, Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stock J. Signaling across membranes: a one and a two and a …. Science. 1996;274:370–371. doi: 10.1126/science.274.5286.370. [DOI] [PubMed] [Google Scholar]
- 51.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 52.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Utsumi R, Brissette R E, Rampersaud A, Forst S A, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 54.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 55.Wolfe A J, Berg H C. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolff C. Diploma thesis. Konstanz, Germany: University of Konstanz; 1983. [Google Scholar]
- 57.Wu J, Li G, Li D, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 58.Zagursky R J, Berman M L. Cloning vectors that yield high levels of single-stranded DNA for rapid DNA sequencing. Gene. 1984;27:183–191. doi: 10.1016/0378-1119(84)90139-2. [DOI] [PubMed] [Google Scholar]