Abstract
Valproic acid (VPA) is an anti-epileptic medication that increases the risk of neural tube defect (NTD) outcomes in infants exposed during gestation. Previous studies into VPA’s mechanism of action have focused on alterations in gene expression and metabolism but have failed to consider how exposure changes the abundance of critical developmental proteins over time. This study evaluates the effects of VPA on protein abundance in the developmentally distinct tissues of the mouse visceral yolk sac (VYS) and embryo proper (EMB) using mouse whole embryo culture. Embryos were exposed to 600μM VPA at 2 hr intervals over 10 hr during early organogenesis with the aim of identifying protein pathways relevant to VPA’s mechanism of action in failed NTC. Protein abundance was measured through tandem mass tag (TMT) labeling followed by liquid chromatography and mass spectrometry. Overall, there were over 1,500 proteins with altered abundance after VPA exposure in the EMB or VYS with 428 of these proteins showing previous gene expression associations with VPA exposure. Limited overlap of significant proteins between tissues supported the conclusion of independent roles for the VYS and EMB in response to VPA. Pathway analysis of proteins with increased or decreased abundance identified multiple pathways with mechanistic relevance to NTC and embryonic development including convergent extension, Wnt Signaling/planar cell polarity, cellular migration, cellular proliferation, cell death, and cytoskeletal organization processes as targets of VPA. Clustering of co-regulated proteins to identify shared patterns of protein abundance over time highlighted 4 hr and 6/10 hr as periods of divergent protein abundance between control and VPA-treated samples in the VYS and EMB, respectively. Overall, this study demonstrated that VPA temporally alters protein content in critical developmental pathways in the VYS and the EMB during early organogenesis in mice.
Keywords: Proteomics, Valproic Acid, Protein Abundance, Organogenesis, Neural Tube Defects
1: Introduction
Valproic acid (VPA) is an anti-epileptic medication that, when taken during pregnancy, increases the risk of having offspring with a neural tube defect (NTD) (Ornoy, 2009; Jentink et al., 2010). NTDs arise due to failed neural tube closure (NTC) during early organogenesis. This failed closure can lead to malformations of the brain and spine resulting in conditions of varying severity depending on the timing and location of the failed closure (Sakai, 1989). The mechanism through which VPA increases the risk of NTDs is not known; however, several downstream targets of VPA may be implicated in incomplete NTC. These mechanisms include disruption of redox balance, histone deacetylase (HDAC) inhibition, blocking voltage gated ion channels, and increasing γ-aminobutyric acid (GABA) levels (Phiel et al., 2001; Ghodke-Puranik et al., 2013; Lapehn et al., 2021). Through these mechanisms, particularly alteration of redox balance and HDAC inhibition, VPA could exert changes in the developmental proteome which could, in turn, have functional consequences for the mechanisms of NTC.
The genetic and histone codes provide the necessary blueprint for making the proteins necessary for every phase of biological function with many factors affecting protein abundance across an individual’s lifespan. Despite the connection between mRNA expression and protein translation, it has been frequently demonstrated that there is not a strong correlation between mRNA and protein abundance using both standard and high-throughput experimental techniques (Gygi et al., 1999; Wang, 2008; Vogel and Marcotte, 2012). Instead, only about 40% of protein abundance is explained by mRNA expression, highlighting the importance of separately assessing and not conflating the transcriptome and proteome (Vogel and Marcotte, 2012). Due to these many variables that can affect protein abundance, the greater complexity of the proteome compared to the genome, and the slower technological advances in the field of proteomics, there is less research on protein abundance compared to mRNA expression (Sidoli et al., 2017). Understanding protein dynamics and the role of the proteome in biological function, however, remains critically important since proteins are the active molecular contributors to cellular activity and morphological/functional development.
Genetic mutations and gene expression have been extensively studied in the context of NTDs with over 200 genes identified in mice as potential targets in the process of NTC (Copp et al., 2013). These genes largely belong to pathway categories which include: disturbance of the cytoskeleton, disturbance of cell proliferation or neuronal differentiation, neuroepithelial cell death, transcriptional regulation and chromatin dynamics, and dysregulation of the sonic hedgehog pathway (Copp and Greene, 2010; Copp et al., 2013). To date, most studies investigating the role of these genes and proteins in the context of NTC have utilized a candidate gene or protein approach which limits discovery to a pre-defined hypothesis and limits understanding of the interplay between genes or proteins and their biological pathways.
The advancement of omics technologies has altered the way hypotheses related to gene and protein expression are developed and tested. There have only been a few studies looking at the proteome of mouse or rat embryos, but each of these studies has indicated that the proteome is dynamic across stages of embryonic development (Gao et al., 2017), and that the embryo proper (EMB) and visceral yolk sac (VYS) have unique patterns of protein expression with protein abundance changing over time (Usami et al., 2007). There are currently no known global proteome studies evaluating changes protein abundance throughout mouse organogenesis, despite this stage being highly dynamic due to the myriad of morphological and physiological processes operating in the embryo. This study evaluates changes in global protein quantity following VPA exposure in organogenesis stage mouse embryos to determine patterns of protein abundance in early development and generate hypotheses related to VPA’s mechanism of failed NTC. Pathway analysis of proteins affected by VPA exposure will aid in determining protein networks and biological processes that may be part of VPA’s mechanism in causing NTDs. This will be accomplished through a time and tissue specific assessment of protein abundance following VPA exposure during the period of NTC using 10-plex tandem mass tags (TMT) with protein identification through liquid chromatography and mass spectrometry (Thompson et al., 2003; McAlister et al., 2012). Through these methods it is hypothesized that NTC relevant patterns of tissue and time specific protein abundance following VPA exposure will differ from the patterns in control conceptuses revealing potential pathways involved in mouse NTC and implicated in VPA’s mechanism of teratogenesis.
2: Materials and Methods
2.1: Animals
Experiments were conducted using mouse whole embryo culture (mWEC) to culture conceptuses from gestational days (GD) 8–9 in time-mated pregnant CD-1 mice (Charles River Laboratories, Raleigh, NC). Gestational day 0 was designated as the morning following mating with the presence of a positive vaginal plug. Animals were housed in groups of 6 or fewer in ventilated cages. All animal methodology was approved by the University of Michigan Institutional Animal Care and Use Committee.
2.2: Culture Conditions
Pregnant GD 8 dams were euthanized by CO2 asphyxiation and uteri were removed and placed in Tyrode’s solution (pH 7.4, HiMedia; Mumbai, India). Implantation sites were dissected with watchmaker’s forceps and iridectomy scissors. Maternal tissues including the decidua and Reichert’s membrane were removed to reveal intact conceptuses. Conceptuses were cultured in groups of 2–6 per bottle of 2mL of immediately centrifuged female rat serum with 4.3 μl/mL penicillin, streptomycin (10,000 units penicillin and 10 mg streptomycin per mL, Sigma Aldrich; St. Louis, MO). All culture conditions were at 37 °C with gas concentrations at 5% O2, 5% CO2, 90% N2 for 6 hr then 20% O2, 5% CO2, 75% N2 for the remainder of the culture period (Harris, 2012).
2.3: Time Course Exposures and Sampling for Tandem Mass Tag Labeling
VPA (600 μM in H2O) was added in vitro to the female rat serum two hours following the start of culture. Samples were collected 2, 4, 6, 8, and 10 hr following the addition of VPA. For each time point and treatment condition (Ctl, VPA) two biological replicates were collected. At the designated end of culture, conceptuses were rinsed 3 times with 1x Hank’s Balanced Salt Solution (HBSS, pH 7.4, Thermo Fisher Scientific; Waltham, MA). The ectoplacental cone was removed and discarded followed by separation of the remaining conceptus into visceral yolk sac (VYS) and embryo proper (EMB). At each designated time point, VYS and EMB were collected and pooled into separate samples consisting of tissue from up to 10 conceptuses to ensure high enough protein content for further analysis. Samples were collected in 100 μL RIPA buffer (pH 7.4; 50 mM Tris-HCl, 1% NP-40 v/v, 1:100 HALT Protease/Phosphatase Inhibitor, 150 mM NaCl, 1mM EGTA, 1mM NaF, 0.25% Na Deoxycholate w/v, and 0.1% SDS w/v), then sonicated to disrupt tissue and homogenize the solution. Homogenized samples were centrifuged for 10 min at 12,000 × g before being frozen at −80 °C.
2.4: Tandem Mass Tag Labeling
A bicinchoninic acid (BCA) assay was performed to determine protein concentration of samples. Based on the BCA results, 100 μg of protein was added to a new microfuge tube with volume brought up to 100 μL with 1x Tetraethylammonium bromide (TEAB, Thermo Fisher Scientific; Waltham, MA). Samples were reduced with 5 μL 200 mM tris(2-carboxyethyl)phosphine (TCEP, Thermo Fisher Scientific; Waltham, MA) for 1 hr at 55 °C then alkylated with 5 μL 375 mM iodoacetamide (Sigma Aldrich; St. Louis, MO) and incubated at room temperature in the dark for 30 minutes. Protein was then precipitated overnight in ice-cold acetone at 4 °C then resuspended in 100 μL 1x TEAB. 2.5 μg trypsin was added to each sample for overnight protein digestion at 37 °C. The next day, 41 μL anhydrous acetonitrile was added to each of the 10-plex TMT labels (Thermo Fisher Scientific; Waltham, MA) and vortexed and centrifuged. Each label was then combined with the corresponding sample (Ctl 2 hr = 126, VPA 2 hr= 127N, Ctl 4 hr= 127C, VPA 4 hr= 128N, Ctl 6 hr=128C, VPA 6 hr= 129N, Ctl 8 hr=129C, VPA 8 hr=130N, Ctl 10 hr=130C, VPA 10 hr=131) and incubated for 1 hr at room temperature. The labeling reaction was quenched with 8 μL hydroxylamine (Thermo Fisher Scientific; Waltham, MA) for 15 minutes. Following labeling, all 10-plex labels were combined into one sample and dried in a centrifugal vacuum concentrator. Pierce reverse-phase high pH fractionation was performed following manufacturer’s instructions to split each 10-plex labelled sample into 8 fractions (Thermo Scientific; Waltham, MA).
2.5: Liquid Chromatography, Mass Spectrometry
Liquid chromatography, mass spectrometry, and protein identification were performed by the Proteomics Resource Facility at the University of Michigan. Multinotch-MS3 was utilized to obtain superior accuracy which minimizes the reporter ion ratio distortion resulting from fragmentation of co-isolated peptides during MS analysis (McAlister et al., 2014). Orbitrap Fusion (Thermo Fisher Scientific; Waltham, MA) and RSLC Ultimate 3000 nano-UPLC (Dionex; Sunnyvale, CA) was used to acquire the data. 2 μl of the sample was resolved on a PepMap RSLC C18 column (75 μm i.d. x 50 cm; Thermo Scientific) at a flow rate of 300 nl/min using a 0.1% formic acid/acetonitrile gradient system (2–22% acetonitrile in 150 min; 22–32% acetonitrile in 40 min; 20 min wash at 90% followed by 50 min re-equilibration) and directly sprayed into the mass spectrometer using an EasySpray source (Thermo Fisher Scientific; Waltham, MA). The mass spectrometer was set to collect one MS1 scan (Orbitrap; 120K resolution; AGC target 2×105; max IT 100ms) followed by data-dependent, “Top Speed” (3 sec) MS2 scans (collision induced dissociation; ion trap; NCE 35; AGC 5×103; max IT 100ms). For multinotch-MS3, the top 10 precursors from each MS2 were fragmented by HCD followed by Orbitrap analysis (NCE 55; 60K resolution; AGC 5×104; max IT 120ms, 100–500 m/z scan range).
2.6: Protein identification in Proteome Discoverer
Proteome Discoverer (v2.4; Thermo Fisher Scientific; Waltham, MA) was used for data analysis. MS2 spectra were searched against the Uniprot mouse protein database (reviewed and unreviewed; 94044 entries; downloaded on 06/20/2019) using the following search parameters: MS1 and MS2 tolerance were set to 10 ppm and 0.6 Da, respectively; carbamidomethylation of cysteines (57.02146 Da) and TMT labeling of lysine and N-termini of peptides (229.16293 Da) were considered static modifications; oxidation of methionine (15.9949 Da) and deamidation of asparagine and glutamine (0.98401 Da) were considered variable. Identified proteins and peptides were filtered to retain only those that passed ≤1% false discovery rate (FDR) threshold. Quantitation was performed using high-quality MS3 spectra (Average signal-to-noise ratio of 10 and <30% isolation interference). P-values were calculated by ANOVA in Proteome Discoverer.
2.7: Protein Inclusion Criteria
Proteins were included in downstream analysis when they were identified with high FDR confidence in both replicates and showed labeling with all 10 TMT labels. We calculated the ratio of protein abundance between the VPA and control conditions (VPA/Ctl) using normalized abundance values from the appropriate TMT channels. Proteins with a p<0.05 were considered significant with those having a log2 abundance ratio greater than 0 being increased and less than 0 being decreased.
2.8: Co-regulation Clustering
Co-regulated protein clustering was performed using Clust (Abu-Jamous and Kelly, 2018). Clustering was completed separately for EMB protein abundances and VYS protein abundances. Default Clust settings were applied for this analysis.
2.9: Pathway Analysis
Pathway analysis was conducted for each list of proteins that met the cut-off criteria for inclusion as well as each set of proteins within a co-regulated Clust protein cluster. DAVID Bioinformatics Database functional annotation tool was used to identify enriched Biological Process Gene Ontology (GO) terms (Huang et al., 2009b, 2009a). Protein lists were searched against the full list of proteins identified for the specific tissue as background to adjust for the presence of higher abundance proteins being overly represented in mass spectrometry data. GO-terms were deemed significant if the p-value was less than 0.05.
2.10: Transcription Factor Enrichment
Enrichment of transcription factors was measured using Enrichr (Chen et al., 2013; Kuleshov et al., 2016). Uniprot IDs were converted to Entrez gene names which were compared against the database ENCODE and ChEA Consensus TFs from ChIP-X (ENCODE Project Consortium, 2012; Davis et al., 2018). An adjusted p-value less than 0.05 was used as the cut-off for inclusion in the results.
2.11: Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was performed to identify pathways that showed differential enrichment between proteins with higher or lower abundance following VPA exposure without the need to use the pre-determined threshold inclusion criteria (Mootha et al., 2003; Subramanian et al., 2005). Data were input into the GSEA software (v. 4.1.0) as a .rnk file containing Uniprot accession numbers and VPA/Ctl abundance ratios and run as a pre-ranked dataset with 1000 permutations and collapsing redundant accession numbers to a single gene name. Proteins were searched against the Biological Process Gene Ontology (GO) term database (v 7.2). Pathways with an FDR < 0.25 were considered significant. These pathways also had a normalized enrichment score with an absolute value greater than 1 and p<0.05.
2.12. Comparative Toxicogenomics Database Enrichment Test
The Comparative Toxicogenomics Database (CTD) is a curated, publicly available database of gene-environment interactions with a wide-breadth of environmental and pharmaceutical exposures across multiple species (Davis et al., 2021). We used the CTD to perform a Fisher’s Exact Overrepresentation Test to evaluate if the proteins associated with VPA in our analysis were overrepresented in the mouse gene lists associated with VPA in the CTD. To perform the analysis, we downloaded: 1) a list of all Mus musculus genes in the CTD, 2) Genes associated with VPA exposure in Mus musculus in the CTD. These lists in combination with the significant proteins from each of the 2–10 hr timepoints in EMB and VYS, were utilized to run a one-sided Fisher’s Exact Test. The list of proteins within a timepoint were considered significantly enriched when p<0.05.
3: Results
3.1: Data Summary
In the VYS 4,815 proteins were identified across both replicates with high FDR confidence across all 10 treatment groups, whereas in the EMB there were 4,897 proteins that met these criteria. Comparing the two tissue datasets, there were 4,238 proteins that appeared in both VYS and EMB.
3.2: Variability
Each tissue (EMB and VYS) protein determination and comparison were based on two biological replicates. Variability of the VPA/control (Ctl) abundance ratio for each tissue and time point is summarized in Supplemental Figure 1. In the EMB, median variability in the abundance ratios across all proteins ranged from 8.7% to 34.1%, while the VYS variability median is between 5.9% and 22.6%. In the EMB, the most variable time points across the two replicates were at 6 and 10 hr, while in the VYS, the most variable time points were at 2 and 4 hr.
3.3: Embryo Time Series
There were 4,897 proteins identified in the EMB at all five time points. Within each of these time points significant proteins (p<0.05) were categorized by increased or decreased abundance following VPA exposure (Figure 1A). 2, 4, and 8 hr time points contained the highest number of affected proteins. At 2 and 10 hr there were more proteins with increased abundance, while at 4, 6, and 8 hr there were more decreased proteins. For all time points, the majority of significant proteins were specific to that time point (Figure 1B). Across all EMB time points only 42 proteins were shared within at least two time points (Figure 1B).
Figure 1:
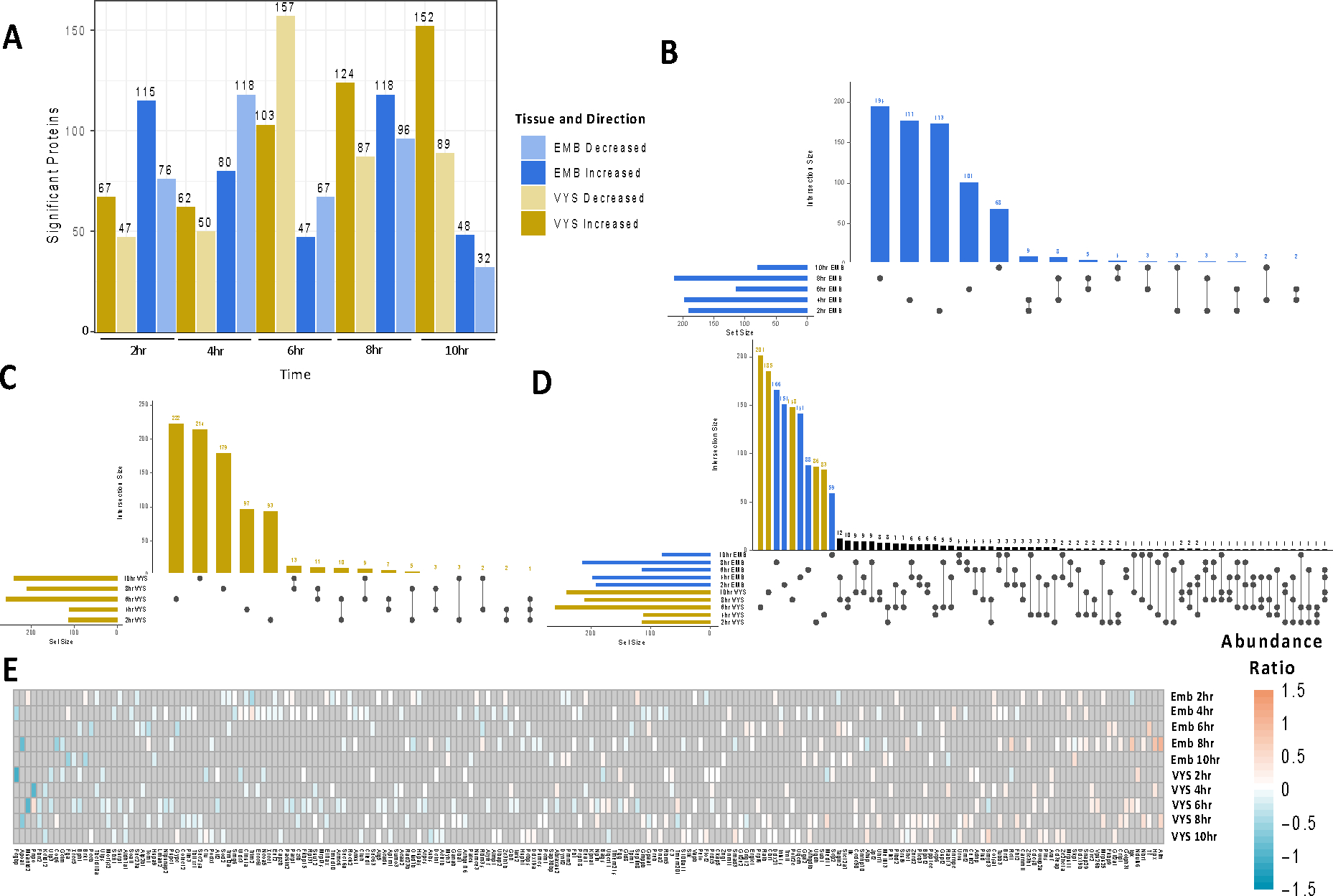
Summary of significant protein changes in EMB and VYS after 2–10hr treatment with VPA compared to Controls. A) Number of significant proteins (p<0.05) for each tissue and time B) Overlap of significant proteins across time points in the EMB C) Overlap of significant proteins across time points in the VYS D) Overlap of significant proteins across time points in the VYS and EMB. E) Abundance ratio directionality for significant proteins shared by at least 2 tissues or time points. Gray shading indicates a lack of significance for a given protein and treatment group.
Pathway analysis was carried out for each time-specific group of increased or decreased proteins and enrichment of biological process GO-terms was identified for all groups (Table 1). In the EMB, pathways with relevance to NTC were most prevalent for proteins that were increased in abundance at 2 hr and included pathways such as Wnt signaling and planar cell polarity, NTC, and convergent extension. Significant transcription factor (TFs) enrichment was identified for all 5 EMB time points with the highest number of TF’s identified at the time points with the greatest number of significant proteins (Table 2). In the EMB developmentally relevant TFs were identified in all 5 time points for decreased proteins and in 3 time points for increased proteins.
Table 1:
Significant biological process gene ontology terms identified through DAVID pathway analysis of significant (p<0.05) proteins that were increased or decreased for VYS and EMB.
| Visceral Yolk Sac | ||
|---|---|---|
| Decreased | Increased | |
|
| ||
| 2 hr | Ribosome biogenesis | Short-term memory |
|
| ||
| Negative regulation of ATPase activity | ||
| 4 hr | Positive regulation of protein ubiquitination involved in ubiquitin-dependent | Cell migration |
| protein catabolic process | ||
|
| ||
| 6 hr | Cellular response to hydrogen peroxide | |
| Regulation of cell proliferation | ||
| - | Proteolysis | |
| DNA methylation | ||
| Negative regulation of canonical Wnt signaling | ||
| pathway | ||
|
| ||
| 8 hr | Regulation of centrosome duplication | - |
| Cellular response to interleukin-1 | ||
| NLS-bearing protein import into nucleus | ||
|
| ||
| 10 hr | Negative regulation of gene expression | |
| Transport | Thymus development | |
| Lipoprotein transport | Thyroid gland development | |
| Bone development | Regulation of cell proliferation | |
| Heart development | Transcription, DNA-templated | |
| Actin cytoskeleton organization | Positive regulation of cell migration | |
| Protein stabilization | ||
|
| ||
| Embryo | ||
|
| ||
| Wnt signaling pathway, planar cell polarity | ||
| 2 hr | - | pathway |
| Neural tube closure | ||
| Convergent extension involved in | ||
| organogenesis | ||
|
| ||
| 4 hr | Regulation of transcription, DNA-templated | Methylation |
| Cellular response to DNA damage stimulus | Ribosomal small subunit assembly | |
| Rac protein signal transduction | S-adenosylmethioninamine metabolic process | |
| Barbed-end actin filament capping | Translation | |
| Regulation of protein kinase C signaling | Cell-matrix adhesion | |
| Immune system process | Cellular response to amino acid stimulus | |
| Cellular response to transforming growth factor | ||
| beta stimulus | ||
|
| ||
| 6 hr | Bicellular tight junction assembly | |
| - | Myelin assembly | |
| Attachment of GPI anchor to protein | ||
|
| ||
| 8 hr | Response to amino acid | Ribosome biogenesis |
| DNA replication | rRNA processing | |
| Positive regulation of macroautophagy | Single organismal cell-cell adhesion | |
| DNA replication initiation | Ribosomal small subunit assembly | |
| Response to cadmium ion | Ventricular cardiac muscle cell action potential | |
| Post-translational protein acetylation | rRNA methylation | |
| Positive regulation of stress fiber assembly | ||
|
| ||
| 10 hr | Alpha-beta T cell differentiation | Metabolic Process |
| Negative regulation of endothelial cell | ||
| apoptotic process | ||
| Cell motility | ||
In addition to pathway analysis based on categorization of proteins into groups increased and decreased by VPA exposure, pre-ranked Gene Set Enrichment Analysis (GSEA) was also performed. In the EMB, GSEA identified 46 developmentally relevant pathways that were more enriched in proteins that increased in abundance following VPA exposure and seven pathways that were more enriched in decreased proteins (Supplemental Table 2). There was no enrichment of developmental pathways at 4 hr for increased proteins, or 6, 8, and 10 hr for decreased proteins.
Table 2:
Transcription factor enrichment of increased and decreased proteins within each tissue and time point from the ENCODE and CHEA consensus transcription factors in Enrichr. Listed TFs are significant (adjusted p<0.05). Bolded TFs are relevant to developmental processes based on GeneCard summaries.
| Visceral Yolk Sac | Embryo | |||
|---|---|---|---|---|
| Decreased | Increased | Decreased | Increased | |
| 2 hr | MYC, MAX, E2F6 | IRF1 | NELFE, BRCA1, ZMIZ1, CREB1, YY1, CHD1 | NRF1, UBTF, TAF1, ATF2, CHD1, ELF1, YY1, SPI1, CTCF, RCOR1, CREB1, GABPA, BRCA1, TCF3, MAX, ZNF384 |
| 4 hr | - | - | E2F1, ZNF384, RFX5, E2F4, NRF1, TCF3, NANOG | MYC, TAF1, GABPA, SIN3A, E2F1, FLI1, MAX, ZNF384, SPI1, BRCA1, ELF1, PML, ZBTB7A, TCF3, KLF4, YY1 |
| 6 hr | TAF1, MAX, MYC, ZMIZ1, CREB1, FLI1, BRCA1, NRF1, ELF1, USF2, ZNF384, SIN3A, GABPA, ATF2, YY1, ZBTB33, USF1, NFYB, PBX3, CEBPD, CHD1, NFYA, SPI1, NELFE, HNF4A, TAF7 | NFYB, NFYA, MAX, MYC, GABPA, CTCF, E2F1, SP2, BRCA1, NRF1, PBX3, IRF3, RFX5, SPI1, UBTF, CREB1, TAF1, E2F6, FLI1, ZBTB7A, ATF2 | NR2C2, MYC, YY1, ZNF384, MAX, GABPA | - |
| 8 hr | TAF1, MYC, YY1, UBTF, ATF2, PML, SIN3A, MAX, ZNF384, BRCA1, CREB1, KLF4, E2F1, TCF3, GABPA, NFIC, STAT3, USF2, PPARG, E2F4 | BRCA1, ELF1, NFYB, CREB1, TAF1, NFYA, CHD1, SIX5, GABPA, YY1, EGR1 | TAF1, E2F4, NRF1, E2F6, MYC, GABPA SIN3A, YY1, MAX, TAF7, ATF2, E2F1, CREB1, ELF1, BRCA1, CHD1, PML, ZBTB33, RUNX1, ZNF384 | TAF1, YY1, NRF1, ATF2, MAX, SIN3A, PML, E2F6, ELF1, MYC, USF2, TAF7, E2F1, GABPA, RFX5, SPI1 |
| 10 hr | USF2, USF1, HNF4A, ELF1, SPI1, BRCA1 | TAF1, CREB1, YY1, UBTF, MYC, BRCA1, SIN3A, MAX, NRF1, E2F4, NFYA, ELF1, TCF3, E2F1, ZMIZ1, EGR1, PML, E2F6, USF1, FOXP2, ATF2 | YY1, KLF4, MYC, TAF1, MAX | ZNF384 |
In addition to pathway analysis based on categorization of proteins into groups increased and decreased by VPA exposure, pre-ranked Gene Set Enrichment Analysis (GSEA) was also performed. In the EMB, GSEA identified 46 developmentally relevant pathways that were more enriched in proteins that increased in abundance following VPA exposure and seven pathways that were more enriched in decreased proteins (Supplemental Table 2). There was no enrichment of developmental pathways at 4 hr for increased proteins, or 6, 8, and 10 hr for decreased proteins.
3.4: Visceral Yolk Sac Time Series
There were 4,815 proteins identified in the VYS at all 5 time points. Within each of these timepoints proteins were categorized by increased or decreased abundance following VPA exposure using the inclusion criteria described in the methods (Figure 1A). 6, 8, and 10 hr showed the highest number of proteins that were affected by VPA exposure. At all the time points, except for 6 hr, there were more proteins with increased abundance than decreased abundance. Similar to the EMB, most significant proteins in the VYS were specific to a single time point with 65 proteins shared across two time points and one protein shared across three time points (Figure 1C).
Pathway analysis identified significantly enriched GO-terms for all time points and directions except 6 hr decreased and 8 hr increased (Table 1). Pathways with the greatest relevance to NTC included, negative regulation of canonical Wnt signaling (6 hr increased), regulation of cell proliferation (10 hr increased), positive regulation of cell migration (10 hr increased), and actin cytoskeleton organization (10 hr decreased). Significant transcription factor enrichment was primarily identified between 6–10 hr timepoints in the VYS (Table 2). For decreased VYS proteins 4 time points identified developmentally relevant TFs, while increased VYS proteins had 3 time points with developmentally relevant TFs.
Pre-ranked GSEA in the VYS identified 28 developmentally relevant pathways that were enriched in proteins with increased abundance following VPA exposure and 12 pathways that were enriched in proteins with decreased abundance (Supplemental Table 2). There was no developmental enrichment at 6 and 8 hr for increased abundance proteins and at 2 and 4 hr for decreased abundance proteins.
3.5: Tissue Comparison Summary- Embryo vs Visceral Yolk Sac
4,238 proteins were identified and labeled in both biological replicates in the EMB and VYS datasets with high FDR confidence. Supplemental Figure 2 compares the VPA/Ctl abundance ratios for each protein between the EMB and VYS. The correlation between the Log2 VPA/Ctl abundance fold change was highest at 2 hr and 4 hr at 0.23 and 0.22 respectively. The correlations for 6, 8, and 10 hr were 0.06, 0.06, and 0.09. Overall, these correlations indicate that protein abundance in the VYS and EMB are affected by VPA independently. Most of the significant proteins in the EMB and VYS were unique to a specific tissue and time point (Figure 1D). There were 202 proteins that were significant for at least two tissues or time points including 180 shared by two groups and 21 shared by three groups (Figures 1D, 1E). A single protein (Gpaa1) was significant for four groups (EMB 4hr and 6hr, VYS 8hr and 10hr) (Figure 1E). It showed decreased abundance compared to controls in the EMB at 4hr and VYS at 8hr, followed by increased abundance compared to controls in the EMB at 6hr and the VYS at 10hr.
3.6: Co-regulated protein clustering
Clustering of EMB abundance data lead to two co-regulated protein clusters of 624 (EMB Cluster 0) and 536 (EMB Cluster 1) proteins each (Figure 2). The main time points of divergence between control and VPA in these clusters are at 6 and 10 hr. Pathway analysis of enriched biological process gene ontology (GO) terms was conducted for each EMB and VYS co-regulated abundance cluster. EMB Cluster 0 showed significant enrichment for 11 biological process GO-terms (Figure 3). Of these 11 terms, there was enrichment for two developmentally relevant pathways including, epidermal growth factor signaling and forebrain development. EMB Cluster 1 was enriched for 21 biological process GO-terms (Figure 3). None of these EMB Cluster 1 pathways were specifically related to development, but negative regulation of apoptotic process and oxidation-reduction process are still noteworthy due to apoptosis serving as an important mechanism of programmed cell death during development and VPA’s association with oxidative endpoints.
Figure 2:
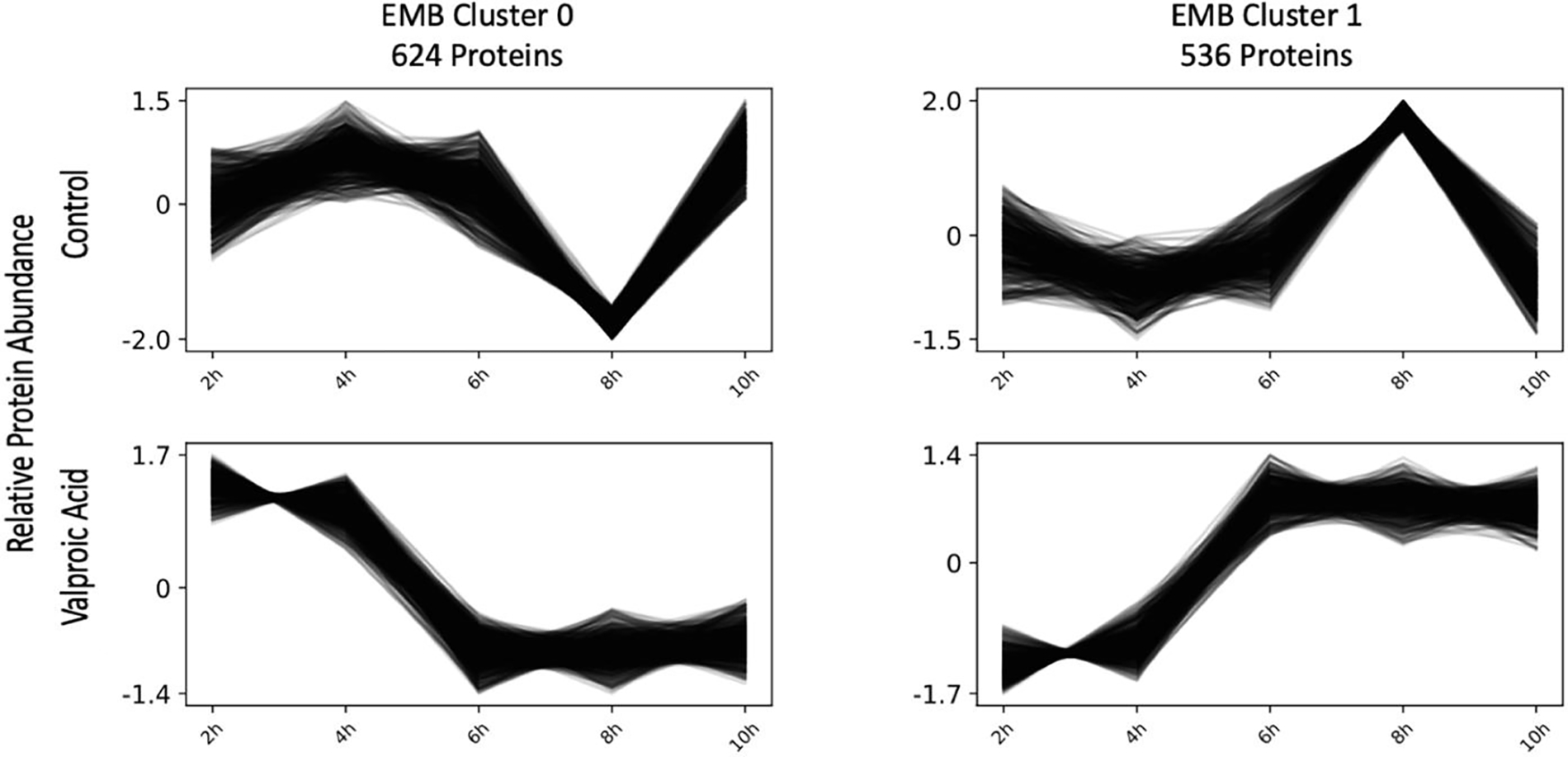
Co-regulated protein clustering of EMB protein abundance values in Clust identified two unique clusters of proteins that showed similar expression patterns within a given treatment group. A full list of proteins in each cluster is available in Supplemental Table 3.
Figure 3:
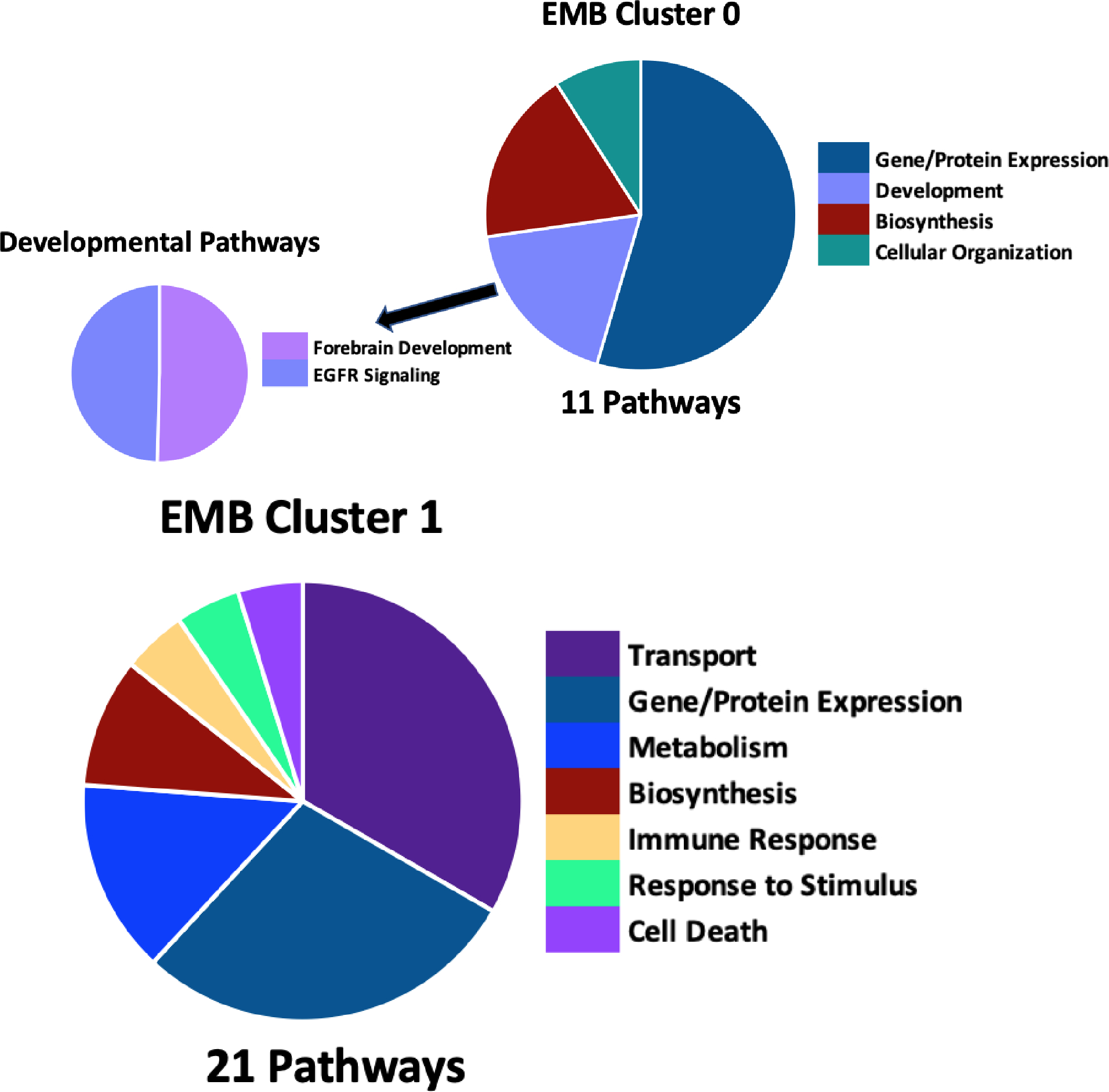
EMB abundance cluster 0 (A) and cluster 1 (B) biological process GO-term pathway enrichment in DAVID identified 11 and 21 enriched pathways, respectively with a p-value < 0.05 using the full EMB protein list as background. Pie chart division is based on the total number of pathways in each category in the main figure with division based on the pathway’s −log10 p-value in the highlighted developmental pathways. A full list of pathways is available in Supplemental Table 3.
Clustering of VYS data also lead to two distinct clusters of 738 (VYS Cluster 0) and 601 (VYS Cluster 1) proteins respectively (Figure 4). For the two VYS clusters, the single time point of divergence between the Ctl and VPA samples occurred at 4 hr in both clusters. VYS Cluster 0 had 14 significantly enriched pathways biological process GO-terms (Figure 5). None of these pathways were directly relevant to development, although cardiac muscle cell apoptosis may be partly related to heart development. VYS Cluster 1 showed significant enrichment for 31 biological process pathways with several relevant to development or oxidation (Figure 5). Developmental pathways included the following: skeletal muscle tissue differentiation, cerebral cortex neuron differentiation, and axon extension. Oxidation relevant pathways included oxidation-reduction process, cellular response to hypoxia, and response to reactive oxygen species (ROS).
Figure 4:
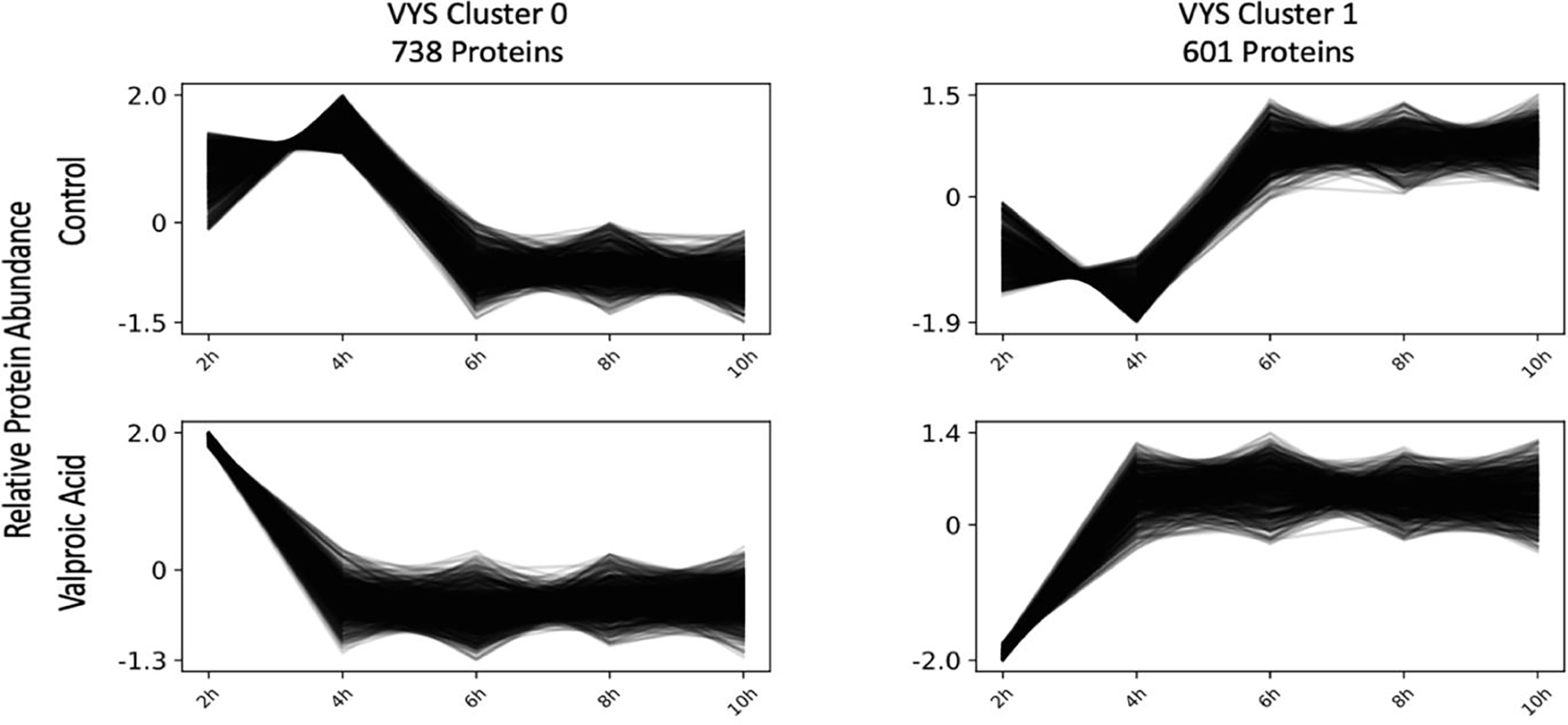
Co-regulated protein clustering of VYS protein abundance values in Clust identified two unique clusters of proteins that showed similar expression patterns within a given treatment group. A full list of proteins in each cluster is available in Supplemental Table 3.
Figure 5:
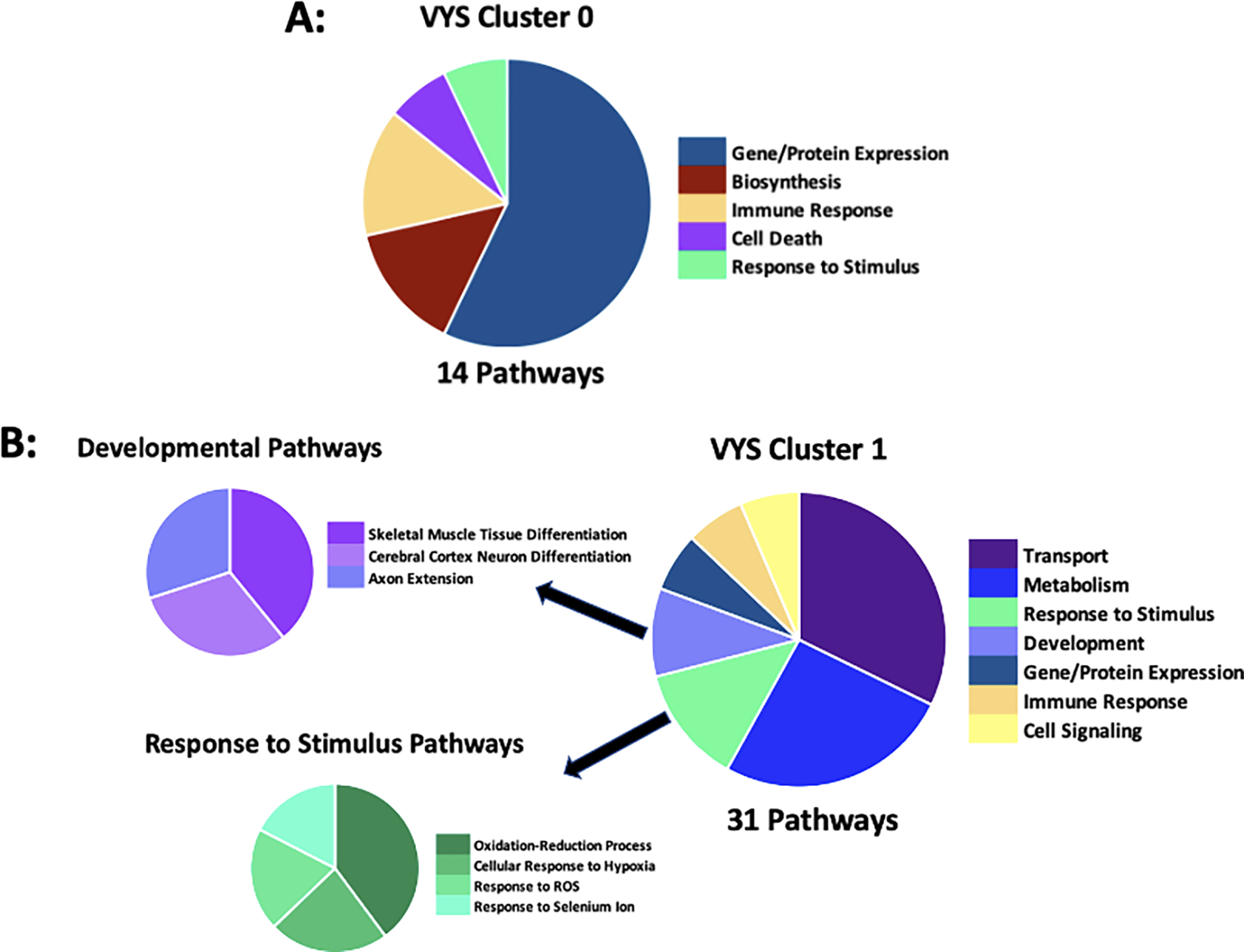
VYS Abundance Cluster 0 (A) and VYS Abundance Cluster 1 (B) biological process GO-term pathway enrichment in DAVID identified 14 and 31 enriched pathways, respectively with a p-value < 0.05 using the full VYS protein list as background. Pie chart division is based on the total number of pathways in each category. A full list of pathways is available in Supplemental Table 3.
EMB Cluster 0 and EMB Cluster 1 were significantly enriched for 65 and 64 transcription factors (TFs), respectively. Of these enriched TFs, 6 were unique to EMB Cluster 0 and 5 were unique to EMB Cluster 1 (Table 3). The TFs unique to EMB Cluster 0 had 4 developmentally relevant TFs based on the NCBI Entrez Gene summary, while EMB Cluster 1 had 2 TFs of developmental relevance. In the VYS abundance clusters, there were 64 significantly enriched TFs in VYS Cluster 0 and 65 in VYS Cluster 1. VYS Cluster 0 was enriched for 7 TFs that were unique compared to 8 unique TFs for VYS Cluster 1. Five of the enriched TFs for VYS Cluster 0 are developmentally relevant and 4 of the enriched TFs for VYS Cluster 1 are developmentally relevant.
Table 3:
Transcription factors (TFs) uniquely enriched in one of the two co-regulated embryo and visceral yolk sac abundance clusters. TFs are listed in order of significance, which required an adjusted p-value less than 0.05. Bolded TFs are related to developmental processes.
| Enriched Cluster-specific Transcription Factors | |
|---|---|
| EMB Cluster 0 | KAT2A, RELA, BCL3, NANOG, POU5F1, FOXM1 |
| EMB Cluster 1 | FOS, RFX5, ZKSCAN1, PPARD, HDAC2 |
| VYS Cluster 0 | KAT2A, FOXM1, NELFE, NANOG, VDR, SOX2, TRIM28 |
| VYS Cluster 1 | ERG, IRF1, ZKSCAN1, NFIC, RFX5, HNF4A, GATA1, CEBPB |
3.7. Comparative Toxicogenomics Database (CTD) Enrichment Test
The CTD contained 4,554 Mus musculus genes with previous associations with VPA. The protein lists from the EMB in our analysis, were significantly overrepresented (p<0.05) in this list of genes at 2, 4, 6, and 8 hr (Table 4). The protein lists from the VYS in our analysis were significantly overrepresented (p<0.05) in this gene list for all 5 time points (Table 4).
Table 4:
Comparative Toxicogenomics Database Enrichment Test p-values and overlap across 2–10 hr timepoints for EMB and VYS.
| 2 hr | 4 hr | 6 hr | 8 hr | 10 hr | ||
|---|---|---|---|---|---|---|
|
| ||||||
| EMB | p-value | 1.46E-10 | 7.70E-11 | 1.08E-09 | 1.15E-12 | 5.30E-02 |
| Overlap | 56/189 | 58/195 | 39/113 | 66/214 | 15/79 | |
|
| ||||||
| VYS | p-value | 4.93E-04 | 1.31E-07 | 6.20E-12 | 1.17E-07 | 9.17E-10 |
| Overlap | 27/112 | 35/110 | 73/258 | 54/209 | 64/239 | |
4: Discussion
Focused research on the role of gene expression during early metazoan development has led to significant insights into the process and control of embryogenesis but has failed to provide a complete understanding that includes the additional layers of intricate control through environmental and chemical cues. Regulatory responses to external and intrinsic signals are mediated, in large part, through the biosynthesis and posttranslational modification of proteins that define growth, differentiation, and morphogenesis, both spatially and temporally. Using VPA as a known embryo toxicant and disruptor of NTC we sought to evaluate changes in protein abundance in the mouse conceptus following VPA exposure that are time and tissue-specific with the goal of identifying proteins that may be involved in VPA’s mechanism of developmental toxicity. The analysis identified over 4,800 proteins in EMB and VYS with hundreds of these proteins exhibiting differential abundance between control and VPA treated samples during at least one time point.
Previous studies of developmental biology and toxicology have frequently focused exclusively on the cells and tissues of the embryo proper, ignoring the shared vascular system and functional dependence of the extraembryonic VYS tissues that make up the intact and viable conceptus. Paucity of EMB tissue and complexities related to developmental dynamics precluded closer inspection of individual EMB tissue types in this analysis but do allow for comparison of EMB and VYS. The observed differences between EMB and VYS in their response to VPA, as interdependent developmental tissues, result from many factors including their germline origins (EMB → female; VYS → male which, through imprinting result in differential gene expression), different types of environmental interface, and overall developmental juxtaposition within the conceptus (Wang et al., 2013). As such, tissue specificity was a key component of the analysis with EMB and VYS protein abundance measured independently. The independent assessment of EMB and VYS is due to the unique roles played by these tissues where the EMB will form into the fetal and eventually adult mouse, and the VYS is a uniquely developmental tissue, vestigial in the human but present in the mouse throughout gestation. The VYS performs many roles that are essential for the development of the EMB proper including germ cell maintenance and release, hematopoiesis (blood islands), vasculogenesis, angiogenesis, nutritional uptake prior to placentation, a source of mesenchymal stem cells and as an essential environmental interface for signaling and regulation (Zohn and Sarkar, 2010; Aires et al., 2015; Halbach et al., 2020; Ross and Boroviak, 2020). Due to these critical roles played by the VYS during early organogenesis, it is essential to include a separate assessment of the EMB and VYS to identify any distinct responses to, and consequences of, VPA exposure even though our downstream stream mechanism of interest of NTC appears historically to lie solely within the EMB proper. This study also extends our knowledge of VPA teratogenesis during early organogenesis by performing a time course analysis of its effects on the mouse VYS and EMB during the process of NTC.
Overall, this study identified 1,510 proteins in the VYS or EMB across five time points that had significantly different abundance in VPA-treated samples compared to controls. Of note, the majority of proteins with significant abundance differences were specific to a given tissue and timepoint (Figure 1B,C,D). This emphasizes the importance of tissue and time specific analyses in developmental toxicology and highlights the need for further analysis to summarize findings at the pathway or regulatory level since the overlap of individual proteins is limited. To ascertain the biological significance of these altered proteins, we performed several subsequent analyses including 1) Pathway Analysis, 2) Transcription Factor Enrichment Analysis, 3) Co-regulated protein clustering, 4) Comparative Toxicogenomics Database Enrichment Test. Together, the findings of these analyses unveil patterns of perturbation by VPA that can be utilized to generate hypotheses for future mechanistic toxicology studies that can more precisely identify mechanisms of VPA teratogenesis.
Pathway analysis was performed using two complementary approaches. DAVID performed an overrepresentation test of proteins that were significantly increased or decreased in abundance (Table 1) while pre-ranked GSEA identified pathways that were significantly enriched for genes with high or low abundance ratios regardless of statistical significance (Supplemental Table 2). The goal of the pathway analysis was to generate hypotheses regarding the molecular mechanisms of VPA’s NTC disruption for each time point and tissue. Due to the large number of significant pathways generated through this analysis, discussion will focus on pathways identified through DAVID analysis that have relevance to NTC (Figure 6). Focusing on these pathways leads to emerging temporal themes with developmental pathways in the EMB being predominately enriched for increased proteins and in earlier stages of exposure (2–6 hr), while in the VYS these pathways are predominately present at later exposure times (4–10 hr). The pathways with the greatest relevance to VPA and NTC are all enriched in the EMB for increased proteins after 2 hr VPA exposure and include Wnt signaling/planar cell polarity, neural tube closure, and convergent extension (CE). The non-canonical Wnt/Planar Cell Polarity (PCP) signaling pathway is a necessity for completion of the CE process involved in NTC (Wang et al., 2019). PCP is responsible for establishing and maintaining cell polarity and movement to establish tissue organization. In the context of CE, the PCP pathway directs the reshaping of the neural epithelium through sequential resolution of the anteroposterior cell junction and extension along the mediolateral junction followed by intercalation of the newly shaped cells guided by actin protrusions (Butler and Wallingford, 2017; Wang et al., 2019). This process of cellular re-shaping and movement is what drives the folding of the neural plate into the neural tube using both directional cell migration and collective cell movement (Muñoz-Soriano et al., 2012; Nikolopoulou et al., 2017). Normally, studies of gene function in cases of developmental toxicity that produce birth defects focus on deletions and mutations that result in loss of function, where the genetic lesion would result in the absence of a critical protein. However, the results of the present study suggest that, irrespective of gene expression, developmental protein abundances can vary dynamically in space and time and the increased abundance of specific critical proteins is also associated with developmental defects. This appears to be the case at 2 hr in the EMB where successful NTC is decreased following VPA exposure, but proteins associated with Wnt signaling, PCP, CE, and NTC pathways exhibit increased abundance. It has been proposed that VPA-induced loss of protein function through posttranslational modification is also occurring, such as through redox mediated oxidation, that is known to result in dynamic changes in cell differentiation and growth (Funato and Miki, 2010; Sandieson et al., 2014).
Figure 6:
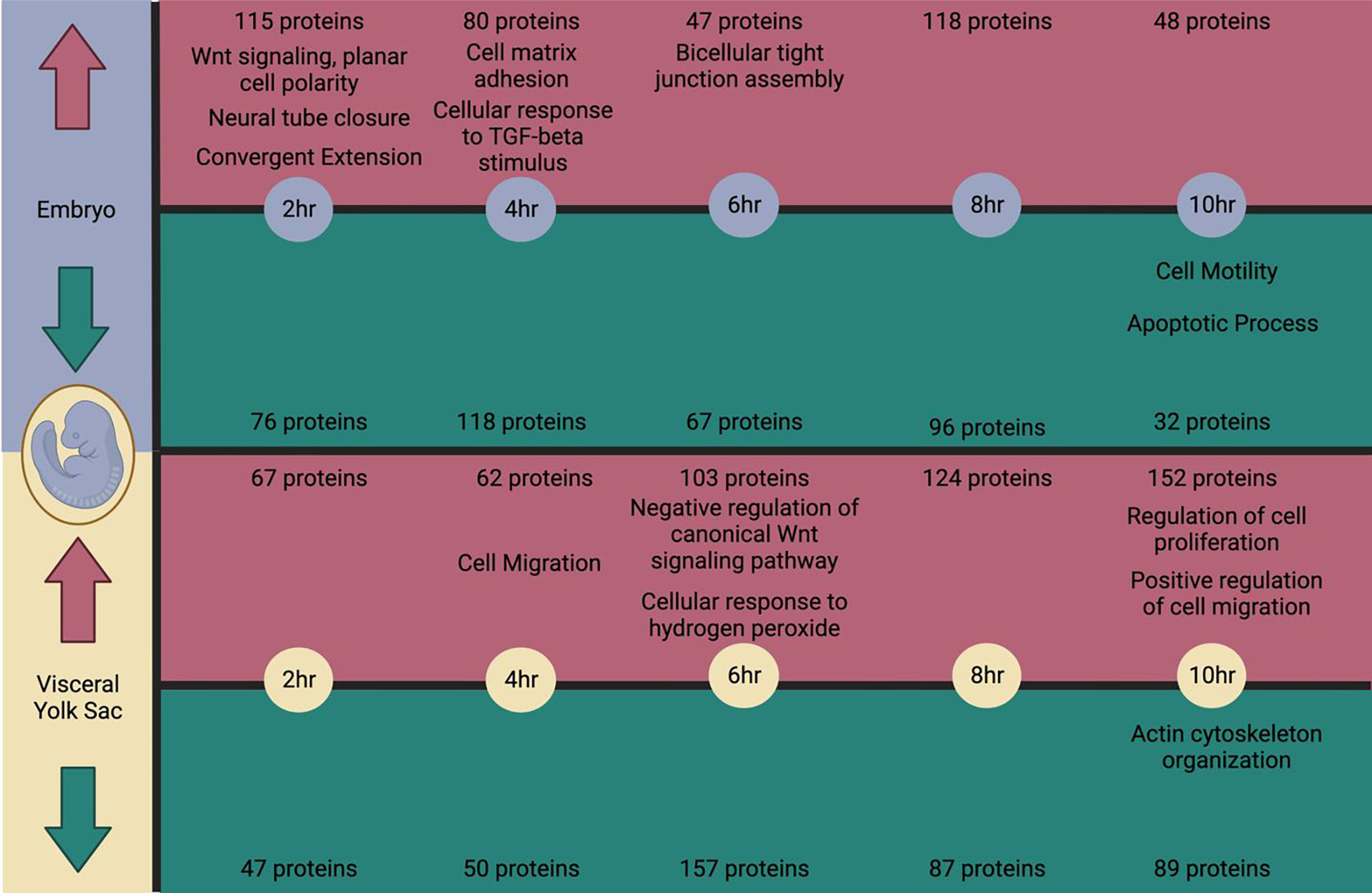
Timeline summary of proteins with significant changes in abundance in the EMB or VYS alongside biological process gene ontology terms identified through DAVID pathway analysis of the subsetted increased or decreased proteins which have relevance to the process of neural tube closure. Created with Biorender.com
Many of the other developmentally relevant pathways identified through DAVID pathway analysis are directly involved in the NTC process through supportive roles that help to orchestrate the cell movement required for CE and guided by Wnt/PCP signaling including cytoskeletal organization, cellular proliferation, cellular motility, and cellular adhesion (Muñoz-Soriano et al., 2012; Nikolopoulou et al., 2017). Early enrichment of these pathways in the EMB may initially be viewed as advantageous for promoting NTC closure. The early enrichment, however, is associated with the failure to complete closure following VPA exposure, suggesting that premature activation of canonical Wnt and other NTC pathways may disrupt the normal developmental sequencing.
Another pathway of interest identified through DAVID for increased proteins in the VYS at 6 hr is cellular response to hydrogen peroxide, which ties in with our previous work that demonstrated spatiotemporal shifts in redox potential in NTC stage mouse conceptuses (Lapehn et al., 2021). In addition to redox shifts being linked to VPA exposure, redox balance is also an essential developmental process that has been linked to signaling mechanisms through post-translational oxidative modifications of protein cysteine residues (Hansen et al., 2020). Changes to the cysteine redox proteome have been measured in aging mice, but more research is needed in this space to understand how VPA and other teratogens with oxidative endpoints may affect the proteome through post-translational modification and redox signaling (Xiao et al., 2020).
Enrichment of redox process pathways was also noted in the DAVID pathway enrichment performed for the cluster analysis. Protein clustering of abundance values in the control and VPA-treated EMB or VYS identified two clusters of proteins (within each tissue) that show co-regulated shifts in abundance over time (Figures 3, 4, 5, 6). Comparing the control and VPA-treated cluster patterns, it is recognized that in the EMB there are distinct differences in abundance of EMB Cluster 0 and EMB Cluster 1 proteins at 6 hr and 10 hr, whereas in the VYS, the time with the greatest abundance difference by treatment was at 4 hr in VYS Cluster 0 and VVYS Cluster 1. The Redox relevant pathways were identified in VYS Cluster 1 with enrichment for oxidation-reduction process, response to reactive oxygen species, and cellular response to hypoxia (Figure 5). These pathways are elevated in VPA-treated VYS at 4 hr compared to control VYS, thereby directly preceding the VYS enrichment for cellular response to hydrogen peroxide in our DAVID analysis of significant VYS proteins at 6 hr.
A transcription factor enrichment analysis was performed to identify possible upstream mechanisms of changes to protein abundance occurring at the transcriptional level. Enrichr performs an overrepresentation test of significant proteins for each time point compared to transcription factor binding sites for the associated genes. Notable findings in the transcription factor enrichment included that there were 22 distinct TFs with functions relevant to developmental processes (bolded in Table 2). Significant proteins in the VYS were enriched for 20 distinct developmental TFs, while the EMB was enriched for 15. There were two developmental TFs that were only enriched in the EMB which included RCOR1 and NANOG and seven developmental TFs that were only enriched in the VYS including: PBX3, HNF4A, NFIC, PPARG, E2F4, SIX5, and FOXP2. Of these tissue-specific developmental TFs, NANOG which was enriched in our data for EMB proteins with decreased abundance at 4 hr has previously been associated with upregulation of TF Gcm1 (under conditions of folate deficiency) which is involved in the regulation of the Wnt signaling pathways that control developmental patterning associated with proper NTC (Li et al., 2021). NFIC was enriched for decreased proteins in the VYS at 8 hr and has previously been shown to be elevated in a mouse model of NTDs (Huang et al., 2021). Further research is needed to understand the potential role of the other enriched TFs in NTD pathogenesis and to understand how the spatiotemporal transcriptome of the VPA-exposed conceptus relates to the proteome.
In total, this study identified 1,510 proteins that were affected by changes in protein abundance in either the VYS or EMB across ten hours of early organogenesis in the mouse conceptus. Due to the large number of proteins, however, we are unable to follow-up on all individual proteins with additional experimental evidence of their role in VPA teratogenesis. The CTD is an online database that contains manually curated associations between genes and environmental or pharmaceutical exposures based on published peer-reviewed literature. In total 428 of the significant proteins identified in our TMT analysis (28.3%) were included in the CTD as genes with previous associations with VPA in mice. To quantitatively assess, if our significant proteins were overrepresented in the CTD at each time point, we leveraged the full list of VPA-associated genes from the CTD to perform a Fisher’s Exact enrichment analysis. The enrichment test concluded that all timepoints in the VYS and the 2–8 hr timepoints in the EMB had greater overlap with the CTD VPA-associated gene list than expected by chance. This conclusion provides strong supporting evidence for the association between these proteins and VPA in mice. Additional evidence supporting the connection of these genes previously associated with VPA is that of the 202 significant proteins that are shared in at least two time points or tissues, there are 53 that are associated with VPA in the CTD (Figure 1E). Of these proteins, there were four that were significant for three time points and/or tissues. These proteins included Tubb3 (4hr EMB Increased, 8hr EMB Increased, 8hr VYS Increased), Fbl (2hr EMB Increased, 8hr EMB Increased, 6hr VYS Decreased), Arhgdlb (4hr EMB Increased, 8hr VYS Increased, 10hr VYS Increased) and Parp1 (2hr EMB Increased, 4hr EMB Increased, 6hr VYS Decreased). Of all 53 of the CTD-overlapping proteins that were significant for at least two time points or tissues, there were 25 that were significant either in both tissues at the same time point, the same tissue at adjacent time points, or different tissues but at adjacent time points. This shared temporality of these proteins in addition to their annotation as VPA-associated genes in the CTD makes them strong candidates for further investigation. Future research should prioritize mechanistic studies of the CTD overlapping proteins (indicated in Supplemental Table 1) for a role in VPA teratogenesis in mice and prioritize investigating their spatiotemporal expression since the CTD enrichment cannot account for tissue of origin or timing of VPA exposure. In addition to the extensive chemical-gene associations in the CTD, there are also a small number of gene-disease associations. Of the genes associated with NTDs in mice within the CTD, there were two that overlap with our proteins with differential abundance in this study. Vangptl2 was increased in the EMB at 2 hr (LogFC=0.17) and associated with NTDs (Wilson et al., 1990) and VPA within the CTD (Jergil et al., 2011). In the VYS, Flor1 was decreased at 10 hr (LogFC= −0.16) and was associated with NTDs (Tang et al., 2005) and VPA within the CTD (Finnell et al., 1997; Jergil et al., 2009, 2011; Kultima et al., 2010). Future research should link the VPA-associated genes and proteins from this study (and the CTD) to phenotypic NTD endpoints to directly confirm the link between VPA-altered gene/protein abundance and the etiology of NTDs.
The findings of this study are the result of a single acute exposure to VPA during an early period of organogenesis. Whole embryo culture experiments conducted in vivo as well as in vitro confirm the ability of VPA to elicit NTDs as a result of a single dose (Tung and Winn, 2011; Piorczynski et al., 2022). Due to the limited exposure window, it is not possible to determine whether our results are specific to the length of exposure or a combination of the timing and length of exposure to VPA. Future studies may benefit from performing earlier in vivo dosing of VPA in dams with measurement of protein abundance at these same time points within organogenesis to determine whether the timing or duration of the exposure is more critical for the response. In vivo dosing provides the benefit of assessing the roles of maternal transport and metabolism to the experimental design, which are lacking in the whole embryo culture model. Despite this, it is evident that proteins exhibit a dynamic response to VPA exposure and do not perpetually remain in a state of increased or decreased abundance, thus emphasizing the importance of temporal measurements when evaluating a compound for safety during pregnancy. An additional future direction for this research would be to separately analyze organ and cell type responses to VPA exposure. By treating the whole EMB proper as a single “tissue,” it is not possible to definitively conclude whether instances of altered differentiation or proliferation for example are directly related to the process of NTC or instead relevant to VPA’s effects on a separate organ or tissue system.
Global quantitative proteomics, and omics technologies in general, are excellent methodologies for generating hypotheses without the need for a priori selection of targets, thus resulting in less biased study design and conclusions. However, despite these advantages to proteomics technology, there are still important considerations for its implementation and interpretation. Unlike RNA sequencing, which has a standardized pipeline, analysis of proteomics is more variable. This is partly due to lower precision in proteomics than RNA sequencing because of limitations in LC-MS/MS methodology resulting in poor quantitation of low abundance proteins and lower certainty in protein identification due to shared peptide sequences (Fonslow et al., 2011; Muth and Renard, 2018). Due to these limitations, it is important to treat initial results as hypothesis generating until samples sizes can be increased and results of important protein targets are validated through additional methods. Despite these limitations, proteomics is a novel methodology in the field of developmental toxicology with the capacity to reveal important patterns of protein network expression that may be involved in structural birth defect development. The presented proteomics results evaluate protein abundance following VPA exposure, however, it is not currently known whether VPA may be affecting the structure, activity, or abundance of these proteins through further post-translational modifications which should be evaluated in future studies.
Herein we have presented evidence to show that the EMB and VYS respond to VPA in a spatially independent and temporally specific manner through altered protein abundance in early organogenesis. The altered abundance of over 1500 proteins following VPA exposure confirms that VPA causes significant alterations in protein abundance within specific regulatory pathways required for normal NTC such as Wnt signaling (planar cell polarity, convergent extension), cytoskeletal dynamics, cellular proliferation, migration, and adhesion, as well as oxidation-reduction processes. In total, 428 proteins of the differentially abundant proteins were coded by genes with previous VPA associations in mice that are highlighted in the CTD. The identification of the other 1,082 proteins affected by VPA without prior evidence of gene expression changes in mice in the CTD, underscores the importance of spatiotemporal proteomics as a discovery tool and the importance of independent investigation of gene expression and protein abundance dynamics in the context of developmental toxicology studies.
Supplementary Material
Early organogenesis stage mouse conceptuses were treated with 600μM VPA using whole embryo culture for 2–10 hr.
Protein abundance was measured through tandem mass tag labeling and LC-MS3 which identified 1,510 proteins with differential abundance in the treated embryo or visceral yolk compared to controls.
Protein abundance changes were dynamic across time and based on tissue (embryo vs. visceral yolk sac).
428 of the significant proteins have previous associations with VPA exposure in mice based on the Comparative Toxicogenomics Database.
Pathway analysis revealed enrichment of multiple pathways relevant to neural tube closure including convergent extension, Wnt Signaling/planar cell polarity, cellular migration, cellular proliferation, cell death, and cytoskeletal organization.
5: Acknowledgements:
We would like to acknowledge our funding sources including NIH T32ES007062, P30ES017885, and R01ES028802, a Rackham Graduate Student Research Grant, a grant from the University of Michigan Office of Research, and the University of Michigan School of Public Health. The authors would like to acknowledge the Proteomics Resource Facility at University of Michigan, including Alexey Nesvizhskii, Venky Basrur, and Kevin Conlon, for assistance in experimental planning and running the LC-MS3 and proteome discover searches. An earlier version of this article appeared as a chapter in the doctoral dissertation of Samantha Lapehn at University of Michigan.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Data Availability
Embryo and Visceral Yolk Sac TMT data has been uploaded to Mendeley Data under DOI: 10.17632/nwf27h76yr.1
References
- Abu-Jamous B, and Kelly S (2018). Clust: automatic extraction of optimal co-expressed gene clusters from gene expression data. Genome Biol. 19, 172. doi: 10.1186/s13059-018-1536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires MB, Santos JRA, Souza KS, Farias PS, Santos ACV, Fioretto ET, et al. (2015). Rat visceral yolk sac cells: viability and expression of cell markers during maternal diabetes. Braz. J. Med. Biol. Res. 48, 676–682. doi: 10.1590/1414-431x20154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MT, and Wallingford JB (2017). Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 18, 375–388. doi: 10.1038/nrm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles G, et al. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, and Greene ND (2010). Genetics and development of neural tube defects: Genetics and development of neural tube defects. J. Pathol. 220, 217–230. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Stanier P, and Greene NDE (2013). Neural tube defects: recent advances, unsolved questions, and controversies. Lancet Neurol. 12, 799–810. doi: 10.1016/S1474-4422(13)70110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC, et al. (2021). Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 49, D1138–D1143. doi: 10.1093/nar/gkaa891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnell RH, Wlodarczyk BC, Craig JC, Piedrahita JA, and Bennett GD (1997). Strain-dependent alterations in the expression of folate pathway genes following teratogenic exposure to valproic acid in a mouse model. Am. J. Med. Genet. 70, 303–311. [PubMed] [Google Scholar]
- Fonslow BR, Carvalho PC, Academia K, Freeby S, Xu T, Nakorchevsky A, et al. (2011). Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J. Proteome Res. 10, 3690–3700. doi: 10.1021/pr200304u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato Y, and Miki H (2010). Redox regulation of Wnt signalling via nucleoredoxin. Free Radic. Res. 44, 379–388. doi: 10.3109/10715761003610745. [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu X, Tang B, Li C, Kou Z, Li L, et al. (2017). Protein Expression Landscape of Mouse Embryos during Pre-implantation Development. Cell Rep. 21, 3957–3969. doi: 10.1016/j.celrep.2017.11.111. [DOI] [PubMed] [Google Scholar]
- Ghodke-Puranik Y, Thorn CF, Lamba JK, Leeder JS, Song W, Birnbaum AK, et al. (2013). Valproic acid pathway: pharmacokinetics and pharmacodynamics. Pharmacogenet. Genomics 23, 236–241. doi: 10.1097/FPC.0b013e32835ea0b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, and Aebersold R (1999). Correlation between Protein and mRNA Abundance in Yeast. Mol. Cell. Biol. 19, 1720–1730. doi: 10.1128/MCB.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach K, Ulrich N, Goss K-U, Seiwert B, Wagner S, Scholz S, et al. (2020). Yolk Sac of Zebrafish Embryos as Backpack for Chemicals? Environ. Sci. Technol. 54, 10159–10169. doi: 10.1021/acs.est.0c02068. [DOI] [PubMed] [Google Scholar]
- Hansen JM, Jones DP, and Harris C (2020). The Redox Theory of Development. Antioxid. Redox Signal. 32, 715–740. doi: 10.1089/ars.2019.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C (2012). “Rodent Whole Embryo Culture,” in Developmental Toxicology Methods in Molecular Biology., eds. Harris C and Hansen JM (Totowa, NJ: Humana Press; ), 215–237. doi: 10.1007/978-1-61779-867-2_13. [DOI] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang W, Huang T, Liu Y, Fu J, Wei X, Liu D, et al. (2021). Nuclear factor I-C disrupts cellular homeostasis between autophagy and apoptosis via miR-200b-Ambra1 in neural tube defects. Cell Death Dis. 13, 17. doi: 10.1038/s41419-021-04473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentink J, Loane MA, Dolk H, Barisic I, Garne E, Morris JK, et al. (2010). Valproic Acid Monotherapy in Pregnancy and Major Congenital Malformations. N. Engl. J. Med. 362, 2185–2193. doi: 10.1056/NEJMoa0907328. [DOI] [PubMed] [Google Scholar]
- Jergil M, Forsberg M, Salter H, Stockling K, Gustafson A-L, Dencker L, et al. (2011). Short-time gene expression response to valproic acid and valproic acid analogs in mouse embryonic stem cells. Toxicol. Sci. Off. J. Soc. Toxicol. 121, 328–342. doi: 10.1093/toxsci/kfr070. [DOI] [PubMed] [Google Scholar]
- Jergil M, Kultima K, Gustafson A-L, Dencker L, and Stigson M (2009). Valproic Acid–Induced Deregulation In Vitro of Genes Associated In Vivo with Neural Tube Defects. Toxicol. Sci. 108, 132–148. doi: 10.1093/toxsci/kfp002. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultima K, Jergil M, Salter H, Gustafson A-L, Dencker L, and Stigson M (2010). Early transcriptional responses in mouse embryos as a basis for selection of molecular markers predictive of valproic acid teratogenicity. Reprod. Toxicol. Elmsford N 30, 457–468. doi: 10.1016/j.reprotox.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Lapehn S, Piorczynski TB, Hansen JM, and Harris C (2021). Spatiotemporal evaluation of the mouse embryonic redox environment and histiotrophic nutrition following treatment with valproic acid and 1,2-dithiole-3-thione during early organogenesis. Reprod. Toxicol., S0890623821000459. doi: 10.1016/j.reprotox.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xie Q, Gao J, Wang F, Bao Y, Wu L, et al. (2021). Aberrant Gcm1 expression mediates Wnt/β-catenin pathway activation in folate deficiency involved in neural tube defects. Cell Death Dis. 12, 234. doi: 10.1038/s41419-020-03313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, et al. (2012). Increasing the Multiplexing Capacity of TMTs Using Reporter Ion Isotopologues with Isobaric Masses. Anal. Chem. 84, 7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, et al. (2014). MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal. Chem. 86, 7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. (2003). PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Muñoz-Soriano V, Belacortu Y, and Paricio N (2012). Planar cell polarity signaling in collective cell movements during morphogenesis and disease. Curr. Genomics 13, 609–622. doi: 10.2174/138920212803759721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth T, and Renard BY (2018). Evaluating de novo sequencing in proteomics: already an accurate alternative to database-driven peptide identification? Brief. Bioinform. 19, 954–970. doi: 10.1093/bib/bbx033. [DOI] [PubMed] [Google Scholar]
- Nikolopoulou E, Galea GL, Rolo A, Greene NDE, and Copp AJ (2017). Neural tube closure: cellular, molecular and biomechanical mechanisms. Development 144, 552–566. doi: 10.1242/dev.145904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornoy A (2009). Valproic acid in pregnancy: How much are we endangering the embryo and fetus? Reprod. Toxicol. 28, 1–10. doi: 10.1016/j.reprotox.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, and Klein PS (2001). Histone Deacetylase Is a Direct Target of Valproic Acid, a Potent Anticonvulsant, Mood Stabilizer, and Teratogen. J. Biol. Chem. 276, 36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- Piorczynski TB, Lapehn S, Ringer KP, Allen SA, Johnson GA, Call K, et al. (2022). NRF2 activation inhibits valproic acid-induced neural tube defects in mice. Neurotoxicol. Teratol. 89, 107039. doi: 10.1016/j.ntt.2021.107039. [DOI] [PubMed] [Google Scholar]
- Ross C, and Boroviak TE (2020). Origin and function of the yolk sac in primate embryogenesis. Nat. Commun. 11, 3760. doi: 10.1038/s41467-020-17575-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y (1989). Neurulation in the mouse: Manner and timing of neural tube closure. Anat. Rec. 223, 194–203. doi: 10.1002/ar.1092230212. [DOI] [PubMed] [Google Scholar]
- Sandieson L, Hwang JTK, and Kelly GM (2014). Redox regulation of canonical Wnt signaling affects extraembryonic endoderm formation. Stem Cells Dev. 23, 1037–1049. doi: 10.1089/scd.2014.0010. [DOI] [PubMed] [Google Scholar]
- Sidoli S, Kulej K, and Garcia BA (2017). Why proteomics is not the new genomics and the future of mass spectrometry in cell biology. J. Cell Biol. 216, 21–24. doi: 10.1083/jcb.201612010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LS, Santillano DR, Wlodarczyk BJ, Miranda RC, and Finnell RH (2005). Role of Folbp1 in the regional regulation of apoptosis and cell proliferation in the developing neural tube and craniofacies. Am. J. Med. Genet. C Semin. Med. Genet. 135C, 48–58. doi: 10.1002/ajmg.c.30053. [DOI] [PubMed] [Google Scholar]
- Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. (2003). Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Anal. Chem. 75, 1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Tung EWY, and Winn LM (2011). Valproic Acid Increases Formation of Reactive Oxygen Species and Induces Apoptosis in Postimplantation Embryos: A Role for Oxidative Stress in Valproic Acid-Induced Neural Tube Defects. Mol. Pharmacol. 80, 979–987. doi: 10.1124/mol.111.072314. [DOI] [PubMed] [Google Scholar]
- Usami M, Mitsunaga K, and Nakazawa K (2007). Comparative proteome analysis of the embryo proper and yolk sac membrane of day 11.5 cultured rat embryos. Birth Defects Res. B. Dev. Reprod. Toxicol. 80, 383–395. doi: 10.1002/bdrb.20127. [DOI] [PubMed] [Google Scholar]
- Vogel C, and Marcotte EM (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D (2008). Discrepancy between mRNA and protein abundance: Insight from information retrieval process in computers. Comput. Biol. Chem. 32, 462–468. doi: 10.1016/j.compbiolchem.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Marco P. de, Capra V, and Kibar Z (2019). Update on the Role of the Non-Canonical Wnt/Planar Cell Polarity Pathway in Neural Tube Defects. Cells 8. doi: 10.3390/cells8101198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Miller DC, Harman R, Antczak DF, and Clark AG (2013). Paternally expressed genes predominate in the placenta. Proc. Natl. Acad. Sci. U. S. A. 110, 10705–10710. doi: 10.1073/pnas.1308998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DB, Wyatt DP, and Gookin JL (1990). Cranial effects of retinoic acid in the loop-tail (Lp) mutant mouse. J. Craniofac. Genet. Dev. Biol. 10, 75–81. [PubMed] [Google Scholar]
- Xiao H, Jedrychowski MP, Schweppe DK, Huttlin EL, Yu Q, Heppner DE, et al. (2020). A Quantitative Tissue-Specific Landscape of Protein Redox Regulation during Aging. Cell 180, 968–983.e24. doi: 10.1016/j.cell.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn IE, and Sarkar AA (2010). The visceral yolk sac endoderm provides for absorption of nutrients to the embryo during neurulation. Birt. Defects Res. A. Clin. Mol. Teratol. 88, 593–600. doi: 10.1002/bdra.20705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Embryo and Visceral Yolk Sac TMT data has been uploaded to Mendeley Data under DOI: 10.17632/nwf27h76yr.1


