Abstract
An inducible middle promoter from the lactococcal bacteriophage φ31 was isolated previously by shotgun cloning an 888-bp fragment (P15A10) upstream of the β-galactosidase (β-Gal) gene (lacZ.st) from Streptococcus thermophilus (D. J. O’Sullivan, S. A. Walker, S. G. West, and T. R. Klaenhammer, Bio/Technology 14:82–87, 1996). The promoter showed low levels of constitutive β-Gal activity which could be induced two- to threefold over baseline levels after phage infection. During this study, the fragment was subcloned and characterized to identify a smaller, tightly regulated promoter fragment which allowed no β-Gal activity until after phage infection. This fragment, defined within nucleotides 566 to 888 (P566–888; also called fragment 566–888), contained tandem, phage-inducible transcription start sites at nucleotides 703 and 744 (703/744 start sites). Consensus −10 regions were present upstream of both start sites, but no consensus −35 regions were identified for either start site. A transcriptional activator, encoded by an open reading frame (ORF2) upstream of the 703/744 start sites, was identified for P566–888. ORF2 activated P566–888 when provided in trans in Escherichia coli. In addition, when combined with pTRK391 (P15A10::lacZ.st) in Lactococcus lactis NCK203, an antisense ORF2 construct was able to retard induction of the phage-inducible promoter as measured by β-Gal activity levels. Finally, gel shift assays showed that ORF2 was able to bind to promoter fragment 566–888. Deletion analysis of the region upstream from the tandem promoters identified a possible binding site for transcriptional activation of the phage promoters. The DNA-binding ability of ORF2 was eliminated upon deletion of part of this region, which lies centered approximately 35 bp upstream of start site 703. Deletion analysis and mutagenesis studies also elucidated a critical region downstream of the 703/744 start sites, where mutagenesis resulted in a two- to threefold increase in β-Gal activity. With these improvements, the level of expression achieved by an explosive-expression strategy was elevated from 3,000 to 11,000 β-Gal units within 120 min after induction.
Lactococcus lactis is an industrially important member of the lactic acid bacteria (LAB). It is used widely in the fermentation of dairy products, including sour cream, buttermilk, and various cheeses such as cheddar. In addition to its role in these important food fermentations, Lactococcus has the potential to play an even greater role in food biotechnology. Its long history of use in the food industry, its generally recognized as safe status, and the ever-expanding knowledge of the molecular genetics of this microorganism make Lactococcus an ideal choice for food-grade production of proteins and enzymes of relevance to the food and pharmaceutical industries.
Relatively few expression systems exist for the food-grade LAB such as lactococci and lactobacilli. In contrast, powerful expression systems for protein and enzyme production exist for some of the more genetically defined microorganisms, such as Escherichia coli. These expression systems are based on transcriptionally regulated bacterial promoters (e.g., lac promoter) or on very specific bacteriophage promoters (e.g., T7 promoter/T7 RNA polymerase [RNAP] [34]). The advancement of molecular techniques for the LAB, including the identification of regulated promoters, has set the stage for the development of expression systems for Lactococcus. For instance, the inducible lacA promoter upstream of the lactose operon in lactococci is one example where induction by lactose resulted in an increase in transcription of downstream genes (36). The regulated lac promoter was utilized in the construction of a T7 RNAP/T7 promoter expression system for use in Lactococcus (38). In this system, T7 RNAP was cloned under control of the lacA promoter, so that shifting the culture to growth on lactose induced expression of the T7 RNAP. By using this system, tetanus toxin fragment C, cloned downstream of the T7 promoter, was expressed to levels up to 22% of the soluble cell protein of L. lactis. Recently, a powerful expression system for Lactococcus has been developed by using the regulatory components of the nisin regulon (7, 22). Two of the three promoters associated with the nisin cluster of genes, the nisA promoter and the nisF promoter, were inducible by the addition of nisin. A combination of the nisA promoter with a strain carrying the components involved in signal transduction, nisR and nisK, allowed efficient expression of proteins or enzymes cloned downstream of the promoter at levels proportional to the amount of nisin added to the medium (6). Other promising, regulated bacterial promoters from Lactococcus include those induced by environmental factors such as NaCl (21) or pH (19). In addition, extensive research into the molecular biology of lactococcal bacteriophages has also provided excellent opportunities to create expression systems based on phage-specific signals. For example, Nauta et al. (25) developed an expression system based on the repressor-operator system isolated from the lactococcal temperate bacteriophage φr1t. By using this system, β-galactosidase (β-Gal) levels were increased 70-fold upon induction of the φr1t prophage with mitomycin. The details of the regulated promoters described above are available in a comprehensive review by Kok (21).
We recently developed a novel expression system which exploits the temporally regulated development of bacteriophage in L. lactis. The expression system consists of two components of the phage genome: a phage origin of replication (ori) and a phage-inducible promoter (27). A phage origin of replication (ori31) was previously cloned from the lytic lactococcal bacteriophage φ31 (26). When present in trans on a vector, ori31 is proposed to act as an alternative target for phage-directed DNA replication. Phage infection of a host carrying ori31 on a low-copy-number replicon results in explosive amplification of the vector, thereby dramatically increasing the copy number of the gene of interest in the host. Recently, the first phage-inducible promoter element from a lactococcal bacteriophage was isolated by shotgun cloning phage φ31 DNA upstream of the β-Gal gene from Streptococcus thermophilus (lacZ.st) (33) in the high-copy-number promoter screening vector pTRK390 (27). The 888-bp phage-inducible promoter (termed P15A10) showed a low level of constitutive activity (200 to 300 β-Gal units) prior to phage φ31 infection. After phage infection of the lactococcal host, β-Gal activity was induced three- to fourfold within 60 min. Combining P15A10::lacZ.st and ori31 to yield the low-copy-number expression vector pTRK392 (27) resulted in negligible β-Gal activity before phage infection. However, following phage infection, an activity level of greater than 2,000 β-Gal units was achieved within 2 h.
In this study, we present the molecular characterization of the phage-inducible promoter P15A10. The goals of the present study were threefold: to identify the essential phage-inducible region, to determine the factors or regions involved in regulation of the promoter, and to improve promoter expression through site-directed mutagenesis.
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. L. lactis subsp. lactis NCK203, the sensitive host for bacteriophage φ31, was propagated in M17 (Difco) supplemented with 0.5% glucose (GM17) at 30°C. Where necessary, erythromycin and/or chloramphenicol was added at 5 or 7.5 μg/ml, respectively. E. coli strains were grown in LB broth at 37°C with shaking or on LB broth supplemented with 1.5% agar. When required, ampicillin was added at 100 μg/ml, chloramphenicol was added at 20 μg/ml, and kanamycin was added at 50 μg/ml. In E. coli, erythromycin resistance was selected for on brain heart infusion agar (Difco) supplemented with 120 μg of erythromycin per ml (29).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains | Descriptiona | Reference or source |
|---|---|---|
| E. coli | ||
| JM110 | E. coli cloning host | 39 |
| DH5α | E. coli cloning host | Gibco-BRL |
| MC1061 | E. coli host for pNZ18 | 4 |
| BIL21 (DE3) | Expression host for pET28a | Novagen |
| L. lactis subsp. lactis NCK203 | Sensitive host for phage φ31 | 15 |
| Plasmids | ||
| pT7Blue | Apr; T-vector for cloning PCR products | Novagen |
| pTRK390 | Emr; lactococcal promoter screening vector | 27 |
| pTRK391 | Emr; pTRK390::P15A10 | 27 |
| pTRK477 | Emr; pTRK390::P566–888 | This study |
| pTRK486 | Emr; pTRK390::P566–862 | This study |
| pTRK481 | Emr; pTRK390::P566–849 | This study |
| pTRK485 | Emr; pTRK390::P566–792 | This study |
| pTRK487 | Emr; pTRK390::P566–732 | This study |
| pTRK483 | Emr; pTRK390::P566–888S | This study |
| pTRK482 | Emr; pTRK390::P566–862S | This study |
| pTRK484 | Emr; pTRK390::P15A10/mutORF2 | This study |
| pET28a | Knr; E. coli expression vector (T7 based) | Novagen |
| pTRK478 | Knr; pET28a::ORF2 | This study |
| pNZ18 | Cmr; high-copy-number shuttle vector | 8 |
| pTRK452 | Cmr; pNZ18::P566–888::ORF2 | This study |
| pTRK479 | Cmr; pNZ18::P6::antiORF2::TT7 | This study |
| pTRK360 | Cmr, Emr; pSA3 + ori31 | 26 |
| pTRK392 | Cmr, Emr; pTRK360::P15A10:: lacZ.st | 27 |
| pTRK480 | Cmr, Emr; pTRK360::P566–862S:: lacZ.st | This study |
| pCITE4a | Apr; for in vitro transcription-translation | Novagen |
Abbreviations: Apr, ampicillin resistance; Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Knr, kanamycin resistance.
Bacteriophage propagation and β-Gal assays.
Phage φ31 is a small, isometric-headed cohesive-ended, lytic lactococcal bacteriophage of the P335 species (1, 20) with a double-stranded DNA genome of 31.9 kb. Phage φ31 was propagated on NCK203 in GM17 supplemented with 10 mM CaCl2 at 30°C. Efficiency of plaquing (EOP) assays were performed as described previously (35). β-Gal assays were performed on NCK203 derivatives at various time points during a phage φ31 lytic cycle by using the O-nitrophenyl-β-d-galactopyranoside (ONPG) assay described by Miller (23), as modified by O’Sullivan et al. (27). β-Gal activity was measured just before phage infection (time, 0 min; optical density at 600 nm [OD600] ≅ 0.5). Phage φ31 was added to a multiplicity of infection of greater than 1 (MOI ≅ 5) so that cell lysis occurred within 60 min. Samples (100 μl) were then assayed for β-Gal levels every 20 min until cell lysis occurred. Lactococcal cells were permeated with chloroform, and β-Gal activity was expressed as units per OD600 of the culture. For β-Gal measurements made after phage-mediated lysis had begun, units were expressed relative to the highest OD600 attained by the culture before lysis. All measurements reported are averages of results from at least three separate assays. At each time point, the assay was performed in duplicate.
DNA isolation.
Small-scale E. coli plasmid preparations were made by using the alkaline-sodium dodecyl sulfate method (31). Large-scale E. coli plasmid preparations were made by using the Qiagen (Chatsworth, Calif.) plasmid kit as described in the manufacturer’s directions. Small-scale isolation of plasmids from L. lactis was as described by O’Sullivan and Klaenhammer (28), except that ethidium bromide was not used prior to phenol-chloroform extraction.
Gene cloning and transformations.
Standard procedures were used for the DNA manipulations described in this study (31). Restriction enzymes and T4 DNA ligase were provided by Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and used as described in manufacturer’s instructions. Ligation products were transformed into RbCl-competent E. coli strains. RbCl-competent E. coli cells were prepared by the procedure of Hanahan (14), modified as described by Dinsmore and Klaenhammer (9). Cells were frozen at −70°C in 100-μl aliquots and transformed by the procedure described for CaCl2-competent cells (31). After screening for the proper insert in E. coli, plasmids were electroporated into L. lactis by using a modified procedure of Holo and Nes (18). L. lactis subsp. lactis NCK203 was propagated in GM17 supplemented with 1% glycine at 30°C. When an OD600 of 0.2 was reached, the cells were washed three times with ice-cold electroporation buffer (0.5 M sucrose, 10% glycerol) and then resuspended in 1/10 original volume with the same buffer. Electroporations were carried out with the Bio-Rad (Richmond, Calif.) gene pulser with 100 μl of cells in a 0.2-cm cuvette under the following conditions: 25 μF, 2.45 kV, and 200 Ω. Recovery was achieved by growing the cells in GM17 supplemented with 10 mM MgCl2 and 1 mM CaCl2 for 2 h at 30°C prior to plating them on selected antibiotic markers.
PCR and DNA sequencing.
PCR was performed with Taq DNA polymerase (Boehringer Mannheim) as described in the manufacturer’s instructions. In each case, 40 cycles were used to amplify the regions of interest. Annealing temperatures were 5 to 10°C below the lowest melting temperature of each primer pair. To facilitate cloning of PCR products, restriction enzyme sites were either inserted into the 5′ ends of the primers or the product was subcloned into the T-vector pT7Blue (Novagen, Madison, Wis.). To ensure the absence of PCR-generated mutations and to confirm the accuracy of site-directed mutations, DNA sequencing was performed on large-scale E. coli plasmid preparations by using the Sequenase 2.0 enzyme and kit (Amersham Life Sciences, Arlington Heights, Ill.) and standard dideoxy sequencing (32) as described by the manufacturers.
Gel retardation assays with ORF2.
The DNA fragments used in the gel retardation assays were fragment 566–888 (spanning nucleotides 566 to 888; also called P566–888), fragment 566–732, and fragment 658–888; they were amplified from P15A10 by using PCR with Taq DNA polymerase (Boehringer Mannheim) so that no 5′ phosphate would be present on any strand. Each fragment was end labeled with 32P by using T4 polynucleotide kinase (Boehringer Mannheim) and [γ-32P]ATP (NEN, Boston, Mass.) as described in the manufacturers’ instructions. The open reading frame ORF2 product (or the control product) was produced by using a single-tube protein system (T7 based; Novagen) as described in Results. DNA binding was achieved in 20-μl reaction mixtures containing the following: 10 μl of the ORF2 product (or control) directly from the single-tube protein system reaction, 2 μl of labeled DNA fragment, and 8 μl of a reaction buffer consisting of 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 100 mM NaCl, and 1 mM dithioerythritol (1× restriction enzyme buffer H; Boehringer Mannheim). Sterile glycerol and sheared salmon sperm were added to levels of 1% and 1 μg/reaction, respectively. To determine the effect of competitive DNA on the ability of ORF2 to bind each specific fragment, 2 μl of unlabeled fragment 566–888 was added to each of the mixtures described above. The concentration of unlabeled fragment used created an approximate 2:1 ratio of unlabeled fragment to labeled fragment. Binding was performed for 40 min at 25°C. Samples were then electrophoresed through a 4% polyacrylamide gel by using 1× standard Tris-borate-EDTA buffer and 75 V. The gels were soaked for 5 min in 7% acetic acid and then rinsed with water before transfer to Whatman 3MM paper. The gels were dried for 15 to 25 min and exposed to Kodak’s Biomax film by using Biomax intensifying screens.
RNA manipulations.
RNA was isolated from L. lactis subsp. lactis NCK203 at various times during the phage infection cycle by using TRIzol reagent (Gibco-BRL, Gaithersburg, Md.) as described by Dinsmore and Klaenhammer (9). Slot blot Northern hybridizations were performed on a Bio-Rad apparatus in accordance with the manufacturer’s protocol. Equivalent amounts of RNA from each time point (approximately 10 μg) were denatured and applied to a Zeta probe membrane (Bio-Rad). The RNA was UV cross-linked to the membrane with the auto-cross-link cycle of the Stratagene (La Jolla, Calif.) Stratalinker and then hybridized to a 32P-labeled probe at 65°C as recommended by Bio-Rad. Probes were 32P labeled by using the multiprime DNA labeling system (Amersham). The lacZ.st-specific probe corresponded to the BamHI/SalI fragment from pTRK390. The ORF2-specific probe was generated by PCR by using primers described in the legend to Fig. 5. Primer extension analysis was performed as described previously (27). The lacZ primer (complementary to nucleotides 189 to 203 of the lacZ.st sequence as provided by Schroeder et al. [33]) was used to determine transcription start sites when appropriate.
FIG. 5.
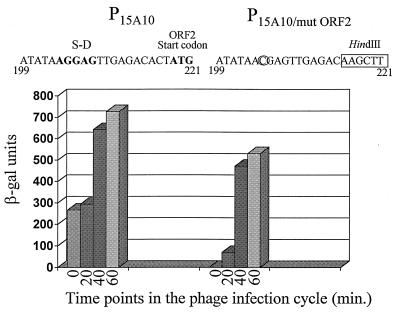
Site-directed mutagenesis of the translational signals of ORF2 on P15A10. P15A10 was amplified by PCR into two separate fragments. Fragment 1–215 was amplified by using the universal −40 primer (on pTRK391) and a primer complementary to nucleotides 194 to 215 (G→C mutation at nucleotide 204). Fragment 222–888 was generated by using a primer consisting of nucleotides 222 to 243 and the lacZ primer (on pTRK391). Nucleotides 216 to 221, containing the ATG start codon for ORF2, were replaced with a HindIII site on both PCR fragments (1–215HindIII and HindIII222–888) to allow fusion. The changes made to ORF2 are indicated above the graph. The graph represents β-Gal results of P15A10 and P15A10/mutORF2. β-Gal assays were performed at least three separate times. For each assay, time point determinations were performed in duplicate.
RESULTS
Subcloning of the phage-inducible promoter element P15A10.
The 888-bp promoter P15A10 represented the first phage-inducible promoter element isolated from a lactococcal bacteriophage. Initial primer extension analysis of P15A10 revealed five putative transcription start sites (Fig. 1) (27). The first three start sites (at nucleotides 167 and 172 and between nucleotides 537 and 550) were phage inducible but were very weak. In contrast, start sites at nucleotides 703 and 744 (start sites 703 and 744) were strongly phage inducible. A weak primer extension product was present before phage infection (time 0) for start site 703 (27).
FIG. 1.
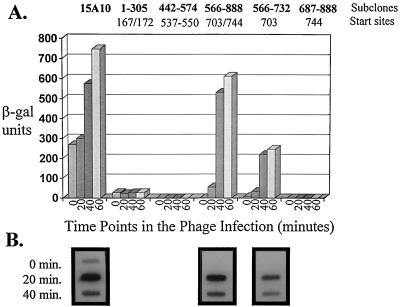
Representation of the phage-inducible promoter fragment P15A10 (27). The five putative transcription start sites determined by O’Sullivan et al. (27) are represented by vertical arrows (numbered 1 to 5). A complete open reading frame (ORF2; nucleotides 219 to 650) located upstream of start sites 4 and 5 is indicated. This fragment showed constitutive activity which was induced three- to fourfold upon phage infection of the host. P15A10 was subcloned by PCR into five different regions, as indicated. Fragment 1–305 was generated by using the universal −40 primer (on pTRK391) and a primer complementary to nucleotides 281 to 306 on P15A10. Fragment 442–574 was amplified by using one primer consisting of nucleotides 442 to 457 and one primer complementary to nucleotides 559 to 574. Subclone 566–888 was generated by using a primer consisting of nucleotides 566 to 582 and the lacZ primer described in Materials and Methods (on pTRK391). Subclone 687–888 was amplified by using a primer consisting of nucleotides 687 to 705 (T→A and A→C mutations at nucleotides 691 and 692, respectively) and the lacZ primer. Subclone 566–732 utilized the nucleotide 566 primer and a primer complementary to nucleotides 714 to 732. Addition of a 5′ BamHI site to at least one primer of each pair facilitated subsequent cloning procedures.
To determine the precise location of the phage-inducible promoter activity, P15A10 was subcloned as five different regions representing various combinations of the transcription start sites identified previously (Fig. 1). Each region, amplified by PCR by using primers described in the legend to Fig. 1, was cloned into the BamHI site of the promoter-screening vector pTRK390 (27). Restriction analysis and DNA sequencing were used to confirm the orientation of the cloned fragments and the absence of PCR-generated mutations, respectively. Tandem start sites at nucleotides 703 and 744 were subcloned both together and separately because they showed the strongest inducibility after phage infection. Start site 703 was isolated by itself on fragment 566–732. Although this subclone contained sequences upstream of start site 744, the primer pair (see legend to Fig. 1) was designed to exclude the putative −10 consensus region for the 744 start site (see Fig. 3B) so that transcription would take place only from the start site 703-associated promoter. Start site 744 was isolated on fragment 687–888. To eliminate activity from start site 703 on this fragment, the upstream primer was designed to introduce a 2-bp mutation in the putative −10 consensus region of start site 703 (TATTAT→ACTTAT; see Fig. 3B) as described in the legend to Fig. 1. Combined with the absence of sequences centered approximately 35 bp upstream of start site 703, the mutations were expected to eliminate any promoter activity associated with start site 703.
FIG. 3.
(A) Sequence of the middle, phage-inducible promoter (P15A10) from the lytic, lactococcal bacteriophage φ31 (27). This fragment was shown to have a baseline level of constitutive activity before phage infection of the host. The amino acid sequence corresponding to the complete open reading frame (ORF2) present on the fragment is shown below the sequence. The Shine-Dalgarno sequence for ORF2 is underlined. The putative helix-turn-helix DNA binding motif is shaded. The PCR primers used in the 5′ and 3′ deletion analysis are marked over the sequence. Primers used in the 5′ deletion analysis are designated by the number of the first nucleotide, while primers used in the 3′ deletion analysis are designated by the number of the final nucleotide. These 3′ primers (3′-792, 3′-826, 3′-849, and 3′-862) are complementary to the sequence shown. (B) Sequence of the tightly regulated phage promoter P566–888. The phage-inducible transcription start sites (703 and 744) are marked by vertical arrows. The consensus −10 promoter sequences are boxed. No consensus −35 sequences were observed for either start site. Instead, inverted or direct repeats were observed in the −35 regions for both start sites. The critical region for activation by phage φ31 (between nucleotides 648 and 658) contained a pair of inverted repeats, marked by solid horizontal arrows. The inverted repeat downstream of the 703/744 start sites is marked by broken horizontal arrows. Small, leftward arrows above the sequence mark the positions of the subclones used to determine the importance of the downstream region (subclones 566–826, 566–849, and 566–862).
These various subclones (fragments) were then tested for their ability to drive β-Gal expression upon phage φ31 infection. The results of β-Gal activity assays performed on L. lactis subsp. lactis NCK203 over the course of a phage φ31 infection cycle are shown in Fig. 2. A very low level of constitutive expression (approximately 20 β-Gal units) was obtained from subclone 1–305. This low level of activity was not affected by infection with phage φ31. As shown in Fig. 2, the phage-inducible promoter activity was fully associated with tandem start sites 703/744. The highest level of activity for the subcloned fragments was obtained with subclone 566–888, which contains both of the tandem 703/744 start sites. In addition, the constitutive activity associated with the original P15A10 fragment (bp 1 to 888) was eliminated in subclone 566–888. A very low level of activity was obtained before phage infection when the fragment with start site 703 was subcloned separately (approximately 10 β-Gal units) (Fig. 2). After phage infection, the fragment with start site 703 alone yielded 33 to 50% of the activity obtained with the fragment with tandem start sites 703/744. No activity was obtained with the fragment with start site 744 alone. Hybridization of RNA isolated from subclones 566–888 and 566–732 at various time points in the phage φ31 infection cycle with 32P-labeled lacZ.st confirmed the absence of lacZ.st mRNA before phage infection (time 0) (Fig. 2). As expected, no lacZ.st mRNA was evident for subclone 687–888 at any time point (data not shown). Primer extension analysis performed on RNA isolated from subclones 566–888 and 566–732 revealed transcription starts at positions 703/744 and at position 703 alone, respectively (data not shown). On the 566–888 subclone, these transcription starts were evident only after phage infection, in contrast to the slight constitutive activity observed previously with start site 703 on P15A10 (27). The subclone 566–732 primer extension results also confirmed that any possible promoter activity from the start site 744 region had been eliminated in subclone 566–732 (data not shown).
FIG. 2.
(A) β-Gal activity results of the five P15A10 regions subcloned into the promoter screening vector pTRK390. Time 0 is immediately before the addition of phage φ31 (cells at OD600 of ≅0.5). β-Gal assays were performed at least three different times. For each assay, time point determinations were performed in duplicate. (B) Northern analysis of RNA hybridized with a 32P-labeled lacZ.st probe. Northern analysis was not performed on subclones 1–305 and 442–574 because phage-inducible activity was associated exclusively with start sites 703 and 744.
Analysis of ORF2 as a transcriptional activator.
The original phage-inducible promoter fragment P15A10 yielded approximately 200 to 250 β-Gal units before phage infection (27). The lack of substantial promoter activity from subclones 1–305 and 442–574 (Fig. 1 and 2), combined with the virtual loss of the constitutive activity associated with start site 703 after deletion of nucleotides 1 to 565, prompted the study of the function of ORF2 (Fig. 1) (present upstream of start sites 703/744) in transcriptional activation of P566–888 ORF2 (coding for 143 amino acids) contains its own ribosome binding site, but analysis of the sequence upstream did not reveal a consensus promoter region (27). A search for amino acid or nucleotide sequence similarities, by using BLAST (2), revealed significant homology between ORF2 and ORF25 on the temperate lactococcal bacteriophage φr1t (99% homology) (37). The function of this ORF in φr1t was not determined previously. A search for a possible helix-turn-helix DNA binding motif was conducted by the method of Dodd and Egan (13). One area of ORF2 (corresponding to amino acid positions 108 to 129) rated a 2.5, indicating that a 25% probability existed for a helix-turn-helix motif (Fig. 3A). The putative helix-turn-helix motif is conserved in both ORF2 (φ31) and ORF25 (φr1t).
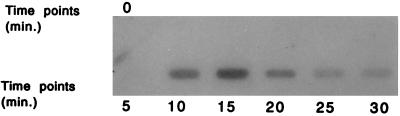
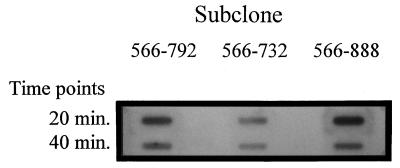
Northern slot blot analysis (Fig. 4) using 32P-labeled ORF2 as a probe showed that mRNA for ORF2 was present 10 min after infection of L. lactis subsp. lactis NCK203 with phage φ31. Levels of ORF2 mRNA peaked at 15 min and were barely detectable 30 min after phage infection. These results correlated very well with the observed induction of lacZ.st mRNA from the phage-inducible promoter. As indicated in Fig. 2, the mRNA level for lacZ.st reached a peak 20 min after phage infection and decreased by 40 min into the lytic cycle. These results suggested that control of promoter activity from P566–888 could be partly regulated by levels of the ORF2 gene product during the phage infection.
FIG. 4.
ORF2 mRNA levels during a phage φ31 lytic cycle of the sensitive host, L. lactis subsp. lactis NCK203. Time 0 is immediately before phage infection (cells at OD600 of ≅0.5).
Mutagenesis of ORF2.
To test the relationship between ORF2 and expression from start sites 703/744, attempts were made to evaluate the activity of ORF2 under control of the llaI constitutive promoter in a high-copy-number vector in L. lactis. However, deletions of the promoter region always occurred in the transformants isolated. Therefore, the Shine-Dalgarno sequence and ATG start codon of ORF2 were modified by site-directed mutagenesis to create P15A10/mutORF2. All changes made to the 5′ region of ORF2 are shown in Fig. 5. DNA sequencing of the entire 888-bp fragment revealed no other mutations. A BamHI fragment containing P15A10/mutORF2 was cloned upstream of lacZ.st in pTRK390, and β-Gal assays were performed on the lactococcal strain containing this clone. The results (Fig. 5) showed that no activity was observed before phage infection after the ORF2 translational signals were eliminated. Following phage infection, P15A10/mutORF2 was still induced, but levels were approximately 25% less than those achieved with P15A10 carrying a functional ORF2.
Effect of an antisense construct of ORF2.
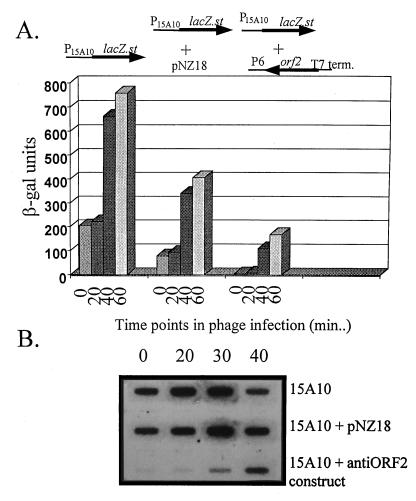
To reverse the effects of ORF2, an antisense construct of ORF2 was assembled and introduced into the host carrying pTRK391 (P15A10::lacZ.st). ORF2 was cloned in an antisense orientation under control of the strong Lactobacillus promoter, P6 (10), in pNZ18. The entire, wild-type ORF2 coding region was used in this construct. To stop transcription and allow production of a small antisense mRNA transcript, the T7 terminator (TT7) was amplified by PCR from the E. coli expression vector pET28a (Novagen) and cloned in a position after the P6::antisense ORF2 (P6::anti-ORF2) cassette in pNZ18, thereby generating pTRK479. pTRK479, containing P6::anti-ORF2::TT7, was combined with pTRK391 in the lactococcal host, NCK203. As a control, pNZ18 was combined with pTRK391 in NCK203. Reverse transcription-PCR confirmed that an antisense ORF2 transcript was produced when the P6::anti-ORF2::TT7 cassette in pTRK479 was present (data not shown).
Results of β-Gal assays are shown in Fig. 6A. Surprisingly, pNZ18 alone had a negative impact on β-Gal activity, reducing levels twofold. Nevertheless, before phage infection, the antisense construct significantly reduced β-Gal activity to levels below detection. After phage infection, β-Gal activity was reduced to about one-third the level obtained with pTRK391 plus pNZ18. By using 32P-labeled lacZ.st probes, Northern dot blot analysis of RNA isolated at various time points in the phage lytic cycle showed a marked reduction in lacZ.st mRNA when the antisense construct was present with pTRK391 (Fig. 6B). Interestingly, the negative effect of pNZ18 alone on β-Gal activity was not at the level of transcription (Fig. 6B).
FIG. 6.
Effect of an antisense construct of ORF2 on activation of P15A10. ORF2 was amplified from P15A10 by using one primer consisting of nucleotides 200 to 221 and one primer complementary to nucleotides 640 to 655 and was cloned behind the strong, constitutive P6 promoter (10) in pNZ18. The T7 terminator was cloned behind the P6::anti-ORF2 cassette. The T7 terminator was amplified from the E. coli expression vector pET28a (Novagen) by using a 5′ primer consisting of 5′-GAGAAGCCCGAAAGGAAGC-3′ and a 3′ primer consisting of 5′-ATCCGGATATAGTTCCTC-3′. (A) β-Gal activity when the antisense construct was combined with pTRK391 (P15A10::lacZ.st) (27) in L. lactis subsp. lactis NCK203 both before and after phage infection. β-Gal levels reported are the average of assays performed at least three separate times. For each assay, time point determinations were performed in duplicate. (B) Slot blot Northern analysis of RNA isolated at various points in the φ31 lytic cycle and probed with 32P-labeled lacZ.st.
The EOP of φ31 was evaluated on the lactococcal host carrying pTRK479 to determine whether the initial reduction in ORF2 could retard phage development. NCK203(pNZ18) was used as the control strain in determining EOP. No decrease in the plaquing ability of phage φ31 on NCK203(pTRK479) was observed (data not shown).
Activation of P566–888 by ORF2 in E. coli.
These data provided strong evidence for the role of ORF2 in transcriptional activation. However, the evidence would be more compelling if ORF2 could activate P566–888 when provided in trans on a compatible vector. As described above, this experiment could not be performed with Lactococcus because intact clones with constitutively expressed ORF2 could not be isolated on a high-copy-number vector. Therefore, the experiment was performed with E. coli. ORF2 was amplified from pTRK391 by using a primer consisting of nucleotides 219 to 243 (5′ NdeI site) and a primer complementary to nucleotides 628 to 650 (5′ HindIII site) and cloned under control of the T7 promoter in the E. coli expression vector pET28a (Novagen). The pET::ORF2 construct was combined with pTRK477, which contains the P566–888::lacZ.st cassette, in the E. coli host BIL21 (DE3). The DE3 lysogen contains T7 RNAP under control of Plac. Induction of T7 RNAP with IPTG (isopropyl-β-d-thiogalactopyranoside) would lead to efficient expression of ORF2 from the T7 promoter. β-Gal assays could then be used to monitor subsequent activation of P566–888 by ORF2. The results (Table 2) showed that induction of ORF2 expression in E. coli resulted in β-Gal levels three times greater than those obtained with the control strain (pET28a plus P566–888::lacZ.st).
TABLE 2.
Activation of P566–888::lacZ.st by ORF2 provided in trans in E. coli
| Constructs (in E. coli) | Time point (min) | Activity (β-Gal units) |
|---|---|---|
| pET28A plus P566–888 (control) | 0 | 69 |
| 60 | 172a | |
| 120 | 185a | |
| pET::ORF2 plus P566–888 | 0 | 73 |
| 60 | 380 | |
| 120 | 684 |
The increase in β-Gal activity in this case represents the stalling of growth caused by induction of E. coli BIL21 (DE3) with IPTG. β-Gal units are expressed per OD600 of the cell culture. Inhibition of growth causes a slight rise in β-Gal levels.
Ability of ORF2 to bind P566–888 and P566–732.
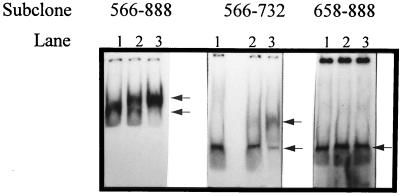
As a confirmation of the role of ORF2 as a transcriptional activator, the ability of ORF2 to bind the promoter element was assessed by gel retardation assays. Due to problems encountered in overproducing and purifying ORF2 with the E. coli expression vector pET28a (Novagen), ORF2 was produced in the single-tube protein system (T7), an in vitro transcription-translation kit available from Novagen. Translation is accomplished in this system by using an extract from rabbit reticulocyte cells. To increase the efficiency of translation of ORF2 in this eukaryotic-based translation system, ORF2 was subcloned from pET28a::ORF2 into pCITE4a (Novagen) and then produced in accordance with the manufacturer’s directions. As a control, the single-tube protein system protocol was used with pCITE4a containing no insert. The ORF2 product (or control) was mixed with 32P-labeled fragment 566–888 or 566–732 and then separated on a polyacrylamide gel as described in Materials and Methods. Results (Fig. 7) showed that the bands for both fragments 566–888 and 566–732 were shifted when ORF2 was present. When competitor DNA was present (unlabeled fragment 566–888), the degree of shifting was altered, confirming that ORF2 was indeed binding specifically to both fragments 566–888 and 566–732.
FIG. 7.
Results of gel retardation assays performed with the ORF2 gene product. The labeled fragments used in the gel retardation assay are indicated at the top of the gels. For each fragment, lane 1 represents the control (no ORF2 added), lane 2 represents the effects of nonlabeled, competitive DNA on DNA binding (unlabeled subclone 588–888 added with ORF2), and lane 3 represents the ability of ORF2 to bind each fragment with no unlabeled 566–888 fragment present. The arrows to the right of each panel indicate the shift in mobility of each fragment upon addition of ORF2. For all three fragments, the ORF2 gene product was added from the same in vitro transcription-translation tube to ensure that the amount was identical between reactions.
Deletion analysis of P566–888.
To determine the minimum sequence required upstream of start sites 703/744 for promoter activation, a series of deletions was made from the 5′ end of fragment 566–888. PCR was used to amplify five new fragments from fragment 566–888, each starting at a different nucleotide (nucleotide 599, 633, 648, 658, or 667) and all ending at the last position, 888. The fragments were cloned into pTRK390 and evaluated for β-Gal activity after phage infection of the lactococcal host, NCK203. Phage inducibility was retained for subclones 599–888, 633–888, and 648–888 and lost in subclone 658–888. Therefore, the critical region was between nucleotides 648 and 658 (data not shown). Gel retardation assays performed on fragment 658–888 as described above showed that ORF2 was not able to bind this fragment (Fig. 7).
A series of deletions was made from the 3′ end of fragment 566–888 to evaluate the importance of sequences downstream of start sites 703/744. Subclones were constructed as described above to incorporate the regions of fragments 566–862, 566–826, and 566–792. Interestingly, a 50 to 65% loss in β-Gal activity levels resulted upon deletion of nucleotides between positions 862 and 826 (Fig. 8). This loss in activity could be explained only partly by a decrease in transcription. While mRNA levels for lacZ.st were reduced in subclones 566–792 and 566–826 when compared to that in subclone 566–888, they were still higher than levels obtained for subclone 566–732 containing only start site 703 (Fig. 9). These data suggest that sequences downstream of start sites 703/744 are required for optimal expression from the phage φ31 promoter.
FIG. 8.
Importance of sequences downstream of start sites 703/744 in promoter function. The β-Gal levels shown are for the time point 60 min after infection with phage φ31, just before cell lysis. The wild-type, downstream inverted repeat (hatched rectangles with inverted arrows) and the mutated, downstream region that disrupted the inverted repeat (solid rectangles) are indicated.
FIG. 9.
Slot blot analysis of RNA isolated at various time points in the phage φ31 lytic cycle of subclones 566–888, 566–792, and 566–732 and probed with 32P-labeled lacZ.st. lacZ.st mRNA was not detectable before phage infection (time 0; see Fig. 2B).
Determination of the importance of downstream sequences on β-Gal activity.
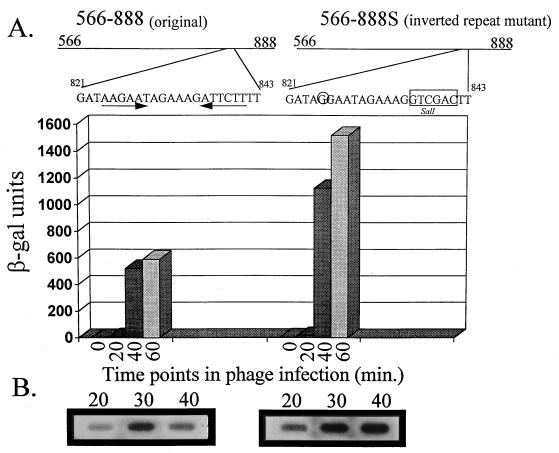
Examination of the sequence between nucleotides 826 and 862 identified an inverted repeat (nucleotides 824 to 841) (Fig. 3B). To determine if this inverted repeat played a role in promoter activity, PCR was used to amplify nucleotides 566 to 849. This PCR product was subcloned into the BamHI site in the promoter screening vector pTRK390. As with subclones 566–826 and 566–792 described above, subclone 566–849 showed a 50 to 65% reduction in β-Gal activity, indicating that the inverted repeat alone did not allow efficient expression of β-Gal (Fig. 8). To gain a better understanding of the importance of this inverted repeat within the context of the whole sequence, the inverted repeat was modified by site-directed mutagenesis to yield a new fragment, designated 566–888S (described in the legend to Fig. 10). Fragment 566–888S was cloned into the BamHI site in pTRK390, and the resulting vector was transformed into L. lactis subsp. lactis NCK203. The mutation did not decrease β-Gal activity as expected but rather caused a twofold increase in activity (Fig. 8 and 10). Northern dot blot analysis of RNA isolated from this clone showed that the increase in enzyme activity was due to an increase in transcription of lacZ.st (Fig. 10).
FIG. 10.
Effects of mutagenizing the downstream inverted repeat (nucleotides 824 to 841) on β-Gal activity and lacZ.st mRNA levels. A site-directed mutation to eliminate the inverted repeat was constructed by amplification of the 566–888 fragment into two separate fragments. Fragment 566–835 was amplified by using the universal −40 primer (on pTRK391) and a primer complementary to nucleotides 817 to 835 (A→G mutation at nucleotide 825). Fragment 842–888 was generated by using a primer consisting of nucleotides 842 to 858 and the lacZ primer (on pTRK391). Nucleotides 836 to 841 were replaced with a SalI site, which allowed fusion of the two fragments (566–835SalI and SalI842–888) to yield fragment 566–888S. Mutations made to the inverted repeat are indicated above the graph. β-Gal levels are the average of at least three separate assays. For each assay, time point determinations were performed in duplicate.
To further evaluate the downstream region, PCR was used to generate two new fragments from the 566–888S subclone: 566–841S and 566–862S. Both fragments contained the mutated inverted repeat, but only 566–862S contained sequences just downstream of the repeat. These fragments were cloned into the BamHI site of pTRK390. β-Gal assays were performed to determine what effect these deletions had on enzyme activity. The results (Fig. 8) showed that when the mutated inverted repeat was included without the regions just downstream (fragment 566–841S), β-Gal activity was increased twofold in comparison to that of subclone 566–849 (wild-type inverted repeat with no downstream sequences). Inclusion of sequences just downstream from the mutated inverted repeat (fragment 566–862S) resulted in an additional two- to threefold increase in enzyme activity.
Construction of an improved expression vector based on P566–862S.
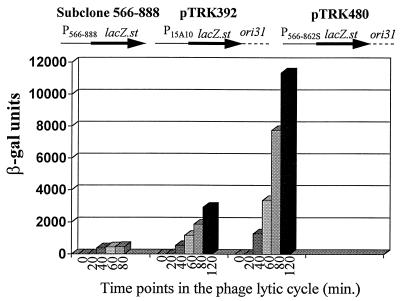
Because of the increased activity from the inverted repeat mutant, a new expression vector was constructed by replacing P15A10 on pTRK392 (27) with promoter fragment 566–862S (P566–862S). Because pTRK392 did not have convenient restriction sites for easy replacement of the promoter, the improved expression vector was constructed by cloning a partial AviII/partial SalI fragment containing P566–862S::lacZ.st into the SalI/NruI site of pTRK360 (pSA3::ori31) (27). The resulting vector (pTRK480) was transformed into L. lactis subsp. lactis NCK203 and tested for β-Gal activity. As shown in Fig. 11, levels of activity were significantly increased to 10,000 β-Gal units and higher, compared with the 3,000 β-Gal units obtained with pTRK392.
FIG. 11.
β-Gal activity of the new expression vector, pTRK480, compared to those of pTRK392 (27) and subclone 566–888 (pTRK477). Subclone 566–888 was constructed in the high-copy-number promoter screening vector, pTRK390. pTRK392 is based on the low-copy-number replicon pSA3 and contains the phage φ31 origin of replication (ori31) (27) and lacZ.st under the control of P15A10. The new expression vector was constructed by replacing P15A10 with P566–862S. The β-Gal results shown represent the averages of at least three different assays, with each time point determination being performed in duplicate.
DISCUSSION
In this study, we have described the molecular characterization of a lactococcal phage-inducible promoter from the lytic phage φ31. The original promoter was isolated as an 888-bp fragment (P15A10) which showed some constitutive β-Gal activity (200 to 300 U) but which was induced three- to fourfold upon infection of the host with phage φ31 (27). Detailed molecular analysis of P15A10 in this study defined a tightly regulated phage promoter (P566–888) which contains two phage-inducible transcription start sites, corresponding to nucleotides 703 and 744. Deletion analysis was used to determine which sequences upstream and downstream of the 703/744 start sites were important for promoter function. In addition, the function of a complete open reading frame (ORF2) upstream of the 703/744 start sites was established as a positive transcriptional regulator of the P566–888 promoter. To our knowledge, this is the first positive transcriptional activator described and characterized in a lactococcal bacteriophage.
Analysis of the P566–888 sequence revealed the presence of −10 consensus promoter regions upstream of both start sites (Fig. 3); however, no strong −35 consensus regions could be identified for either start site. Instead, an inverted repeat and a direct repeat were identified upstream of start sites 703 and 744, respectively (Fig. 3). This lack of a −35 consensus region is not surprising, since many regulated promoters which lack canonical −35 regions have been identified (21, 30, 40). In many cases, inverted or direct repeats present in this region may act as binding sites for transcriptional activators of the promoter element (21, 40). The start site 703 associated promoter proved active on its own (P566–732), yielding approximately 50% of the activity achieved with P566–888. The fragment containing start site 744, however, was not inducible on its own (subclone 687–888). At first glance, it appears from these results that nucleotide 744 may not actually be a transcription start site but is, instead, for example, an RNA processing site. However, Fig. 9 shows that the levels of lacZ.st mRNA obtained during phage φ31 infection of P566–732 are lower than the levels obtained with P566–792 and P566–888. The RNA data suggest that nucleotide 744 does act as a separate transcription start site.
Deletion analysis for the 5′ end of P566–888 showed that the critical region for phage inducibility was between nucleotides 648 and 658 (subclone 648–888 was inducible by phage φ31 while subclone 658–888 was not). Located within this region was part of the 28-bp imperfect inverted repeat centered approximately 35 bp upstream of start site 703 (Fig. 3). It is interesting to note that this region appears important for activation of both transcription start sites and may act as a binding site for activation of P566–888. The inability of the transcriptional activator, ORF2, to bind fragment 658–888 in gel shift assays corroborates this role for the upstream inverted repeat (Fig. 7).
Interestingly, start site 703 alone proved to be slightly leaky, yielding approximately 10 to 15 β-Gal units before phage infection. In contrast, the combination of start sites 703 and 744 was more tightly regulated. β-Gal activity levels from P566–888 were below the limit of detection before phage infection, as was the level of lacZ.st mRNA. The “tightness” of this promoter element was confirmed in a separate study by Djordjevic et al. (11). In this study, a novel phage defense system was constructed by cloning the 703/744 start site promoter (φ31P; corresponding to fragment 566–804) upstream of a lethal gene (the LlaI restriction cassette) in a high-copy-number vector. Upon phage infection, induction of the promoter resulted in production of the restriction enzyme LlaI, killing the host and the phage and preventing phage proliferation. With this system, the EOP of φ31 was reduced to approximately 10−4. However, in the absence of phage infection, the construct was successfully maintained in Lactococcus. Therefore, P566–888 could be an important component of an expression system used to produce proteins or enzymes which may be relatively toxic to L. lactis.
Constitutive activity associated with the original phage-inducible promoter, P15A10, was associated with transcription start site 703. Primer extension analysis showed a faint product initiating at nucleotide 703 before phage infection (27). Loss of the constitutive activity associated with P15A10 upon deletion of nucleotides 1 to 565, coupled with the lack of substantial promoter activity from subclones upstream of start sites 703/744, suggested that some upstream element present on P15A10 was enabling the nucleotide 703-associated promoter to be expressed before phage infection. This element was subsequently identified as ORF2, which contained a putative helix-turn-helix DNA binding motif. Helix-turn-helix motifs were recognized as important DNA recognition motifs for many transcriptional regulators, including the λ cI (17) and λ cII (16) proteins and the Mu C protein, which regulates late transcription of phage Mu (3). In these examples, the promoters utilizing these transcriptional activators lack consensus −35 sequences, and the proteins recognize DNA sequences within or just upstream of the −35 region. Again, the loss of the ability of ORF2 to bind fragment 658–888, which contains the putative −10 region upstream of the 703 start site but lacks part of the upstream inverted repeat centered 35 bp upstream of start site 703 (Fig. 3B), substantiates the role of ORF2 as a transcriptional activator and may suggest that the putative helix-turn-helix motif present in ORF2 is important for its DNA binding function. The importance of this region in ORF2, however, remains to be determined.
In addition to demonstrating the ability of ORF2 to bind the phage promoter element (P566–888 and P566–732) (Fig. 7), three main approaches were used to establish ORF2’s role as a transcriptional activator of P566–888. When translation of ORF2 was eliminated by site-directed mutagenesis of its Shine-Dalgarno sequence and ATG start codon on pTRK391 (P15A10::lacZ.st), the constitutive activity associated with P15A10 was lost. Accordingly, β-Gal activity could be detected only after infection of the lactococcal host with phage φ31. ORF2 was also capable of activating P566–888::lacZ.st when provided in trans in E. coli. These results show that the ORF2 product is able to work, at least on some level, with the host transcription machinery in both L. lactis and E. coli. In addition, an antisense construct of ORF2 significantly reduced both the level of lacZ.st mRNA and the level of β-Gal activity associated with pTRK391 (P15A10::lacZ.st), before and after phage infection. It is important to note that the presence of the T7 terminator cloned in a position after anti-ORF2 was critical to its antisense activity. In the absence of the T7 terminator, the amount of lacZ.st mRNA was only partially decreased and β-Gal activity was not affected at all (data not shown). The terminator causes production of small, antisense transcripts, which may be more likely to target the mRNA of interest with little nonspecific binding to other targets. Along the same line, the presence of a long, nonspecific RNA tail due to lack of proper termination could cause the formation of secondary structure, possibly inhibiting the antisense transcript from functioning properly. Finally, the terminator may stabilize the transcript, making it less susceptible to RNase attack.
Collectively, these data demonstrate that ORF2 is indeed a transcriptional activator of this phage φ31 middle promoter and is designated tac (transcriptional activator). To our knowledge, ORF2 represents the first transcriptional activator isolated from a lactococcal bacteriophage. This important role for ORF2 prompted us to determine whether an antisense construct of ORF2 would have any effect on phage proliferation. It has been shown previously that using antisense technology to target phage structural proteins has little effect on phage proliferation, mainly due to the excess production of these proteins by the phage (5, 24). The use of antisense technology to target a middle transcriptional activator might be more effective if this activator was produced in more limiting amounts. As stated above, the antisense construct significantly inhibited lacZ.st transcription from pTRK391, even after phage infection. Unfortunately, the presence of anti-ORF2 behind a strong, constitutive promoter on a high-copy-number vector had no effect on the proliferation or EOP of phage φ31. Several possible explanations for these results exist. First, the amount of ORF2 mRNA produced by phage φ31 during the infection process may be higher than the amount of anti-ORF2 mRNA expressed, so that the antisense construct was unable to disrupt phage development. Alternatively, if certain phage-encoded factors were able to inhibit some host promoters, transcription of anti-ORF2 may also be affected. Although it was shown through RT-PCR that an anti-ORF2 mRNA was present both before and after phage infection, no effort was made to quantify the amount of transcript present or to measure its stability. It is possible, therefore, that the antisense ORF2 transcript was not maintained at a high enough level to prevent phage proliferation. Finally, as stated above, certain late phage proteins, such as the structural proteins, are produced in excess during the lytic cycle. In this case, a decrease in the levels of ORF2 may not affect the overall ability of phage φ31 to infect L. lactis subsp. lactis NCK203.
A separate study by Djordjevic and Klaenhammer (12) showed that the strength of promoter induction could be affected by mutations in ORF2. In this study, mutant phages were isolated that activated P566–888 to only 50% of the level obtained with wild-type phage φ31, as measured by β-Gal activity assays. Sequence analysis of four of these mutant phages identified an identical amino acid change (F142→L) in the carboxy-terminal region corresponding to ORF2. Although this change appears to have altered the effectiveness of ORF2, no decrease in the ability of the mutant phages to plaque efficiently on the native lactococcal host, NCK203, was observed.
In addition to the importance of sequences upstream of the 703/744 start sites, at least two regions were located downstream of the 703/744 start sites which regulated the level at which the P566–888 promoter was induced. First, an inverted repeat encompassing nucleotides 824 to 841 (Fig. 3B) may function in down-regulation of the promoter element. Site-directed mutagenesis of this inverted repeat to yield P566–888S resulted in a twofold increase in β-Gal activity. This increase was at the level of transcription. The true function of this site is as yet unknown, but it is possible that the inverted repeat may act as a binding site for some phage-borne factor, leading to down-regulation of transcription of this region of the phage genome. A combination of P566–888S::lacZ.st (mutated inverted repeat) with pET28a::ORF2 in E. coli BIL21 (DE3) did not result in increased β-Gal expression in E. coli when compared to the combination of P566–888::lacZ.st and pET28a::ORF2 described in Results (data not shown). These results suggest two things. First, ORF2 does not appear to be involved in the transcriptional regulation observed at this site. If it were, one might expect to see the same type of increase in β-Gal expression in E. coli as that observed during the lytic cycle in L. lactis subsp. lactis NCK203. The inability of ORF2 to bind fragment 658–888 strengthens the view that ORF2 does not act at this downstream inverted repeat. Second, at least in E. coli, the mutated site itself is not responsible for increased β-Gal activity. Indeed, primer extension analysis performed on RNA isolated during a phage infection of NCK203 carrying P566–888S::lacZ.st did not reveal any new transcription start sites that could explain the increase in transcription (data not shown). Therefore, it appears likely that another phage-borne factor is acting at this position.
In addition to the inverted repeat, a second region of importance was identified. At least part of the region downstream of the inverted repeat is required for optimal activation of the phage φ31 promoter. As indicated in Fig. 8, β-Gal activity increased twofold or more when the mutated inverted repeat was combined with adjacent downstream sequences (P566–841S versus P566–862S and P566–888S). Northern slot blot results shown in Fig. 9 indicate that the sequences 3′ to the downstream inverted repeat may act at least partly at the level of transcription. In addition, steady-state lacZ.st mRNA levels for P566–888S and P566–862S were higher than those achieved with P566–841S (data not shown). The increase in activity levels between P566–862S and P566–888S (Fig. 8) were surprising, especially since β-Gal activity levels from P566–862 were slightly lower than those achieved with P566–888 (Fig. 2). The reason for this difference was not determined. Northern analysis revealed that the region downstream of the inverted repeat did not function to increase the stability of the mRNA (data not shown). Therefore, the functions of both the downstream inverted repeat and the sequences immediately downstream remain to be determined.
The increase in promoter activity from the combination of the mutated inverted repeat with the downstream sequences was confirmed in a separate study by Djordjevic and Klaenhammer (12). The improved promoter P566–888S was used to replace P566–804 in the novel phage defense strategy described above. Replacement of the original phage promoter resulted in a further 2.2-fold reduction in EOP and a dramatic reduction in plaque size and appearance.
The promoter element yielding the highest level of activity in the high-copy-number promoter screening vector (P566–862S) was used to replace P15A10 in the expression vector pTRK392 (27). Replacement resulted in a four- to fivefold increase in β-Gal activity. These data reiterate the importance of both phage elements, a phage-inducible promoter and a phage ori, to the expression vector. The importance of the origin of replication (ori31) was previously shown by O’Sullivan et al. (27). A low-copy-number version of P15A10::lacZ.st without ori31 yielded an activity level of only 85 β-Gal units. Inclusion of ori31 resulted in activity levels of 2,500 to 3,000 β-Gal units within 2 h of phage infection of the host. Now, by site-directed improvements in the promoter element, an activity level of close to 11,000 β-Gal units can be obtained after phage infection. Further molecular characterization could possibly lead to improvements in expression levels from the expression vector as well as to greater understanding of gene regulation in lactococcal bacteriophages. We are currently exploring the use of this improved expression system for the production of other heterologous proteins and enzymes.
ACKNOWLEDGMENTS
This research was supported by NRICGP projects 92-37500-8018 and 97-35503-4368, the Southeast Dairy Foods Research Center, Dairy Management, Inc., and Rhone-Poulenc, Inc. Shirley Walker was supported by a USDA-GAANN Biotechnology Fellowship.
We thank Gordana Djordjevic, Evelyn Durmaz, Gwen Allison, and David Mills for helpful discussions and for critical review of the manuscript and Eric Miller and Mark Conkling for their insights into alternative methods for overproduction of ORF2. We also thank Evelyn Durmaz for her technical assistance with the photographs.
Footnotes
Paper FSR 97-32 of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Alatossava T, Klaenhammer T R. Molecular characterization of three small isometric-headed bacteriophages which vary in their sensitivity to the lactococcal phage resistance plasmid pTR2030. Appl Environ Microbiol. 1991;57:1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Bölker M, Wulczyn F G, Kahmann R. Role of bacteriophage Mu C protein in activation of the mom gene promoter. J Bacteriol. 1989;171:2019–2027. doi: 10.1128/jb.171.4.2019-2027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 5.Chung D K, Chung S K, Batt C A. Antisense RNA directed against the major capsid protein of Lactococcus lactis subsp. cremoris bacteriophage F4-1 confers partial resistance to the host. Appl Microbiol Biotechnol. 1992;37:79–83. doi: 10.1007/BF00174207. [DOI] [PubMed] [Google Scholar]
- 6.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vos W M. Gene cloning and expression in lactic streptococci. FEMS Microbiol Rev. 1987;46:281–295. [Google Scholar]
- 9.Dinsmore P K, Klaenhammer T R. Molecular characterization of a genomic region in a Lactococcus bacteriophage that is involved in its sensitivity to the phage defense mechanism AbiA. J Bacteriol. 1997;179:2949–2957. doi: 10.1128/jb.179.9.2949-2957.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djordjevic G M, Bojovic B, Miladinov N, Topisorovic L. Cloning and molecular analysis of promoter sequences isolated from the chromosomal DNA of Lactobacillus acidophilus ATCC4356. Can J Microbiol. 1997;43:61–69. doi: 10.1139/m97-009. [DOI] [PubMed] [Google Scholar]
- 11.Djordjevic G M, O’Sullivan D J, Walker S A, Conkling M A, Klaenhammer T R. Triggered-suicide system designed for bacteriophage defense of Lactococcus lactis. J Bacteriol. 1997;179:6741–6748. doi: 10.1128/jb.179.21.6741-6748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djordjevic G M, Klaenhammer T R. Bacteriophage-triggered defense systems: phage adaptation and design improvements. Appl Environ Microbiol. 1997;63:4370–4376. doi: 10.1128/aem.63.11.4370-4376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Hill C, Pierce K, Klaenhammer T R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction/modification (R/M) and abortive infection (HSP+) Appl Environ Microbiol. 1989;55:2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho Y S, Wulff D L, Rosenberg M. Bacteriophage λ protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature (London) 1983;303:703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- 17.Hochschild A, Irwin N, Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983;32:319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- 18.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Israelsen H, Madsen S M, Vrang A, Hansen E B, Johansen E. Cloning and partial characterization of regulated promoters from Lactococcus lactis TN917-lacZ integrants with the new promoter probe vector, pAK80. Appl Environ Microbiol. 1995;61:2540–2547. doi: 10.1128/aem.61.7.2540-2547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis A W, Klaenhammer T R. Bacteriophage resistance conferred on lactic streptococci by the conjugative plasmid pTR2030: effects on small isometric-, large isometric-, and prolate-headed phages. Appl Environ Microbiol. 1986;51:1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kok J. Inducible gene expression and environmentally regulated genes in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:129–145. doi: 10.1007/BF00395930. [DOI] [PubMed] [Google Scholar]
- 22.Kuipers O P, Beerthuyzen M M, de Ruyter P G G A, Luesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 24.Moineau S, Durmaz E, Pandian S, Klaenhammer T R. Differentiation of two abortive mechanisms by using monoclonal antibodies directed toward lactococcal bacteriophage capsid proteins. Appl Environ Microbiol. 1993;59:208–212. doi: 10.1128/aem.59.1.208-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nauta A, van Sinderen D, Karsens H, Smit E, Venema G, Kok J. Inducible gene expression mediated by a repressor-operator system isolated from Lactococcus lactis bacteriophage r1t. Mol Microbiol. 1996;19:1331–1341. doi: 10.1111/j.1365-2958.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 26.O’Sullivan D J, Hill C, Klaenhammer T R. Effect of increasing the copy number of bacteriophage origins of replication, in trans, on incoming-phage proliferation. Appl Environ Microbiol. 1993;59:2449–2456. doi: 10.1128/aem.59.8.2449-2456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Sullivan D J, Walker S A, West S G, Klaenhammer T R. Development of an expression strategy using a lytic phage to trigger explosive plasmid amplification and gene expression. Bio/Technology. 1996;14:82–87. doi: 10.1038/nbt0196-82. [DOI] [PubMed] [Google Scholar]
- 28.O’Sullivan D J, Klaenhammer T R. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Sullivan D J, Klaenhammer T R. High and low copy number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 30.Parreira R, Valyasevi R, Lerqayer A L S, Ehrlich S D, Chopin M-C. Gene organization and transcription of a late-expressed region of a Lactococcus lactis phage. J Bacteriol. 1996;178:6158–6165. doi: 10.1128/jb.178.21.6158-6165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sanger G, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus β-galactosidase sequences. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 34.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 35.Terzaghi B, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rooijen R J, de Vos W M. Characterization of the Lactococcus lactis lactose operon promoter: contribution of flanking sequences and LacR repressor to promoter activity. J Bacteriol. 1992;174:2273–2280. doi: 10.1128/jb.174.7.2273-2280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Sinderon D, Karsens H, Kok J, Terpstra P, Ruiters M H J, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 38.Wells J M, Wilson P W, Norton P M, Gasson M J, Le Page R W F. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol. 1993;8:1155–1162. doi: 10.1111/j.1365-2958.1993.tb01660.x. [DOI] [PubMed] [Google Scholar]
- 39.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 40.Ye Z-H, Lee C Y. Cloning, sequencing, and genetic characterization of regulatory genes, rinA and rinB, required for the activation of staphylococcal phage φ11 int expression. J Bacteriol. 1993;175:1095–1102. doi: 10.1128/jb.175.4.1095-1102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]