Abstract
The gut microbiome has coevolved with humans to aid in physiologic functions and prevent disease. An increasing prevalence of gut dysbiosis in modern society exists and has strong linkages to multiple disease processes common in the developed world. Mechanisms for microbiome-human interactions that impact host homeostasis include bacterial metabolite/toxin production, biofilm formation with mucous layer infiltration, and host immune system modulation. Most of this crosstalk occurs at the epithelial layer of the gut, and as such the role of these interactions in the induction of colorectal cancer—a highly prevalent disease globally and one undergoing significant epidemiologic shifts—is under increasing scrutiny. Although multiple individual gut bacteria have been hypothesized as possible driver organisms in the oncogenic process, no bacterium has been definitively identified as a causal agent of colorectal cancer, suggesting that host lifestyle factors, microbiome community interactions, and the mucosal and/or systemic immune response may play a critical role in the process. Recent evidence has emerged implicating the ubiquitous human pathogen Clostridioides difficile as a possible promoter of colorectal cancer through chronic toxin-mediated cellular changes. Although much remains to be defined regarding the natural history of infections caused by this pathogen and its potential for oncogenesis, it provides a strong model for the role of both individual bacteria and of the gut microbial community as a whole in the development of colorectal cancer.
Keywords: microbiome, cancer, dysbiosis, Clostridioides difficile, biofilms
The gut microbiome can play a crucial role in the development and prevention of human cancers through a variety of individual bacterial and broader community mechanisms. Emerging evidence suggests that Clostridioides difficile could be an important contributor to this balance.
The communities of bacteria, archaea, fungi, parasites, and viruses colonizing the human body are collectively referred to as the microbiota or microbiome (ie, the living microbes and their gene content, respectively). Functionally, the microbiota, through its virulence factors, cell adhesins, and associated gene products, intimately impacts human physiology, primarily to aid in homeostatic function but also with modulation of local and systemic diseases. The gut contains the highest density and species numbers of bacteria, whereas the skin is a close second [1]. A robust, ever-expanding literature implicates the colonic microbiome as critical to the development of a diverse array of disease processes ranging from local gut disorders (eg, inflammatory bowel disease) to distant pathologies (eg, neuropsychiatric diseases) [2]. In particular, the role of gut dysbiosis in cancer pathogenesis—especially colorectal cancer (CRC)—is considered increasingly important [3], particularly with the emergence of early-onset CRC (EO-CRC, ie, at ages under 45–50 years). In this review, we focus on CRC to introduce key determinants of human gut microbiome composition; whether community structure contributes to colon oncogenesis; and the mechanisms by which individual gut bacteria may sway the balance between health and disease. We close by discussing new evidence for Clostridioides difficile (C. difficile) association with human CRC.
BROAD LINKAGES CONNECTING THE MICROBIOME and CRC
Microbiome Composition and Dysbiosis
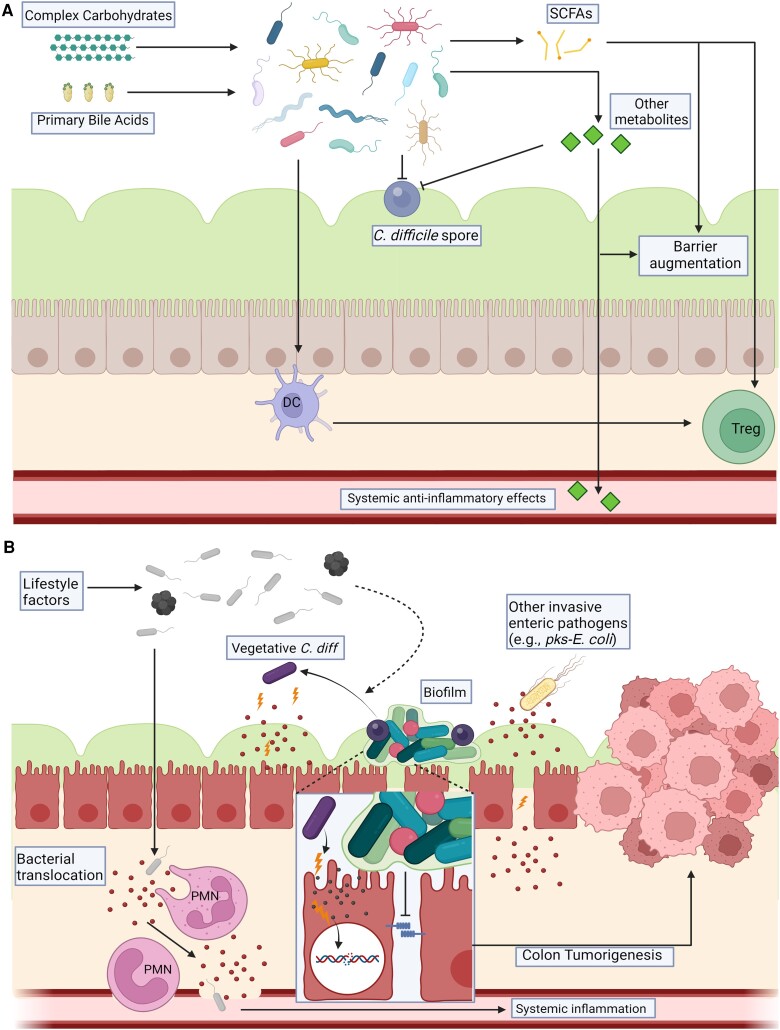
The composition of an individual's microbiome is influenced by many factors. Dietary patterns and social structures (eg, agricultural vs cosmopolitan communities) markedly influence gut microbial signatures [4]. Prior to the advent of modern society, infrequent shifts in lifestyle factors appeared to allow for coevolution on both sides of this interaction that grew into an increasingly dependent symbiosis (ie, an intimate interaction between two distinct organisms, as vs commensals that do not significantly interact with the host). Namely, humans grew dependent on the gene repertoire of their symbionts to fulfill certain physiologic functions (eg, mucosal barrier integrity, food digestion, and immunologic development) and prevent disease processes (eg, infectious diarrhea or auto-inflammation) (Figure 1A) [5]. However, this delicate balance has been immensely disrupted by improved hygiene, antimicrobial medications, and dietary changes that occurred over the past 2 centuries, leading to marked, persistent shifts in gut microbiome composition especially amongst urban populations [4]. Although the implications of these changes still require exploration, the observation that mode of delivery (ie, vaginal vs Caesarian section) impacts the infant microbiome and may be associated with early onset of atopic diseases in childhood is only one example of how microbial context may result in human pathology in a short timeframe [6].
Figure 1.
The Microbiome's role in cancer suppression (A) and development (B). A, A “healthy” gut microbiome is characterized by high species richness and a diverse gene and gene product pool. It is considered to be associated with consumption of a diet high in complex carbohydrates and a lifestyle devoid of high-risk behaviors. In this setting, we propose the microbiome suppresses cancer development through mechanisms such as digestion of complex carbohydrates and primary bile acids to create SCFAs that augment the mucosal barrier and may directly inhibit the growth of cancerous cells. SCFAs and other metabolites produced by healthy microbiomes also can stimulate an anti-inflammatory mucosal immune response (including Treg development partially mediated through interactions with dendritic cells [DC]). Direct competition by endogenous symbionts for nutrients helps protect against invading pro-inflammatory pathogens (eg, C. difficile, shown here in dormant spore phase restricted to the outer mucous layer). Recent data further suggest that the systemic release of certain small molecules synthesized or modulated by gut microbes can serve anti-neoplastic effects at sites distant from the gut. B, In hosts displaying chronic dysbiosis due to lifestyle factors and/or other local or systemic diseases, the species richness and gene pool of the gut microbiome is typically reduced. In these settings, the mucosal barrier is predicted to be more fragile and susceptible to specific pathogen impacts and/or invasion due to release of toxins or other virulence factors (in addition to a relatively reduced production of anti-inflammatory small molecules such as SCFAs) that can contribute to chronic clinical or subclinical intestinal inflammation and further barrier breakdown, predisposing to persistent bacterial translocation. These changes result in the production of pro-inflammatory cytokines and neutrophil (PMN) recruitment, further effecting a cycle of inflammation. A dysbiotic gut microbial community also enables the germination of C. difficile leading to toxin production and mucosal damage (see details in text). Alternatively, microbiota communities such as bacterial biofilms that invade the normally sterile inner mucus layer may activate signaling cascades to additionally impair barrier function and promote chronic mucosal inflammation. These cumulative inflammatory stressors are one potential trigger for colonic stem cell DNA mutation that can foster the development of CRC. The figures in (A) and (B) were created using a paid subscription to BioRender. Abbreviations: C. difficile, Clostridioides difficile; CRC, colorectal cancer; pks-E. coli, Escherichia coli; SCFA, short chain fatty acids; Treg, regulatory T-cell.
The definition of a “healthy microbiome” and, in turn, “dysbiosis’ remain poorly delineated, as functional redundancy allows for astounding inter-individual variability in microbiome composition to accomplish the same physiologic functions. However, there are emerging descriptions of microbiome signatures somewhat consistently associated with specific disease processes, including CRC [7, 8]. The epidemiologic factors associated with sporadic CRC include lifestyle parameters such as obesity, diabetes, a Western diet, and alcohol and tobacco use [9], each alone known to disrupt the microbiota, adding strength to the thesis that the colon microbiota is a crucial environmental risk factor for the development of CRC [10]. Medications, particularly antibiotics, are potent microbiome disrupters with associations now drawn between the quantity and type of oral antibiotic exposure and the development of colonic polyps [11] and colon cancer [12], observations supporting the concept that colon oncogenesis is, at least in part, mediated by disturbances in microbial community structure [13].
Bacteria-Human Interface and Colon Oncogenesis
The mechanisms of microbial impacts on human physiologic function are many. There exists strong evidence demonstrating a causative effect by individual microbes in the induction of human cancers; these organisms range from helminths such as Schistosoma haematobium [14] to viruses such as human papillomavirus [15], and they accomplish oncogenesis in their respective tissues (bladder and cervical or head and neck cancer, respectively) through a wide variety of active and passive mechanisms. In contrast, beyond Helicobacter pylori [16], attempts to definitively implicate gut bacteria in the development of cancer have been less successful, likely in large part due to the sheer number of species, metabolites, and interdependent ecosystems that surround these organisms and confound both controlled experimentation and human studies. These rich communities engage in constant communication amongst themselves and with nearby human cells. Although some microbe:host interactions exhibit far-reaching effects within the human body (eg, trimethylamine [TMA] production in the colon and cardiovascular disease) [17], the majority of crosstalk occurs within the gut. As such, the colon epithelial barrier represents the first line of defense against colon microbiome disruption.
Sporadic CRC pathogenesis including EO-CRC involves slow accumulation of genetic changes in colon epithelial cells (CECs) mediated by repeated, and likely heterogeneous, environmental exposures [9]. Microbial toxins and metabolites that directly damage CEC DNA, bypass cellular controls to activate oncogenic pathways, and/or induce a robust host mucosal immune response likely serve to directly or indirectly contribute to the initiation and/or progression of colon cancer (Figure 1B). Several prominent microecological theories exist regarding the hypothesized community-level mechanisms of colonic tumorigenesis, ranging from the driver-passenger model of more long-term colonization effects to the hit-and-run model of transient impacts that leave permanent genetic changes in CECs [10]. The general concept in many of these models is that, beyond an “instigator” bacterium, microbial community enables “bad actor” bacteria to effect critical human cell changes that lead to the development of disease processes such as CRC.
Despite a preponderance of preclinical evidence supporting these models in colon tumor development, no individual microbes are yet proven as causative in CRC [10]. This underscores the likely importance of community and abiotic factors in the oncogenic process. Nonetheless, understanding the mechanisms by which specific bacteria may contribute to pre-cancerous changes in vitro has potential to inform the development of more efficient screening or effective therapeutics. We will next discuss mechanisms utilized by select bacteria hypothesized to foster colon oncogenesis and will also explore the potential for certain gut bacteria to aid the host in its campaign to counteract cancer development and progression, both colonic and otherwise.
INDIVIDUAL AND COMMUNITY BACTERIAL IMPACTS ON CANCER
Individual Bacterial Implicated in Colon Oncogenesis
Although ubiquitous lifestyle factors contributing to dysbiosis and CRC development may be slowly beginning mitigation in Western society [18], approaches to intervene and disrupt the contributions of individual oncogenic gut bacteria are unknown. Linkages between CRC and specific bacteria trace back decades to the initial recognition that endovascular infection with Streptococcus bovis type I (now Streptococcus gallolyticus) demanded colonoscopy to rule out colon neoplasia [19]. For some time, this and other similar bacterial associations were felt to represent more of a “bystander” effect as opposed to a causal one. However, strong evidence is mounting that individual bacteria (including potentially Streptococcus gallolyticus [20]) may be active in the oncogenic process. A well-studied example is Bacteroides fragilis, a commensal microbe with 2 molecular subgroups, nontoxigenic (NTBF) and toxigenic (ETBF). The latter strains harbor a pathogenicity island and produce a metalloprotease toxin (B. fragilis toxin [BFT]) that is required to induce colonic inflammation and colon tumorigenesis in mouse models through interleukin (IL)-17-dependent and other mechanisms [21]. ETBF is identified at a disproportionally high level in patients with CRC when compared to controls [10]. Similarly, the acquisition of a pathogenicity island (termed the pks island) by strains of another commensal organism, Escherichia coli (pks-E. coli), also drives tumorigenesis in mouse models through DNA-damaging secondary metabolite (colibactin) production [22] and has strong human CRC linkages [10].
Outside of acquired virulence factors, individual bacteria can utilize other mechanisms to cause pro-inflammatory mucosal changes. Some strains of Enterococcus faecalis produce extracellular superoxide that, in certain contexts, leads to genomic instability in nearby human cells [23, 24] and microenvironmental changes favoring “blooms” of additional inflammatory bacteria such as Ruminococcus gnavus, a bug implicated in flares of inflammatory bowel disease (IBD) [25]. More recently, high-throughput techniques revealed detection of microbial metabolites such as small-molecule genotoxins called indolimines, produced by CRC-associated bacteria like Morganella morganii. These bacterial genotoxins may contribute to direct colonocyte DNA damage and increased intestinal inflammation, known precursors to CRC formation [26]. Both proteomics and metabolomics of colon bacteria involve a staggering amount of data analysis and, as such, the field remains in its infancy but with significant potential for paradigm-shifting discoveries. This list is not comprehensive; detailed and updated reviews of individual bacteria implicated in colon oncogenesis are catalogued elsewhere [10].
Community Structure and Communication
Although the activities of individual bacteria are anticipated to be important in the pathogenesis of CRC, the role of community structure and environmental factors in enabling bacterial behavior may be just as crucial. It is now recognized that at least 50% of human CRC are covered with mucus-invasive bacterial biofilms, bringing dense polymicrobial communities in direct CEC contact in both the cancers and normal tissue [7, 27]. Bacterial biofilms on normal tissue diminish barrier function and induce inflammatory signals and Stat3 activation that contribute to the onset of pro-carcinogenic IL-17 mucosal inflammation [27]. Diminished barrier function is considered an early step in colon carcinogenesis that is permissive to bacterial translocation and activation of innate immune receptors creating a positive feedback loop to further inflammatory cell infiltration as well as dysbiosis [28]. Which communities consistently foster pro-carcinogenesis versus anti-carcinogenesis is unclear. However, increasing attention is focused on the metabolic output of both individual bacteria and of communities in colon in systemic disease pathogenesis linked to the microbiota. For example, early analysis identified N1, N12-diacetylspermine, a polyamine product, as associated with colon biofilm formation and cancer with contributions by both the bacterial biofilm and host cells [29]. However, further investigation is needed to, for example, contrast biofilm community function on normal tissue versus CRC. Such analyses might provide insight into pro-carcinogenic microbial community functions that could assist in biomarker development for CRC prevention.
Quorum sensing (QS) is a likely mechanism governing both biofilm and fecal microbial community function in this context. QS refers to complex molecular circuits that mediate communication between bacteria within the same community but not necessarily in physical contact with one another. QS acts via small-molecule autoinducers that diffuse through the cell membranes of other bacteria—frequently, but not exclusively, of the same species—and can influence gene expression designed to enhance microbial fitness, increase virulence factor production, and/or otherwise modify function [30, 31]. Additional mechanisms by which bacteria influence bacterial and host cell functions are extracellular vesicles [32] and miRNA production [33].
Anti-Carcinogenic Activity of Gut Bacteria
Although much focus has been on identifying microbes contributing to cancer development risk, the human microbiome has immense genetic reservoir that has been toned over millennia to coexist with and/or facilitate human development and physiology. Even with recent drastic shifts in human gut microbiome composition, there undoubtedly remains significant potential in gut microbial communities to buffer against disease development. For example, most data available on bacterial metabolites such as short-chain fatty acids (SCFAs) suggest these small molecules largely reinforce the colon mucosal epithelial barrier and act to limit colonic/systemic inflammation and cancer risk. Specifically, the SCFA butyrate, the key substrate for epithelial cell energy and present in micromolar concentrations in human blood, is synthesized by commensal microbes as a product of dietary complex carbohydrate metabolism (eg, fiber) with the capacity to inhibit the proliferation of colorectal cancer cells [34] and induce cancer cell apoptosis [35]. Thus, dietary approaches to augment intracolonic/systemic SCFAs may impact cancer biology, including both CRC [35] and melanoma [36].
The colon microbiome may also promote anti-tumor effects outside of its local environment, especially through modulation of the immune response. A recent study [37] demonstrated that the production of trimethylamine by colon microbes with systemic release of the metabolite trimethylamine N-oxide (TMAO) potentiated the response to immune checkpoint inhibitor (ICI) therapy in patients with pancreatic ductal adenocarcinoma through the induction of type I interferon pathways and the subsequent activation of effector T-cell responses within the tumor microenvironment (TiME). Similarly, the translocation and subsequent tumor microenvironmental habitation of gut-derived Lactobacillus reuteri in melanoma patients was recently described [38]; this microbe released a dietary tryptophan catabolite into the TiME, leading to the stimulation of interferon-γ-producing CD8 T cells, response to immune checkpoint inhibitor therapy, and increased survival in advanced melanoma patients.
The potential for linkages between specific bacteria, bacterial products and impacts on cancer, colorectal or otherwise, is vast. Teasing apart the relative contributions that individual bacteria have to cancer development or other disease processes versus the role of that bacterium's local environment and community effects is challenging. Nonetheless, it is anticipated that these types of investigations will inform our understanding of disease genesis with the potential of identifying microbes, pathways, and molecules to guide new prevention and therapy approaches.
INVESTIGATING THE MICROBIOME: CLOSTRIDIOIDES DIFFICILE AS A MODEL BACTERIUM FOR DISCERNING MICROBIOME-CANCER INTERACTIONS
C. difficile and Colorectal Cancer
C. difficile is a gram-positive, spore-forming anaerobic bacterium that colonizes the large intestine. Toxigenic strains release protein exotoxins (TcdA, TcdB) that induce local inflammation leading to the clinical signs of diarrhea and/or colitis in susceptible individuals [39]. Its hardy spores are immensely difficult to eradicate in the colon as evidenced by disease recurrence rates ranging from 20% to 65% even after current standard-of-care treatments [40]. Since the early 2000s, there has been a dramatic increase in rates of severe C. difficile infection [40], as well as community-acquired infection [41]. This trend is not limited to the developed world, as comparable or even elevated prevalence rates have been reported in epidemiologic studies of countries such as India [42] and Indonesia [43].
Along roughly the same time period as this rise in C. difficile cases, the global epidemiology of CRC has undergone significant shifts. Specifically, the incidence of CRC has been increasing in individuals under the age of 50 since the early 1990s [44] as well as in areas of the world previously considered to have low CRC risk [45]. This is especially true for Asian countries, which now account for nearly 50% of the worldwide CRC mortality as they have increasingly adopted a “Westernized” lifestyle [46]. Despite these parallel trends in evolving epidemiology and the data supporting that antibiotic exposure is a risk factor for both diseases, no strong mechanistic link between C. difficile infection and CRC had been drawn prior to a recent study [3].
In this study, the slurry or polymicrobial cultures of biofilm-positive Stage 1 colon cancers induced colon tumors in a murine model. Notably, removal of C. difficile from the cultured bacterial community demonstrated that C. difficile was both necessary and sufficient to convert the bacterial community from non-tumorigenic to tumorigenic, raising the specter that C. difficile could act, at least in a mouse model, as a colon pro-carcinogenic bacterium. In fact, 5 of 6 colon cancers evaluated in the study were positive for C. difficile, and 2 were toxigenic, tumor-inducing strains. Important nuances included the fact that prolonged mucosal colonization with exposure to TcdB (but not TcdA) appeared necessary for colon tumor induction; pro-carcinogenic C. difficile strains were recovered from both biofilm-positive and biofilm-negative CRCs; and limited C. difficile abundance was tumor-inducing (alluding to a possible keystone role in gut ecology [10]). At present, human data on longitudinal colonization in stool or the mucosa with C. difficile strains are lacking. In addition, C. difficile is genetically a very pleomorphic bacterium [47], and preliminary data suggest that not all C. difficile strains possess procarcinogenic properties. In consideration of this possibility, understanding the “life cycle” of C. difficile has merit.
C. difficile Microbiology and Microecology
C. difficile lives a complex, binary life cycle that remains sub-optimally understood; a variety of environmental stimuli induce genetic regulatory factors that force switches between the incredibly durable spore phase and a metabolically active vegetative phase, the latter of which is capable of the toxin production necessary for human disease [48]. Although much remains to be defined regarding the inducers and repressors of the C. difficile life cycle, local environmental factors likely play a key role. The cascade of sporulation, which transforms the bacterium into the aerotolerant, transmissible form, appears driven by stimuli such as nutrient depletion and QS as it is in other anaerobic gram-positive bacteria [49, 50]. The spore phase “dormant” state is highly resistant to elimination due to its low metabolic activity. A germination phase transforming the bacterium into a more active vegetative form completes the life cycle. In the case of C. difficile, it is thought that the key germinants are primary bile acids produced by the host [48]. Interestingly, secondary bile acids synthesized by other commensal gut bacteria inhibit the germination of C. difficile and may yield colonization resistance against this bacterium [51], providing one of the main mechanistic hypotheses regarding how the endogenous microbiota protects against C. difficile infection.
Germination is a critical step required for the synthesis of the toxins crucial to human disease and, potentially, CRC promotion [3]. A recent study [52] suggests that toxin production may be designed to induce host inflammation to alter the bacterium's nutrient landscape through targeted degradation of collagen and other connective tissue components; this shredding of the extracellular matrix and the obligatory severe host inflammatory response that follows may liberate key nutrients to sustain long-term C. difficile colonization and toxin production while also selecting against certain competitor species, such as members of the Bacteroidaceae family. Host inflammation may further impact toxin production by C. difficile, as instability in iron balance, common in inflammatory conditions, can impact the bacterium's synthesis of toxins [53] and augment its defense against host antimicrobial compounds [54]. QS also appears to be an important variable in toxin production, as autoinducers and toxins are synthesized during the same window of the C. difficile life cycle [31]. Although biofilms are a feature of many CRCs (discussed above), a study of C. difficile-dominant biofilms suggested a predominance of highly resistant spores with reduced germination potential and relatively low toxin production, overall projecting a “hunker-down” as opposed to a symptomatic disease phenotype [55]. However, based on Drewes, Chen et al [3]., we would hypothesize that low level, persistent TcdB release in proximity to CECs, whether within biofilms or not, is critical to colon neoplasia development in this context.
Gut Microbial Evolution and Niche-Seeking Behavior
Establishment of C. difficile colonization in the host colon appears to require particular microecological criteria—potentially including but not limited to macro- and micro-nutrient disturbances, host primary bile acid synthesis, an absence of secondary bile acid production by commensal gut microbes, and an appropriate autoinducer milieu—to facilitate germination and the subsequent toxin production critical for human disease and, possibly, the induction of colonic neoplasia. In line with ubiquitous clinical observations, depletion of the endogenous microbiota and stimulation of colon mucosal inflammation are the triggers for symptomatic C. difficile infection. To this end, the idea that toxigenic C. difficile may be obligately pathogenic in human hosts to support its life style [52] is intriguing. It is interesting to consider whether this hypothesis may extend to all other gut bacteria suspected to play a role in CRC development, such as ETBF and pks-E. coli.
CRC development is a chronic process—estimated ≥10 years from onset of neoplastic clonal expansion in mutated CECs to visible neoplasia observable at colonoscopy [56]. At present, little is known about the durability of fecal or mucosal colonic C. difficile colonization and the mechanisms that may promote chronic pathogenicity. A recent publication described a model of divergent within-host evolution of certain gut bacteria (ie, Enterococcus gallinarum and Lactobacillus reuteri) to adapt to fill distinct microecological niches within the host gastrointestinal tract [57]. In a murine model, isolates evolved into 2 broad, coexisting phenotypes, mucosal and luminal; the former developed a heightened ability to translocate through the gut wall, induce hepatoenteric inflammation, and evade immune clearance when compared with its counterpart, hypothesized to be equipped for long-term colonization and higher transmission potential. This identification of apparent targeted, time-dependent functional advantages achieved through within-host evolution suggests that bacterial phenotype may be strongly influenced by the opening of ecologic niches within the host, a finding with potential consequences for other enteric bacteria capable of developing pathogenicity. Importantly, the data suggested that the mucosal strain phenotype was also influenced by host genetics and the neighboring microbiome.
Although within-host evolution of C difficile remains to be investigated, studies of the broad evolutionary signatures on its genome suggest that it is designed for survival within gut niches and thus is primed for long-term co-habitation within a host [47], although notably it is unclear whether the forces of selection on its genome imply an obligately pathogenic relationship [58]. Additionally, the genetic architecture of C. difficile's QS circuit(s) and its ability to uptake genetic information readily from neighboring organisms may be important factors in its toxin production capability and virulence evolution potential, suggesting a predilection for prolonged community interactions in settings such as the microbiome [59].
Pursuing the C. difficile Pro-carcinogenesis Hypothesis
Murine model data support that persistent colonization and production of TcdB in the colon lumen are required for C. difficile procarcinogenesis. However, critical gaps in knowledge exist including a dearth of information regarding persistent human C. difficile fecal or mucosal colonization, the duration of colon TcdB production, the impact of differing C. difficile toxinotypes, and whether persistent colon mucosal inflammation (a critical contributor to oncogenesis) occurs. Prior studies suggest that asymptomatic C difficile colonization may occur in 5%–15% of community adults [60], but linkages to inflammation and/or toxin production are unknown. A competent adaptive immune response to TcdA and TcdB through antibody production seems important to diminish the likelihood of clinical disease but does little to protect against colonization, suggesting that a relative loss of humoral immunity may impact bacterial toxin production and the inflammation that results [61]. Key steps to study the oncogenic potential of C. difficile will involve detailing epidemiological links between C. difficile infection and the rates of colonic neoplasia in humans, as well as determining the frequency of and factors contributing to persistent colonization and toxin production in certain individuals. In addition, mapping the within-host evolution of C. difficile in humans deemed persistent carriers may help shed light on how intimate the associations between colonization, toxin production, gut inflammation, and the immune response may be.
CONCLUDING THOUGHTS AND FUTURE DIRECTIONS
As yet, unlike the causal role of Helicobacter pylori in gastric cancer [16], no causal linkages between gut bacteria and the development of human CRC are proven. However, there is a preponderance of evidence connecting disruptions in microbiome community structure and function with the induction of an oncogenic environment at the crucial interface of the microbe-human crosstalk, the epithelial layer of the colon. Possible mechanistic explanations for these findings, both at the level of bacterial pathogenicity and community dysfunction, are rapidly emerging. Despite this, the potential for the gut microbiome to buffer against and even reverse the development of neoplasia, within the colon or distant from it, remains a tantalizing area for future research; after all, only a small number of colon polyps progress to CRC [56]. Although many different individual bacteria have been implicated as potential promoters in the colon oncogenic process, few have the clinical ubiquity and resilience of toxigenic C. difficile, a bacterium whose evolving epidemiology, at least in part, mirrors that of the epidemic of EO-CRC. Further studies aimed at defining the true burden and persistence of C. difficile following treatment in humans; identifying and manipulating the ecological factors contributing to this bacterium's ability to proliferate and produce pathogenic toxins; and identifying approaches to either durably eliminate and/or neutralize the bacterial-mediated induction of colonic inflammation may inform the approach to understanding colon bacterial oncogenesis with resounding consequences for unraveling microbiota-cancer connections. More importantly, as microbiota-associated disease mechanisms emerge, clinically relevant approaches to disease prevention and therapy will be introduced. There are now two FDA-approved targeted microbiota therapies (Rebiota® and Vowst®) based on studies of C. difficile disease with many additional products in development. Although the field of microbiota-host interactions is still young, clinicians can anticipate the introduction of microbiota biomarkers for disease detection and monitoring and microbiota-modifying approaches to assist in disease prevention and therapy.
Contributor Information
Sean M Anderson, Division of Infectious Diseases, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Cynthia L Sears, Division of Infectious Diseases, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Notes
Author Contributions. S. M. A. and C. L. S. both contributed to composition of the body of the text and to subsequent edits, figure creation, and final drafting.
Supplement sponsorship. This article appears as part of the supplement “The Microbiome and Human Health Perspective,” sponsored by Ferring Pharmaceuticals Inc., Seres Therapeutics Inc., and Nestlé Health Science.
Financial support. S. M. A. is funded by the National Institutes of Health (grant number T32-AI007291) and Johns Hopkins University Division of Infectious Diseases for salary support. The work by C. L. S. is supported by the Bloomberg∼Kimmel Institute for Immunotherapy, Cancer Grand Challenges OPTIMISTICC team grant number (A27140) funded by Cancer Research UK, Johns Hopkins University Department of Medicine; and National Institutes of Health grant number R01CA196845 (C. L. S.).
References
- 1. Domingue JC, Drewes JL, Merlo CA, Housseau F, Sears CL. Host responses to mucosal biofilms in the lung and gut. Mucosal Immunol 2020; 13:413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Golofast B, Vales K. The connection between microbiome and schizophrenia. Neurosci Biobehav Rev 2020; 108:712–31. [DOI] [PubMed] [Google Scholar]
- 3. Drewes JL, Chen J, Markham NO, et al. Human colon cancer-derived Clostridioides difficile strains drive colonic tumorigenesis in mice. Cancer Discov 2022; 12:1873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tito RY, Knights D, Metcalf J, et al. Insights from characterizing extinct human gut microbiomes. PLoS One 2012; 7:e51146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oliveira RA, Pamer EG. Assembling symbiotic bacterial species into live therapeutic consortia that reconstitute microbiome functions. Cell Host Microbe 2023; 31:472–84. [DOI] [PubMed] [Google Scholar]
- 6. Sandall J, Tribe RM, Avery L, et al. Short-term and long-term effects of caesarean section on the health of women and children. Lancet 2018; 392:1349–57. [DOI] [PubMed] [Google Scholar]
- 7. Drewes JL, White JR, Dejea CM, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017; 3:34. Erratum in: NPJ Biofilms Microbiomes. 2019 Jan 9; 5(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Young C, Wood HM, Fuentes Balaguer A, et al. Microbiome analysis of more than 2,000 NHS bowel cancer screening programme samples shows the potential to improve screening accuracy. Clin Cancer Res 2021; 27:2246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Venugopal A, Carethers JM. Epidemiology and biology of early onset colorectal cancer. EXCLI J 2022; 21:162–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knippel RJ, Drewes JL, Sears CL. The cancer microbiome: recent highlights and knowledge gaps. Cancer Discov 2021; 11:2378–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao Y, Wu K, Mehta R, et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut 2018; 67:672–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J, Haines C, Watson AJM, et al. Oral antibiotic use and risk of colorectal cancer in the United Kingdom, 1989–2012: a matched case-control study. Gut 2019; 68:1971–8. [DOI] [PubMed] [Google Scholar]
- 13. Perez-Cobas AE, Artacho A, Knecht H, et al. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One 2013; 8:e80201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mostafa MH, Sheweita SA, O’Connor PJ. Relationship between schistosomiasis and bladder cancer. Clin Microbiol Rev 1999; 12:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 2010; 11:1048–56. [DOI] [PubMed] [Google Scholar]
- 16. Marshall BJ, Windsor HM. The relation of Helicobacter pylori to gastric adenocarcinoma and lymphoma: pathophysiology, epidemiology, screening, clinical presentation, treatment, and prevention. Med Clin North Am 2005; 89:313–44, viii. [DOI] [PubMed] [Google Scholar]
- 17. Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013; 368:1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ouakrim D A, Pizot C, Boniol M, et al. Trends in colorectal cancer mortality in Europe: retrospective analysis of the WHO mortality database. BMJ 2015; 351:h4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Association of Streptococcus bovis with carcinoma of the colon. N Engl J Med 1977; 297:800–2. [DOI] [PubMed] [Google Scholar]
- 20. Kumar R, Herold JL, Schady D, et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog 2017; 13:e1006440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valguarnera E, Wardenburg JB. Good gone bad: one toxin away from disease for Bacteroides fragilis. J Mol Biol 2020; 432:765–85. [DOI] [PubMed] [Google Scholar]
- 22. Boussuet-Greif N, Vignard J, Taieb F, et al. The colibactin genotoxin generates dna interstrand cross-links in infected cells. mBio 2018; 9:e02393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 2007; 132:551–61. [DOI] [PubMed] [Google Scholar]
- 24. Huycke MM, Moore D, Joyce W, et al. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol Microbiol 2001; 42:729–40. [DOI] [PubMed] [Google Scholar]
- 25. Hall AB, Yassour M, Sauk J, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 2017; 9:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao Y, Oh J, Xue M, et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 2022; 378:eabm3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A 2014; 111:18321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanaka T, Kohno H, Suzuki R, et al. Dextran sodium sulfate strongly promotes colorectal carcinogenesis in Apc(Min/+) mice: inflammatory stimuli by dextran sodium sulfate results in development of multiple colonic neoplasms. Int J Cancer 2006; 118:25–34. [DOI] [PubMed] [Google Scholar]
- 29. Johnson CH, Dejea CM, Edler D, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab 2015; 21:891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uhlig F, Hyland NP. Making sense of quorum sensing at the intestinal mucosal interface. Cells 2022; 11:1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gunaratnam S, Millette M, McFarland LV, DuPont HL, Lacroix M. Potential role of probiotics in reducing Clostridioides difficile virulence: interference with quorum sensing systems. Microb Pathog 2021; 153:104798. [DOI] [PubMed] [Google Scholar]
- 32. Xie J, Haesebrouck F, Van Hoecke L, Vandenbroucke RE. Bacterial extracellular vesicles: an emerging avenue to tackle diseases. Trends Microbiol 2023:S0966-842X(23)00163-4. [DOI] [PubMed] [Google Scholar]
- 33. Nikolaieva N, Sevcikova A, Omelka R, Martiniakova M, Mego M, Ciernikova S. Gut microbiota-microRNA interactions in intestinal homeostasis and cancer development. Microorganisms 2022; 11:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zeng H, Umar S, Rust B, Lazarova D, Bordonaro M. Secondary bile acids and short chain fatty acids in the colon: a focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int J Mol Sci 2019; 20:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Donohoe DR, Holley D, Collins LB, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov 2014; 4:1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simpson RC, Shanahan ER, Batten M, et al. Diet-driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome. Nat Med 2022; 28:2344–52. [DOI] [PubMed] [Google Scholar]
- 37. Mirji G, Worth A, Bhat SA, et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci Immunol 2022; 7:eabn0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bender MJ, McPherson AC, Phelps CM, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell 2023; 186:1846–1862.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010; 467:711–3. [DOI] [PubMed] [Google Scholar]
- 40. Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med 2015; 372:1539–48. [DOI] [PubMed] [Google Scholar]
- 41. Guh AY, Mu Y, Winston LG, et al. Trends in U.S. Burden of Clostridioides difficile infection and outcomes. N Engl J Med 2020; 382:1320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Monaghan TM, Biswas R, Satav A, Ambalkar S, Kashyap RS. Clostridioides difficile epidemiology in India. Anaerobe 2022; 74:102517. [DOI] [PubMed] [Google Scholar]
- 43. Collins DA, Gasem MH, Habibie TH, et al. Prevalence and molecular epidemiology of Clostridium difficile infection in Indonesia. New Microbes New Infect 2017; 18:34–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siegel RL, Fedewa SA, Anderson WF, et al. Colorectal cancer incidence patterns in the United States, 1974–2013. J Natl Cancer Inst 2023; 109:djw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Douaiher J, Ravipati A, Grams B, Chowdhury S, Alatise O, Are C. Colorectal cancer-global burden, trends, and geographical variations. J Surg Oncol 2017; 115:619–30. [DOI] [PubMed] [Google Scholar]
- 46. Kono S. Secular trend of colon cancer incidence and mortality in relation to fat and meat intake in Japan. Eur J Cancer Prev 2004; 13:127–32. [DOI] [PubMed] [Google Scholar]
- 47. Sebaihia M, Wren BW, Mullany P, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 2006; 38:779–86. [DOI] [PubMed] [Google Scholar]
- 48. Zhu D, Sorg JA, Sun X. Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect Microbiol 2018; 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 2012; 36:131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Darkoh C, Odo C, DuPont HL. Accessory gene regulator-1 locus is essential for virulence and pathogenesis of Clostridium difficile. mBio 2016; 7:e01237-16. Erratum in: MBio. 2017 Oct 31; 8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winston JA, Theriot CM. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016; 41:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fletcher JR, Pike CM, Parsons RJ, et al. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat Commun 2021; 12:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamaki J, Chawla S, Tong S, Lozada KA, Yang S. Iron effects on Clostridioides difficile toxin production and antimicrobial susceptibilities. Antibiotics (Basel) 2022; 11:537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Knippel RJ, Wexler AG, Miller JM, et al. Clostridioides difficile senses and hijacks host heme for incorporation into an oxidative stress defense system. Cell Host Microbe 2020; 28:411–421.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Semenyuk EG, Laning ML, Foley J, et al. Spore formation and toxin production in Clostridium difficile biofilms. PLoS One 2014; 9:e87757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Winawer SJ, Fletcher RH, Miller L, et al. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology 1997; 112:594–642. Erratum in: Gastroenterology 1997 Mar; 112(3):1060. Erratum in: Gastroenterology 1998 Mar; 114(3):625. [DOI] [PubMed] [Google Scholar]
- 57. Yang Y, Nguyen M, Khetrapal V, et al. Within-host evolution of a gut pathobiont facilitates liver translocation. Nature 2022; 607:563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sachs JL, Essenberg CJ, Turcotte MM. New paradigms for the evolution of beneficial infections. Trends Ecol Evol 2011; 26:202–9. [DOI] [PubMed] [Google Scholar]
- 59. Knight DR, Elliott B, Chang BJ, Perkins TT, Riley TV. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev 2015; 28:721–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crobach MJT, Vernon JJ, Loo VG, et al. Understanding Clostridium difficile colonization. Clin Microbiol Rev 2018; 31:e00021-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kyne L, Warny M, Qamar A, Kelly CP. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 2000; 342:390–7. [DOI] [PubMed] [Google Scholar]