Abstract
Introduction
Liquid biopsies are increasingly being adopted in the care of patients with cancer. Not only in patients with metastatic disease but the utility is also being recognized in earlier phases of the journey of a patient with cancer. More recently, methylated platforms are offering another lens of looking at the same question more so in minimal residual disease (MRD) and early detection settings. While false positives secondary to clonal hematopoiesis of indeterminate potential (CHIP) are recognized as one entity to consider when interpreting these assays, and advanced CHIP filtering bioinformatics platforms can prevent this, false positives secondary to aberrant methylation are not described.
Case Presentation
Herein, we report a case of a patient with hepatitis C-related viremia and a very high viral load that had a false-positive plasma-only colorectal MRD assay. The colorectal MRD assay spontaneously cleared on hepatitis C virus therapy which led to clearance of the virus.
Conclusion
As these assays are increasingly applied in real-world settings, it would be of value to consider non-cancer chronic disease states that may lead to aberrant methylation that could lead to a false-positive assay.
Keywords: Circulating tumor DNA, Methylation, Liquid biopsy, Hepatitis C virus, Colorectal cancer, False positive
Introduction
Minimal residual disease (MRD) assays, which can be divided into (i) plasma-only and (ii) tumor-informed circulating tumor DNA (ctDNA) assays, are a recent development in cancer care [1]. Colorectal cancer has been the poster child in terms of the number and types of assays. In patients with resected colorectal cancer, they have made a significant impact in monitoring disease recurrence and tailoring therapy and are currently being evaluated as predictive markers in numerous ongoing clinical trials [2]. Even outside of clinical trials, physicians use these tests to aid in therapeutic decision-making and initiation of treatment. There is increasing evidence of how these tests have profoundly changed clinical practice by moving from tissue to peripheral blood as a source of information [3].
However, plasma-only and tumor-informed assays have individual limitations to themselves, and there are pros and cons to each approach [4]. With increasing utility, we are understanding better both the analytical and pre-analytical variables that affect sensitivity and specificity of these assays, and also instances where they might be false negative or false positive. Herein, we report our experience with a patient who had a moderately differentiated resected sigmoid adenocarcinoma with concurrent hepatitis C virus (HCV) infection and a false-positive plasma-only assay on the methylation portion. This cleared without any anti-cancer treatment as the HCV viremia resolved on anti-HCV-based therapy.
Case Presentation
A 62-year-old male with a medical history significant for type 2 diabetes mellitus and HCV, presented with irregular bowel movements and a positive stool DNA test. He subsequently underwent a colonoscopy that revealed a partially obstructing suspicious sigmoid mass and was biopsied, confirming a moderately differentiated adenocarcinoma that was mismatch repair proficient/microsatellite stable. His staging scans were negative for any distant metastases. Based on his findings, he underwent a laparoscopic sigmoid colectomy, and a 5.5 × 5.6 × 1.3 cm mass was removed, with metastases to one of twenty-one lymph nodes. Surgical staging classified this as stage IIIB (tumor, node, metastasis pT3N1a) with positive lymphovascular invasion, negative perineural invasion, and pMMR/microsatellite stable as noted earlier. RAS/RAF-based testing that was done as part of the evaluation of an MRD-based clinical trial showed that the tumor was extended RAS/RAF-wildtype. Tumor-informed ctDNA assay was sent by the surgical team in the postoperative setting, which was negative. The patient received 3 months (4 cycles) of capecitabine and oxaliplatin as adjuvant chemotherapy with routine restaging scans and MRD-based ctDNA detection.
However, at 19 weeks post-surgery, the patient had both a plasma-only as well as a tumor-informed assay for evaluation of eligibility for some of our MRD-based clinical trials. Intriguingly, the plasma-only test revealed that MRD was detected, whereas the tumor-informed assay was negative. Further evaluation and discussion revealed that this was only for the methylation component and not the mutational component of the composite assay. Concurrent imaging of the CT chest, abdomen, and pelvis showed no evidence of disease.
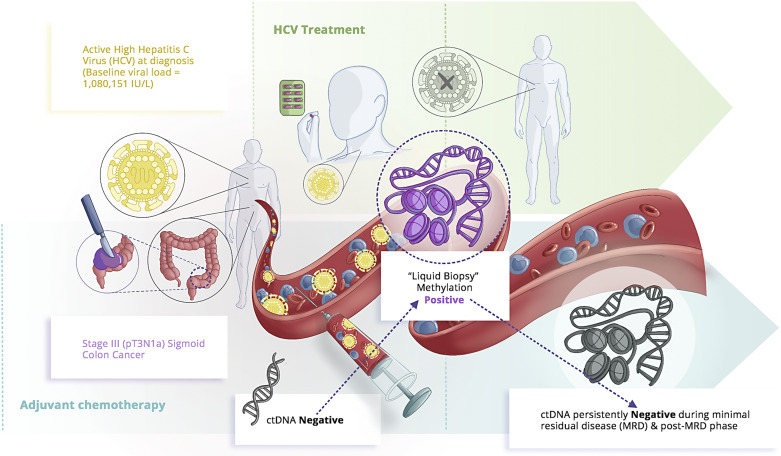
While workup and investigations were being done to evaluate options given the MRD-positive plasma-only assay, the patient got treatment for the very high HCV viral load without signs of cirrhosis, which was planned after he had completed his adjuvant therapy. He had been earlier referred to hepatology and subsequently started on anti-HCV therapy with glecaprevir/pibrentasvir, a combination of HCV NS3/4A protease inhibitor and an HCV NS5A inhibitor. Figure 1 summarizes the timeline of events and treatment received with respect to sigmoid colon cancer and HCV.
Fig. 1.
Simplistic schema summarizing the sequence of events and therapy for patient’s diagnosis of stage Ill colon cancer and associated HCV treatment.
As options for further chemotherapy were discussed given the MRD-positive assay, the patient opted for no more systemic chemotherapy and more time to think about this. After completion of his anti-HCV therapy, patient reestablished care and new baseline plasma-only and tumor-informed assays were repeated along with imaging.
Surprisingly, repeat testing at 33 weeks post-surgery revealed that ctDNA was not detected by either of the plasma-only assay and tumor-informed assay tests. Multiple subsequent follow-ups and more than 2 years into the surveillance setting, the assays have continued to remain negative with no evidence of disease recurrence. In this case, our working hypothesis is that the HCV viral load of over a million copies per mL likely caused the false-positive methylation signal. The treatment for HCV, which was curative, leads to clearance of the virus and the methylation signal on the colorectal cancer plasma-only assay. The fact that the mutational component of the plasma-only assay as well as an independent tumor-informed ctDNA assay continued to be negative, and clinically, this being relatively low risk, argues against the presence of cancer in this patient. The CARE Checklist has been completed by the authors for this case report and is attached as online supplementary material (for all online suppl. material, see https://doi.org/10.1159/000535174).
Discussion
This is an intriguing case and potentially the first report of any false-positive case described in the real-world setting pertaining to a methylated assay that is secondary to a non-cancer disease state (in this case, the very high HCV viremia). As these liquid biopsies increasingly get adopted in MRD and earlier detection settings, it is important to realize that there could be potential instances of overlapping methylation signals with other cancers and/or chronic non-cancer conditions.
According to the time points, the patient’s plasma-only assay was only positive when the HCV viral load was high. After completion of anti-HCV therapy and no detectable HCV viral load, the plasma-only assay had become negative. Not surprisingly, the mutational component on both the plasma-only as well as the tumor-informed assay detected no ctDNA. Our concern is that the plasma-only assay may have had a false MRD signal due to the active HCV viremia in our patient. To hypothesize, it might be possible that after HCV treatment, the background signal might have died out and hence no longer interfered with MRD detection. As we learn more about these assays, not only in the MRD setting but also now in early detection/diagnosis where multiple assays are in development and available for commercial use, these kinds of instances could potentially result in false positives.
This observation might also potentially explain some of the patients who on long-term follow-up still do not have a diagnosis of cancer despite a positive signal in some of these multi-cancer early detection tests. Most of the earlier detection assays are looking at some component of epigenomics/methylated markers. The National Cancer Institute has released a white paper report on factors that could influence these results. These include and are not limited to certain pre-analytical variables, the characteristic of the ctDNA, sequencing methods, storage of plasma, or PCR-related errors [5]. But non-cancer disease states that could influence these assays is a novel finding.
With more use of these assays and learning, some guidelines and/or limitations have been put forward. E.g., in situations where there could be a lot of background noise such as pregnancy, bone marrow, or solid organ transplant patients, and/or if someone has had a recent blood transfusion, the sensitivity or specificity could be impacted. As noted earlier, CHIPs are a consideration in those with older age. Someone with another malignancy (hematological or solid tumors) may cause a false-positive signal. And finally, as noted in our case, another non-cancer disease state potentially may impact methylation. The sensitivity of this test is also influenced by the amount of ctDNA shed by different cancer types, especially low-shedding cancers [6]. However, real-world data and studies with positive controls (other malignancies, and in this case, non-cancer disease states) would be crucial factors to take into account for any assay in development in clinical use. The false-positive rates (FPRs) with ctDNA and CHIPs (clonal hematopoiesis of indeterminate potential) are decreasing with better databases and bioinformatics filtering [7]. But we are probably still on the learning curve in the field of these epigenetic/methylation signals.
Building upon this, an interesting lens to see this issue is to recognize the introduction of Multi-Cancer Early Detection (MCED) tests that can detect cancer-origin signals by using cell-free DNA-based targeted methylation assays [8]. These are now commercially available in clinics and trials to screen the general population and identify site- and type-specific cancer signals. The hope is that it will help physicians in ordering particular imaging or blood markers to confirm cancer findings much before the disease can be picked up by conventional tools. However, the flipside to this is that cancer signals could be picked up by the test with no evidence of cancer, despite frustrating and meticulous detection attempts. The thought process here is that it could be a potential false positive or very early-stage cancer that even with the best of imaging and diagnostics is still not detectable and needs more follow-up. In such cases, cancer site origin FPR was dependent on whether the signal was from a solid tumor or hematological cancer, as the latter was associated with a higher FPR. To counteract this, the detection classifier was adjusted according to hematological cancer and solid tumors [9]. In the case of a false-positive signal, it could be possible that (1) either the test was truly a false positive or (2) there was the presence of background signals caused by other comorbid conditions or diseases which may lead to false-positive signals. Therefore, this screening test has gone through multiple iterations, including modification, of the hematological signal threshold to avoid false positives [10].
Building upon this, it is likely that the HCV infection with high viremia acted as a possible aberrant methylation signal that was cleared with anti-HCV therapy [11–13]. For any assay utilizing methylation-based platforms, we will eventually learn with time about the non-hematological, non-cancer group of illnesses causing aberrant methylation patterns. Recognition of CHIP also took years. Additionally, these assays can help identify certain mutations that can act as predictive biomarkers, especially in the setting of systemic immunotherapy. The profound effect of this is seen in the colorectal space for microsatellite instability-high (dMMR-MSI-H) cancers, where immunotherapy has significantly improved disease outcomes [14].
Our report has several limitations. First, it is a case report. To make sure this was not premature, we waited 2+ years of follow-up and multiple plasma-only and tumor-informed ctDNA-based MRD tests and clinical follow-up/imaging that all continue to be negative to date. The strength of the report is this being the first of this novel finding. As noted earlier, the strength of our findings is the multiplicity of time points and long-term follow-up. The serendipitous timing of the methylation component of the plasma-only assay being positive and turning immediately thereafter once HCV viremia resolved supports our hypothesis.
Conclusion
There is a paucity of literature regarding false-positive liquid biopsy assays, particularly from non-cancer disease states. Our report is probably the first and an important addition to the literature that needs to evolve more in this subject. It would be worthwhile going back to patients who have had methylation-based earlier detection assays and still do not have any obvious evidence of cancer to see if they have any concomitant/chronic non-cancer disease states to help explain this finding.
Acknowledgments
We are also deeply indebted to Emma Vidal at DrawImpacts for her work on the illustration with the author accompanying the article on social media.
Statement of Ethics
Written informed consent was obtained from the patient for publication of the details of their medical case and any accompanying images. We would also like to extend our gratitude to him for allowing us to present his information. This retrospective review of patient data did not require ethical approval according to hospital/national guidelines. A copy of the written informed consent will be made available for review to the journal editor.
Conflict of Interest Statement
P.M.K. reports grants paid to the institution by Merck, Agenus Bio, Novartis, Advanced Accelerator Applications, TerSera, and Boston Scientific; a consultancy and advisory board relationship with Elicio (scientific advisory board member/shares/stock ownership); co-founder of Precision BioSensors Inc.; consultancy/advisory board fees from Guardant Health, Natera, Foundation Medicine, Illumina, BostonGene, Merck/MSD Oncology, Tempus, Bayer, Lilly, Delcath Systems, IPBA, QED Therapeutics, Boston Healthcare Associates, Servier, Taiho Oncology, Exact Sciences, Daiichi Sankyo/AstraZeneca, Eisai, Saga Diagnostics, NeoGenomics, DoMore Diagnostics AS, and Seattle Genetics; consulting fees paid to the institution by Taiho Pharmaceutical and Ipsen; receiving travel support from AstraZeneca for presentation of an investigator initiated trial. All other authors have no conflicts of interest to report.
Funding Sources
No funding was received for this article.
Author Contributions
A.L., M.K.A., and P.M.K. were responsible for drafting the manuscript and providing clinical information. P.M.K. and B.S. were responsible for the final manuscript review. All authors have read and approved the final version of this study.
Funding Statement
No funding was received for this article.
Data Availability Statement
This is a case report, and all data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.
Supplementary Material
References
- 1. Kasi PM. Tumor-informed versus plasma-only liquid biopsy assay in a patient with multiple primary malignancies. JCO Precis Oncol. 2022;6:e2100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan HT, Nagayama S, Otaki M, Chin YM, Fukunaga Y, Ueno M, et al. Tumor-informed or tumor-agnostic circulating tumor DNA as a biomarker for risk of recurrence in resected colorectal cancer patients. Front Oncol. 2022;12:1055968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rizzo A, Ricci AD, Tavolari S, Brandi G. Circulating tumor DNA in biliary tract cancer: current evidence and future perspectives. Cancer Genom Proteom. 2020;17(5):441–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mauri G, Vitiello PP, Sogari A, Crisafulli G, Sartore-Bianchi A, Marsoni S, et al. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br J Cancer. 2022;127(3):394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB 3rd, Boland P, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal–Anal Task Forces whitepaper. Nat Rev Clin Oncol. 2020;17(12):757–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kotani D, Oki E, Nakamura Y, Yukami H, Mishima S, Bando H, et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat Med. 2023;29(1):127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2018;8(2):164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexander GE, Lin W, Ortega FE, Ramaiah M, Jung B, Ji L, et al. Analytical validation of a multi-cancer early detection test with cancer signal origin using a cell-free DNA–based targeted methylation assay. PLoS One. 2023;18(4):e0283001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klein EA, Richards D, Cohn A, Tummala M, Lapham R, Cosgrove D, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol. 2021;32(9):1167–77. [DOI] [PubMed] [Google Scholar]
- 10. Beer TM, McDonnell CH, Nadauld L, Liu MC, Klein EA, Reid RL, et al. A prespecified interim analysis of the PATHFINDER study: performance of a multicancer early detection test in support of clinical implementation. J Clin Oncol. 2021;39(15_Suppl l):3070. [Google Scholar]
- 11. Zheng Y, Hlady RA, Joyce BT, Robertson KD, He C, Nannini DR, et al. DNA methylation of individual repetitive elements in hepatitis C virus infection-induced hepatocellular carcinoma. Clin Epigenetics. 2019;11(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El-Bendary M, Nour D, Arafa M, Neamatallah M. Methylation of tumour suppressor genes RUNX3, RASSF1A and E-Cadherin in HCV-related liver cirrhosis and hepatocellular carcinoma. Br J Biomed Sci. 2020;77(1):35–40. [DOI] [PubMed] [Google Scholar]
- 13. Zekri AERN, Nassar AAM, El-Din El-Rouby MN, Shousha HI, Barakat AB, El-Desouky ED, et al. Disease progression from chronic hepatitis C to cirrhosis and hepatocellular carcinoma is associated with increasing DNA promoter methylation. Asian Pac J Cancer Prev. 2014;14(11):6721–6. [DOI] [PubMed] [Google Scholar]
- 14. Fan A, Wang B, Wang X, Nie Y, Fan D, Zhao X, et al. Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci. 2021;17(14):3837–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This is a case report, and all data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.