Abstract
Overproduction of the capsular polysaccharide alginate appears to confer a selective advantage for Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. The regulators AlgB and AlgR, which are both required as positive activators in alginate overproduction, have homology with the regulator class of two-component environmental responsive proteins which coordinate gene expression through signal transduction mechanisms. Signal transduction in this class of proteins generally occurs via autophosphorylation of the sensor kinase protein and phosphotransfer from the sensor to a conserved aspartate residue, which is present in the amino terminus of the response regulator. Recently, kinB was identified downstream of algB and was shown to encode the cognate histidine protein kinase that efficiently phosphorylates AlgB. However, we show here that a null mutation in kinB in a mucoid cystic fibrosis isolate, P. aeruginosa FRD1, did not block alginate production. The role of the conserved aspartate residue in the phosphorylation of AlgB was examined. The predicted phosphorylation site of AlgB (D59) was mutated to asparagine (N), and a derivative of an AlgB lacking the entire amino-terminal phosphorylation domain (AlgBΔ1-145) was constructed. A hexahistidine tag was included at the amino terminus of the wild-type (H-AlgB), H-AlgBΔ1-145, and mutant (H-AlgB.59N) AlgB proteins. These derivatives were purified by Ni2+ affinity chromatography and examined for in vitro phosphorylation by the purified sensor kinase protein, KinB. The results indicated that while KinB efficiently phosphorylated H-AlgB, no phosphorylation of H-AlgBΔ1-145 or H-AlgB.D59N was apparent. An allelic exchange system was developed to transfer mutant algB alleles onto the chromosome of a P. aeruginosa algB mutant to examine the effect on alginate production. Despite the defect in AlgB phosphorylation, P. aeruginosa strains expressing AlgB.D59N or H-AlgBΔ1-145 remained mucoid. The roles of the conserved aspartate residues in the phosphorylation of AlgR were also examined. As seen with AlgB, mutations in the predicted phosphorylation site of AlgR (AlgR.D54N and AlgR.D85N) did not affect alginate production. These results indicate that in vivo phosphorylation of AlgB and AlgR are not required for their roles in alginate production. Thus, the mechanism by which these response regulators activate alginate genes in mucoid P. aeruginosa appears not to be mediated by conventional phosphorylation-dependent signal transduction.
Cystic fibrosis (CF) is a common, serious, and often fatal genetic disease characterized by oversecretion of pulmonary mucus, bacterial infections, respiratory congestion, and, in many cases, death due to respiratory failure. Although the lungs of CF patients are colonized by several microorganisms, infections by Pseudomonas aeruginosa are the most common, are usually chronic, and are the most serious in terms of clinical prognosis (15, 23). P. aeruginosa isolates from such chronic infections often have a mucoid colony appearance. This phenotype is due to the overproduction and secretion of a capsular polysaccharide called alginate which plays an important role in chronic P. aeruginosa infections in CF patients (for a review, see reference 23).
Alginate production is controlled by a complex regulatory hierarchy involving several genes (65). A key element in alginate gene regulation is the alternative sigma factor ς22 (alternatively known as AlgT and AlgU), which is a member of the RpoE family of extracytoplasmic function sigma factors (13, 30). The activity of ς22 appears to be modulated by the mucABCD gene products, which are encoded by the algT gene operon at 68 min on the P. aeruginosa chromosome (23, 34, 41). Many mucoid P. aeruginosa isolates derived from CF patients harbor mutations in mucA (32), and inactivation of mucA or mucB (also referred to as algN) in wild-type nonmucoid P. aeruginosa strains causes induction of alginate synthesis (20, 31, 32). A membrane complex formed by MucA-MucB may be involved in regulating the stability of ς22 in the cell (34). Biochemical data show that MucA has an affinity for ς22 (50, 66). Active ς22 induces the expression of at least four genes or operons which are required for alginate synthesis. These include the algT operon (13, 33), the algD operon encoding most of the genes required for alginate synthesis (8, 11, 65), algR (33, 65), and the algB operon (29, 64, 65).
The algB and algR genes encode proteins that have homology to response regulators of the two-component superfamily (44). Both AlgR and AlgB control alginate levels by activating transcription of algD, the first gene of the alginate biosynthetic operon located at 34 min on the P. aeruginosa chromosome (8, 11, 65). AlgR activates algD expression directly by binding to three sites, two of which are located unusually far upstream of the algD transcription start site (25, 37).
The mechanism by which AlgB stimulates algD transcription and alginate production is unclear. AlgB shows homology to response regulators of the NtrC subfamily (63). Although AlgR contains a conserved amino-terminal phosphorylation domain typical of response regulators, its output domain does not appear to fall into a known subfamily (10).
Response regulators generally have a cognate sensor kinase protein that responds to environmental stimuli and undergoes autophosphorylation at a histidine residue. The phosphate is then transferred to an aspartate residue in the amino-terminal domain of the response regulator. This phosphorylation usually activates the response regulator leading to an adaptive response. This kind of phosphorelay is a general mechanism for the activation of response regulators of two-component regulatory systems (for a review, see reference 43). Recently, a gene downstream of algB, designated kinB, was identified to encode the cognate sensor kinase for AlgB (29). The KinB protein was localized to the membrane, and a purified carboxyl terminus of KinB was able to undergo autophosphorylation and to phosphorylate AlgB (29). Upstream of algR is fimS, a gene involved in type 4 pilus-mediated twitching motility that encodes an atypical sensor protein (62); it has also been termed algZ (68).
Despite the evidence that KinB-AlgB and FimS-AlgR are cognate sensor response regulator pairs, it has not been established that phosphorylation of AlgB or AlgR is required for alginate production in vivo. This is a clinically important question, since it has been proposed that inhibitors of two-component signal transduction systems might have therapeutic value for CF patients colonized with P. aeruginosa (46). In the present study, P. aeruginosa strains with mutations in kinB, algB, and algR were constructed to test the role of phosphorylation and signal transduction in alginate synthesis. These studies showed that the response regulators AlgB and AlgR did not require phosphorylation in order to promote alginate production in mucoid P. aeruginosa. This suggests that an alternative and unusual mechanism may be used by these response regulators to activate gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The P. aeruginosa strains used in this study are listed in Table 1. Escherichia coli JM109 (Promega) and XL1-Blue (Stratagene) were used for most plasmid manipulations. Bacteria were cultured in L broth (10.0 g of tryptone, 5.0 g of yeast extract, 5.0 g of sodium chloride per liter [pH 7.5]) or on L agar (Difco) plates. The media used for selection of P. aeruginosa and counterselection of E. coli following triparental mating were either a 1:1 mix of L agar and Pseudomonas Isolation Agar (Difco) or L agar lacking sodium chloride plus irgasan (Irgasan DP300; Ciba Geigy) at a final concentration of 25 μg/ml. Sucrose plates (for sacB-mediated counterselection) contained sucrose at a concentration of 5% (wt/vol) in L agar lacking sodium chloride, and the cultures were incubated at 30°C for 24 h. Selective antibiotics were used at the following concentrations for P. aeruginosa: carbenicillin, 300 μg/ml; gentamicin, 100 μg/ml; and tetracycline, 100 μg/ml. For E. coli, the concentrations were as follows: ampicillin, 100 μg/ml; gentamicin, 15 μg/ml; and tetracycline, 15 μg/ml. Mercuric chloride was used at 18 μg/ml for both P. aeruginosa and E. coli. Chemicals were purchased from Sigma unless stated otherwise.
TABLE 1.
P. aeruginosa strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Characteristic(s) or sequence | Source or reference |
|---|---|---|
| E. coli | ||
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Promega |
| XL1-Blue | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10] | Stratagene |
| BMH71-18 mutS | thi supE lac-proAB mutS::Tn10 [F′ proAB lacIqZΔM15] | Promega |
| TB1 | ara Δ(lac-proAB) rspL φ80 lacZΔM15 hsdR | Bethesda Research Laboratories |
| BL21 (λDE3) | F−ompT [lon] hsdSB(rB− mB−; B strain); λDE3 lysogen with T7 RNA polymerase | Novagen |
| P. aeruginosa | ||
| FRD1 | CF isolate; Alg+ (due to mucA22 mutation) | 40 |
| FRD2 | algT18 Alg− | 13, 40 |
| FRD440 | algT::Tn501 | 18 |
| FRD444 | algB::Tn501 | 22 |
| FRD831 | ΔalgR::ΩaacC1 | This study |
| FRD833 | algR+ derivative of FRD831 | This study |
| FRD836 | algR7 (encoding AlgR.D54N) | This study |
| FRD838 | algR10 (encoding AlgR.D85N) | This study |
| FRD839 | algR11 (encoding AlgR.D54N.D85N) | This study |
| FRD840 | algBΔ::ΩaacC1 | This study |
| FRD842 | algB48 (encoding AlgB.R442E) | This study |
| FRD844 | algB45 (encoding AlgB.D59N) | This study |
| FRD846 | algB+ derivative of FRD840 | This study |
| FRD848 | algB22 (encoding H-AlgB) | This study |
| FRD850 | algB23 (encoding H-AlgBΔ1-145) | This study |
| FRD1049 | kinB::Tn501 | This study |
| Plasmids | ||
| pALTER-1 | ColE1 Tcr Amps f1ori | Promega |
| pRK2013 | ColE1-Tra(RK2)+ Kmr | 17 |
| pUC18 | ColE1 Ampr | 57 |
| pLAFR3 | IncP1 Tcr λcos oriT | 54 |
| pUC4K | ColE1 Ampr Kmr | Pharmacia |
| pKS− | ColE1 Ampr | Stratagene |
| pTrcHisA | ColE1 AmprlacIq Ptrc 6x His tag vector | Invitrogen |
| pEX100T | ColE1 AmproriT sacB | 53 |
| pEMR-ST | ColE1 Tcr Amps f1ori cos oriT sacB | 36 |
| pUCGM | ColE1 Ampr GmrΩaacC1 | 52 |
| pAB2001 | ColE1 Ampr Gmr | 4 |
| pSM14 | pJG222 + 0.8-kb HindIII fragment of cat; kinB-cat transcriptional fusion | This study |
| pSM50 | pSM14 with KpnI-AgeI fragment deleted | This study |
| pSM53 | pSM50 + 1.6-kb HindIII fragment containing oriT | This study |
| pSM95 | ColE1 Kmr pT7 His-KinB | 29 |
| pAL1 | IncP1 Tcr pCP13 + 20-kb HindIII fragment; algR+ | 65 |
| pJG1 | IncP1 Tcr pLAFR1 + 20-kb EcoRI fragment; algB+ kinB′ | 21 |
| pJG1::Tn501-49 | pJG1 with kinB::Tn501 | 22 |
| pDJW15 | pKS− + 3.9-kb HindIII-XhoI fragment; algB+ | 64 |
| pDJW17 | pALTER-1 + 4.3-kb HindIII-EcoRI fragment; algB+ | This study |
| pDJW52 | pKK223-3 + ptac-algB; Ampr | 63 |
| pDJW130 | pKS− + 0.9-kb XhoI-EcoRI fragment of pJG1; ′kinB | This study |
| pDJW148 | pALTER-1 + 3.9-kb HindIII-XhoI fragment; algB+ | This study |
| pDJW385 | pDJW106 mutagenized with algR5 and algR6 | This study |
| pDJW387 | pDJW385 ΔBglII + 1.5-kb BamHI of pUCGM; ΔalgR::ΩaacC1 | This study |
| pDJW389 | pEMR-ST + 3.7-kb SalI of pDJW387; ΔalgR::ΩaacC1 | This study |
| pDJW400 | pUC18 + 1.3-kb BamHI-EcoRI PCR fragment; algB22 | This study |
| pDJW403 | pTrcHisA + 1.3-kb BamHI-EcoRI fragment of pDJW400; algB22 | This study |
| pDJW406 | pUC18 + 800-bp BamHI-EcoRI PCR fragment; algB23 | This study |
| pDJW408 | pTrcHisA + 800-bp BamHI-EcoRI PCR fragment; algB23 | This study |
| pDJW430 | pALTER-1 + 3.0-kb SphI fragment of pDJW403; algB22 | This study |
| pDJW433 | pALTER-1 + 2.6-kb SphI of pDJW408; algB23 | This study |
| pDJW437 | pAB2001 + 3.0-kb SphI fragment of pDJW430; algB22 | This study |
| pDJW438 | pAB2001 + 2.6-kb SphI of pDJW433; algB23 | This study |
| pDJW470 | pUS68 + 3.1-kb KpnI-XhoI of pDJW437; algB22 | This study |
| pDJW471 | pUS68 + 2.7-kb KpnI-XhoI of pDJW438; algB23 | This study |
| pDJW525 | ColE1 Ampr KmroriT sacB | This study |
| pDJW527 | ColE1 Ampr GmroriT sacB | This study |
| pUS4 | pDJW148 mutagenized with oligonucleotide algB45 | This study |
| pUS5 | pDJW148 mutagenized with oligonucleotide algB48 | This study |
| pUS14 | pLAFR3 + HindIII-BamHI fragment of pUS4; algB45 | This study |
| pUS50 | pEX100T + 3.9-kb HindIII-XhoI fragment of pUS4; algB45 | This study |
| pUS51 | pEX100T + 3.9-kb HindIII-XhoI fragment of pUS5; algB48 | This study |
| pUS55 | pUC18 + 1.3-kb BamHI-EcoRI PCR fragment; algB45 | This study |
| pUS56 | pTrcHisA + 1.3-kb BamHI-EcoRI fragment of pUS55; algB45 | This study |
| pUS61 | pDJW17 mutagenized with oligonucleotides algB50 and algB51 | This study |
| pUS63 | pUS66 + 1.5-kb BamHI fragment of pUCGM; ΔalgB::ΩaacC1 | This study |
| pUS65 | pEX100T + 5.1-kb EcoRI-ClaI fragment of pUS63; ΔalgB::ΩaacC1 | This study |
| pUS66 | pUS61 cleaved with BglII and the 1.8-kb algB fragment deleted; ΔalgB | This study |
| pUS68 | pEX100T + 2.9-kb HindIII-EcoRI fragment of pUS66; ΔalgB | This study |
| pUS69 | pUS68 + 2.1-kb KpnI-XhoI fragment of pDJW17; algB+ | This study |
| pUS150 | pEX100T + 2.7-kb SalI fragment from pDJW106; algR | This study |
| pUS152 | pDJW106 mutagenized with oligonucleotide algR7 | This study |
| pUS157 | pEX100T + 2.7-kb SalI fragment from pUS152; algR7 | This study |
| pUS164 | pDJW106 mutagenized with oligonucleotide algR10 | This study |
| pUS165 | pDJW106 mutagenized with oligonucleotides algR7 and algR10; algR11 | This study |
| pUS166 | pDJW525 + 2.7-kb SalI fragment from pUS164; algR10 | This study |
| pUS168 | pDJW525 + 2.7-kb SalI fragment from pUS165; algR11 | This study |
| Oligonucleotides | ||
| algB45 | GGCGCAGGTTGAGGAAGCAa | This study |
| algB48 | TGCTTGCGCTTCTCGTACAGGGTCGAa | This study |
| algB50 | TTCGCTACCCAGATCTGCCTGGCCCCa | This study |
| algB51 | ACCGGGTCCAGATCTTCATCGGa | This study |
| algB52 | CGGGATCCATGGAAACCACTTCC | This study |
| algB53 | CGGAATTCATCGGCAGCGGC | This study |
| algB54 | CGGGATCCTGGAATCGCACAGGCC | This study |
| algR5 | CGGCACTAGATCTGTCTACGa | This study |
| algR6 | GCATCAGATCTGACGGCa | This study |
| algR7 | GTCCTGCTGAATATCCGCAa | This study |
| algR10 | ACGGCCCATAACGAATTCGa | This study |
| algR11 | GAAGCGCTGACGCTGAT | This study |
| algR12 | TCGCTGCGCACCGGCTTGA | This study |
| kinBP7 | CTCGACAGCATCGACGACGG | This study |
| kinBP10 | CGCTATGCCCGCCGGGATGG | This study |
Mutagenic oligonucleotide (underlined residue[s] represents altered nucleotide).
Plasmids and DNA manipulations.
The plasmids and oligonucleotides used in the study are listed in Table 1. Restriction enzymes were purchased from Boehringer Mannheim, Promega, or New England Biolabs. Protocols for routine cloning were described elsewhere (1, 63). Triparental matings as described previously (21, 65) were used to mobilize plasmids into P. aeruginosa. DNA sequences from plasmid DNA were determined by the dideoxy chain-termination method as described previously (63) with minor modifications. PCRs were performed as described elsewhere (3). Oligonucleotide-directed mutagenesis was performed with the Altered Sites Mutagenesis system (Promega), pALTER-1, and mutagenic oligonucleotides (Table 1) as described by the manufacturer. Plasmid pDJW148 was mutagenized with oligonucleotides algB45 and algB48 to generate the algB45 and algB48 alleles, respectively, and the resulting plasmids were designated pUS4 (algB45) and pUS5 (algB48). To place BglII sites flanking algB, pDJW17 was mutagenized with oligonucleotides algB50 and algB51 to generate pUS61. To place BglII sites flanking algR, pDJW106 was mutagenized with oligonucleotides algR5 and algR6 to generate pDJW385.
Determination of the sites of Tn501 insertion in algB and kinB.
The exact position of the algB::Tn501-2 in FRD444 was determined by sequence analysis and shown to be inserted following bp 1034 of the algB open reading frame. By restriction analysis and Southern hybridization, pJG1::Tn501-49 (22) was shown to have a Tn501 insertion ∼300 bp into the kinB open reading frame. P. aeruginosa genomic DNAs from FRD1 and FRD1049 (with Tn501-49 in the chromosome) were isolated as described previously (21). Tn501 has EcoRI sites at both its termini, which were used for mapping. The DNAs were digested with EcoRI and ClaI, subjected to electrophoresis on 0.7% agarose (SeaKem; FMC), and transferred to a nylon membrane (Boehringer Mannheim) by the capillary transfer procedure described elsewhere (1). A 280-bp digoxigenin-labeled probe, matching kinB sequences 5′ to the EcoRI site, was synthesized from pDJW130 by PCR with oligonucleotide primers P7 and P10 (Table 1) following a prior protocol (27, 29). Hybridization and detection were performed with the Genius system (Boehringer Mannheim), which revealed 1.2- and 0.9-kb bands in FRD1 and FRD1049, respectively.
Analysis of cat transcriptional fusions.
Extracts of P. aeruginosa containing pSM53 (kinB-cat) were obtained as previously described (63). Extracts were assayed for protein concentrations by the Bradford method (7) and were assayed for chloramphenicol acetyltransferase (CAT) levels by an enzyme-linked immunosorbent assay as indicated by the manufacturer (5 Prime→3 Prime, Inc., Boulder, Colo.). CAT levels in dilutions of the cell extracts were determined by extrapolation from a standard curve and were normalized for protein content. The values were expressed as picograms of CAT per microgram of extract protein and are averages from three independent experiments.
Allelic exchange techniques.
A kinB::Tn501-49 mutant of FRD1 was generated with PAO1(pJG1::Tn501-49) and phage F116L to transfer plasmid DNA fragments by a transduction-mediated gene replacement technique as previously described (42). Mutants with altered algB alleles were generated by gene replacement with suicide plasmids containing sacB for counterselection. A schematic representation of the allele replacement technique is illustrated in Fig. 5. In order to generate the intermediate strain FRD840 (ΔalgB::ΩaacC1) used as a recipient for most gene replacements at algB, single-stranded DNA from JM109/pDJW17 was subjected to site-directed mutagenesis with oligonucleotides algB50 and algB51. This was performed to introduce BglII cloning sites 5′ and 3′ of the algB coding sequence due to the lack of convenient restriction sites. The positions of these sites were important, since all desired algB mutations had to be contained within the BglII restriction sites (see below). The resulting plasmid (pUS61) was cleaved with BglII, and a 1.5-kb ΩaacC1 cassette (encoding resistance to gentamicin [Gmr]) derived from pUCGM by treatment with BamHI was used to replace algB to form pUS63. The ΔalgB::ΩaacC1 allele with flanking sequences was subcloned into pEX100T, a ColE1 carbenicillin resistance (Cbr) vector used for allelic exchange in P. aeruginosa (53). pEX100T can be propagated in E. coli but cannot replicate in P. aeruginosa. This vector has an oriT sequence which allows for pRK2013-mediated transfer from E. coli to P. aeruginosa. In addition, pEX100T contains the sacB gene, allowing for counterselection when P. aeruginosa strains containing sacB are cultured in the presence of sucrose (51). The subsequent plasmid (pUS65) was transferred to P. aeruginosa FRD1 (Alg+), and colonies were selected for Gmr (see Fig. 5A). Since pEX100T cannot replicate in P. aeruginosa, the only way in which a Gmr colony can arise is through homologous recombination between sequences on the chromosome and sequences flanking algB on pUS65. Most Gmr colonies were also Cbr and contained both wild-type and ΔalgB::ΩaacC1 alleles, indicating single recombination events (merodiploids). To generate the second recombination, a Gmr merodiploid strain was cultured overnight and aliquots were plated on media containing gentamicin (selectable marker) and sucrose (counterselectable marker). These sucrose-resistant, Gmr bacteria were screened for loss of Cbr, and introduction of the ΔalgB::ΩaacC1 mutant allele was verified by PCR and Southern hybridizations of chromosomal DNA (data not shown). Techniques similar to those outlined above were used to generate FRD831 (ΔalgR::ΩaacC1), an intermediate strain used for algR allele replacements, except that the gene replacement plasmid pDJW389 used to create the intermediate strain was derived from pEMR-ST (36) rather than pEX100T. To introduce specific algB alleles (e.g., mutation algB45 [see Fig. 5B]), the reverse procedure was utilized, relying on regions of homology flanking algB. algB alleles (plus flanking sequences, e.g., pUS50 [see Fig. 5B]) were subcloned into pEX100T and transferred to FRD840, and the transconjugants were plated on carbenicillin plates to select for merodiploids. These Cbr Gmr, sucrose-sensitive bacteria were plated on sucrose media to select for the second recombination. This provided a direct selection for allele replacement and introduction of the algB mutation into the chromosome. As a second screen, the sucrose-resistant bacteria were tested for sensitivity to carbenicillin and gentamicin. Thus, the final strain contained the desired single-copy allele at the algB locus and did not require antibiotic selection. Introduction of each mutation was verified by PCR amplification of mutant chromosomal DNA followed by DNA sequence analysis of the PCR product (data not shown).
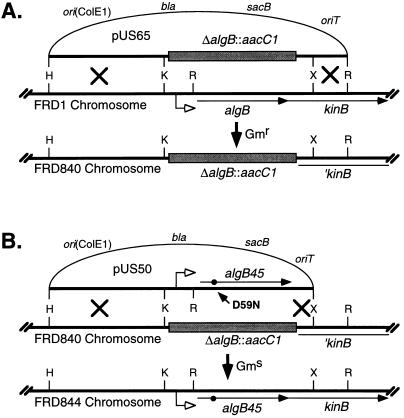
FIG. 5.
Depiction of the two-step procedure used to transfer algB alleles to the P. aeruginosa chromosome by allelic exchange. (A) Generation of ΔalgB::ΩaacC1 intermediate strain FRD840. Plasmid pUS65 (pEX100T plus ΔalgB::ΩaacC1), which cannot replicate in P. aeruginosa, was mobilized into strain FRD1 and single-crossover recombinants were isolated by selection for Gmr. Most Gmr bacteria were also Cbr, indicating integration of the entire plasmid (single-crossover events). Double recombinants were isolated by plating Gmr bacteria on agar containing 5% sucrose. The desired (double) recombinants were Gmr Cbs (marker on pUS65). (B) Allele replacements. To perform allele replacements, pEX100T containing a specific algB allele (algB45 in this example) was mobilized into the intermediate strain FRD840, and single recombinants were selected by resistance to carbenicillin. The desired allele replacements (Gms Cbs) were then obtained by counterselection on sucrose-containing medium. Abbreviations: ori, ColE1 origin; sacB, gene encoding levansucrase; oriT, transfer origin; bla, resistance to carbenicillin; aacC1, resistance to gentamicin; s, antibiotic sensitivity; Cb, carbenicillin; Gm, gentamicin; H, HindIII; K, KpnI; R, EcoRI; X, XhoI.
Most pEX100T-derived plasmids were generated from pALTER-1 derivatives (Table 1). Plasmid pUS50 was derived by cleaving pUS4 with HindIII-BamHI treatment with T4 polymerase and subcloning the 3.9-kb fragment into the unique SmaI site of pEX100T. This plasmid was mobilized into P. aeruginosa FRD1, and allele replacements were performed as above, generating strain FRD844. Similar approaches were used to produce strain FRD842 from pUS51 (algB48). Some allele replacements were performed with derivatives of pUS68. Plasmid pUS69 (wild-type algB) was constructed simply by subcloning the 2.1-kb KpnI-XhoI fragment from DJW17 into similarly digested pUS68. pUS69 was used to generate P. aeruginosa FRD846. P. aeruginosa FRD848 (algB22 [H-AlgB]) and FRD850 (algB23 [H-AlgBΔ1-145]) were constructed by similar allelic exchange techniques with plasmids pDJW470 and pDJW471, respectively. For gene replacements involving algR, the intermediate strain FRD831 (ΔalgR::ΩaacC1) was utilized with pEX100T- or pDJW525-derived plasmids harboring wild-type or mutant algR alleles. These plasmids, which included pUS150 (wild-type algR), pUS157 (algR7), pUS166 (algR10), and pUS168 (algR11), were used in allele replacements of FRD831 to generate P. aeruginosa FRD833, FRD836, FRD838, and FRD839, respectively.
Purification of AlgB and H-AlgB proteins.
AlgB was overproduced in E. coli XL1-Blue(pDJW52) and purified by streptomycin sulfate precipitation, precipitation with 30% ammonium sulfate, and DEAE anion-exchange chromatography essentially as previously described (29). Approximately 40 μg of this AlgB preparation (>90% pure) was concentrated with an Applied Biosystems Pro Spin Sample Preparation Cartridge, and the polyvinylidene difluoride membrane containing AlgB was subjected to direct amino-terminal sequence analysis. The sequence was found to be Glu-Thr-Thr-Ser-Glu-Lys-Gln-Gly-Arg-Ile-Leu, which is the same as that deduced from previous DNA sequence analysis of algB (63).
H-AlgB fusion proteins were expressed and purified from E. coli JM109 containing either pDJW403 (H-AlgB), pUS56 (H-AlgB.D59N), or pDJW408 (H-AlgBΔ1-145). DNA encoding H-AlgB or H-AlgB.D59N was obtained by PCR amplification of plasmids containing wild-type algB (pDJW148) or algB45 (pUS4) with primers algB52 and algB53. The 1.3-kb fragments from the PCR amplification products were cloned into pUC18 as BamHI-EcoRI fragments resulting in pDJW400 (wild-type algB) or pUS55 (algB45), and the DNA sequences of the PCR-generated fragments were determined and shown to be identical to pDJW148 or pUS4 sequences, respectively (data not shown). The BamHI-EcoRI fragments of pDJW400 or pUS5 were subcloned into pTrcHisA (Invitrogen) to generate pDJW403 and pUS56, respectively. Similar approaches were used to clone DNA expressing H-AlgBΔ1-145, except that oligonucleotide algB54 was substituted for algB52 in the PCR of pDJW148. The 0.8-kb BamHI-EcoRI PCR fragment was cloned into pUC18 (pDJW406), and the sequence was determined to be identical to that of pDJW148. The BamHI-EcoRI fragment of pDJW406 was subcloned into pTrcHisA to generate pDJW408. pTrcHisA is an expression vector which contains the Ptac promoter with a lac operator sequence, the lacIq gene, and a multicloning site. When the BamHI-EcoRI fragments described above were cloned into pTrcHisA, the resulting plasmids expressed fusion proteins consisting of an amino-terminal (∼3-kDa) peptide sequence derived from bacteriophage T7 coat protein and an additional stretch of six histidine residues. The hexahistidine tag allowed for purification of the H-AlgB proteins by nickel agarose chromatography (Qiagen). For purification of the H-AlgB proteins, 500-ml cultures of JM109 harboring pDJW403, pUS56, or pDJW408 were cultured in L broth plus ampicillin to an A600 of 0.3. Isopropyl-β-d-thiogalactopyranoside was added to a concentration of 1 mM, and the cells were cultured for an additional 3 h, harvested by centrifugation, and suspended in 5 ml of fractionation buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM MgCl2). Cell extracts were prepared by subjecting the mixture to a French press (15,000 lb/in2) followed by centrifugation. H-AlgB proteins were purified under native conditions from the supernatant fraction by nickel agarose chromatography as outlined by the manufacturer of the Ni-nitrilotriacetic acid agarose resin (Qiagen). Approximately 1 mg of pure H-AlgB was obtained per 500 ml of culture.
Immunoblot analysis.
Polyclonal antisera against AlgB were elicited in New Zealand White rabbits (Immunodynamics, Inc.) with purified AlgB protein (0.75 mg). Anti-AlgB antibodies were used in immunoblots at a dilution of 1:20,000 with chemiluminescent reagents by procedures outlined by the manufacturer (Amersham), and film was exposed for 30 s prior to development.
In vitro phosphorylation assays.
The conditions used in the autophosphorylation of KinB and phosphotransfer from KinB to AlgB have been described previously (29). Briefly, the cytoplasmic carboxy terminus of KinB (1.3 μM) was incubated with 33.3 μM [γ-32P]ATP for 60 min at room temperature in a buffer containing 50 mM KCl and 5 mM MgCl2. H-AlgB protein (3.0 μM) was added to the mixture, which was further incubated for 60 s. The reaction was terminated by adding sodium dodecyl sulfate (SDS) sample buffer (60 mM Tris hydrochloride [pH 6.8], 2% SDS, 10% glycerol, 0.1 mg/ml bromphenol blue, 5% 2-mercaptoethanol), unincorporated label was removed, and the products were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography. For competition assays, kinase reactions were performed as described above, except that H-AlgB.D59N or H-AlgBΔ1-145 was included in the mixture at concentrations of 1.3, 5.2, or 13.0 μM.
Alginate assays.
Alginates were collected from cultures grown in L broth with rapid aeration at 37°C for 22 h, and levels were determined as previously outlined (26), with modifications (19). Briefly, samples (5 ml) of cultures were mixed with 5 ml of saline, and the cells were removed by centrifugation (12,000 × g for 30 min). The culture supernatant was mixed with 5 ml of 2% cetyl pyridinium chloride, and the precipitated alginate was collected by centrifugation (12,000 × g for 10 min at room temperature). The pellet was dissolved in 10 ml of 1 M NaCl, precipitated again with 10 ml of cold (−20°C) isopropanol, and dissolved in 10 ml of saline. The concentration of alginate in solution was determined by the carbazole method described by Knutson and Jeanes (26), in which a solution of alginate (30 μl) was mixed with 1.0 ml of borate-sulfuric acid reagent (10 mM H3BO3 in concentrated H2SO4) and 30 μl of carbazole reagent (0.1% in ethanol). The mixture was then incubated in a 55°C bath for 30 min, and absorbance at 530 nm was determined spectrophotometrically. The alginate concentration was determined by extrapolation from a standard curve with various concentrations (0 to 50 μg/ml) of alginate (high viscosity from Macrocystis pyrifera).
RESULTS
The algB and kinB genes form an operon.
AlgB is a two-component regulator that is required for expression of the alginate biosynthetic operon, and the level of algB expression is elevated in mucoid strains (64). The kinB gene, downstream and adjacent to algB, encodes a cognate histidine kinase that efficiently phosphorylates AlgB (29). To determine if kinB was part of the alginate regulon under ς22 (algT/algU) control, we examined the expression of kinB in Alg+ and Alg− P. aeruginosa. A kinB-cat transcriptional fusion in a suicide vector was constructed and integrated into the chromosomes of P. aeruginosa strains by single-crossover homologous recombination (Fig. 1A). Alg+ P. aeruginosa FRD1 carrying kinB-cat (pSM53) contained CAT levels that were approximately threefold higher than that seen in the Alg− algT18 mutant strain, FRD2 (Table 2). These results were similar to the expression levels of a plasmid-borne algB-cat fusion in these Alg+ and Alg− strains (64). This was not unexpected, since sequence analysis showed that the predicted translational start (ATG) for kinB overlaps the stop codon for algB, suggesting that they form an operon (29). This was further tested by examining whether the algB transposon insertion (algB::Tn501-2) in FRD444 was polar on the downstream kinB gene. The position of Tn501-2 in algB was determined by sequence analysis and was shown to be inserted following bp 1034 of the algB open reading frame. Analysis of kinB-cat expression in Alg− FRD444::pSM53 (where kinB-cat was positioned downstream of the polar Tn501-2 insertion [Fig. 1B]) revealed dramatically reduced kinB levels (Table 2). This further suggested that algB and kinB formed an operon. Furthermore, providing FRD444::pSM53 with algB in trans on pJG1 did not restore kinB-cat expression, indicating that kinB did not have an AlgB-dependent promoter. Interestingly, providing Alg− FRD444::pSM53 with algB in trans did restore the Alg+ phenotype, even though one would predict this strain to be kinB defective.
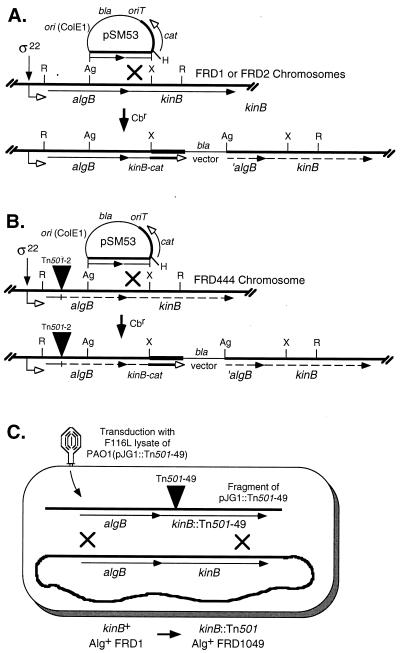
FIG. 1.
Diagram of genetic constructions used to modify the chromosomal kinB gene in P. aeruginosa FRD. (A) Construction of kinB-cat transcriptional fusion in Alg+ strain FRD1 and Alg− strain FRD2. A promoterless cat gene cassette (0.8-kb HindIII fragment) was cloned to form a kinB-cat transcriptional fusion in pSM53. This plasmid has a ColE1 origin, which cannot replicate autonomously in P. aeruginosa, and was integrated into the chromosomes of FRD1 and FRD2 (algT18) by homologous recombination via selection for carbenicillin resistance encoded by bla. Dashed arrows indicate genes that are not transcribed due to the polar upstream insertion of the vector. (B) Construction of kinB-cat transcriptional fusion in Alg− strain FRD444 (algB::Tn501-2). The Tn501 insertion in algB (closed triangle) is polar on downstream genes, as indicated by dashed arrows. (C) Construction of the kinB mutant FRD1049. A gene replacement technique (42) was used to transfer an kinB::Tn501 allele into the chromosome of P. aeruginosa FRD1. Briefly, a lysate of phage F116L was generated on P. aeruginosa PAO1(pJG1::Tn501-49) and used to transduce FRD1. Mercury resistance encoded by Tn501 was used to select for double-crossover events of kinB::Tn501 with the chromosome, and the strain was scored for loss of plasmid-borne tetracycline resistance. Abbreviations: Ag, AgeI; R, EcoRI; H, HindIII; X, XhoI; bla, gene encoding carbenicillin resistance; cat, gene encoding CAT.
TABLE 2.
Analysis of kinB-cat expression in P. aeruginosa strains
| Strain | Genotype | Alginate pheno- type | Amt of CAT (pg/μg of protein) from kinB-cata |
|---|---|---|---|
| FRD1 | Wild type | Alg+ | <10 |
| FRD1::pSM53 | kinB-cat | Alg+ | 438 ± 31 |
| FRD2::pSM53 | algT18 kinB-cat | Alg− | 146 ± 21 |
| FRD444::pSM53 | algB::Tn501 kinB-cat | Alg− | <10 |
| FRD444::pSM53 (pJG1) | algB::Tn501 kinB-cat (algB+) | Alg+ | <10 |
P. aeruginosa FRD strains containing a chromosomally integrated pSM53 (kinB-cat) were cultured to the same density in logarithmic phase and were harvested. As a measure of relative kinB transcription, cell extracts of these strains were assayed for CAT levels relative to the protein concentrations.
A kinB null mutation did not affect alginate production.
The genetic data above suggested that KinB was not essential for alginate production, even though kinB was in an operon with algB and encoded its cognate kinase, which has been shown to efficiently phosphorylate AlgB in vitro (29). To directly test the role of KinB in alginate production, a kinB::Tn501 mutant of Alg+ FRD1 was constructed by gene replacement. In a prior study, plasmid pJG1 was subjected to Tn501 transposon mutagenesis in an attempt to localize the algB gene (22). One of these plasmids, pJG1::Tn501-49, was found to carry a Tn501 insertion within the first 300 bp of the kinB open reading frame and was used to generate the kinB::Tn501 null mutant, FRD1049 (Fig. 1C). Interestingly, the colony morphology of FRD1049 on L agar (following incubation for 18 h at 37°C) was mucoid, and this strain synthesized alginate at levels comparable to those of the parental strain FRD1 (Fig. 2). Other mutants generated by insertional disruption of kinB in the FRD1 background also remained Alg+ (data not shown). As controls, strains FRD840 (ΔalgB::ΩaacC1, see below) and FRD440 (algT::Tn501) were nonmucoid and produced little if any detectable alginate (Fig. 2). Thus, a null mutation in kinB appeared to have no obvious effect on alginate production under the conditions tested here. This suggested that phosphorylation of AlgB by KinB was not required for alginate overproduction in mucoid P. aeruginosa.
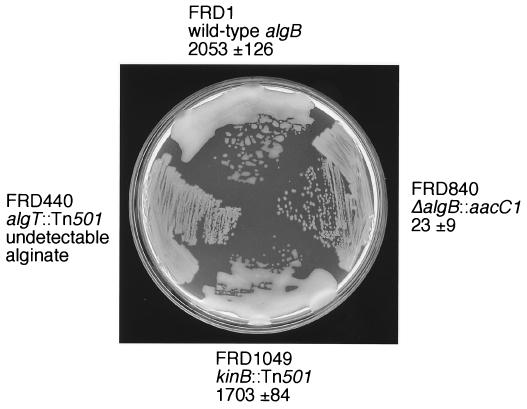
FIG. 2.
Plate phenotypes of P. aeruginosa strains carrying wild-type or mutant kinB alleles. FRD1, wild type; FRD840, ΔalgB::ΩaacC1; FRD1049, kinB::Tn501; and FRD440, algT::Tn501 on L agar. Numbers are levels (in micrograms per milliliter) of alginate produced as determined by the carbazole assay (19, 26). Note that a kinB null mutation (FRD1049) did not affect alginate production.
Purification of AlgB derivatives predicted to have phosphorylation defects.
The data above did not negate the possibility that AlgB was phosphorylated by another kinase. We then tested the possibility that AlgB activity for alginate production may require phosphorylation by a process independent of KinB kinase activity. Another histidine kinase (i.e., a cross-talk mechanism) or a small-molecular-weight phosphodonor may be sufficient for this phosphorylation reaction, and this has been proposed for other response regulators (35, 59). To address this, algB alleles that were predicted to encode AlgB proteins defective in phosphorylation were constructed. Based on its close relatedness to the well-studied NtrC subfamily of response regulators (38, 55, 63), AlgB was predicted to contain three functional domains (Fig. 3A): (i) an amino-terminal phosphorylation domain that is conserved across families of response regulators, (ii) a central nucleotide-binding domain that is required for facilitating transcription initiation by RNA polymerase containing ς54, and (iii) a carboxy-terminal helix-turn-helix motif that is presumably involved in binding DNA sequences that are often located far upstream of the target promoter.
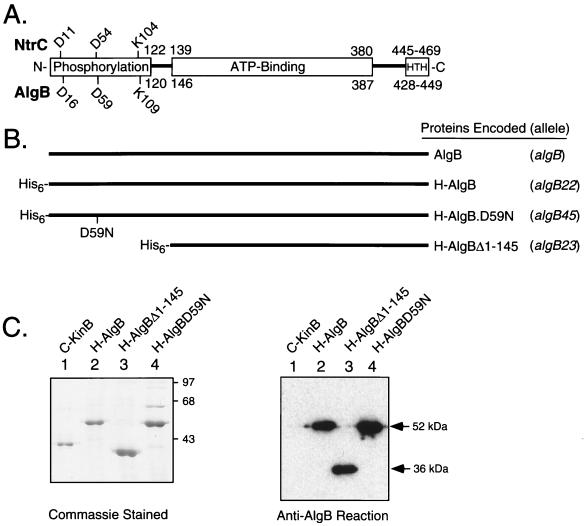
FIG. 3.
(A) The modular structures of the AlgB protein and related NtrC protein are depicted. AlgB and NtrC are homologous over the entire lengths of the proteins, including a central domain with consensus ATP binding sites and the helix-turn-helix (HTH) DNA binding domain (63). Numbers above and beneath the boxes indicate positions of amino acid residues in the respective proteins. Both proteins contain a highly conserved amino-terminal domain (residues 1 to 120) which in NtrC, CheY and PhoB, OmpR, and VirG (and likely AlgB and AlgR) represents the phosphorylation domain (56). Residues corresponding to Asp13, Asp57, and Lys109 in CheY are the most highly conserved among response regulators, since these residues cluster together around the site of phosphorylation. The phosphorylation site of NtrC (Asp54 in the center of the N-terminal domain) aligns with Asp57 of AlgB and Asp54 of AlgR. (B) Depiction of AlgB proteins used in this study. The algB alleles which encode these proteins are indicated on the right and include algB (wild-type protein), algB22 (H-AlgB), algB45 (H-AlgB.D59N), and algB23 (H-AlgBΔ1-145). (C) Purification of H-tagged wild-type and mutant AlgB proteins. (Left) H-tagged proteins were purified and subjected to SDS-PAGE followed by Commassie blue staining. Each lane contains 1.5 μg of protein purified from E. coli BL21(λDE3, pSM95) (lane 1) or JM109 containing pDJW403 (lane 2), pDJW408 (lane 3), or pUS56 (lane 4). Purified C-KinB (lane 1) was included to show its relative size compared to those of AlgB protein and derivatives. Positions of protein size markers (97, 68, and 43 kDa) are indicated. (Right) Immunoblot assay of purified H-AlgB proteins. Lanes 1 to 4, preparations identical to those described for the left gel except that 200 ng of protein/lane was used. Immunoblots were performed with rabbit anti-AlgB serum, and antigen-antibody complexes were detected with enhanced chemiluminescence reagents (Amersham).
The phosphorylation domain was targeted here for site-directed mutagenesis. In the well-characterized response regulator CheY, three essential residues in this domain, including one lysine and two aspartate residues, form an acid pocket (56, 58), and Asp-57 within this pocket is the site of phosphorylation (48). The aspartate residue represented by Asp-57 in CheY is also the primary site of phosphorylation in NtrC, VirG, and OmpR (9, 24, 47). The three highly conserved residues in the NtrC phosphorylation domain (Asp-11, Asp-54, and Lys-104) correspond to Asp-16, Asp-59, and Lys-109, respectively, in AlgB (Fig. 3A). To investigate the site of AlgB phosphorylation, the algB45 allele was constructed by oligonucleotide-directed mutagenesis to encode AlgB.D59N, in which the predicted phosphorylated residue Asp-59 was changed to asparagine (Fig. 3B). Also, the algB23 allele was constructed encoding AlgBΔ1-145, in which the entire phosphorylation domain of AlgB (residues 1 to 145) was deleted. To facilitate purification by nickel affinity chromatography, the wild-type and two mutant AlgB proteins were produced as fusion proteins with amino-terminal tags (∼3 kDa) consisting of six histidine residues and a peptide sequence derived from bacteriophage T7 coat protein. The purified His-tagged AlgB (H-AlgB) proteins showed the following expected mobilities on SDS-PAGE: 52 kDa for H-AlgB, 36 kDa for H-AlgBΔ1-145, and 52 kDa for H-AlgB.D59N (Fig. 3C-left, lanes 2, 3, and 4, respectively). For comparison, a previously described (29) 39-kDa soluble derivative of KinB (C-KinB) that lacked the amino-terminal membrane hydrophobic sequence yet retained kinase activity is shown (lane 1). All of the AlgB derivatives reacted with a polyclonal antiserum specific for AlgB in an immunoblot assay (Fig. 3C, right). The amino-terminal tags were not removed, because they did not appear to affect AlgB function (see below).
AlgB.D59N and AlgBΔ1-145 show defects in phosphorylation.
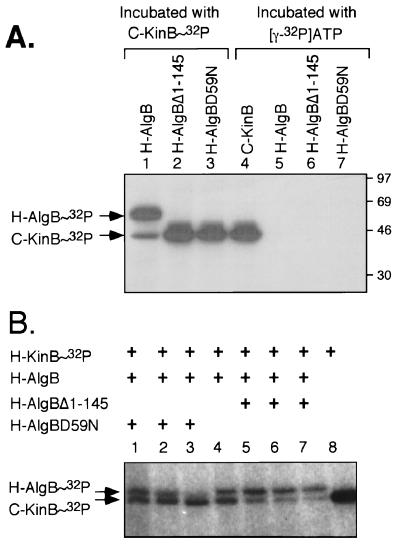
An in vitro reaction was used to determine whether the wild-type and mutant forms of H-AlgB were capable of being phosphorylated by KinB, its cognate histidine protein kinase. KinB is a membrane protein, but the soluble and readily purified carboxyl-terminal fragment of KinB (C-KinB) has been shown to rapidly phosphorylate AlgB (29) and was used here. When C-KinB (1.3 μM) was incubated with excess [γ-32P]ATP (33.3 μM), it underwent autophosphorylation (C-KinB∼P; Fig. 4A, lane 4) as previously described (29). None of the other H-AlgB proteins alone showed any autophosphorylation activity (lanes 5 to 7). When purified H-AlgB protein (3.0 μM) was incubated for 60 s with C-KinB∼P, most of the label transferred to H-AlgB (Fig. 4A, lane 1). Thus, the amino-terminal tag on H-AlgB did not block its phosphorylation by C-KinB∼P. However, phosphorylation of H-AlgBΔ1-145 (Fig. 4A, lane 2) or H-AlgB.D59N (lane 3) was not detected. This phosphotransfer procedure was performed with a wide range of H-AlgB.D59N and H-AlgBΔ1-145 protein concentrations, yet phosphorylation of these proteins was still not observed (data not shown).
FIG. 4.
Assays of in vitro phosphorylation of purified H-AlgB and derivatives by C-KinB∼32P. (A) C-KinB (1.3 μM) showed autophosphorylation following incubation with 33.3 μM [γ-32P]ATP for 60 min (lane 4) and was then incubated for 60 s with 3.0 μM purified H-AlgB (lane 1), H-AlgBΔ1-145 (lane 2), or H-AlgB.D59N (lane 3), followed by termination of the reaction with SDS sample buffer. Unincorporated label was removed, and the samples were analyzed by SDS–10% PAGE, followed by autoradiography. As controls, H-AlgB (lane 5), H-AlgBΔ1-145 (lane 6), and H-AlgB.D59N (lane 7) were incubated with [γ-32P]ATP under identical conditions for 60 s. The positions of the phosphorylated forms of C-KinB (C-KinB∼P) and H-AlgB (His-AlgB∼P) are indicated on the side. Note that C-KinB can phosphorylate H-AlgB but not H-AlgB.D59N or H-AlgBΔ1-145. (B) Demonstration that H-AlgB.D59N can compete with H-AlgB for interaction with C-KinB∼32P. (Top) schematic diagram of H-AlgB proteins used in the competition assay (+, addition of that protein). C-KinB was autophosphorylated as described above, and aliquots (1.3 μM) were removed and added to SDS-PAGE sample buffer (lane 8) or to a sample containing H-AlgB (3.0 μM) either alone (lane 4) or with increasing amounts of H-AlgB.D59N (lane 1, 1.3 μM; lane 2, 5.2 μM; lane 3, 13.0 μM H-AlgB.D59N addition) or H-AlgBΔ1-145 (lane 5, 1.3 μM; lane 6, 5.2 μM; lane 7, 13.0 μM H-AlgBΔ1-145 addition). Kinase reactions were allowed to proceed and analyzed as described for A.
A competition assay was also performed. Compared to the phosphorylation of H-AlgB in the presence of C-KinB∼P (Fig. 4B, lane 4), the addition of H-AlgB.D59N (10-fold molar excess) nearly eliminated H-AlgB phosphorylation (Fig. 4B, lane 3). However, this effect was not observed when H-AlgBΔ1-145 was used as a competitor (Fig. 4B, lanes 5 to 7). The ability of H-AlgB.D59N to inhibit the phosphorylation of H-AlgB suggests that this mutant protein still retained the ability to interact with C-KinB∼P even though it could not be phosphorylated. These data provide experimental evidence that Asp-59 represents the site of AlgB phosphorylation and fulfills the prediction based on sequence homology to NtrC. Thus, substitution of this residue or deletion of the phosphorylation domain was predicted to block phosphorylation of AlgB in vivo.
AlgB derivatives blocked in phosphorylation still promoted alginate production in mucoid P. aeruginosa.
To test the potential role of AlgB phosphorylation in alginate gene activation, the algB45 allele (encoding AlgB.D59N but lacking a His tag) was cloned onto pLAFR3, a low-copy-number, broad-host-range plasmid, to form pUS14, which was transferred to Alg− FRD444, an algB::Tn501 mutant. Although AlgB.D59N was shown above to be defective in phosphorylation, the transconjugates obtained were complemented and displayed the Alg+ phenotype (data not shown). This suggested that AlgB may function to promote alginate overproduction without phosphorylation. However, we could discount the possibility that this result was due to the multiple copies of plasmid-borne algB alleles in the cell.
To avoid the possibility of gene dosage effects, gene replacement was used to place mutant alleles onto the P. aeruginosa FRD1 chromosome so that they could be tested in single copy under native transcriptional and translational controls (see Materials and Methods). To test the role of AlgB phosphorylation, we constructed strain FRD844 through a two-step process in which the wild-type algB allele was exchanged for a ΩaacC1 cassette encoding Gmr and then by the algB45 allele, which encoded AlgB.D59N (Fig. 5). As a control, FRD846 was constructed by the same two-step process to restore the wild-type algB allele. FRD842 was constructed with the algB48 allele, which encoded AlgB.R442E. In addition, FRD848 (algB22 encoding H-AlgB) and FRD850 (algB23 encoding H-AlgBΔ1-145), in which the algB alleles were integrated into the chromosome, expressed under the vector’s promoter, and produced His-tagged proteins, were constructed.
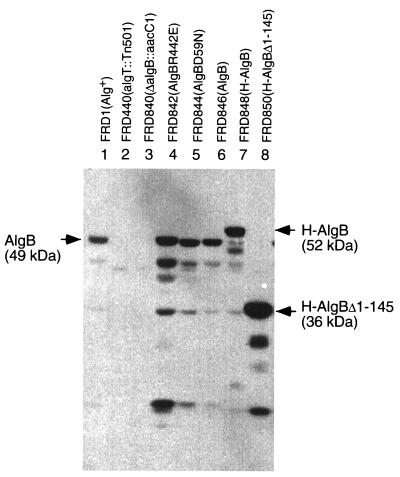
The presence of a single copy of algB in the chromosomes of each of these strains was confirmed by Southern hybridization (data not shown). An immunoblot analysis demonstrated AlgB-reactive bands of anticipated sizes in all of these strains, and proteins appeared to be at a level similar to that in wild-type FRD1 when expressed under the native algB promoter (Fig. 6). Also, the mutant alleles introduced encoded proteins that appeared to have approximately the same susceptibilities to endogenous proteolytic degradation in the cell extracts as wild-type AlgB, which suggests that the substitutions had little effect on the overall structures of the AlgB proteins (Fig. 6).
FIG. 6.
Immunoblot detection of AlgB in extracts derived from P. aeruginosa strains. Extracts were prepared from each strain (see Materials and Methods), and 35 μg was applied in each lane. The immunoblot was performed with a 1/20,000 dilution of rabbit anti-AlgB serum. Antigen-antibody complexes were detected with enhanced chemiluminescence reagents (Amersham). Positions (and molecular masses) of H-AlgB and wild-type AlgB proteins are indicated.
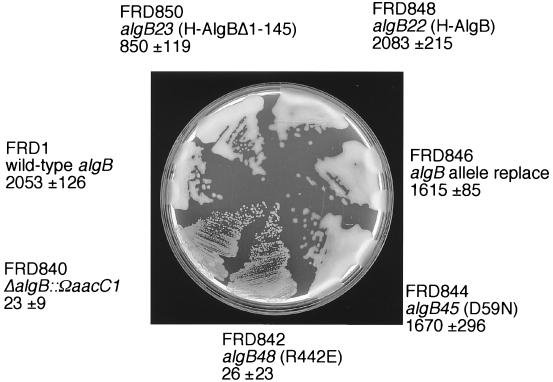
The colony morphologies of these strains were examined on L agar following incubation overnight at 37°C (Fig. 7). The ΔalgB::ΩaacC1 mutant FRD840 had an Alg− phenotype as expected, because algB is required for high-level alginate production (22). Replacing the ΔalgB::ΩaacC1 marker with wild-type algB restored the Alg+ phenotype in FRD846. However, FRD844 (AlgB.D59N) and FRD850 (AlgBΔ1-145) also had an Alg+ phenotype similar to that of FRD1, indicating that AlgB can promote high-level alginate production without phosphorylation. In contrast, the algB48 mutant FRD842, which produced AlgB.R442E with an altered DNA binding domain, was Alg− (Fig. 7). A plasmid-borne algB48 allele also failed to complement an algB::Tn501 mutant in trans. This suggests that AlgB functions as a DNA binding protein in its role in alginate production. The alginate levels produced by these strains in L broth (after 22 h of incubation at 37°C) was determined, and the values obtained were consistent with their colony morphologies; Alg− FRD840 (AlgB−) and FRD842 (AlgB.R442E) cultures synthesized only about 3% of the alginate made by the parental strain FRD1, whereas Alg+ FRD846 (AlgB+), FRD844 (AlgB.D59N), and FRD848 (H-AlgB) produced alginate levels that were similar to that of FRD1 (Fig. 7). Even FRD850, in which the entire phosphorylation domain of AlgB was deleted, still produced about 45% of wild-type alginate levels (Fig. 7). Since the mutant AlgB proteins in FRD844 and FRD850 could not be phosphorylated in vitro, these results suggest that AlgB functions in a phosphorylation-independent, DNA-binding-dependent manner to promote alginate production in mucoid P. aeruginosa.
FIG. 7.
Plate phenotypes of P. aeruginosa strains carrying wild-type or mutant algB alleles. The strains were constructed as outlined in Materials and Methods. L agar plates with the indicated strains indicated were incubated at 37°C for 20 h. The numbers are levels of alginate (in micrograms per milliliter) produced as quantitated by the carbazole assay (19, 26). The results are the means ± standard deviations for three independent experiments. Note that the AlgB.D59N-producing strain (FRD846), which is AlgB phosphorylation defective, still produced alginate; however, the AlgB.R442E-producing strain (FRD842), with an AlgB protein defective for DNA binding, lost the alginate-producing phenotype.
A mutation in the predicted phosphorylation sites of AlgR did not affect alginate production in mucoid P. aeruginosa.
Prior in vitro studies showed that the alginate response regulator AlgR could be phosphorylated by the enteric chemotaxis histidine protein kinase CheA and the small phosphodonor molecule acetyl phosphate (12). Similarly to AlgB and other well-characterized response regulators, AlgR contains a conserved aspartate residue (D54) which is likely the site of phosphorylation. This is supported by prior studies which demonstrated that the phosphorylated form of AlgR had biochemical properties characteristic of response regulators phosphorylated at aspartate side chains (12). More recent studies which identified fimS, an atypical sensor located adjacent to algR in P. aeruginosa, suggested that Asp85 might represent a second phosphorylation site unique to the AlgR subfamily of response regulators (62). To address whether phosphorylation of AlgR is required for alginate production, Asp54 and Asp85 were individually changed to asparagine residues (AlgR.D54N and AlgR.D85N). In addition, an algR allele expressing both alterations (AlgR.D54N.D85N) was generated. These mutations were introduced into the FRD1 chromosomal background via allelic exchange as described above (except that the ΔalgR::ΩaacC1 strain FRD831 was used as an intermediate), so that the algR alleles were in single copy under native control. In a control gene replacement experiment, wild-type algR restored alginate production to the FRD831 intermediate strain, forming FRD833. Interestingly, strains with mutant algR alleles, expressing AlgR with substitutions in the predicted sites of AlgR phosphorylation, were not affected in alginate production levels (Table 3). Strains FRD836 encoding AlgR.D54N, FRD838 encoding AlgR.D85N, and FRD839 encoding AlgR.D54N.D85N also synthesized wild-type levels of alginate (Table 3). Similarly to the results with AlgB, these data suggest that phosphorylation of AlgR does not appear to be required for activation of algD and alginate synthesis in mucoid P. aeruginosa.
TABLE 3.
Levels of alginate production by P. aeruginosa algR strains
| Strain | Genotype | Alginate pheno- typea | Alginate levelb |
|---|---|---|---|
| FRD1 | Wild type | Alg+ | 1,905 ± 313 |
| FRD831 | ΔalgR::aacC1 | Alg− | 43 ± 2 |
| FRD833 | algR+ (by allele replacement) | Alg+ | 1,607 ± 157 |
| FRD836 | algR7 (AlgR.D54N) | Alg+ | 1,775 ± 157 |
| FRD838 | algR10 (AlgR.D85N) | Alg+ | 2,002 ± 240 |
| FRD839 | algR11 (AlgR.D54N.D85N) | Alg+ | 2,238 ± 324 |
Alginate phenotypes of P. aeruginosa strains containing the indicated algR alleles were scored as mucoid (Alg+) or nonmucoid (Alg−) after growth for 20 h on L agar plates.
DISCUSSION
In this study, we have examined the requirement for phosphorylation of the P. aeruginosa AlgB and AlgR alginate response regulators. Three derivatives of AlgB (H-AlgB, H-AlgB.D59N, and H-AlgBΔ1-145) were purified and examined for in vitro phosphorylation activity with purified C-KinB. The results indicated that although C-KinB could efficiently phosphorylate H-AlgB, no KinB-mediated phosphorylation of H-AlgB.D59N or H-AlgBΔ1-145 was observed under any conditions. The inability of H-AlgB.D59N to undergo phosphorylation by C-KinB was apparently not due to a lack of interaction between these two proteins, since H-AlgB.D59N (but not H-AlgBΔ1-145) was able to compete with wild-type H-AlgB in an in vitro phosphorylation assay. To evaluate the in vivo requirement for AlgB phosphorylation, a kinB mutant strain (FRD1049) was generated. Surprisingly, FRD1049 exhibited a mucoid phenotype and produced amounts of alginate similar to those of the parental strain FRD1. Since FRD1049 contained a null kinB allele, it was highly unlikely that AlgB activity in this strain was due to phosphorylation by KinB. However, AlgB could have been phosphorylated by another histidine kinase via cross-talk or with small-molecular-weight phosphodonors such as acetyl phosphate (28, 35). To address this, an algB allele replacement strategy was developed to examine the activities of wild-type, AlgB.D59N, and AlgBΔ1-145 derivatives in vivo. Alginate levels from strains expressing AlgB.D59N were similar to amounts produced from wild-type FRD1, and strains containing the algB23 allele encoding AlgBΔ1-145 synthesized 45% of the wild-type levels of alginate. These results imply that there is little if any requirement for AlgB phosphorylation associated with its role in alginate overproduction in mucoid P. aeruginosa.
Alginate synthesis requires two independent signal transduction systems involving AlgB and AlgR (65). The data described in the present study also raise the question about a requirement for AlgR phosphorylation in alginate production. Recent work which identified a gene upstream of algR called fimS, encoding an atypical sensor kinase required for twitching motility, may shed some light on this. In these studies, which utilized P. aeruginosa PAK strains overexpressing the alternative sigma factor AlgU (AlgT), a mutation in fimS did not appear to affect alginate production whereas a mutation in algR resulted in a substantial reduction in alginate synthesis (62). These observations indicate that FimS and AlgR have different effects on twitching motility and alginate production. Subsequent to these studies, others reported that fimS (designated algZ in these studies) played a negative regulatory role in alginate production, since inactivation of algZ in a mucA2 genetic background resulted in an approximately twofold increase in alginate synthesis (68). Our studies with algR alleles encoding proteins with mutations in the predicted AlgR phosphorylation site(s) indicated little if any requirement for AlgR phosphorylation in alginate production. One plausible hypothesis is that FimS modulates the phosphorylation state of AlgR; the nonphosphorylated form of AlgR may be involved in alginate production, while the phosphorylated form of AlgR may play a role in other cellular functions such as twitching motility.
During signal transduction, response regulators are usually phosphorylated at a conserved aspartate residue in the receiver module. This phosphorylation results in the activation of a nonconserved output domain culminating in an adaptive response. Response regulators have been classified into two broad categories based on the mechanism by which they are activated by phosphorylation (16). In one class of response regulators (exemplified by NtrC, ArcA, OmpR, and PhoB), the receiver and output domains interact in the unphosphorylated form, and this interaction leads to inhibition of dimerization of the receiver domain. This inhibition is relieved either by phosphorylation of the input domain or by deletion of the output domain. In the second class (characterized by CheB and FixJ), interaction between the receiver and output domains results in inhibition of the output domain, and this inhibition can be reversed by either phosphorylation or removal of the input domain. In both classes of response regulators, mutations in the conserved aspartate residue within the phosphorylation domain are almost always deleterious to function, although there are notable exceptions. For example, in Rhizobium meliloti, transcription of nitrogen fixation genes is induced under microaerobic conditions, and this control is modulated by the response regulator FixJ and a hemoprotein kinase, FixL. When a mutant FixJ protein, FixJ.D54N, was analyzed in heterologous host E. coli, it was able to activate transcription of fixK at levels similar to those of wild-type FixJ, and this activation required FixL (45). FixL stimulation of FixJ.D54N activity was due to phosphorylation of an alternate FixJ residue (45). Phosphorylation at alternate residues in other response regulators such as CheY and OmpR has also been observed (5, 9), albeit the efficiencies of these phosphorylation reactions are much lower than those observed for the wild-type proteins. Although alternate phosphorylation of AlgB.D59N cannot be ruled out with the present data, two lines of evidence indicate that alternate phosphorylation is not likely to be the reason why AlgB.D59N retains wild-type activity in promoting alginate synthesis. First, despite numerous attempts, in vitro phosphorylation of H-AlgB.D59N or H-AlgBΔ1-145 was never observed. Second, in the cases in which alternate phosphorylation of response regulators has been demonstrated, this phosphorylation was confined to the highly conserved amino-terminal phosphorylation domain (5, 9, 45). However, FRD850 cells which expressed an AlgB derivative lacking the amino-terminal phosphorylation domain (H-AlgBΔ1-145) had a mucoid phenotype and synthesized high levels of alginate. Although it remains a possibility that the activity of H-AlgBΔ1-145 could be due to relief of the inhibitory effect imposed by the phosphorylation domain as observed in FixJ, it is more difficult to reconcile the in vivo activity of H-AlgB.D59N via this mechanism.
In Bacillus subtilis, the DegS-DegU two-component system controls the expression of a wide variety of genes that encode degradative enzymes and late-competence proteins (39). In this system, phosphorylated DegU was shown to be required for the expression of genes encoding degradative enzymes, as well as degQ, degR, and srfA, whereas nonphosphorylated DegU was capable of activating the late-competence genes comC and comG (39). Expression of genes encoding degradative enzymes was abolished in B. subtilis mutants which synthesized a DegU derivative that could not be phosphorylated (DegU.D56N), whereas the competence pathway was not affected. Thus, phosphorylation of the DegU response regulator apparently acts as a molecular switch controlling the expression of either the degradative-enzyme or late-competence gene. By analogy with the DegS-DegU two-component system, the KinB-AlgB pair may also have dual function in P. aeruginosa, whereby nonphosphorylated AlgB is required for alginate production but the phosphorylated form has another unidentified role(s) in the cell. This is supported by the observation that algB is expressed at low but clearly detectable levels in nonmucoid strains (64).
Strains of P. aeruginosa recovered from CF patients with chronic lung infections are mucoid and synthesize copious amounts of alginate. The molecular mechanism underlying overproduction of alginate in these strains has been elucidated and suggests that the activity of the alternative sigma factor ς22 is negatively controlled by accessory elements encoded by adjacent muc genes (20, 23, 31, 32, 34, 50, 66). Whereas the activity of ς22 in wild-type P. aeruginosa strains appears to be modulated by the anti-sigma factors MucA and MucB, most CF isolates including FRD1 used in our study harbor mutations in mucA and synthesize high levels of active ς22. The levels of expression of algB and algR have been shown to be increased in mucoid strains, and this activation requires ς22 (64, 65) (Fig. 6 [compare lanes 1 and 2]). An attractive hypothesis to explain a lack of requirement for KinB or AlgB and AlgR phosphorylation in the control of alginate synthesis is that elevated levels of these response regulators in the cell may bypass the need for phosphorylation. If phosphorylation controls an equilibrium between active and inactive response regulators, overexpression of AlgB.D59N or AlgR.D54N or wild-type AlgB in a kinB mutant may lead to levels of active protein which are sufficient to promote alginate synthesis. This mechanism was proposed to account for the observation that overexpression of the P. aeruginosa response regulator PilR in the absence of the PilS sensor allowed for transcription of pilA (6). The ComA (response regulator) and ComP (sensor) proteins control competence in B. subtilis. Overexpression of comA can overcome mutations in comP, restoring ComA activity which is insensitive to environmental signals (14). Another example is the UhpA protein of E. coli, which is a response regulator required for the transcription of uphT, encoding the sugar phosphate transport system. UhpA activity is modulated by two membrane proteins, UhpB and UhpC. Overexpression of wild-type UhpA in a uhpBC mutant or high-level expression of a UhpA.D54N variant leads to wild-type activation of uhpT. This was not observed when uhpA was in single copy (60, 61). At the onset of sporulation in B. subtilis, levels of the response regulator Spo0A increase significantly. It has been proposed that the increase of Spo0A alone, independently of phosphorylation, mediates some regulatory interactions between Spo0A and selected promoters with high-affinity Spo0A binding sites (2). It is possible that constitutive AlgB and AlgR activities in FRD1 may be due to “runaway” ς22 synthesis which results in elevated levels of these response regulators. This apparently novel natural mechanism, which may represent an adaptive response to allow P. aeruginosa to survive in the CF lung environment, will be addressed in future studies.
The CF lung represents a unique environment for P. aeruginosa. Under strong selective pressure, an accumulation of mutations such as those in mucA occurs, leading to breakdown of the regulatory circuit of ς22. It is not entirely clear what specific function of ς22 is selected for in the CF lung, since ς22 is involved in regulating alginate production, twitching motility, stress response, heat shock, and likely other unknown cellular processes (23, 49, 62, 67). Nevertheless, studies have suggested that alginate provides P. aeruginosa with a selective advantage in the CF lung (see reference 23 and references therein). It is possible that there are signals present in the CF lung that promote low-level alginate production and that, under these conditions, sensor proteins such as KinB and FimS are required for phosphorylation of AlgB and AlgR, respectively. While this possibility remains to be investigated, removing or blocking such signals and/or inhibiting the KinB-AlgB or FimS-AlgR signal-transducing pathways may prevent low-level alginate production by the initially colonizing P. aeruginosa strains. Since P. aeruginosa is a ubiquitous organism commonly found in soil and water, it is likely that the AlgB and AlgR signal transduction systems evolved to monitor conditions in these environments rather than in the CF lung. The development of algB and algR alleles defective in the signal transduction pathways will also allow us to investigate the natural roles of these response regulators and specific environmental cues in the production of alginate and other properties associated with P. aeruginosa.
ACKNOWLEDGMENTS
We thank I. Blomfield and B. Bourret for helpful discussions. We acknowledge H Schweizer for advice about allele replacement experiments and for providing us with pEX100T and pUCGM.
This work was supported by Public Health Service grants AI-35177 (D.J.W.) and AI-19146 (D.E.O.) from the National Institutes of Allergy and Infectious Diseases and in part by a Forsyth County United Way NIH grant RR-0504 (D.J.W.) and Veterans Administration Medical Research funds (D.E.O.). Oligonucleotide synthesis and amino-terminal analyses were performed in the DNA and Protein Synthesis Core Laboratories of the Cancer Center of Wake Forest University, which is supported in part by NIH grant CA-12197.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates and John Wiley-Interscience; 1987. [Google Scholar]
- 2.Baldus J M, Green B D, Youngman P, Moran C P., Jr Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J Bacteriol. 1994;176:296–306. doi: 10.1128/jb.176.2.296-306.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baynham P J, Wozniak D J. Identification and characterization of AlgZ, an AlgT-dependent DNA binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 4.Becker A, Schmidt M, Jager W, Puhler A. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene. 1995;162:37–39. doi: 10.1016/0378-1119(95)00313-u. [DOI] [PubMed] [Google Scholar]
- 5.Bourret R B, Hess J F, Simon M I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd J, Lory S. Dual function of PilS during transcriptional activation of the Pseudomonas aeruginosa pilin subunit gene. J Bacteriol. 1996;178:831–839. doi: 10.1128/jb.178.3.831-839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Chitnis C E, Ohman D E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence for an operonic structure. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 9.Delgado J, Forst S, Harlocker S, Inouye M. Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol Microbiol. 1993;10:1037–1047. doi: 10.1111/j.1365-2958.1993.tb00974.x. [DOI] [PubMed] [Google Scholar]
- 10.Deretic V, Dikshit R, Konyecsni M, Chakrabarty A M, Misra T K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989;171:1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deretic V, Gill J F, Chakrabarty A M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic V, Leveau H J, Mohr C D, Hibler N S. In vitro phosphorylation of AlgR, a regulator of mucoidy in Pseudomonas aeruginosa, by a histidine protein kinase and effects of small phospho-donor molecules. Mol Microbiol. 1992;6:2761–2767. doi: 10.1111/j.1365-2958.1992.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 13.Devries C A, Ohman D E. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubnau D, Hahn J, Roggiani M, Piazza F, Weinrauch Y. Two-component regulators and genetic competence in Bacillus subtilis. Res Microbiol. 1994;145:403–411. doi: 10.1016/0923-2508(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 15.Fick R B J, Sonoda F, Hornick D B. Emergence and persistence of Pseudomonas aeruginosa in the cystic fibrosis airway. Semin Respir Infect. 1992;7:168–178. [PubMed] [Google Scholar]
- 16.Fiedler U, Weiss V. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 1995;14:3696–3705. doi: 10.1002/j.1460-2075.1995.tb00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flynn J L, Ohman D E. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J Bacteriol. 1988;170:1452–1460. doi: 10.1128/jb.170.4.1452-1460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frankin M J, Chitnis C E, Gacesa P, Sonesson A, White D C, Ohman D E. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994;176:1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg J B, Gorman W L, Flynn J L, Ohman D E. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol. 1993;175:1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg J B, Ohman D E. Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J Bacteriol. 1987;169:1593–1602. doi: 10.1128/jb.169.4.1593-1602.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin S, Prusti R K, Roitsch T, Ankenbauer R G, Nester E W. Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J Bacteriol. 1990;172:4945–4950. doi: 10.1128/jb.172.9.4945-4950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato J, Chakrabarty A M. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1991;88:1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knutson C A, Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 27.Lion T, Haas O A. Nonradioactive labeling of probe with digoxigenin by polymerase chain reaction. Anal Biochem. 1990;188:335–337. doi: 10.1016/0003-2697(90)90616-h. [DOI] [PubMed] [Google Scholar]
- 28.Lukat G S, McCleary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma S, Wozniak D J, Ohman D E. Identification of the histidine protein kinase KinB in Pseudomonas aeruginosa and its phosphorylation of the alginate regulator AlgB. J Biol Chem. 1997;272:17952–17960. doi: 10.1074/jbc.272.29.17952. [DOI] [PubMed] [Google Scholar]
- 30.Martin D W, Holloway B W, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin D W, Schurr M J, Mudd M H, Deretic V. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol Microbiol. 1993;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 32.Martin D W, Schurr M J, Mudd M H, Govan J R W, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to ςe and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathee K, McPherson C J, Ohman D E. Posttranslational control of the algT (algU)-encoded ς22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 36.McIver K S, Kessler E, Olson J C, Ohman D E. The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol Microbiol. 1995;18:877–889. doi: 10.1111/j.1365-2958.1995.18050877.x. [DOI] [PubMed] [Google Scholar]
- 37.Mohr C D, Leveau J H J, Krieg D P, Hibler N S, Deretic V. AlgR-Binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Msadek T, Kunst F, Rapoport G. A signal transduction network in Bacillus subtilis includes the DegS/DegU and ComP/ComA two-component systems. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 447–471. [Google Scholar]
- 40.Ohman D E, Chakrabarty A M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohman D E, Mathee K, McPherson C J, DeVries C A, Ma S, Wozniak D J, Franklin M J. Regulation of the alginate (algD) operon in Pseudomonas aeruginosa. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1996. pp. 472–483. [Google Scholar]
- 42.Ohman D E, West M A, Flynn J L, Goldberg J B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985;162:1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 44.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 45.Reyrat J M, David M, Batut J, Boistard P. FixL of Rhizobium meliloti enhances the transcriptional activity of a mutant FixJD54N protein by phosphorylation of an alternate residue. J Bacteriol. 1994;176:1969–1976. doi: 10.1128/jb.176.7.1969-1976.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roychoudhury S, Zielinski N A, Ninfa A J, Allen N E, Jungheim L N, Nicas T I, Chakrabarty A M. Inhibitors of two-component signal transduction systems: inhibition of alginate gene activation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1993;90:965–969. doi: 10.1073/pnas.90.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanders D A, Gillece-Castro B L, Burlingame A L, Koshland D E. Phosphorylation site of NtrC, a protein phosphatase whose covalent intermediate activates transcription. J Bacteriol. 1992;174:5117–5122. doi: 10.1128/jb.174.15.5117-5122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanders D A, Gillece-Castro B L, Stock A M. Identification of the site of phosphorylation of the chemotaxis response regulator protein CheY. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 49.Schurr M J, Deretic V. Microbial pathogenesis in cystic fibrosis: coordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:411–420. doi: 10.1046/j.1365-2958.1997.3411711.x. [DOI] [PubMed] [Google Scholar]
- 50.Schurr M J, Yu H, Martinez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the ςE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 52.Schweizer H P. Small broad-host-range gentamycin resistance cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 53.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 54.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race-0 and race-1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 57.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 58.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7 Å resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 59.Wanner B L. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webber C A, Kadner R J. Action of receiver and activator modules of UhpA in transcriptional control of the Escherichia coli sugar phosphate transport system. Mol Microbiol. 1995;15:883–893. doi: 10.1111/j.1365-2958.1995.tb02358.x. [DOI] [PubMed] [Google Scholar]
- 61.Webber C A, Kadner R J. Involvement of the amino-terminal phosphorylation module of UhpA in activation of uhpT transcription in Escherichia coli. Mol Microbiol. 1997;24:1039–1048. doi: 10.1046/j.1365-2958.1997.4021765.x. [DOI] [PubMed] [Google Scholar]
- 62.Whitchurch C B, Alm R A, Mattick J S. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1996;93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wozniak D J, Ohman D E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC-family, is required for algD transcription. J Bacteriol. 1991;173:1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wozniak D J, Ohman D E. Involvement of the alginate algT gene and integration host factor in the regulation of the Pseudomonas aeruginosa algB Gene. J Bacteriol. 1993;175:4145–4153. doi: 10.1128/jb.175.13.4145-4153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie Z, Hershberger D, Shankar S, Ye R W, Chakrabarty A M. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu H, Boucher J C, Hibler N S, Deretic V. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (sigma E) Infect Immun. 1996;64:2774–2781. doi: 10.1128/iai.64.7.2774-2781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu H, Mudd M, Boucher J C, Schurr M J, Deretic V. Identification of the algZ gene upstream of the response regulator algR and its participation in control of alginate production in Pseudomonas aeruginosa. J Bacteriol. 1997;179:187–193. doi: 10.1128/jb.179.1.187-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]