Abstract
The heterochronic genes of Caenorhabditis elegans comprise the best-studied pathway controlling the timing of tissue and organ formation in an animal. To begin to understand the evolution of this pathway and the significance of the relationships among its components, we characterized 11 Caenorhabditis briggsae orthologs of C. elegans heterochronic genes. Using CRISPR/Cas9, we made a variety of alleles and found that several mutant phenotypes differ in significant ways from those of C. elegans. Although most mutant orthologs displayed defects in developmental timing, their phenotypes could differ in which stages were affected, the penetrance and expressivity of the phenotypes, or by having additional pleiotropies that were not obviously connected to developmental timing. However, when examining pairwise epistasis and synergistic relationships, we found those paralleled the known relationships between their C. elegans orthologs, suggesting that the arrangements of these genes in functional modules are conserved, but the modules’ relationships to each other and/or to their targets has drifted since the time of the species’ last common ancestor. Furthermore, our investigation has revealed a relationship between this pathway to other aspects of the animal's growth and development, including gonad development, which is relevant to both species.
Keywords: Caenorhabditis elegans, Caenorhabditis briggsae, heterochronic genes, developmental timing, CRISPR/Cas9, comparative genetics, evolutionary conservation, nematode development
Introduction
A developmental regulatory system performs its function in part due to the specific activities of its components and in part due to the manner in which these components interact. It has been found through comparative analysis that as these systems evolves, components may be replaced or their relationships may change. Such investigations can illuminate important features of a developmental regulatory system and how it performs its function (True and Haag 2001; Hill et al. 2006; Sommer 2012; Ellis 2022).
The heterochronic pathway of the nematode Caenorhabditis elegans is the most thoroughly characterized developmental regulatory system controlling the timing of tissue and organ development in an animal (Rougvie and Moss 2013). The components of the core pathway include transcription factors, RNA-binding proteins, and several microRNAs (miRNAs). This is the pathway in which miRNAs were discovered, and they play an important role in how it works: several regulatory factors are down-regulated at specific times during larval development by the miRNAs. Furthermore, the transcription and processing of miRNAs are temporally regulated and, in some cases, under the control of other heterochronic regulators.
Mutations in heterochronic genes alter the relative timing of diverse developmental events independent of spatial or cell type-specific regulation. Similar animal-wide timing pathways have not been characterized in other species. The core heterochronic pathway includes the protein-coding genes lin-14, lin-28, lin-29, lin-41, lin-46 and hbl-1, and the miRNA-encoding lin-4, let-7, mir-241, mir-48, and mir-84 (Rougvie and Moss 2013). (Several other genes with heterochronic effects are not considered here.)
Most of the proteins encoded by heterochronic genes are expressed at the beginning of postembryonic development whereas the miRNAs are not. The levels of the miRNAs rise during the larval stages and block the expression of proteins whose activities promote stage-specific developmental events. In general, when the miRNAs are missing or defective, developing mutant animals repeat some stage-specific events and postpone later events, which is called a reiterative phenotype. Mutations that delete miRNA binding sites from the 3′UTRs of their heterochronic gene targets also cause reiterative phenotypes. By contrast, when target genes are defective due to loss-of-function mutations, stage-specific events are skipped, which is called a precocious phenotype.
The core heterochronic genes of C. elegans have one-to-one orthologs in Caenorhabditis briggsae. Some have orthologs in other phyla, such as the miRNAs and lin-28, and others belong to conserved gene families, such as hbl-1, lin-29, and lin-41 (Rougvie and Moss 2013). lin-14 and lin-46 are found only in the Caenorhabditis genus of nematodes. The degree of conservation does not correlate with how important a gene is in the regulation of development: lin-14 has a key role in controlling L1 and L2 cell fates, and in many ways, it is the paradigmatic heterochronic gene (Ambros and Horvitz 1987).
C. elegans and C. briggsae are remarkably similar nematodes despite being separated evolutionarily by 5–30 million years (Cutter 2008). They both occupy the same ecological niche and have nearly identical development down to their cell lineages (Zhao et al. 2008; Félix and Duveau 2012). Although they are nearly indistinguishable anatomically, only 60% of their loci are clear orthologs (Stein et al. 2003).
Comparative developmental studies—especially of the sex determination pathway in C. elegans, C. briggsae, and other Caenorhabditis species—have revealed that many alterations, shifts, and substitutions of components and their relationships are possible while preserving morphology, life history, and behavior (Ellis 2017, 2022). Such evolution in developmental pathways while the resulting morphology remains unchanged is a phenomenon called developmental systems drift (True and Haag 2001). Random mutations that do not dramatically decrease fitness may linger for several generations and become suppressed by other mutations. Over time, many genetic differences can accumulate, causing the roles of individual genes to change and components of developmental pathways to be replaced or change their relationships to their targets.
Our goal was to investigate the functional organization of the heterochronic pathway by seeing how much of its composition and arrangement are the same across a short evolutionary time—short enough so that orthologs are identifiable for all components, but where sufficient time has passed for developmental systems drift to have occurred. We began by mutating each C. briggsae ortholog of the 11 core heterochronic genes and characterizing their phenotypes, as well as comparing some well-characterized epistasis relationships. To study genes expected to have early embryonic lethal, pleiotropic, or infertile null phenotypes we used the auxin-inducible degron (AID) system in C. briggsae (Zhang et al. 2015; Hills-Muckey et al. 2021).
Materials and methods
Sequence analysis
C. briggsae orthologs of C. elegans heterochronic genes were identified previously in Wormbase (wormbase.org) and miRBase (mirbase.org) and were confirmed by reciprocal Basic Local Alignment Search Tool (BLAST) on each genome. Links to the database entry for each gene are given in Supplementary Table 1.
Strains and culture conditions
Nematodes were grown at 20°C on standard nematode growth medium (NGM) plates seeded with E. coli AMA1004 unless otherwise indicated.
Strains used
C. elegans strains:
RG733 (wIs78 [SCMp::GFP + ajm-1p::GFP + F58E10 (cosmid) + unc-119(+)]) (wild type for this study),
HML1029 cshIs140[rps-28pro::TIR1(F79G)_P2A mCherry-His-11; Cbr-unc-119(+)] LGII,
ME502 cshIs140[rps-28pro::TIR1(F79G)_P2A mCherry-His-11; Cbr-unc-119(+)] LGII; hbl-1(aeIs8[hbl-1::AID]); wIs78,
ME504 cshIs140[rps-28pro::TIR1(F79G)_P2A mCherry-His-11; Cbr-unc-119(+)] LGII; lin-41(aeIs10[lin-41::AID]); wIs78,
ME507 cshIs140[rps-28pro::TIR1(F79G)_P2A mCherry-His-11; Cbr-unc-119(+)] LGII; lin-14(aeIs5[lin-14::AID]); wIs78.
C. briggsae strains:
AF16 (wild type),
ME421 Cbr-lin-28(ae25),
ME444 Cbr-dpy-5(v234) +/+ Cbr-lin-28(ae35),
ME449 Cbr-lin-46(ae38),
ME450 Cbr-lin-28(ae39),
ME451 Cbr-lin-46(ae38); Cbr-lin-28(ae39),
ME454 Cbr-dpy-5 (v234) +/+ Cbr-lin-28(ae39),
ME480 Cbr-lin-46(ae43),
ME482 Cbr-lin-46(ae44),
ME486 Cbr-let-7(ae47),
ME487 Cbr-let-7(ae48),
ME489 Cbr-lin-14(ae51) Cbr-let-7(ae50),
ME493 Cel-lin-4(ae53),
ME494 Cbr-lin-28(ae39); Cbr-let-7(ae47),
ME497 Cbr-lin-4(ae54),
ME500 Cbr-lin-4(ae55),
ME511 Cbr-hbl-1(aeIs12[Cbr-hbl-1::AID]),
ME514 Cbr-lin-14(ae58),
ME515 Cbr-lin-14(ae59),
ME519 Cbr-dpy-8(v262) + Cbr-unc-7(v271)/+ Cbr-lin-14(ae62) Cbr-unc-7(v271),
ME520 Cbr-dpy-8(v262) + Cbr-unc-7(v271)/+ Cbr-lin-14(ae63) Cbr-unc-7(v271),
ME526 Cbr-mir-241(ae64),
ME527 Cbr-mir-48(ae65),
ME529 Cbr-mir-84(ae68),
ME530 Cbr-mir-84(ae69),
ME531 Cbr-mir-241(ae64); Cbr-mir-84(ae70),
ME533 Cbr-mir-241(ae64); Cbr-mir-84(ae69),
ME534 Cbr-lin-4(ae71); Cbr-dpy-8(v262)+Cbr-unc-7(v271)/+ Cbr-lin-14(ae62) Cbr-unc-7(v271),
ME535 Cbr-lin-4(ae72); Cbr-dpy-8(v262)+Cbr-unc-7(v271)/+ Cbr-lin-14(ae62) Cbr-unc-7(v271),
ME538 Cbr-mir-241 Cbr-mir-48(ae73),
ME541 Cbr-mir-241 Cbr-mir-48(ae73)/+; Cbr-mir-84(ae70),
ME544 Cbr-lin-41(ae76),
ME545 Cbr-spe-8 (v142) +/+ Cbr-lin-41(ae77),
ME547 Cbr-lin-41(aeIs14[Cbr-lin-41::AID]),
ME548 Cbr-trr-1(v76) +/+ Cbr-lin-29(ae75),
ME549 aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)]; Cbr-hbl-1(aeIs12[Cbr-hbl-1::AID]),
ME550 aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)],
ME552 aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)]; Cbr-lin-41(aeIs14[Cbr-lin-41::AID]),
ME553 aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)]; Cbr-mir-241 Cbr-mir-48(ae73); Cbr-hbl-1(aeIs12[Cbr-hbl-1::AID]) Cbr-mir-84(ae70),
ME554 aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)]; Cbr-lin-28(aeIs13[Cbr-lin-28::AID]); Cbr-hbl-1(aeIs12[Cbr-hbl-1::AID]),
ME555 aeEx44[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)],
ME556 aeEx45[Cbr-lin-28::GFP],
ME558 Cbr-lin-41(ae76); Cbr-let-7(ae48),
ME559 aeEx46[Cbr-lin-28::GFP(Y35R F37A C127R C137A)],
ME561 aeEx48[Cbr-lin-28::GFP(Y35R F37A C127R C137A) 3′-UTR deletion],
ME562 Cbr-lin-41(ae76); Cbr-lin-29(ae75)/+,
ME563 Cbr-lin-28(ae39); Cbr-lin-29(ae75)/+,
ME564 aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)] Cbr-lin-4(ae79); Cbr-mir-241 Cbr-mir-48(ae73); Cbr-hbl-1(aeIs12[Cbr-hbl-1::AID]) Cbr-mir-84 (ae70),
ME565 Cbr-lin-41(ae76); aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)]; Cbr-hbl-1(aeIs12[Cbr-hbl-1::AID]),
ME566 Cbr-hbl-1(aeIs12[Cbr-hbl-1::AID]) Cbr-let-7(ae48); aeIs15[rps-28pro>atTIR1(F79G)::P2A::GFP::His-11; Cbr-unc-119(+)]
ME567 Cbr-lin-28(ae39); Cbr-mir-48(ae65)
ME568 Cbr-dpy-5(v234) +/+ Cbr-lin-28(ae80)
Development synchronization
To generate developmentally synchronized populations of worms, gravid adults (filled with eggs) were washed from crowded plates into 15 ml tubes, spinned down, the excess liquid then was removed leaving a worm pellet. 500–1000 µl of household bleach solution was added to the tube and vortexed every 2 minutes until the cuticles of worms were dissolved enough to release eggs. Tubes then were filled with sterilized distilled water to slow the bleaching activity and centrifuged at 400 g for 3 minutes, then the supernatant was discarded and eggs were washed twice with sterilized distilled water and then twice with M9. Then eggs were transferred into M9 and left on a shaker for 20–48 hours at room temperature. Then the liquid with larvae was centrifuged to concentrate larvae and they were transferred to plates with food in a small amount of M9.
Microscopy
Animals were examined using differential interference contrast (DIC) and fluorescence microscopy on a Zeiss Axioplan2 microscope with Zeiss objectives: Plan-NEOFLUAR 5x, Plan-NEOFLUAR 16x, Plan-NEOFLUAR 63x, and alpha Plan-FLUAR 100x. Images were acquired using AxioCam with AxioVision software.
Seam cell and gonad phenotype scoring
Fluorescent seam cell markers to facilitate the counting of seam cells are not yet available for C. briggsae as for C. elegans. Therefore, there was some variability in seam cell counts for animals of apparently identical age due to occasional difficulty distinguishing seam cells from lateral hypodermal nuclei by DIC microscopy. Seam cells were confidently identified when located along the lateral midline or slightly off and having a visible oval-shaped outline. Errors in counting may occur when seam cell nuclei or hypodermal syncytial nuclei lay away from or close to the apparent midline, respectively.
A disorganized gonad was scored if gonad or oocyte contents leaked into the pseudocoelom or oocytes or spermatozoa were found outside the gonad.
CRISPR/Cas9
We followed general protocols for the usage of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 ribonucleoprotein complexes in genome editing described in Paix et al. (2017). gRNA was synthesized from PCR-amplified templates using Invitrogen MEGAshortscript T7 Transcription Kit (Catalog# AM1354) and purified with Invitrogen MEGAclear Transcription Clean-Up Kit (Catalog# AM1908). Purified gRNAs were mixed with Cas9 (EnGen Spy Cas9 NLS, Catalog# M0646T) and used in microinjections. Typical concentrations of the components in the injection mix: gRNA (up to 200 ng/µl), Cas9 (250 ng/µl), co-injection Cbr-myo::GFP plasmid (35 ng/µl).
When making insertions, a hybrid dsDNA repair template was used as described in (Dokshin et al. 2018). Repair templates were melted and cooled before injections (Ghanta and Mello 2020). The repair template then was added to the CRISPR mix to the final concentration of 100–500 ng/µl of DNA.
RNAi
RNA interference used to knockdown Cbr-hbl-1 expression was performed as described (Hammell and Hannon 2016). Part of the Cbr-hbl-1 ORF flanked by T7 promoters was amplified using these primers: 5′-GCGCGCTAATACGACTCACTATAGGTCCCAGCACCCCTACCACCAC-3′, 5′-GCGCGCTAATACGACTCACTATAGTGGTGACGCCGGCTCTCCTTT-3′
RNA was synthesized and purified with the in-vitro transcription kit mentioned above. Purified RNA was diluted with sterile distilled water to 200 ng/µl concentration and injected into gonads.
Auxin-inducible degron system
We used a modified auxin-inducible system with TIR1(F79G), using 5-Ph-IAA as the auxin analog (Zhang et al. 2015; Hills-Muckey et al. 2021). To express TIR1, wildtype C. briggsae were injected with a modified pCMH2074 plasmid (C. Hammell, pers. comm.) containing TIR1(F79G) mutation and GFP in place of mCherry. Bright fluorescing animals carried a stable array with a high inheritance rate (50–75%) were selected and a strain with stable extrachromosomal expression was established (aeEx44).
To integrate TIR1(F79G) into the genome, fluorescent L4 and young adult animals were transferred to a plate without bacteria and irradiated with UV in a UV-crosslinker set to an energy level of 13 kJ/cm2 to generate chromosomal breaks and attach the array to a chromosome. Fluorescent F1 animals were isolated and then plates were screened for 100 or 75% transmission rates or animals displaying uniform (vs mosaic) fluorescence. Stable integrants were identified as animals with uniform fluorescence with 100% penetrance. This strain then was outcrossed at least 3 times.
To assess phenotypes using the AID system, animals carrying alleles with fused degron tag and expressing TIR1(F79G) were grown on plates containing 50 µmol of 5-Ph-IAA that was spread on standard NGM plates to approximately 0.005 µM concentration in the agar.
Cbr-lin-28::GFP plasmid
The Cbr-lin-28::GFP expression plasmid was produced by PCR and restriction digestion and ligation techniques. Q5 High-Fidelity DNA Polymerase (NEB #M0491S) and Phusion High-Fidelity DNA Polymerase (NEB #M0530S) were used for amplification. Cbr-lin-28 with promoter region and 3′UTR were amplified from the C. briggsae (AF16) genome, and the C. elegans-optimized GFP sequence with introns was amplified from pVT221 (Moss et al. 1997). To mutate the plasmid, a Q5 Site-Directed Mutagenesis Kit (NEB #E0552S) was used. The plasmid scheme and mutation sequences are shown in Supplementary Fig. 10.
Plots and statistics
Data was analyzed using Prism software. P-values were calculated using unpaired Welch's t-tests for absolute values (seam cell and intestinal nuclei count averages) and Fisher’s exact tests for fractions (percents). Error bars in plots indicate 95% CI, asterisks indicate the following: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, ns—not significant (P > 0.05).
Results
Wild-type seam and intestinal cell fates are similar in C. briggsae and C. elegans
Heterochronic phenotypes of C. elegans can be reliably observed in the postembryonic lineages of the lateral hypodermal seam cells (Ambros and Horvitz 1984, 1987; Ambros 1989). Seam cells are located along each side of the newly hatched larva, dividing at each larval stage and differentiating at adulthood (Sulston and Horvitz 1977). We counted seam cells at each larval stage and observed their divisions in wildtype C. briggsae to see if the lineage patterns resembled those of C. elegans.
We found that C. briggsae L1 larvae had 10 seam cells within 3 hours of hatching (n = 10, Supplementary Fig. 1). Seam cells were observed to divide in L1 larvae within 6 hours after hatching, with one of the daughters staying at the midline while the other moved dorsal or ventral to join the hypodermal syncytium. As a result, we saw that molting L1 larvae still had 10 seam cells (n = 10). Seam cells H1, V1-V4, and V6 were divided symmetrically in L2 larvae, and late L2 or early L3 larvae had 15.5±0.5 seam cells (n = 16). Both L3 and L4 larvae still had asymmetrical divisions like those in the L1. L4 larvae and adult worms had 15–16 seam cells (Fig. 1a). All seam cells aligned along the midline and produced cuticular alae in adulthood. These observations indicate that the numbers and division patterns of seam cells in C. briggsae are like those of C. elegans at each larval stage: 10 in the L1 and 16 in the L2 and later.
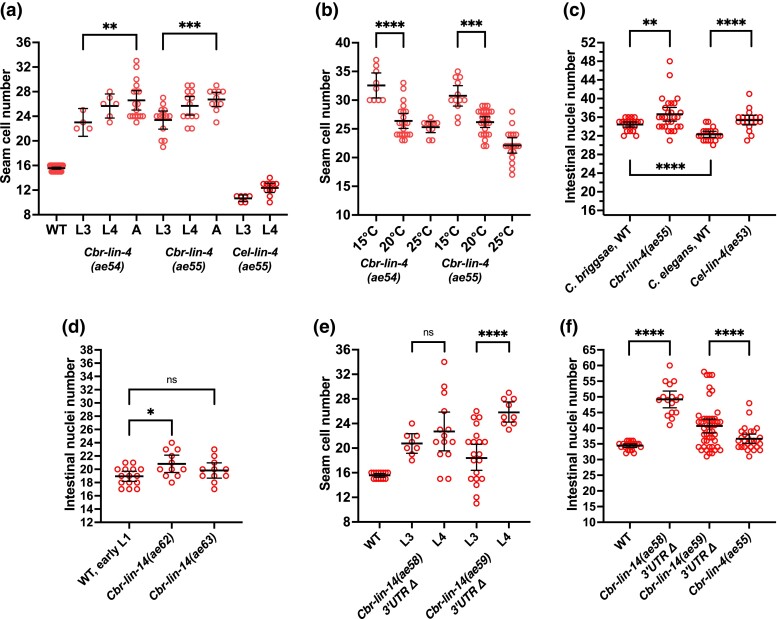
Fig. 1.
Seam cell and intestinal nuclei changes in Cbr-lin-4(0) and Cbr-lin-14(0) or (gf) mutants. Plots show seam cell and intestinal nuclei counts in L4 and young adults (unless otherwise specified) at 20°C (unless otherwise specified) for the strains indicated. a) The number of seam cells is higher in Cbr-lin-4(0) mutants than in the wild type, and increases between the L3 and adult stages. On the contrary, Cel-lin-4(0) mutants have a reduced number of seam cells. b) The number of seam cells in Cbr-lin-4(0) mutants increases at 15°C. c) Intestinal nuclei numbers increase in Cbr-lin-4(0) mutants. d) Intestinal nuclei numbers in the Cbr-lin-14(0) mutants are similar to those in early L1 larvae. e) Cbr-lin-14 3′UTR deletion mutants have an increased number of seam cells. f) Cbr-lin-14 3′UTR deletion mutants have even higher numbers of intestinal nuclei than Cbr-lin-4(0) mutants. The data set for Cbr-lin-4(ae55) is the same as in panel C. The reason for the difference between ae58 and ae59 is unclear as both deletions had approximately the same length and location. Statistical analysis is described in Materials and Methods.
Also like C. elegans, newly hatched C. briggsae had 20 intestinal nuclei. In C. elegans, 10 to 14 of 20 intestinal nuclei divide at the beginning of L1 lethargus (Sulston and Horvitz 1977; Hedgecock and White 1985). In C. briggsae, some intestinal cell nuclei also divided during the L1 lethargus and molt, with some divisions coinciding with the first round of the L2 seam cell divisions. In addition, we observed a slight but statistically significant increase in the average number of intestinal nuclei between the L3 and L4 stages. It is possible that some intestinal nuclei in C. briggsae divide during stages after the L1, which does not occur in C. elegans (Supplementary Fig. 2).
We used seam cell and intestinal nuclei, as well as vulval lineages, which have been previously documented (Brown 2001), to characterize developmental timing phenotypes in C. briggsae mutants. However, we also observed phenotypic changes in other tissues, as described below.
Cbr-lin-4(0) mutants reiterate L2 stages
C. elegans lin-4 is the first heterochronic gene to be identified and encodes the first miRNA to be discovered (Chalfie et al. 1981; Ambros and Horvitz 1984; Lee et al. 1993). Cel-lin-4(0) mutants have a profound reiterative phenotype where L1-specific events are repeated causing adult animals to lack both vulvae and alae.
We isolated 2 Cbr-lin-4(0) mutant alleles with 6 (allele ae54) and 8 bp (allele ae55) deletions that remove most or all of the miRNA seed sequence (Supplementary Table 1). Grossly, these animals were egg-laying defective for lacking a vulva (Supplementary Fig. 3a and b).
To learn if Cbr-lin-4(0) mutants reiterated stage-specific fates like C. elegans, we examined seam cells, molts, adult alae, and intestinal nuclei. Based on seam cell counts, Cbr-lin-4(0) mutants appeared to re-iterate the symmetric divisions characteristic of the L2 stage during later larval stages (Fig. 1a, Supplementary Fig. 4). Seam cells divided symmetrically at L2, and most larvae had 15.2±0.7 seam cells (n = 30) after the L2 divisions. Thus, L1 stages were not reiterated by most seam cells. This effect on seam cell lineages was also cold-sensitive: L4 larvae and young adults had more seam cells at 15°C than at 20°C or 25°C (Fig. 1b).
In contrast, both the C. elegans reference allele Cel-lin-4(e912) and Cel-lin-4(ae53), a mutant generated using the same single guide RNA used for the Cbr-lin-4(0) mutant alleles, reiterated mostly L1 stage seam cell fates. As a result, these animals had 10.7±0.5 seam cells before the L4 stage (n = 6) and 12.7±1.7 seam cells after the L4 (n = 13).
Like C. elegans lin-4(0) mutants, Cbr-lin-4(0) mutant animals also have extra molts. Five L4 larvae from the Cbr-lin-4(ae55) strain were placed together on a plate at 20°C, and the next day, 7 shed cuticles were found in the bacterial lawn. The animals were adults (carried eggs), and 4 of them were stuck while shedding extra cuticles. Adult egg-producing wild-type C. briggsae were never observed shedding cuticles. Thus, Cbr-lin-4(0) mutants have at least 1 additional molt after reaching adulthood, and possibly as many as 2. Unlike Cel-lin-4(0) mutants, whose cuticles are devoid of adult alae, Cbr-lin-4(0) mutants developed partial adult alae at the end of the development, most young adults (adults without embryos) had alae “patches”, meaning that more than 0% but less than 50% of their seam cells generated adult alae (50%, n = 12), while most adult worms with embryos had “gapped” alae, meaning that more than 50%, but less than 100%, of the seam cells, formed alae (78%, n = 18, Supplementary Fig. 3c and Table 2). Thus, most seam cells differentiate at the end of the development in C. briggsae lin-4 mutants, while they fail to differentiate in C. elegans.
In wild-type C. elegans and C. briggsae, dauer larvae represent an alternative developmental stage that forms in unfavorable environmental conditions like overcrowding or lack of nutrients. When heterochronic mutants reiterate L1 stages and do not transition to L2 they cannot form dauer larvae, which was observed for Cel-lin-4(0) mutants (Liu and Ambros 1989). Surprisingly, Cbr-lin-4(0) mutants could enter the dauer developmental pathway, reinforcing the conclusion that they enter the L2 stage. However, Cbr-lin-4(0) dauers had gapped dauer alae, and some segments of their bodies looked expanded which was not observed in wild-type dauers (Supplementary Fig. 5). We suspect that those worms transitioned to the dauer stage incompletely.
Interestingly, despite these differences, both Cbr-lin-4(0) and Cel-lin-4(0) mutants had a slightly increased number of intestinal nuclei compared to wild-type (Fig. 1c). The number of intestinal nuclei in Cbr-lin-4(ae55) mutant was not significantly different (P > 0.05, unpaired Welch's t-test) at 15°C (36.9±3.1, n = 28) compared to the same mutant at 20°C. So, whereas the phenotype of Cel-lin-4(0) mutants is interpreted as a reiteration of L1 stage-specific fates, we find some ambiguity with Cbr-lin-4(0) mutants, since they appear to reiterate L1 fates in the intestine and L2 fates in the hypodermis.
C. briggsae lin-14(0) mutants resemble C. elegans lin-14(0) mutants
C. elegans lin-14 encodes a transcription factor unique to this genus (Ruvkun and Giusto 1989; Hristova et al. 2005). Alleles are of 2 general types: loss-of-function (lf) and null (0), which cause a precocious phenotype, and gain-of-function (gf), which lack miRNA binding sites in the 3′UTR and cause a reiterative phenotype (Ambros and Horvitz 1987; Wightman et al. 1991).
We made mutant alleles ae62 and ae63 with frameshift mutations that create premature stop codons in Cbr-lin-14 by targeting the first exon shared by all isoforms (Supplementary Table 1 and Fig. 6). The mutants were often infertile, so we balanced them with a mutation that caused a visible phenotype: Cbr-dpy-8(v262) (Wei et al. 2014). Cbr-lin-14(0) progeny from the balanced strain resembled Cel-lin-14(0) mutants in several key features, including a protruding vulva, and shared a similar overall morphology (Supplementary Fig. 7).
To see if these mutants displayed a precocious phenotype, we examined the L4 cuticle, seam cell divisions, and intestinal nuclei number. Cbr-lin-14(0) mutants developed full adult alae by the L4 stage (100% had precocious alae, n = 20). As occurs in C. elegans lin-14(0) mutants, seam cell counts of Cbr-lin-14(0) mutants are close to albeit slightly below the wild-type number by the L4 (Supplementary Fig. 8). This is consistent with most seam cells of Cbr-lin-14(0) animals dividing symmetrically during the L1 stage, as occurs in C. elegans lin-14(0) mutants (Ambros and Horvitz 1984). Also like Cel-lin-14(0), the number of intestinal nuclei in later development was reduced in Cbr-lin-14(0) mutants, although some divisions did occur (Fig. 1d). Overall, our observations suggest Cbr-lin-14 is required for stage-appropriate expression of L1-specific fates and that the gene function is largely conserved between the 2 species.
The Cbr-lin-4(0) reiterative phenotype requires functional Cbr-lin-14
In C. elegans, lin-4 mutations that lead to a reiterative phenotype do so because they cause prolonged expression of lin-14 (Ruvkun and Giusto 1989; Lee et al. 1993; Wightman et al. 1993). As a result, the phenotype of a loss-of-function Cel-lin-14(0) mutation is completely epistatic to that of Cel-lin-4(0) (Ambros 1989).
To test whether the temporal fate reiterations that occur in Cbr-lin-4(0) mutants occurred due to the elevated Cbr-lin-14 function, we made Cbr-lin-4(0); Cbr-lin-14(0) double mutants by disrupting the Cbr-lin-4 gene in a Cbr-lin-14(ae62) balanced strain using CRISPR/Cas9 and then isolating double homozygotes from among the progeny. We made strains with 2 different Cbr-lin-4 alleles, ae71, and ae72 (Supplementary Table 1).
The Cbr-lin-4(0); Cbr-lin-14(0) double mutant phenotype mostly resembled the Cbr-lin-14(0) single mutant phenotype—the number of seam cells by the L4 was 14.7±0.9 (n = 16, combined data from 2 strains) and most of the intestinal nuclei did not divide; after the L1 stage, the number of intestinal nuclei was 19.6±1.5 (n = 16, combined data from 2 strains). Surprisingly, however, precocious alae were not always observed in double mutant worms: 2 of the observed L4 larvae did not have precocious alae and the other 2 had full precocious alae. This differs significantly from the C. elegans double mutant, in which alae appear precociously in all worms (Ambros 1989). This observation indicates a difference between the species in the relationships of Cbr-lin-4 and Cbr-lin-14 to downstream regulators controlling the timing of alae formation. But as in C. elegans, Cbr-lin-14 appears to be required for the reiterative and vulvaless phenotypes of Cbr-lin-4(0).
Cbr-lin-14(gf) mutants resemble weak Cel-lin-14(gf) alleles
In C. elegans, 2 mutants with deletions in the 3′UTR of lin-14 display a reiterative phenotype: an allele with nearly all miRNA sites removed reiterates L1 stages and lacks a vulva and adult alae, closely resembling lin-4(0), and a weaker allele with a few intact miRNA sites reiterates both L1 and L2 stage events, also lacks a vulva, and develops some alae (Ambros and Horvitz 1987; Wightman et al. 1993).
Cbr-lin-14 3′UTR deletions (alleles ae58 and ae59) were generated with CRISPR/Cas9, removing approximately 1.3 kb that includes all predicted miRNA binding sites (Supplementary Table 1 and Fig. 6). As adults, the mutants lacked vulvae, had extra seam cells and intestinal nuclei, and had alae patches (52%, n = 21) or lacked alae completely (48%, Supplementary Table 2). The patches of adult alae were more transparent and thinner than wild-type alae.
The number of seam cells in Cbr-lin-14(gf) mutants was slightly lower and more variable at late stages than in Cbr-lin-4(0) mutants (Fig. 1a and e). Additionally, late L2 and early L3 larvae had fewer seam cells than expected if Cbr-lin-14(gf) phenocopied Cbr-lin-4(0) (mean = 11±1, n = 11). This difference would be explained by most seam cells reiterating L1 cell fates before they reiterate L2 fates. Also, the number of intestinal nuclei was higher than in Cbr-lin-4(0) mutants—a sign of reiteration of L1 fates in this tissue—which supports the interpretation that these animals reiterate L1 stage events to some degree. Thus, Cbr-lin-14(gf) mutants resemble the weaker Cel-lin-14(gf) allele.
Cbr-lin-28(0) mutants have minor heterochronic defects and arrest development at the L4 stage
C. elegans lin-28 encodes an RNA-binding protein that is widely conserved in animals (Moss et al. 1997; Moss and Tang 2003; Vadla et al. 2012). Cel-lin-28(0) mutants display a completely penetrant precocious phenotype where they skip cell fates of the L2 and a less penetrant defect of skipping L3 fates (Ambros and Horvitz 1984; Vadla et al. 2012). They also show an incompletely penetrant fertility problem as a result of spermathecal defects (Choi and Ambros 2019).
We made Cbr-lin-28 mutant alleles ae25, ae35, and ae39 by targeting the second exon to generate frameshifts with premature stop codons (Supplementary Table 1 and Fig. 6). Cbr-lin-28 mutant animals were strikingly different from Cel-lin-28(0) animals: many arrested their development during the late L4 stage, did not undergo the final molt and lacked adult alae. These arrested animals retained features characteristic of mid- to late- wild-type L4 animals: the reflexed gonad arms stopped developing toward each other, and the vulva ceased development during morphogenesis (Fig. 2a). Observing a synchronized population, we found that Cbr-lin-28 mutants developed at the same rate and produced oocytes at the same time as wild-type animals, except that wild type proceeded to the last molt and developed a vulva and alae (n ≍ 20).
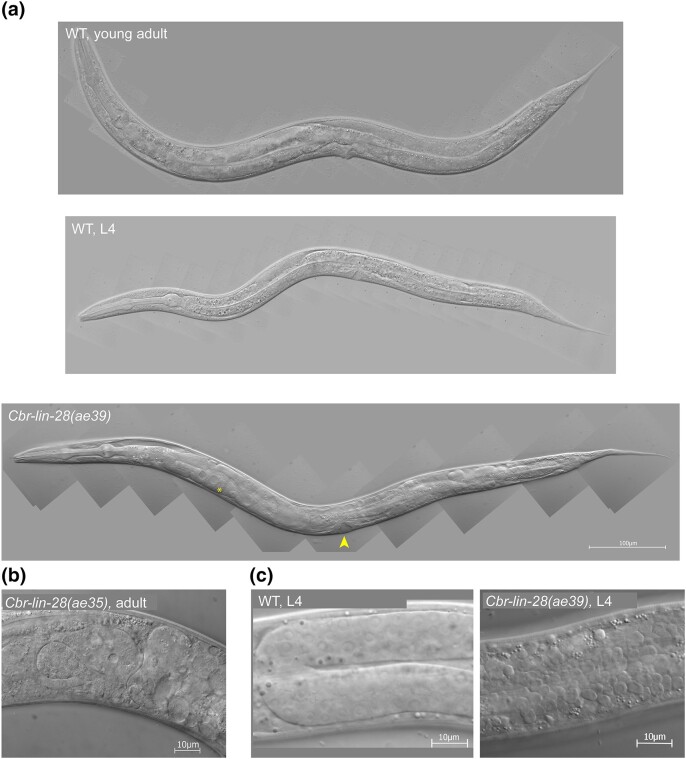
Fig. 2.
Developmental timing defects in Cbr-lin-28(0) mutants cause an L4 arrest and gonad disintegration. DIC photomicrographs of animals grown at 20°C. a) Wild-type (AF16) L4 larva and young adult compared to Cbr-lin-28(ae39) with “arrested L4” phenotype. Arrowhead indicates L4-like vulva (larval trait) and asterisk indicates oocytes (adult trait). b) A closer view at the disorganized gonad of a Cbr-lin-28(ae35) adult. c) An earlier stage gonad disorganization that occurs in some Cbr-lin-28(ae39) mutant L4 larvae compared to a normal gonad. All animals are oriented anterior end left, dorsal side up.
The gonads of Cbr-lin-28 mutants became disorganized or disintegrated after animals were arrested in L4 or reached adulthood. Gonad contents leaked into the pseudocoelom and sometimes the gonad fell apart into separate cells (Fig. 2b). In some cases, gonad disorganization was not visible at first but manifested later (Supplementary Fig. 9). The underlying defect is unknown. Some mutants developed through the L4 stage and had normal vulvae and adult alae, but became stuck in the L4 cuticle during molting. Those few that completed the L4 molt (“successfully molted”) were not morphologically different from the wild type, although sometimes they had disorganized gonads.
In contrast to what happens in C. elegans, Cbr-lin-28 mutants had only weak heterochronic defects: most animals had a small patch of precocious alae near the pharynx at the L4 stage, and the number of seam cells was similar to the wild type (Fig. 3a). Undergoing dauer development suppressed precocious alae, but did not suppress L4 arrest (Fig. 3b and c). By contrast, dauer development completely suppresses C. elegans lin-28(0) heterochronic defects (Liu and Ambros 1991). Some other observed phenotypes included rolling (less than 10%), protruding vulvae (10–20%), larvae stuck at L2 and L3 molts, incompletely shed cuticles (“belts”), and sometimes gonads losing their structure during the L4 stage (Fig. 2c).
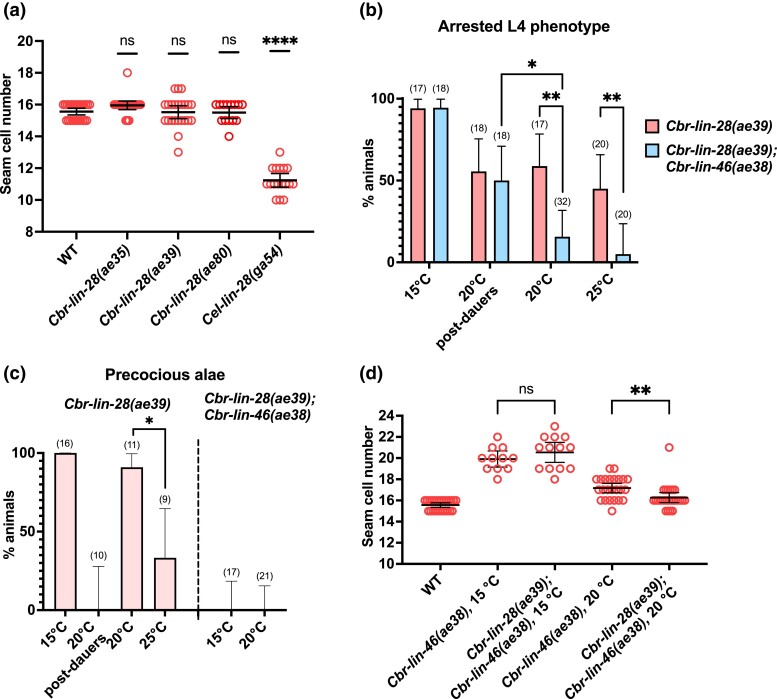
Fig. 3.
Cbr-lin-28(0) mutant phenotypes and their suppression by Cbr-lin-46(0) allele. a) Cbr-lin-28(0) mutants do not have a reduced number of seam cells at 20°C, unlike Cel-lin-28(0). b) L4 developmental arrest was more penetrant in the Cbr-lin-28(0) mutant at 15°C than at 20°C (both dauers and postdauers, P ≤ 0.05). It also was not suppressed by Cbr-lin-46(0) mutation at 15°C in contrast to 20°C and 25°C. Those animals that did not arrest their development at L4, had “successfully molted” or intermediate phenotypes. Sample sizes are specified in parenthesis above the bars. c) Precocious alae of Cbr-lin-28(0) occurred less often at 25°C and were suppressed by dauer pathway and Cbr-lin-46(0) mutation at 15°C and 20°C. d) Increased seam cell number of Cbr-lin-46(0) mutant was suppressed by Cbr-lin-28(0) mutation at 20°C but not at 15°C. Statistical analysis is described in Materials and Methods.
Cbr-lin-28 's mutant phenotype was cold-sensitive. At 15°C, most animals became arrested L4s and all had precocious alae (n = 16) (Fig. 3b and c). Moreover, patches of precocious alae were longer and some animals had complete precocious alae. Additionally, the strain could not be maintained at 15°C. Some animals produced eggs at this temperature but they did not hatch, although eggs placed at 15°C after the mothers were grown at 25°C were viable and did hatch, suggesting a maternal effect embryonic problem at cold temperatures.
To confirm that the Cbr-lin-28 mutant phenotypes that we observed are those of null alleles, we generated a deletion (ae80) that removed 78% of the 206 amino-acid coding region. This deletion starts within the cold-shock RNA-binding domain (CSD) and deletes both CCHC zinc knuckles so that the 46 remaining amino acids contain none of Cbr-lin-28's known functional domains.
Cbr-lin-28(ae80) L4 larvae had 15.5±0.7 (n = 14) seam cells, and 42% (n = 12) had precocious alae patches. After 24 hours, all 30 L4 larvae picked to a separate plate had vulvae arrested at early L4 stages (L4.2-L4.3; Mok et al. 2015), produced oocytes (most of them also had embryos), had L4 cuticles (60% had precocious alae patches, n = 15), and disorganized gonads.
Therefore, the phenotypes of Cbr-lin-28(ae80) worms were not significantly different from those of ae35 and ae39. Heterochronic defects remained weak, and L2 seam cell divisions were not skipped. The penetrance of the “arrested L4” phenotype might be higher in this strain since no “successfully molted” worms were observed among the 30 isolated L4, although some advanced-stage worms (with adult-like vulvaе) were rarely observed on plates as well as worms with protruding vulvaе.
Overall, Cbr-lin-28(0) has only a minor resemblance to Cel-lin-28(0), pleiotropic effects, and variable penetrance and expressivity for most phenotypes.
Cbr-lin-28 is expressed at all stages in C. briggsae and down-regulated in seam cells after the L1 and L3 stages
In C. elegans, lin-28 shows a characteristic “on early, off late” expression pattern that parallels its function in controlling L2 fates, and this temporal down-regulation is a consequence of miRNAs acting via its 3′UTR (Moss et al. 1997; Tsialikas et al. 2017). Because the phenotype of mutant Cbr-lin-28 differed from that of its C. elegans ortholog, we examined the expression of Cbr-lin-28 to see if that was different as well.
We employed a transgenic approach that had been successfully used in C. elegans which creates multicopy extrachromosomal arrays of plasmids (Stinchcomb et al. 1985; Mello et al. 1991). A full-length translational fusion with GFP that included intact 5′ and 3′ regulatory regions was constructed (Supplementary Fig. 10) The construct was injected into wild-type C. briggsae, producing a stable extrachromosomal array aeEx45.
In transgenic animals, GFP fluorescence was observed in head and tail neurons, motor neurons, muscles (including the pharynx), intestinal cells, and seam cells (Fig. 4), which is similar to C. elegans, except that Cel-lin-28::GFP had obvious expression throughout the hypodermis (Moss et al. 1997). In contrast to C. elegans, most GFP expression did not appreciably decline with age.
Fig. 4.
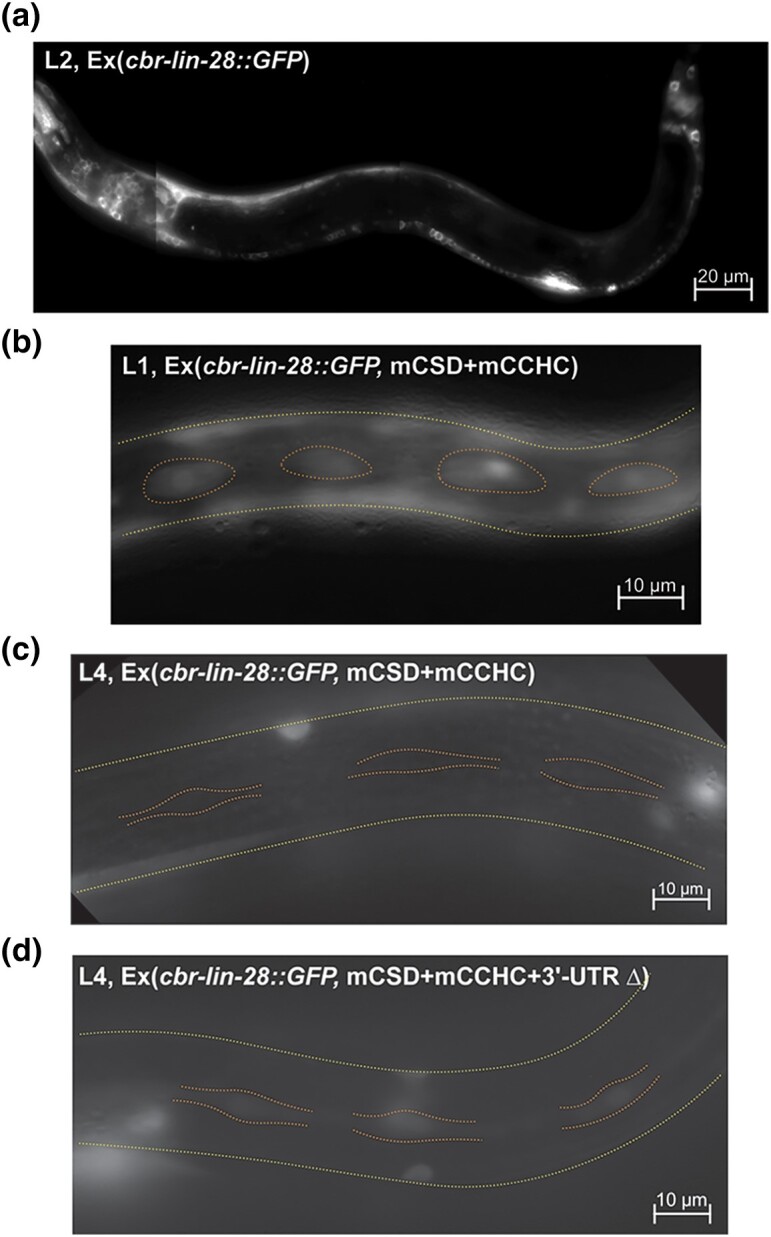
The expression of LIN-28 is down-regulated during C. briggsae development. Fluorescent microscopy images, green channel. All animals are oriented anterior end left, dorsal side up. Yellow dotted line indicates sides of animals, orange dotted line outlines seam cells. a) An AF16 L2 larva from a brood of animals carrying Cbr-lin-28::GFP on an extrachromosomal array. The expression is visible in neurons, pharynx, and P-cells. b) Fluorescing seam cells in an L1 larva expressing Cbr-lin-28::GFP with mutated CSD and CCHC domains. c) Seam cells not glowing in an L4 larva expressing Cbr-lin-28::GFP array with mutated CSD and CCHC domains and an intact 3′UTR. d) Seam cells glowing in an L4 larva with Cbr-lin-28::GFP array with mutated CSD and CCHC domains and a 3′UTR deletion.
About 30% of fluorescing animals also had alae gaps. Extrachromosomal arrays might exceed wild-type levels of expression since stable transgenes contain several copies of the gene (Mello et al. 1991). Thus, it is possible that our construct resulted in overexpression of Cbr-lin-28—possibly by overcoming miRNA repression—to cause a weak reiterative phenotype. Furthermore, we observed that embryos showing very bright GFP fluorescence failed to hatch or died soon after hatching; only 2 larvae hatched out of 34 brightly fluorescing eggs, suggesting that very high Cbr-lin-28 expression causes an embryonic lethal phenotype.
To address the possibility that overexpression of the transgene affected Cbr-lin-28 regulation, we generated aeEx46 transgene with mutations in the lin-28 protein's 2 functional domains, the CSD (Y35A, F37A) and the CCHC zinc fingers (C127A, C137A) (Supplementary Fig. 10). The GFP fluorescence was still observed at all stages and did not decline except for seam cells. In seam cells, robust fluorescence was observed mostly at the L1 stage. Weaker fluorescence was observed at the L2 and L3 stages and occurred less often than at the L1 stage, and no fluorescence was observed at the L4 stage (Table 1).
Table 1.
Percent’s of animals with fluorescent seam cells in strains with a Cbr-lin-28::GFP extrachromosomal array.
| Array | Stage | |||||
|---|---|---|---|---|---|---|
| L1 | L2 | L3 | L4 | Young adult | ||
| Cbr-lin-28-GFP mCSD + mCCHC | 1 | 73% (15) | 42% (12) | 28% (18) | 0% (24) | 0% (11) |
| 2 | 70% (20) | 18% (17) | 11% (19) | 0% (19) | 0% (5) | |
| 3 | 40% (15) | 41% (17) | 31% (13) | 0% (28) | 0% (2) | |
| Cbr-lin-28-GFP mCSD + mCCHC + 3′-UTR del | 75% (16) | 50% (14) | 35% (20) | 17% (23) | 11% (18) | |
The number in parentheses is the total number of fluorescent animals observed; percents are those with at least 1 fluorescent seam cell. mCSD, mutations in the Cold Shock Domain; mCCHC, mutations in the CCHC zinc fingers (see Supplementary Fig. 10).
We further modified the transgene construct to contain a deletion in the 3′UTR to remove miRNA sites (Supplementary Fig. 10). Animals with aeEx48 transgene encoding a mutant protein and a 3′UTR deletion did not show any differences in the place and timing of Cbr-lin-28 expression, except for seam cells. Fluorescing seam cells were observed less often after the L1 stage, similar to arrays with an intact 3′UTR. Nevertheless, they were observed in L4 and young adult animals in this strain, in contrast to 3 independently generated strains bearing the CSD/CCHC mutant with an intact 3′UTR (Table 1, Fig. 4b–d). Thus, Cbr-lin-28 is down-regulated in seam cells in late larval development in part via its 3′UTR.
Overall, the marked difference in expression between C. elegans and C. briggsae parallels the differences in phenotype, where it seems Cbr-lin-28 has a broader role in the animal than Cel-lin-28. Nevertheless, 3′UTR-dependent down-regulation in seam cells occurs in both species.
Cbr-lin-46(0) mutants are similar to Cel-lin-46(0) mutants
In C. elegans, lin-46 was discovered as a suppressor of lin-28(0) phenotype (Pepper et al. 2004). lin-46(0); lin-28(0) double mutants appear mostly as wild type, whereas lin-46(0) single mutants have alae gaps and an increased number of seam cells, with both defects being cold-sensitive. lin-46 encodes an unusual protein with protein-protein interaction activity.
A Cbr-lin-46(0) allele was generated by targeting the second exon: the Cbr-lin-46(ae38) mutation is an insertion causing a frameshift that results in a premature stop codon. A second allele, Cbr-lin-46(ae44), has a deletion removing the start-codon (Supplementary Table 1 and Fig. 6).
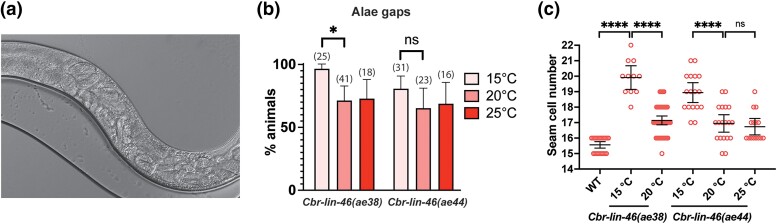
The null mutant has a weak reiterative phenotype similar to Cel-lin-46(0) mutants. In C. briggsae, around 65% of the animals had alae gaps at either 20°C or 25°C, and over 80% at 15°C (Fig. 5b). The penetrance at higher temperatures is higher than is seen for Cel-lin-46(0) animals, but the cold sensitivity is shared (Pepper et al. 2004). In both species, the number of seam cells was significantly higher at 15°C (Fig. 5c). There were also slight egg-laying defects, and vulvae were often abnormally shaped in C. briggsae (Fig. 5a, Supplementary Fig. 11). Thus, Cbr-lin-46(0) mutants have similar defects and cold sensitivity as Cel-lin-46(0) mutants, suggesting that the orthologs have similar functions. Furthermore, the fact that null alleles are cold sensitive in both species implies that a process that is exposed by the loss of lin-46 has inherent cold sensitivity.
Fig. 5.
Null mutants of Cbr-lin-46 have a reiterative phenotype. a) A DIC photomicrograph of a cbr-lin-46(ae38) adult animal with an egg-laying defect and accumulated late stage embryos at 20°C can be seen. The worm is oriented anterior end left, dorsal side up. b) Alae gaps in Cbr-lin-46(0) adults are slightly more frequent at 15°C. Sample sizes are specified in parenthesis above the bars. c) Cbr-lin-46(0) L4 and young adult animals have increased numbers of seam cells that are also cold-sensitive.
Cbr-lin-46(0) partially suppresses the Cbr-lin-28(0) phenotype
To see whether the relationship between Cbr-lin-28 and Cbr-lin-46 is conserved despite the drift in lin-28's role, we constructed a Cbr-lin-46(ae38); Cbr-lin-28(ae39) double null mutant. Surprisingly, we found that Cbr-lin-46(0) suppressed not only the precocious alae defect of Cbr-lin-28(0) mutants, but that the L4 developmental arrest was partly suppressed (although not at 15°C), and that the gonad disorganization was partly suppressed at all temperatures (Fig. 3b and c, Supplementary Figs. 9 and 12).
Interestingly, Cbr-lin-46(0) did not suppress the L4 arrest phenotype when the double-mutant passed through dauer: the fraction of L4-arrested animals at 20°C in Cbr-lin-28(0); Cbr-lin-46(0) double mutants that had passed through dauer was comparable to the fraction of “arrested L4” animals in Cbr-lin-28(0) postdauers at 20°C (Fig. 3b). This suggests that different downstream effectors exist for Cbr-lin-28 in continuous development and dauer development.
We also observed some reciprocal suppression: the increased number of seam cells in Cbr-lin-46(0) mutants was suppressed by the Cbr-lin-28(0) mutation at 20°C, although not at 15°C (Fig. 3d). By contrast, successfully molted double mutants had alae gaps at comparable rates to the Cbr-lin-46(0) single mutants (63.1%, n = 19 at 20°C and 61.5%, n = 13 at 25°C, Fig. 5b). Thus, some of the reiterative traits of Cbr-lin-46(0) were not suppressed by Cbr-lin-28(0), which is in contrast to what occurs in C. elegans (Pepper et al. 2004).
The Cbr-lin-46 5′UTR mutant phenotype differs from that of a Cbr-lin-28(0) mutant
In C. elegans, the lin-46 5′UTR is a regulatory region through which lin-28 acts to inhibit lin-46 expression; small deletions in this sequence cause a phenotype that resembles the Cel-lin-28(lf) phenotype (Ilbay et al. 2021). This 36-nt 5′UTR is conserved among all species of Caenorhabditis and is identical between C. elegans and C. briggsae. We created a 6-bp deletion (allele ae43) in this region using CRISPR/Cas9 (Supplementary Table 1 and Fig. 13a). The Cbr-lin-46 5′UTR mutants had protruding vulvae and either full or gapped precocious alae at the L4 stage; however, they had the same number of seam cells as the wild type (Supplementary Fig. 13b–d). This phenotype resembles the heterochronic traits of Cbr-lin-28(0), however, the penetrance and expressivity of the alae defect are more severe in the Cbr-lin-46(gf) mutant. Significantly, the Cbr-lin-46 5′UTR mutant lacks the larval arrest and gonad disintegration defects of the Cbr-lin-28(0) mutants. Thus, the role of Cbr-lin-46 in developmental timing resembles that of Cel-lin-46, but the fact that the phenotype of the Cbr-lin-46 5′UTR deletion differs substantially from that of Cbr-lin-28(0) suggests that the relationship between lin-28 and lin-46 has drifted as these species evolved.
mir-241, mir-48, and mir-84 have a conserved function in C. elegans and C. briggsae
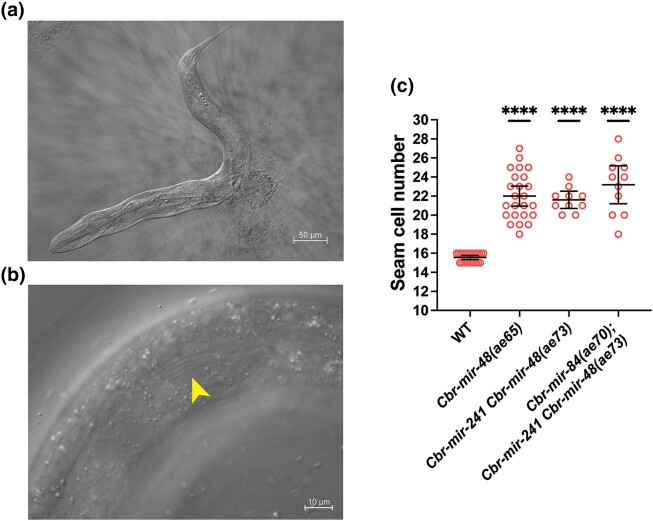
In C. elegans, 3 let-7-family miRNAs, mir-48, mir-84, and mir-241 (the “3let-7s”), redundantly control hbl-1 and lin-28: a strong phenotype appears when all 3 are knocked out, causing reiteration of L2-specific cell fates, whereas single mutants have little or no effect (Abbott et al. 2005; Tsialikas et al. 2017). By contrast, in C. briggsae, a Cbr-mir-48(0) mutation alone yielded a strong phenotype: Cbr-mir-48(ae65) mutant burst at the vulva at the end of the L4 molt, had an increased number of seam cells, and incomplete adult alae (Supplementary Table 1, Fig. 6a and b). We saw that 95% of mutant adults had less than half of the normal amount of adult alae, and 11% lacked alae entirely (n = 44).
Fig. 6.
Cbr-3let-7s mutants have reiterative phenotypes. a) DIC micrograph of a Cbr-mir-48(ae65) mutant that has burst at vulva after reaching adulthood. b) Cbr-mir-48(ae65) young adults develop alae patches (indicated by an arrowhead). c) Cbr-mir-48(ae65) have an increased number of seam cells, but adding Cbr-mir-241(0) and Cbr-mir-84(0) mutations does not cause further increase in the seam cell number. The difference between mutant groups was not statistically significant (unpaired Welch’s t-test, P > 0.05).
The Cbr-mir-48 and Cbr-241 genes are within 3 kb of each other in linkage group V, and we obtained a deletion allele that removed both (Supplementary Table 1; ae73). Deletion of both miRNAs resulted in a more severe phenotype than Cbr-mir-48(ae65) alone: 92% of adult animals lacked alae altogether (n = 13), compared to 11% for the single mutant. However, the double mutants that had developed through the dauer pathway did not burst at the vulva and appeared wild-type, indicating that these mutations, like their C. elegans counterparts, are suppressed by the dauer developmental pathway (data not shown).
Deleting all 3 miRNAs resulted in animals that could not be maintained as homozygotes because most were sterile. However, the frequency of alae patches in Cbr-mir-48 Cbr-mir-241(ae73); Cbr-mir-84(ae70) animals segregating from heterozygotes was similar to that of the double mutant: 92% lacked alae (n = 13). Furthermore, this triple mutant did not show an increase in the number of seam cells compared to the Cbr-mir-48(0) single mutant (Fig. 6c).
In other aspects, the Cbr-mir-241(0) and Cbr-mir-84(0) single null mutants and the Cbr-mir-241(0); Cbr-mir-84(0) double null mutants had few differences from the wild type. There were no alae gaps and the number of seam cells was close to normal at both 20°C and 15°C (Supplementary Fig. 14). A few animals had egg-laying defects and abnormal vulvae (Supplementary Fig. 15), and some of these egg-laying defective animals appeared to be stuck in lethargus (not pumping). Finally, a small percentage of sterile animals was observed in these strains (Table 2).
Table 2.
Phenotypes of Cbr-mir-241(0) and Cbr-mir-84(0) mutants.
| Egl | Sterile | Lethargic | N | |
|---|---|---|---|---|
| Wild type | 2.5% (7) | 0 | 0.7% (2) | 283 |
| Wild type | 0 | 0 | 0 | 174 |
| Cbr-mir-241(ae64) | 3% (11) | 1.1% (4) | 1.4% (5) | 363 |
| Cbr-mir-84(ae68) | 6% (11) | 0 | 1.6% (3) | 182 |
| Cbr-mir-84(ae69) | 8% (25) | 0.32% (1) | 1.3% (4) | 313 |
| Cbr-mir-241(ae64); Cbr-mir-84(ae69) | 13.1% (23) | 0 | 1.7% (3) | 175 |
| Cbr-mir-241(ae64); Cbr-mir-84(ae70) | 10.5% (17) | 0 | 1.85% (3) | 162 |
Egg-laying defective (Egl) animals accumulated eggs in the uterus. Lethargic animals had no pharyngeal pumping. Animals with both traits were included in both columns. Wild-type animals were observed twice. On less crowded plates, no Egl animals were observed, on a more crowded plate 2.5% were Egl.
In C. elegans, mutations in lin-4 and the 3 let-7-related miRNAs mir-48, mir-84, and mir-241, have distinct phenotypes: deletion of lin-4 causes reiteration of L1-specific fates and deletion of all of the 3let-7s causes reiteration of L2-specific fates (Chalfie et al. 1981; Abbott et al. 2005). All 4 of these miRNA genes have been shown to act together in stage-specifically down-regulating lin-28 to ensure appropriate expression of L2 fates (Tsialikas et al. 2017). Because we found that mutations in Cbr-lin-4 and Cbr-3let-7s both cause reiteration of L2 fates, we tested whether these mutations enhanced one another, leading to a reiteration of the earlier L1 fates. To our knowledge, the equivalent mutant of C. elegans has not been reported. A Cbr-lin-4 deletion was introduced into Cbr-mir-241(0), mir-48(0), mir-84(0) mutant background (also containing Cbr-hbl-1::AID; TIR1(F79G); see below). The quadruple mutants were vulvaless with gapped alae at adulthood (data not shown). However, in contrast to Cbr-lin-4(0) mutants, late L2 larvae of the quadruple mutant had a lower number of seam cells (mean = 12.4±1.3, n = 14), suggesting that some seam cells reiterated L1 fates. The phenotype resembles the Cbr-lin-14 3′UTR deletion, suggesting that Cbr-3let-7s participate in the down-regulation of Cbr-lin-14, which contains let-7 sites in its 3′UTR.
Auxin-inducible degron system in C. briggsae
In C. elegans, certain heterochronic genes have pleiotropic phenotypes that include embryonic lethality or infertility. Anticipating that these genes might have similar pleiotropies in C. briggsae, we used the AID system to generate conditional alleles. We produced lines of C. briggsae expressing TIR1(F79G) from extrachromosomal (aeEx44) and attached (aeIs15) arrays (Hills-Muckey et al. 2021). The attached transgene (aeIs15) was located by crossing with marker strains and found to be on LGII.
Unexpectedly, both the extrachromosomal and attached TIR1(F79G) arrays caused a reduction in the number of intestinal nuclei (Supplementary Table 3). However, no other abnormalities were observed and the animals appeared to develop normally and be healthy. The reason for this reduction in intestinal nuclei is unclear. The intestinal nuclei glowed brightly, indicative of high array expression.
Cbr-hbl-1(lf) causes a precocious phenotype like Cel-hbl-1(lf)
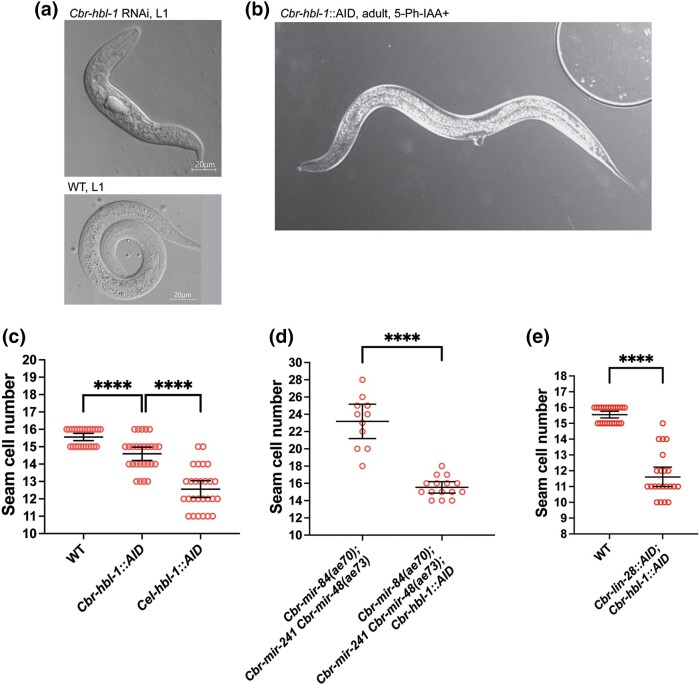
In C. elegans, hbl-1 encodes an Ikaros-family transcription factor involved in hypodermis development where null alleles are embryonic lethal and weak alleles have a heterochronic phenotype with a reduced number of seam cells, precocious alae, and a protruding vulva, resembling lin-28(0) (Fay et al. 1999; Abrahante et al. 2003; Lin et al. 2003). To study Cbr-hbl-1 loss-of-function while avoiding potential embryonic lethality, the locus was tagged with an AID using CRISPR/Cas9 (Supplementary Table 1). The hbl-1::AID strain was then crossed with lines bearing the TIR1(F59G) transgene to generate lines in which Cbr-hbl-1 activity could be reduced in response to the auxin analog 5-Ph-IAA (Hills-Muckey et al. 2021).
Adult animals were placed on plates with 0.01–0.02 µmol of 5-Ph-IAA, and the phenotypes of the next generation were characterized. We saw that 47% of animals on those plates had fully precocious alae, and the remainder had gapped precocious alae (n = 30). There was no embryonic lethality. However, 82% of adult animals remained stuck in the L4 molt, and 27% of mutants had a precocious vulval differentiation (reaching the “Christmas tree”, or L4.4-L4.5 according to Mok et al. (2015), stage of morphogenesis by the end of the L3 stage) or a protruding vulva (Fig. 7b). As in C. elegans hbl-1(lf), the protruding vulva developed during the L4 stage. Other animals had normal vulval development and a functional vulva. There was a slight reduction in seam cell number in some Cbr-hbl-1(lf) animals. However, the reduction was not as significant as in Cel-hbl-1::AID under similar conditions (Fig. 7c). These observations show that the functions of hbl-1 are at least partly conserved between C. elegans and C. briggsae.
Fig. 7.
Cbr-hbl-1 is required for early development and promotes L2 seam cell fates. a) DIC micrograph of a wild-type L1 larva and deformed L1 larva from Cbr-hbl-1 dsRNA injections. These animals do not survive and develop into adults. b) A Cbr-hbl-1::AID young adult grown on 5-Ph-IAA. Notice a protruding vulva. c) Some Cbr-hbl-1::AID mutants have a slightly reduced number of seam cells in the presence of 5-Ph-IAA, but the reduction is not as large as for Cel-hbl-1::AID, which may due to differences in hbl-1 orthologs functions or a difference in the degradation efficiency. d) Cbr-hbl-1 depletion on 5-Ph-IAA suppresses increased seam cell numbers in the Cbr-3let-7s triple mutant. Cbr-3let-7s seam cell counts are the same data set as in Fig. 6c. e) Combined depletion of Cbr-lin-28::AID and Cbr-hbl-1::AID on 5-Ph-IAA causes a reduced number of seam cells.
To test whether a more drastic reduction of Cbr-hbl-1 activity could cause embryonic lethality like that seen in C. elegans, we performed RNAi. A dsRNA representing 668 bp from exon 4 of the Cbr-hbl-1 ORF was injected into wildtype C. briggsae, and we observed dead L1 larvae in the next generation (Fig. 7a). Their terminal phenotype appeared to be more developmentally advanced than that observed in similar C. elegans experiments with none of the eggs failing to hatch (Fay et al. 1999). No older larvae or adults with heterochronic phenotypes were observed, potentially indicating that all animals receiving RNAi failed to proceed with development after hatching. This result suggests that Cbr-hbl-1 may have a role in embryonic development but one that differs slightly from Cel-hbl-1.
Reiterative phenotype of Cbr-3let-7s is suppressed by Cbr-lin-28(0) and Cbr-hbl-1(lf)
The 3let-7s play an important role in down-regulating lin-28 and hbl-1 in C. elegans, a conclusion supported by the fact that lin-28(0) and hbl-1(lf) are epistatic to loss of the 3let-7s (Abbott et al. 2005). Likewise, we found that Cbr-lin-28(0) and Cbr-hbl-1(lf) were epistatic to the reiterative phenotype caused by Cbr-3let-7s(0). The Cbr-hbl-1::AID; Cbr-3let-7s(0); TIR1(F79G) strain had a number of seam cells close to normal (Fig. 7d), precocious alae (100%, n = 14), and protruding vulvae (43%, n = 14) when grown on 5-Ph-IAA plates. Cbr-lin-28(ae39); Cbr-mir-48(ae65) double mutants had a normal number of seam cells (15.8±0.6, n = 13), precocious alae patches (66.7%, n = 12) at L4, nearly complete adult alae, although some had a gap (25%, n = 8), and protruding vulvae (24%, n = 21).
These observations suggest that both Cbr-lin-28 and Cbr-hbl-1 act downstream of the Cbr-3let-7s and are necessary for the reiteration of the L2 stage caused by these 3 mutants, as in C. elegans. This is surprising since Cbr-lin-28(0) and Cbr-hbl-1(0) single mutants mostly did not show a reduction in seam cell numbers.
Simultaneous reduction of Cbr-lin-28 and Cbr-hbl-1 activities shows that Cbr-lin-28 acts in the L2
In C. elegans, both lin-28 and hbl-1 are needed for L2 fates to occur. Because C. briggsae lin-28(0) mutants showed no L2 defect, we investigated whether it might still be involved at this stage by testing whether mutations would enhance the precocious phenotype caused by loss of Cbr-hbl-1 activity. To do this, we generated a Cbr-lin-28::AID; Cbr-hbl-1::AID strain in a TIR1(F79G) background. When grown on 5-Ph-IAA plates, these animals had a reduction in seam cell numbers that was more severe than Cbr-hbl-1:aid alone (Fig. 7e; compare with Fig. 7c). They also developed gapped alae at the L3 stage (100% of animals had some precocious alae at L3), and complete alae by the L4. Thus, the reduction of both Cbr-lin-28 and Cbr-hbl-1 activity resembled the Cel-lin-28(0) phenotype. These double mutants also had a prolonged L3 stage and became stuck in the L3 molt: 24 hours after L3 animals were selected, some still had L3 cuticles with gapped alae and nonreflexing gonads (Supplementary Fig. 16). In other animals, the gonads migrated closer to the pharynx and anus than normal before reflexing, and occasionally, the distal tips cells leading the gonad arms migrated in unexpected directions. Finally, the vulvae were either protruding or stuck in an L4-like (pre-“Christmas tree”, or L4.2-L4.3 according to Mok et al. 2015) shape. Interestingly, all of the Cbr-lin-28::AID; Cbr-hbl-1::AID animals grown on 5-Ph-IAA were sterile and had disorganized gonads.
These observations show that Cbr-lin-28 is involved with Cbr-hbl-1 in promoting L2 cell fates, as in C. elegans. But in C. elegans, both genes are necessary, and in C. briggsae they are partially redundant.
Cbr-let-7(0) mutants have additional molts but no heterochronic defects
C. elegans let-7(0) mutants have delayed adult alae formation due to reiteration of L3-specific developmental events, and they burst at the vulva upon reaching adulthood (Reinhart et al. 2000; Vadla et al. 2012). Two mutant alleles of Cbr-let-7 were generated: an insertion (ae47) and deletion (ae48), both of which eliminate Cbr-let-7 activity (Supplementary Table 1).
Both Cbr-let-7(0) mutants displayed egg-laying defects, slightly abnormal vulvae shapes, and at least 1 extra molt (Supplementary Fig. 17). The vulvae of these animals were slightly protruding, and adults burst at the vulvae on microscope slides, suggesting defects in vulval development or structure. In contrast to Cel-let-7(0), the Cbr-let-7(0) mutants developed normal adult alae at the L4 molt. The alae looked thinner than the wild type, perhaps due to a cuticle defect or the formation of an overlying cuticle during an extra molt (Supplementary Fig. 18). Some Cbr-let-7(0) mutants may have 2 extra molts: A plate containing 4 L4 larvae were placed at 20°C and the number of shed cuticles were counted the next day: 6 cuticles were found and 2 adult animals (with eggs) were stuck in cuticles (Supplementary Fig. 19). The Cbr-let-7(0) mutants had a slightly increased number of seam cells at low temperatures, but it is unclear whether this is a heterochronic defect (Supplementary Fig. 20).
Cbr-let-7(0) mutation suppresses later defects of Cbr-lin-14(0) and Cbr-lin-28(0) mutants
Despite Cbr-let-7(0) mutants not reiterating late larval stage seam cell fates (as assessed by alae formation), we tested whether they could nevertheless suppress the precocious alae defect of Cbr-lin-28(0), as occurs in C. elegans (Slack et al. 2000; Vadla et al. 2012). Examining Cbr-lin-28(ae39); Cbr-let-7(ae47) animals, we found that Cbr-let-7(0) mutation completely suppressed the “arrested L4” phenotype and gonad disorganization of the Cbr-lin-28(0) mutant, which implies that this aspect of the Cbr-lin-28(0) phenotype is in part due to the inappropriate upregulation of Cbr-let-7 and presumably the subsequent silencing of the miRNA's targets.
Some Cbr-lin-28(0); Cbr-let-7(0) animals looked wild-type, whereas others had egg-laying defects and resembled Cbr-let-7(0) mutants. The double mutants had slightly abnormal vulvae similar to Cbr-let-7(0) single mutants, and sometimes developed protruding vulvae (data not shown). The double mutants also underwent extra molts, however, unlike the Cbr-let-7(0) single mutants, they usually did not complete those molts and remained stuck in cuticles (Supplementary Fig. 21). Four L4 larvae from the Cbr-lin-28(0); Cbr-let-7(0) strain were isolated on a separate plate, and cuticles were scored next day; 4 cuticles were found and all of the animals were adults stuck in the cuticle while molting. Thus, the loss of Cbr-lin-28 slightly mitigates this Cbr-let-7(0) phenotype, suggesting that some functions of Cbr-let-7 depend on the activity of Cbr-lin-28.
Interestingly, although some Cbr-lin-28(0); Cbr-let-7(0) double mutants developed precocious alae at the L4 stage at both 15°C (84.2%, n = 38) and at 20°C (25%, n = 8), the frequencies were lower than in Cbr-lin-28(0) single mutant (compare to Fig. 3c). Alae patches were usually located on the head and just behind the pharynx, but short patches were also observed in other areas. The results suggest that precocious alae formation in Cbr-lin-28(0) mutants is in part caused by premature Cbr-let-7 upregulation.
Finally, we examined a Cbr-lin-14(ae51) Cbr-let-7(ae50) double mutant. The Cbr-let-7(0) mutation restored fertility to Cbr-lin-14(0) mutants and suppressed precocious alae formation. This observation suggests that sterility and precocious alae occur in Cbr-lin-14(0) mutants because of the inappropriate Cbr-let-7 expression. In C. elegans, lin-14 acts in part through lin-28 to control late-stage events, so the sterility and precocious alae of Cbr-lin-14(0) could be due to the down-regulation of Cbr-lin-28, although we have not tested that hypothesis here (Seggerson et al. 2002; Tsialikas et al. 2017).
The Cbr-hbl-1(lf) phenotype is partly epistatic to that of Cbr-let-7(0)
Work in C. elegans suggests that hbl-1 acts downstream of let-7 (Abrahante et al. 2003; Lin et al. 2003; Abbott et al. 2005; Vadla et al. 2012). We tested the ability of Cbr-hbl-1::AID to suppress Cbr-let-7(0) phenotype by constructing the double mutant. Of double mutant animals grown on 5-Ph-IAA plates, 81% (n = 21) developed patches of precocious alae at the L3 molt, although no animals were stuck in the L4 molt. These results indicate that the Cbr-hbl-1(lf) mutant phenotype is partly epistatic to that of Cbr-let-7(0), as seen in C. elegans (Abrahante et al. 2003).
A Cbr-lin-41(lf) mutation causes a developmental arrest at the L4 stage
C. elegans lin-41 has multiple roles in the animal: null mutants are sterile (due to a germline defect) and hypomorphs have a heterochronic phenotype (Slack et al. 2000; Tocchini et al. 2014). We chose to tag the C. briggsae ortholog with the AID so its level of activity could be controlled. We inserted the aid sequence near the start codon of Cbr-lin-41 with CRISPR/Cas9. In this process, we also generated 2 loss-of-function alleles, including a potential null allele (Supplementary Table 1). This allele, Cbr-lin-41(ae77), is an incorrect aid insertion creating a false ORF that did not contain any Cbr-lin-41 exons. A potential start codon that was in-frame with the remaining Cbr-lin-41 ORF was located 181 bp downstream of the original start. Because homozygous Cbr-lin-41(ae77) animals were sterile, the allele was balanced with the marker Cbr-spe-8(v142) which is also sterile when homozygous (R. Ellis, pers. comm.). Thus, only heterozygous animals reproduced. Cbr-lin-41(ae77) segregating from the heterozygous strain was identified as having a shorter body (Dpy-like phenotype).
Homozygous Cbr-lin-41(ae77) animals had a developmental delay at the end of the L4 stage similar to that observed in Cbr-lin-28(0) mutants. Around 52 hours posthatching, when wild-type animals complete the L4 molt, develop alae, adult vulva, and both types of gametes, the Cbr-lin-41(ae77) animals also had oocytes and spermatozoa but did not have alae, and their vulvae were stuck at the pre-“Christmas tree” stage of morphogenesis (Fig. 8). Sometimes they had disorganized gonads similar to Cbr-lin-28(0) mutants. However, older Cbr-lin-41(ae77) animals had alae and adult-shaped vulvae in contrast to Cbr-lin-28(0) mutants, which sometimes failed to continue development after the L4 arrest. This suggests that the “arrested L4” state in Cbr-lin-28(0) mutants cannot be due solely to Cbr-lin-41 down-regulation. Notably, in contrast to Cel-lin-41 mutants, Cbr-lin-41(ae77) animals did not have precocious alae.
Fig. 8.
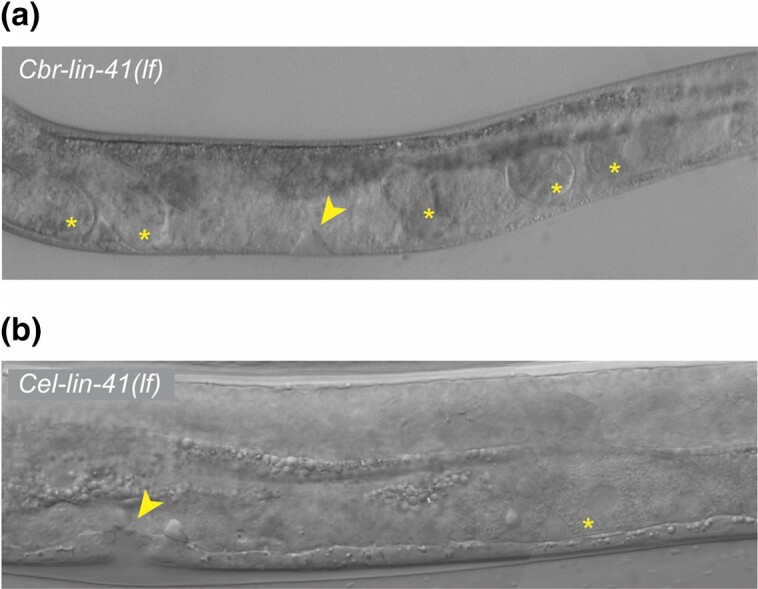
Both Cel-lin-41(lf) and Cbr-lin-41(lf) have L4 developmental delay or arrest. DIC micrographs of (top) Cbr-lin-41(ae76) and (bottom) Cel-lin-41::AID(aeIs10) animals grown on 5-Ph-IAA. In both, the vulva is arrested at an L4 stage of development (arrowheads) and the germ line is producing embryos or oocytes (asterisks). Animals are oriented anterior end left, dorsal side up.
The Cbr-lin-41(ae76) allele is a small insertion that creates a frameshift and early stop codon. Another in-frame start codon occurred 170 bp downstream of the original, which might allow some expression of the functional protein (Supplementary Table 1). This allele caused a weaker phenotype than ae77 and could be maintained as a homozygous line. The Cbr-lin-41(ae76) mutants had these additional defects: 17.9% successfully molted into adults, 67.9% were stuck in the L4 molt, and 14.2% had an “arrested L4” phenotype with disorganized gonads (n = 28). None had precocious alae.
Animals with a degron-tagged Cbr-lin-41 and TIR1(F79G) expressed from an attached array grown on 5-Ph-IAA produced oocytes and sperm before the soma completed development: the animals had vulvae still undergoing morphogenesis and no alae. The Cbr-lin-41::AID animals also had slightly abnormal vulvae, with asymmetric shapes and protrusions, and had egg-laying defects. There were no sterile animals on 5-Ph-IAA, and no precocious alae. The observations reveal similar drifts in the roles of lin-28 and lin-41 during the evolution of the 2 species, with more explicit control of developmental timing per se in C. elegans and less in C. briggsae.
Depletion of C. elegans lin-41 causes L4 developmental arrest
Although a variety of defects caused by loss of Cel-lin-41 activity have been observed (Slack et al. 2000; Tocchini et al. 2014), to our knowledge, L4 arrest like we observed in Cbr-lin-41(lf) mutants, have not been reported. To test whether C. elegans lin-41(lf) mutants can arrest in the L4 stage, we generated a Cel-lin-41::AID strain and crossed it with a strain expressing TIR1(F79G) allele (Hills-Muckey et al. 2021). Synchronized L1 larvae of wild-type C. elegans and Cel-lin-41::AID were placed on plates with or without 5-Ph-IAA. After 57 hours of development, 71% (n = 14) of wild-type animals were molting or had already shed cuticles, and the remainder were still late L4 larvae. Most molting animals had mature vulvae and were still producing sperm. Only 14% of the animals were already producing oocytes (1 young adult and 1 molting animal). Of the Cel-lin-41::AID animals, 40% (n = 15) were molting, only 13% had mature vulvae (2 of the molting animals), and the others had L4-shaped vulvae. Surprisingly, 73% of the animals already had both sperm and oocytes, including animals that appeared like “late L4” larvae. Moreover, when we looked at Cel-lin-41::AID animals on 5-Ph-IAA at 72 hours of development, 13% of animals had an arrested L4 phenotype characterized by an L4-like vulva (“Christmas tree”-like morphology), lack of adult alae, the absence of the final molt, and the presence of oocytes (Fig. 8). These observations indicate that reduction of Cel-lin-41 activity can produce an L4 developmental delay or arrest similar to that observed for both Cbr-lin-41(lf) and Cbr-lin-28(0), resulting in asynchrony between the germline and soma.
The Cbr-lin-41 gene acts downstream of Cbr-let-7 in the heterochronic pathway
In C. elegans, lin-41 is a primary target of let-7 in the heterochronic pathway (Slack et al. 2000). To test whether Cbr-lin-41 acts downstream of Cbr-let-7, we made the Cbr-lin-41(ae76); Cbr-let-7(ae48) double mutant. Of the double mutants observed (n = 33), 97% of the animals were stuck in the L4 molt and 3% had an “arrested L4” phenotype (L4-shaped vulva, no alae, and produced embryos). The difference in the penetrance of the “arrested L4” phenotype was not significant compared to the Cbr-lin-41(ae76) single mutant (P > 0.05, Fisher's exact test). Because the double mutant displayed the Cbr-lin-41(lf) mutant characteristics and none of the Cbr-let-7(0) mutant features were displayed, the Cbr-lin-41(lf) phenotype was epistatic to that of Cbr-let-7(0), suggesting that Cbr-lin-41 acts downstream of Cbr-let-7, as in C. elegans.
A Cbr-lin-41(0) mutation enhances the Cbr-hbl-1(lf) phenotype
In C. elegans, lin-41 and hbl-1 both appear to control L3 cell fates, and the animals lacking the activities of both genes generate some precocious alae at the L2 molt (Abrahante et al. 2003; Vadla et al. 2012). To test whether depletion of both these genes would cause a more severe phenotype in C. briggsae, a Cbr-lin-41(ae76); Cbr-hbl-1::AID double mutant was generated. Double mutants carrying the TIR1(F79G) unattached array were placed on 5-Ph-IAA-containing plates, and their phenotypes were analyzed. Double mutants resembled Cbr-lin-28::AID; Cbr-hbl-1::AID animals grown on 5-Ph-IAA: they had an L3 developmental delay, gonads with delayed reflexion, and abnormal distal tip cells migration after the reflection. Moreover, precocious alae patches appeared at the L2 molt in 69% of animals (n = 26), and 100% of animals had gapped (68.4%) or complete (31.6%) precocious alae at the L3 molt (n = 19). The double mutants also failed to shed L3 cuticles. The number of seam cells was slightly reduced (14.5±0.8, n = 26) compared to wild type (15.6±0.5, n = 25), but was similar to that observed in Cbr-hbl-1::AID strain grown on 5-Ph-IAA (14.8±0.8, n = 23, Fig. 7c). Thus, these counts do not show an effect of Cbr-lin-41(lf) on seam cell number. Our observations suggest that Cbr-lin-41 and Cbr-hbl-1 also control L3 hypodermal cell fates redundantly, but the fact that seam cell duplication occurs normally shows that lin-41 does not control L2 fates.
The Cbr-lin-29(0) phenotype resembles the Cel-lin-29(0) phenotype
The lin-29 gene encodes a zinc finger transcription factor that directly regulates the larval-to-adult adult switch in the C. elegans hypodermis; in its absence, the seam cells fail to differentiate, whereas precocious alae are formed because of early lin-29 activity (Ambros and Horvitz 1984; Ambros 1989; Rougvie and Ambros 1995; Azzi et al. 2020).
A Cbr-lin-29(0) mutant allele was made by targeting the 6th exon, where orthologous Cel-lin-29 mutations are located (Rougvie and Ambros 1995). The deletion ae75 is a frameshift which leads to a premature stop codon (Supplementary Table 1). The Cbr-lin-29(ae75) mutants did not develop alae and had delayed vulval development that caused them to burst at the adult stage (n = 15, Supplementary Fig. 22). Some animals retained a patch of L4 cuticle around the vulva, which occasionally prevented bursting and allowed some animals to survive and produce eggs. Cbr-lin-29(0) rarely produced larvae and could not be maintained as homozygotes. We, therefore, balanced Cbr-lin-29(ae75) with Cbr-trr-1(v76) (Guo et al. 2013). Overall, the phenotypes of Cel-lin-29(0) and Cbr-lin-29(0) mutants are very similar, suggesting conserved function and relationship to targets.
Cbr-lin-28 and Cbr-lin-41 act through Cbr-lin-29
In C. elegans, lin-29 acts at the end of the heterochronic gene hierarchy and is necessary for the late-stage phenotypes of earlier-acting heterochronic genes (Ambros 1989). To test whether the L4 developmental arrest of Cbr-lin-28(0) and Cbr-lin-41(lf) mutants required Cbr-lin-29 activity, we made the Cbr-lin-28(ae39); Cbr-lin-29(ae75) and Cbr-lin-41(ae76); Cbr-lin-29(ae75) double mutants.
Double homozygotes of these alleles could not be maintained because they burst at the vulva in adulthood and had very few progeny. Strains that were heterozygous for Cbr-lin-29(ae75) and homozygous for Cbr-lin-28(ae39) or Cbr-lin-41(ae76) (determined by PCR genotyping) segregated mostly worms (more than 50%) that lacked developmental arrest and disorganized gonads, suggesting that the loss of 1 copy of Cbr-lin-29 is sufficient to suppress these phenotypes. These strains segregated worms phenotypically similar to Cbr-lin-29(0), which lacked adult alae and burst at the vulva (Supplementary Fig. 23), as well as worms that phenotypically resembled Cbr-lin-41(lf) or Cbr-lin-28(0), respectively. Among Cbr-lin-41(ae76); Cbr-lin-29(ae75)/+ animals, 60% of worms resembled wild type (a normal L4 molt, vulva and adult alae), 19% were stuck in the L4 molt and looked like Cbr-lin-41(ae76) single mutants, and 21% looked like Cbr-lin-29(ae75) single mutants (n = 48). Cbr-lin-28(ae39); Cbr-lin-29(ae75)/+ animals segregated L4 larvae that had precocious alae patches (12 of 14 animals examined), and adults that had the Cbr-lin-29(0) phenotype (burst vulvae) lacked alae completely (n = 11). These observations suggest that loss of Cbr-lin-29 is epistatic to loss of either Cbr-lin-28 or Cbr-lin-41. Thus, Cbr-lin-28 and Cbr-lin-41 act through Cbr-lin-29, as they do in C. elegans, and interestingly, the arrested L4 phenotype also depends on Cbr-lin-29.
Discussion
We characterized 11 C. briggsae orthologs of C. elegans heterochronic genes using a total of 35 genetic lesions and 18 double and triple mutants and found that their mutant phenotypes differ in significant ways from those of C. elegans. Although most orthologs displayed defects in developmental timing, some of the phenotypes differed in which stages were affected, the penetrance and expressivity of the phenotypes, or by having pleiotropic effects that were not obviously connected to developmental timing. However, when examining pairwise epistasis and synergistic relationships, we found those reflected the relationships between their C. elegans orthologs, suggesting that the arrangements of these genes in functional modules are conserved, but the modules’ relationships to each other and/or to their targets has drifted since the time of the species’ last common ancestor.
A previous comparison of C. elegans and C. briggsae orthologs by RNA-interference (RNAi) showed that only a small fraction (91 of 1,333 orthologs) have significantly different loss-of-function phenotypes in the 2 species (Verster et al. 2014). This study included the protein-coding heterochronic gene orthologs, except Cbr-lin-28, but in general did not detect the degree of divergence that we observed. However, the level of analysis was limited and included only larval or embryonic lethality, growth rate, morphology, and fertility.
Our observations suggest that the level of functional divergence in the heterochronic gene orthologs is greater than previously thought. Despite this, the fundamental structure of the heterochronic pathway is largely conserved between C. elegans and C. briggsae. Since these species have nearly indistinguishable larval development and postembryonic cell lineages, the differences in single-gene mutant phenotypes and some pairwise relationships indicate a significant degree of developmental systems drift has occurred while giving rise to essentially the same anatomy and life history.
Conservation of key regulatory modules
As has been done in the analysis of other complex developmental pathways, the heterochronic pathway can be divided into subcircuits or modules that govern individual aspects of the phenotype (Supplementary Fig. 24, Verd et al. 2019). The modules are defined by enhancement or epistasis relationships among genes that act either together or in opposition to control cell fates at specific larval stages. Our findings suggest that these modules are largely conserved between C. elegans and C. briggsae.
The lin-4/lin-14 module in C. elegans specifies L1 cell fates and controls the transition to L2 fates (Chalfie et al. 1981; Ambros and Horvitz 1987; Wightman et al. 1993). lin-14 acts to specify L1 cell fates then is down-regulated by lin-4, allowing L2 and later fates to occur. We found the same is true in C. briggsae: Cbr-lin-14 has nearly the same role and is required for the reiterative mutant phenotype of mutant Cbr-lin-4. Taken with the fact that the miRNA sites in Cbr-lin-14's 3′UTR are conserved, and their deletion also causes a reiterative phenotype, we find that the regulatory relationship between lin-4 and lin-14 is conserved between the 2 species.
The genes lin-28, lin-46, 3let-7s, and hbl-1 comprise a complex regulatory module that, in C. elegans, specifies L2 cell fates and the transition to the L3 (Pepper et al. 2004; Vadla et al. 2012; Tsialikas et al. 2017; Ilbay and Ambros 2019; Ilbay et al. 2021). In C. briggsae, the relationships among these genes are conserved, although relative roles within the module have drifted. As in C. elegans, in C. briggsae both lin-28 and hbl-1 are required for L2 fates, lin-46 acts downstream of lin-28 while lin-28 also has lin-46-independent activity, the 3let-7s act upstream of hbl-1, and the 3let-7s are needed for the transition to L3 fates.
The transition from L3 to L4 and subsequently to adult cell fates is regulated by the let-7/lin-41/lin-29 module in C. elegans (Slack et al. 2000; Vadla et al. 2012; Azzi et al. 2020). Like lin-28, let-7's individual role has drifted, but its relationship to other genes is largely conserved: In both species, let-7 generally acts downstream of lin-14, lin-28, and hbl-1 and upstream of lin-41. lin-41's role has also drifted, but it appears to be a regulatory target of let-7 and acts to negatively regulate lin-29 in both species. lin-29 is the furthest downstream of all the genes in both species, at least with regard to the larva-adult transition. Thus, the core relationships in the let-7/lin-41/lin-29 regulatory module are conserved.
It is also worth noting the high phenotypic variability that we observed, which is greater in C. briggsae than in C. elegans for some mutants. This fact speaks to the high degree of genetic buffering that these modules provide when fully intact, a condition that would accommodate considerable developmental systems drift.
Evolutionary drift in regulatory relationships
Despite the fact that key regulatory relationships are conserved between the species, the single-gene phenotypes and some double mutant effects reveal substantial drift in the relationships of these genes to downstream targets. This drift is manifested in 2 ways: a shift in the role of the gene in the developmental timing of cell fates (their heterochronic roles), and the uncovering of additional roles that are not obviously related to cell fate timing, specifically the completion of larval development and gonad integrity.
Two aspects of the role of the lin-4/lin-14 module in C. briggsae compared to C. elegans are significant. First, the fact that Cbr-lin-4 primarily affects the transition from L2 to L3 suggests either that it alone is not sufficient to repress Cbr-lin-14, or that Cbr-lin-14 is not sufficient to specify L1 fates. Second, the fact that Cbr-lin-4(0) suppresses—but is not epistatic to—the precocious Cbr-lin-14(0) indicates drift in lin-4's relationship to its targets. Specifically, whereas in C. elegans lin-4's primary target is lin-14 and secondarily the L2 regulators lin-28 and hbl-1, in C. briggsae lin-4 may play a more significant role in regulating lin-28 and hbl-1 than it does with lin-14.
In both species, both lin-14 and lin-28 have sites for lin-4 and let-7 family miRNAs in their 3′UTRs. The regulation of lin-28 by both miRNA families was described previously and it was shown that the lin-4/lin-14 module controls the expression of the 3let-7s (Tsialikas et al. 2017). The drift we see in the role of lin-4 may reflect differences in the relative strengths of multiple miRNAs from the lin-4 and let-7 families in down-regulating their multiple targets. Developmental systems drift arises due to the accumulation of small changes—in this case, perhaps, in miRNA expression, abundance, or target sensitivity—and how those changes are compensated for by other small changes that keep developmental outcomes the same (True and Haag 2001).
Such subtle shifts in relative strengths are more apparent among the 3let-7s. In C. elegans, mutations in mir-48, mir-84, and mir-241 individually have very weak or undetectable phenotypes but when combined cause a strong L2 reiterative defect (Abbott et al. 2005; Li et al. 2005). These miRNAs all resemble let-7 in having the same 8-nt seed sequence and therefore have the potential to regulate the same targets (Lau et al. 2001; Lim et al. 2003). In C. elegans, mir-48 is largely redundant with the other 2 genes, but in C. briggsae mir-48 is mostly responsible for the work of all 3 genes for this phenotype (loss of mir-48 has nearly the same heterochronic effect as that of the triple mutant). Thus, small evolutionary changes in the relative levels of these miRNAs, or differences in the regulation caused by changes in 3′UTRs of their target genes, may be offset by compensatory changes in other miRNAs or target sites, leading to identical outputs of the regulatory module for the different species.
A reciprocal example of drift involving redundancy is lin-28 and hbl-1. In C. elegans, each gene is needed to specify L2 cell fates (Ambros and Horvitz 1984; Abrahante et al. 2003; Lin et al. 2003). By our observations, the regulatory module involving these genes is conserved, but in C. briggsae, lin-28's role in specifying L2 fates is barely detectable until Cbr-lin-28(lf) is combined with Cbr-hbl-1(lf), indicating its redundancy with Cbr-hbl-1. The substantial difference in lin-28's solo role in the 2 species may reflect a drift in 2 parallel components of this regulatory module, which are the negative regulators of hbl-1: the 3let-7s and lin-28's direct target, lin-46. In C. elegans, the effect of deleting lin-46's 5′UTR is similar to, although weaker than, lin-28's null phenotype (Ilbay et al. 2021). It is, therefore, surprising that deletion of the presumed Cbr-lin-28 regulatory site in the 5′UTR of Cbr-lin-46 does not phenocopy Cbr-lin-28(0) at all, but rather looks more like Cbr-hbl-1(lf). Furthermore, the role of lin-46 appears not to have drifted during the divergence of the 2 species. It is possible that Cbr-lin-28 may not be the only gene acting via the 5′UTR of Cbr-lin-46, but that alone would not explain Cbr-lin-28's redundancy with Cbr-hbl-1. A shift in the relative strength of lin-28's regulation of let-7 or another target may be responsible.
Significant drift was also found in the let-7/lin-41/lin-29 module. On their own, neither Cbr-let-7 nor Cbr-lin-41 influences the developmental timing of cell fates, in striking contrast to what happens in C. elegans. However as in C. elegans, in C. briggsae, lin-41 shows some redundancy with hbl-1 in controlling seam cell differentiation, demonstrating that its regulatory relationships with other heterochronic regulators are conserved. let-7, on the other hand, does not have the same suppressor interactions with the early-acting heterochronic genes lin-14 and lin-28 in C. briggsae as it has in C. elegans, although it does have a role in cessation of the molting cycle in both species. This “split” phenotype is consistent with the analysis of Azzi and colleagues who showed a branching of the pathway in the control of lin-29 isoforms that affect different aspects of terminal differentiation in the hypodermis (Azzi et al. 2020).
By far, the most significant difference between C. elegans and C. briggsae is lin-28's phenotype, which shows only minor heterochronic defects in C. briggsae while at the same time displaying significant late larval arrest and gonad integrity problems, 2 unexpected phenomena that are not well understood even in C. elegans. This was surprising given lin-28's broad conservation and role in timing among animals (Moss and Tang 2003; Balzer et al. 2010; Romer-Seibert et al. 2019). We have demonstrated here that a reduction in lin-41 activity can also lead to L4 arrest in C. elegans. Perhaps a difference in lin-28's regulatory relationship with lin-41 (possibly via its direct regulation of let-7) is responsible for lin-28's different influence on the completion of larval development in the 2 species. Additionally, we found significant differences in the time and place of lin-28's expression between the 2 species—including different degrees of significance of 3′UTR regulation—which may also account for the drift in pleiotropies over evolutionary time.
Our comparison of the heterochronic genes of C. elegans to those of their orthologs in C. briggsae revealed less drastic changes than has been seen in the sex determination pathways. Sex determination pathways evolve rapidly: C. elegans and C. briggsae developed hermaphroditism independently, since their common ancestor was dioecious (Ellis 2017; Haag et al. 2018). Some sex-determination genes have conserved roles in Caenorhabditis species, such as tra-1, tra-2, the fem genes, and fog-3 whereas others have quite different functions, including genes involved in sex determination in 1 species but not the other. Whether there exist genes in C. briggsae that have primary roles in developmental timing but are not among the orthologs studied here can be determined by forward genetic screens in these species for mutants with developmental timing defects.
Insights into the heterochronic pathway of C. elegans
Our investigation of the heterochronic gene orthologs of C. briggsae revealed new relationships between this pathway and other aspects of the animal's growth that may be relevant to both species. In particular, we found that a Cel-lin-41 mutation can cause a developmental arrest similar to that caused by Cbr-lin-28 and Cbr-lin-41 mutations. It is possible that redundancy that has not yet been uncovered in C. elegans could connect the heterochronic genes to the completion of larval development and gonad integrity. Given the number of heterochronic gene orthologs directly involved in gonad integrity—either because mutations in them cause disintegration (Cbr-lin-28, Cbr-hbl-1, Cbr-lin-41) or mutations suppress that disintegration (Cbr-lin-46, Cbr-let-7)—it would not be surprising to find that further investigation of heterochronic genes in C. elegans uncovers such a connection.
The majority of developmental systems drift in the heterochronic pathway appears to have occurred in redundant and parallel components of regulatory modules. Parallel regulatory branches may have different “weights” in determining phenotypic outcomes in different species. Even in C. elegans, we do not yet fully understand the nature of these parallel branches—such as non-lin-4 regulation of lin-14, lin-46-independent regulation of hbl-1 by lin-28, and the different contributions of the 3 let-7 miRNAs to the repression of multiple heterochronic genes. Further investigation using multiple Caenorhabditis species may reveal why the pathway is organized as it is and why it has evolved the ways it has.
Supplementary Material
Acknowledgments
We would like to thank members of Ellis lab and Kevin Kemper for valuable comments on the manuscript, Ronald Ellis for comments, advice and strains, Chris Hammell for TIR1 strains, and plasmid. We thank WormBase for genetic sequences and WormAtlas for reference information on nematode biology and development. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Contributor Information
Maria Ivanova, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering and Sciences, Rowan University, Stratford, NJ 08084, USA.
Eric G Moss, Department of Molecular Biology, Rowan University, Stratford, NJ 08084, USA.
Data availability
Strains and plasmids are available upon request. Figures and supplemental files available at figshare; https://doi.org/10.25386/genetics.24116676. Sequence data are available at Wormbase and Mirbase, links to database entries for each gene are given in Supplementary Table 1.
Funding
This work was fully funded by Rowan University.
Literature cited
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. 2005. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 9(3):403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 4(5):625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ambros V. 1989. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 57(1):49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 226(4673):409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. 1987. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1(4):398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- Azzi C, Aeschimann F, Neagu A, Großhans H. 2020. A branched heterochronic pathway directs juvenile-to-adult transition through two LIN-29 isoforms. Elife. 9:e53387. doi: 10.7554/eLife.53387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. 2010. LIN28 Alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Dev Camb Engl. 137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- Brown K., 2001. Comparative Studies of Vulva Development in C. briggsae. CalTech PhD thesis.
- Chalfie M, Horvitz HR, Sulston JE. 1981. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 24(1):59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- Choi S, Ambros V. 2019. The C. elegans heterochronic gene lin-28 coordinates the timing of hypodermal and somatic gonadal programs for hermaphrodite reproductive system morphogenesis. Dev Camb Engl. 146(5):dev164293. doi: 10.1242/dev.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 25(4):778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC. 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics. 210(3):781–787. doi: 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE. 2017. “The persistence of memory”—hermaphroditism in nematodes. Mol Reprod Dev. 84(2):144–157. doi: 10.1002/mrd.22668. [DOI] [PubMed] [Google Scholar]
- Ellis RE. 2022. Sex determination in nematode germ cells. Sex Dev. 16(5-6):305–322. doi: 10.1159/000520872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB. 1999. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 205(2):240–253. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- Félix M-A, Duveau F. 2012. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanta KS, Mello CC. 2020. Melting dsDNA donor molecules greatly improves precision genome editing in Caenorhabditis elegans. Genetics. 216(3):643–650. doi: 10.1534/genetics.120.303564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chen X, Ellis RE. 2013. Evolutionary change within a bipotential switch shaped the sperm/oocyte decision in hermaphroditic nematodes. PLoS Genet. 9(10):e1003850. doi: 10.1371/journal.pgen.1003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag ES, Fitch DH, Delattre M. 2018. From “the worm” to “the worms” and back again: the evolutionary developmental biology of nematodes. Genetics. 210(2):397–433. doi: 10.1534/genetics.118.300243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Hannon GJ. 2016. Inducing RNAi in Caenorhabditis elegans by injection of dsRNA. Cold Spring Harb. Protoc. 2016(1):pdb.prot086306. doi: 10.1101/pdb.prot086306. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, White JG. 1985. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol. 107(1):128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, Haag ES. 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell. 10(4):531–538. doi: 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hills-Muckey K, Martinez MAQ, Stec N, Hebbar S, Saldanha J, Medwig-Kinney TN, Moore FEQ, Ivanova M, Morao A, Ward JD, et al. 2021. An engineered, orthogonal auxin analog/AtTIR1(F79G) pairing improves both specificity and efficacy of the auxin degradation system in Caenorhabditis elegans. Genetics. 220(2):iyab174. doi: 10.1093/genetics/iyab174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova M, Birse D, Hong Y, Ambros V. 2005. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol Cell Biol. 25(24):11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbay O, Ambros V. 2019. Regulation of nuclear-cytoplasmic partitioning by the lin-28-lin-46 pathway reinforces microRNA repression of HBL-1 to confer robust cell-fate progression in C. elegans. Dev Camb Engl. 146(21):dev183111. doi: 10.1242/dev.183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbay O, Nelson C, Ambros V. 2021. C. elegans LIN-28 controls temporal cell fate progression by regulating LIN-46 expression via the 5′ UTR of lin-46 mRNA. Cell Rep. 36(10):109670. doi: 10.1016/j.celrep.2021.109670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE. 2005. Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev Cell. 9(3):415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. 2003. The microRNAs of Caenorhabditis elegans. Genes Dev. 17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-Y, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ, 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 4(5):639–650. doi: 10.1016/S1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Liu ZC, Ambros V. 1989. Heterochronic genes control the stage-specific initiation and expression of the dauer larva developmental program in Caenorhabditis elegans. Genes Dev. 3(12b):2039–2049. doi: 10.1101/gad.3.12b.2039. [DOI] [PubMed] [Google Scholar]
- Liu Z, Ambros V. 1991. Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature. 350(6314):162–165. doi: 10.1038/350162a0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DZ, Sternberg PW, Inoue T. 2015. Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev Biol. 15(1):26. doi: 10.1186/s12861-015-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 88(5):637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. 2003. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 258(2):432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Seydoux G. 2017. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods. 121-122:86–93. doi: 10.1016/j.ymeth.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AS-R, McCane JE, Kemper K, Yeung DA, Lee RC, Ambros V, Moss EG, 2004. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Dev Camb Engl. 131:2049–2059. doi: 10.1242/dev.01098. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G, 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Romer-Seibert JS, Hartman NW, Moss EG. 2019. The RNA-binding protein LIN28 controls progenitor and neuronal cell fate during postnatal neurogenesis. FASEB J Off Publ Fed Am Soc Exp Biol. 33:3291–3303. doi: 10.1096/fj.201801118R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Ambros V. 1995. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 121(8):2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Moss EG. 2013. Developmental transitions in C. elegans larval stages. Curr Top Dev Biol. 105:153–180. doi: 10.1016/B978-0-12-396968-2.00006-3. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Giusto J. 1989. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 338(6213):313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 243(2):215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 5(4):659–669. doi: 10.1016/S1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Sommer RJ. 2012. Evolutionary Systems Biology. 1st edn. New York (NY): Springer New York. p. 79–91. 10.1007/978-1-4614-3567-9. [DOI] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1(2):e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. 1985. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 5(12):3484–3496. doi: 10.1128/mcb.5.12.3484-3496.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Tocchini C, Keusch JJ, Miller SB, Finger S, Gut H, Stadler MB, Ciosk R. 2014. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet. 10(8):e1004533. doi: 10.1371/journal.pgen.1004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Haag ES. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 3(2):109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Tsialikas J, Romens MA, Abbott A, Moss EG. 2017. Stage-Specific timing of the microRNA regulation of lin-28 by the heterochronic gene lin-14 in Caenorhabditis elegans. Genetics. 205(1):251–262. doi: 10.1534/genetics.116.195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadla B, Kemper K, Alaimo J, Heine C, Moss EG. 2012. lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 8(3):e1002588. doi: 10.1371/journal.pgen.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verd B, Monk NA, Jaeger J. 2019. Modularity, criticality, and evolvability of a developmental gene regulatory network. Elife. 8:e42832. doi: 10.7554/eLife.42832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster AJ, Ramani AK, McKay SJ, Fraser AG. 2014. Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function. PLoS Genet. 10(2):e1004077. doi: 10.1371/journal.pgen.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Shen Y, Chen X, Shifman Y, Ellis RE. 2014. Rapid creation of forward-genetics tools for C. briggsae using TALENs: lessons for nonmodel organisms. Mol Biol Evol. 31(2):468–473. doi: 10.1093/molbev/mst213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Bürglin TR, Gatto J, Arasu P, Ruvkun G. 1991. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 5(10):1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 142(24):4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Boyle TJ, Bao Z, Murray JI, Mericle B, Waterston RH. 2008. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev Biol. 314(1):93–99. doi: 10.1016/j.ydbio.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. Figures and supplemental files available at figshare; https://doi.org/10.25386/genetics.24116676. Sequence data are available at Wormbase and Mirbase, links to database entries for each gene are given in Supplementary Table 1.