Summary
Background
Malaria, a widespread parasitic disease caused by Plasmodium species, remains a significant global health concern. Rapid and accurate detection, as well as species genotyping, are critical for effective malaria control.
Methods
We have developed a Flexible, Robust, Equipment-free Microfluidic (FREM) platform, which integrates recombinase polymerase amplification (RPA) and clustered regularly interspaced short palindromic repeats (CRISPR)-based detection, enabling simultaneous malaria infection screening and Plasmodium species genotyping. The microfluidic chip enabled the parallel detection of multiple Plasmodium species, each amplified by universal RPA primers and genotyped by specific crRNAs. The inclusion of a sucrose solution effectively created spatial separation between the RPA and CRISPR assays within a one-pot system, effectively resolving compatibility issues.
Findings
Clinical assessment of DNA extracts from patients with suspected malaria demonstrates the FREM platform's superior sensitivity (98.41%) and specificity (92.86%), yielding consistent results with PCR-sequencing for malaria detection, which achieved a positive predictive agreement of 98.41% and a negative predictive agreement of 92.86%. Additionally, the accuracy of species genotyping was validated through concordance rates of 90.91% between the FREM platform and PCR-sequencing.
Interpretation
The FREM platform offers a promising solution for point-of-care malaria screening and Plasmodium species genotyping. It highlights the possibility of improving malaria control efforts and expanding its applicability to address other infectious diseases.
Funding
This work was financially supported by International Joint Laboratory on Tropical Diseases Control in Greater Mekong Subregion, National Natural Science Foundation of China, the Natural Science Foundation of Shanghai, Bill & Melinda Gates Foundation and National Research and Development Plan of China.
Keywords: Malaria, Point-of-care testing, Microfluidic, CRISPR, Surveillance
Research in context.
Evidence before this study
Nucleic acid amplification tests represent the predominant diagnostic approach worldwide. However, their wide-scale implementation faces significant challenges, particularly in resource-limited settings, due to the need for well-equipped laboratories and trained personnel. This has led to a critical challenge in malaria-affected regions, where the absence of field-deployable diagnostics capable of detecting malaria infection impedes elimination efforts. The advent of CRISPR/Cas systems has brought about a revolution in point-of-care molecular diagnostics. These systems, known for their precise and programmable nucleic acid cleavage capabilities, have been successfully applied in infectious disease detection, including malaria. It's important to note that most CRISPR-based assays typically involve two separate operational steps: isothermal amplification and CRISPR-based nucleic acid detection. This dual-step process complicates diagnostic procedures and hampers their widespread adoption. However, the idea of performing both reactions directly within a confined system raises concerns regarding biocompatibility and potential damage to detection efficiency. Additionally, malaria, caused by various Plasmodium species, necessitates accurate genetic typing for guiding treatment and control efforts. However, challenges remain in achieving CRISPR-based multiplex biosensing due to indiscriminate collateral cleavage of unrelated probes.
Added value of this study
In response to these challenges, we introduce the Flexible, Robust, Equipment-free Microfluidic (FREM) platform, presenting a novel approach for simultaneous malaria screening and Plasmodium species genotyping. Our innovative incorporation of a sucrose solution strategically addresses the compatibility issues associated with combining recombinase polymerase amplification (RPA) and CRISPR assays within a one-pot system. This breakthrough not only simplifies the diagnostic process but also ensures high efficiency. Furthermore, the integration of RPA and CRISPR-based detection within a microfluidic chip offers a versatile and reliable solution for five Plasmodium species identification. This innovation represents a substantial advancement in point-of-care diagnostics for malaria, particularly in resource-limited settings. Our research extends the boundaries of molecular diagnostics, providing a user-friendly, rapid, and precise tool for healthcare professionals.
Implications of all the available evidence
Our study demonstrates the effectiveness of the FREM platform in screening malaria infection and genotyping Plasmodium species. This platform holds the potential to enhance epidemiological surveillance and inform treatment decisions, especially in regions with limited access to advanced laboratory facilities. By enabling species-specific identification of Plasmodium parasites, the FREM platform supports the tracking of different species, facilitating more targeted interventions and resource optimization. Furthermore, the FREM platform has the capacity to revolutionize healthcare delivery, aligning with the broader goal of eradicating malaria by 2040. Beyond its immediate application in malaria diagnostics, our work sheds light on innovative approaches that can be applied to combat other infectious diseases, extending the reach of our research beyond malaria alone.
Introduction
Malaria, one of the most serious mosquito-borne infectious diseases in tropical and subtropical regions,1,2 remains a major global challenge. It is caused by five species of unicellular eukaryotic Plasmodium parasites,3 namely Plasmodium falciparum, P. knowlesi, P. malariae, P. ovale, and P. vivax. Despite increased global elimination efforts over the past decade, malaria continues to impose a substantial burden on affected communities. In 2021, there were more than 247 million reported malaria cases in 84 endemic countries, leading to an estimated 619,000 deaths (https://www.who.int/publications/i/item/9789240064898).
Rapid and sensitive clinical diagnosis is critical for malaria control and elimination. Many of the remaining malaria cases receive diagnoses without confirmatory tests, leading to potential misdiagnosis and unnecessary or ineffective treatment. It is also crucial to distinguish between the various Plasmodium species, as they exhibit different drug–resistance profiles and responses to medications.4 Accurate identification of Plasmodium species is essential to guide the selection of species-specific drugs for effective treatment, which can significantly reduce the parasite reservoir. Given the global efforts shift from controlling malaria to pursuing elimination and eradication,5 there is a growing interest in tracking the prevalence and distribution of each species in a region,6,7 which is important for targeted public health interventions and resource allocation. In this context, the development of rapid, user-friendly and field-deployable diagnostic tests is an urgent priority for screening asymptomatic carriers and supporting elimination campaigns.
Microscopic examination has been considered the gold standard diagnostic method for confirming malaria infection due to its ease of operation and cost-effectiveness. However, solely relying on Giemsa-stained blood smears presents challenges in distinguishing atypical malarial morphology and identifying multiple species co-infections.8 Rapid diagnostic tests (RDTs) based on immunochromatography serve as an adjunct to microscopy, offering rapid and relatively straightforward performance and interpretation. Nevertheless, RDTs have their limitations in sensitivity and may not effectively detect infections with low parasite density, which is a common situation encountered in region with low transmission rates. Currently, nucleic acid amplification tests (NAATs) are the most sensitive diagnostic approach accessible globally. However, their wide-scale implementation is hampered by the requirement for well-equipped laboratories and trained personnel, especially in resource-limited settings. Consequently, the lack of field-deployable diagnostics capable of detecting malaria infection in endemic zones has posed a significant challenge to malaria elimination efforts.9
To overcome these limitations, there is a pressing need for an ideal diagnostic tool capable of performing on-site surveillance, promptly confirming suspected cases in resource-limited settings, and simultaneously distinguish between multiple Plasmodium species. Current research efforts have focused on clustered regularly interspaced short palindromic repeats (CRISPR)-based diagnostic tools10,11 due to their reliability, high specificity, and sensitivity when combined with isothermal amplification methods such as recombinase polymerase amplification (RPA).12 However, most CRISPR-based assays require two separate operational steps, which complicate the process and hamper widespread adoption.13 Also, challenges remain in achieving CRISPR-based multiplex biosensing due to indiscriminate collateral cleavage of unrelated probes.14,15 To address these challenges, recent developments in CRISPR-based microfluidic systems, featuring a “sample-to-answer” capability, have shown great potential in the fields of infectious diseases diagnosis.16,17 The manipulation of spatial separation within microfluidic systems, facilitated by a multi-channel design, provides a practical and effective solution for multiplex detection, while preserving the advantages of CRISPR and averting interference.
In this study, we present a Flexible, Robust, Equipment-free Microfluidic integrated platform, referred to FREM, which combines RPA and CRISPR-associated (Cas) 12a detection system within a microfluidic chip. This point-of-care testing (POCT) platform is designed for the accessible and comprehensive monitoring of critical malaria infection, including the identification of all five Plasmodium species. Our strategy utilizes a sucrose solution to successfully integrate RPA and CRISPR reactions into a single process, simplifying the workflow and reducing the risk of aerosol contamination. By utilizing microfluidic technology in conjugation, with species-specific CRISPR RNAs (crRNAs), we enabled parallel testing of six targets, enhancing both the efficiency and robustness of the platform. The outstanding performance of diagnostic platform shows the great potential for rapid malaria detection, which is invaluable not only for countries and regions seeking to increase their detection capacity in response to rising malaria cases but also for those requiring surveillance measures to monitor imported malaria cases caused by different species.
Methods
Primers and crRNAs design
The full-length sequences of the 18S rRNA gene for each Plasmodium species were downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). A multiple sequence alignment of all sequences was conducted using Geneious software 11.1.3 (Biomatters Ltd, Auckland, New Zealand), and highly conserved regions were visually identified with a threshold of 95%. Primer candidates for RPA pre-amplification were designed to be reactive across all Plasmodium species, following the instructions of the TwistAmp Assay Design Manual. Universal and species-specific crRNA candidates for the five Plasmodium species, each incorporating a protospacer adjacent motif (PAM) for Cas12a recognition, were designed following the common principles18 of Cas12a crRNA design (https://www.neb.com/en/faqs/2018/05/03/how-do-i-design-a-guide-rna-for-use-with-engen-lba-cas12a). All primers and crRNAs were synthesized by Tsingke Biotech (Beijing, China) and GenScript Biotech (Nanjing, China), respectively. The sequences of the primers and crRNAs used in this study are provided in Supplementary Data.
Specimen collection and processing
To confirm the proposed FREM method, clinical dry blood spot (DBS) samples of malaria infection were collected from imported cases to China from African and Southeast Asian countries. Six circles were punched out from a DBS sample and then incubated in 180 μL Buffer ATL at 85 °C for 10 min. After incubation, DNA was extracted using a QIAamp® DNA Mini Kit (Qiagen, Valencia, USA) according to the manufacturer's protocol.
RPA and CRISPR reactions in tubes
The RPA reaction was first carried out at 38 °C for 20 min using the TwistAmp Basic Kit (TwistDx), following the manufacturer's instructions. In brief, a 50 μL RPA mixture was prepared by resuspending one RPA pellet with 29.5 μL rehydration buffer, 0.48 μM forward and reverse primers, 14 mM magnesium acetate and 4 μL of the target sample. For the two-step detection process, 1.5 μL of the RPA product was then transferred to a Cas12a-mediated assay containing 100 nM LbCas12a, 100 nM crRNA, 400 nM ssDNA reporter, and 1 × r2.1 buffer, making a total reaction volume of 15 μL. The reaction was incubated at 38 °C for 30 min with fluorescence measurements taken every 30 s. For the one-step detection process, 10 μL of RPA system, without prior incubation, was immediately transferred to Cas12a-mediated assay containing 100 nM LbCas12a, 100 nM crRNA, 400 nM ssDNA reporter, 1 × r2.1 buffer and 10% sucrose solution to reach a total volume of 20 μL. The reaction was incubated at 38 °C for 60 or 90 min with fluorescence measurements taken every 1 min. For malaria infection testing, the crRNA targeting the conserved sequence of Plasmodium species was used. For the specific Plasmodium species detection, five crRNAs targeting distinguished locus of P. falciparum, P. malariae, P. ovale, P. vivax and P. knowlesi were used in each Cas12a-mediated assay, respectively. For a negative control, the target sample used for the RPA reaction was replaced by nuclease-free water.
Fabrication of microfluidic chip
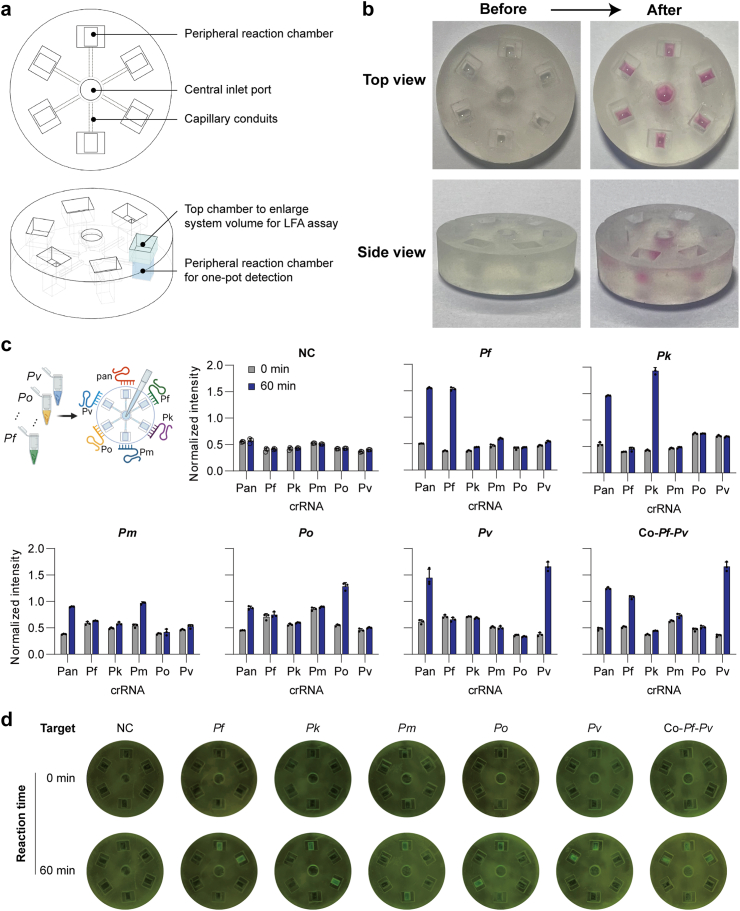
To achieve multiplex detection, the microfluidic chip measures 26 mm in diameter and 7.2 mm in height. It comprises six peripheral reaction chambers for multiple Cas12a systems, six connecting capillary conduits, and a central RPA inlet reactor. Each peripheral reaction chamber consists of two interconnected parts: the bottom chamber measures 3 mm in length, 2.1 mm in width, and 3.8 mm in height, the top chamber measures 4.5 mm in length, 3.5 mm in width, and 2.4 mm in height. The central inlet port has dimensions of 4.2 mm in diameter and 3.5 mm in depth, with six surrounding channels measuring 0.55 mm in width, connecting to the Cas12a reaction chambers.
The microfluidic chip was designed using SolidWorks 2021 (DS SolidWorks™, Waltham, USA) and fabricated using a 3D printer, Form 3 (Formlabs™, Boston, USA). After printing using clear resin FLGPCL04 (Formlabs™, Boston, USA), the printed objects were separated from the supporting platform and thoroughly washed for 15 min using isopropanol under ultrasonication conditions to remove any uncured resin, followed by a 15 min post-wash with deionized water. Finally, the microchannels and reaction chambers were statically coated with 2% polyethylene glycol (PEG) 3350 aqueous solution at room temperature for 30 min to reduce reagent absorbance.19,20 To mimic the aliquoting of solution from the central inlet port to the peripheral reaction chambers, sucrose solution and dye solution were pre-loaded into the chambers, and their distribution was recorded using a smartphone camera.
RPA and CRISPR reactions on microfluidic chip
Initially, each peripheral reaction chamber was loaded with 10 μL of Cas12a reaction components, containing 100 nM Cas12a, 100 nM crRNA, 400 nM ssDNA reporter, 1 × r2.1 buffer and 10% sucrose solution. To achieve multiplex detection, we prepared six Cas12a systems, each containing specific crRNAs for Plasmodium and different species, were introduced them into their corresponding chambers. Then, 90 μL of RPA solution was introduced into the central inlet port, effectively distributing it evenly across the peripheral chambers through capillary channels. The RPA solution was prepared as described above in the tube-based assay. Finally, the chip was incubated at 38 °C on a heat block for 60 min.
Fluorescent signal and lateral flow readouts
To readout the fluorescent signal, the chip was exposed to blue LED irradiation and imaged using a smartphone camera. The acquired fluorescent images were further processed and analyzed using Image J 1.8.0 software. For lateral flow strip readout, 200 μL deionized water was injected through the central inlet port to dilute solutions within the peripheral reactors. A lateral flow strip (LeSun Bio, China) was then added to each peripheral reactors to detect the presence of targets and a result was visualized after approximately 5 min. Lateral flow strips can be interpreted visually or were quantified using Image J gel analyzer tool and signal was normalized to the max signal intensity.
PCR, bidirectional sequencing analysis and qPCR
To screen and initially classify Plasmodium species in clinical DBS samples, the same nucleic acid extracts employed in the FREM assay were also utilized for PCR and sequencing. The PCR reaction mixture consisted of a total volume of 25 μL, with the following components: 12.5 μL 2 × Taq Master Mix (Vazyme, Nanjing, China), 0.4 μM of forward and reverse primers (details in Supplementary Data), and 2 μL of DNA template. The thermal cycling program included an initial step at 95 °C for 3 min, followed by 30 cycles at 95 °C for 15 s, 51 °C for 15 s, and 72 °C for 30 s. PCR products were analyzed by performing 2% agarose gel electrophoresis, and the DL2000 ladder was used to determine the position of PCR fragments. For positive samples, bidirectional sequencing was performed at Tsingke Biotechnology (Beijing, China). The sequence data were analyzed using BioEdit software version 7.0.9 (Ibis Bioscience, Inc., USA) and Geneious software (Biomatters Ltd, Auckland, New Zealand).
The qPCR reaction was performed to further analyze samples that had discrepancies between our work and PCR-sequencing. The qPCR reaction mixture consists of a total volume of 20 μL, with the following components: 10 μL Fast SYBR™ Green Master Mix (Applied Biosciences, Foster City, USA), 0.5 μM of forward and reverse primers (details in Supplementary Data), and 2 μL of DNA template. The thermal cycling program included an initial step at 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.
Statistical analysis
Unless otherwise indicated, each experiment was conducted with three technical replicates, one biological replicate. Two-tailed Student's t-test was used to evaluate statistical differences between two groups and one-way ANOVA was used to evaluate statistical differences among three or more groups using GraphPad Prism 8, with statistical significance defined as P < 0.05.
Ethical statement
This study was conducted in accordance with the principles of the Declaration of Helsinki. Prior to blood collection, participants were informed of the study protocol and the potential risks and benefits, and the written informed consent was obtained. Blood collected was performed in accordance with the institutional ethical guidelines reviewed and approved by the Ethics Committee of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention (20181203).
Role of funders
The funders were not involved in the study design, data collection, data analysis, interpretation or writing of the manuscript.
Results
Overview of the FREM platform
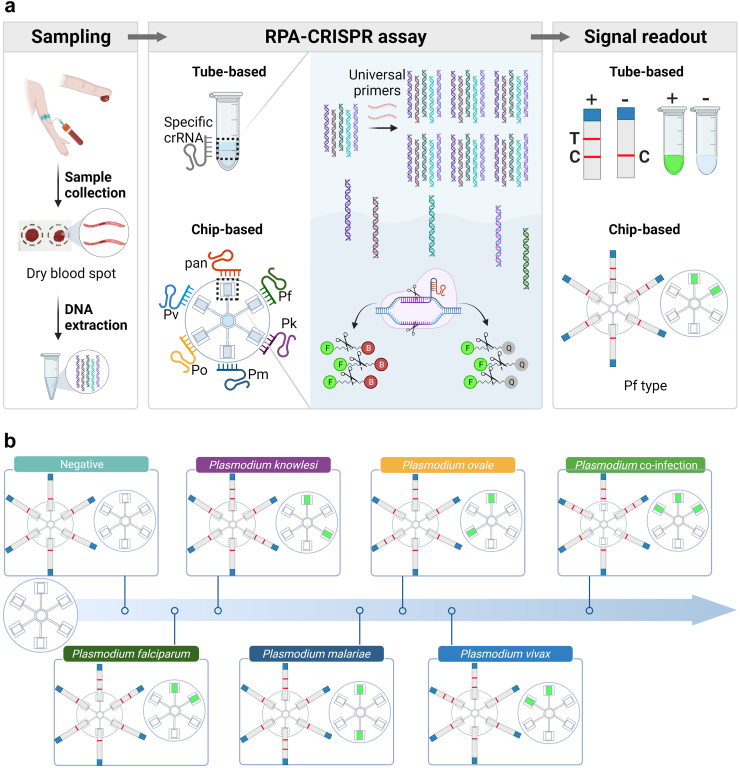
We developed a Flexible, Robust, Equipment-free Microfluidic platform (FREM) for comprehensive screening of malaria and Plasmodium species genotyping (Fig. 1). The FREM platform integrates RPA and CRISPR technologies, enabling the efficient detection of target nucleic acids. The sucrose-assisted one-pot assay design ensures spatial separation of the two systems while streamlining the workflow (Fig. 1a). After sample preparation and DNA extraction, RPA reagents are added to the CRISPR systems with sucrose solution in one tube. The sucrose solution enables density-based phases separation of the two systems, allowing the target nucleic acids to be pre-amplified through universal RPA reaction and then dynamically diffused to CRISPR detection system. Upon specific recognition of the crRNA to RPA amplicons, the parallel cleavage activity of Cas12a will be activated, generating a colorimetric or fluorescent signal to distinguish positive malaria cases. To achieve comprehensive malaria diagnosis, we have incorporated a microfluidic device capable of testing six targets in parallel, achieving effective stratification and increasing the robustness of the platform. The procedure and working principle for on-chip Plasmodium detection are shown in Figure S1. According to the results interpretation criteria for the FREM platform (Fig. 1b), each Plasmodium species can be clearly differentiated.
Fig. 1.
Schematic workflow of malaria infection screening and Plasmodium species genotyping in FERM platform. (a) Overview of the distinct steps involved in the detection process in FREM; (b) Interpretation of the representative genotyping results of multiple Plasmodium species in FERM platform, visual detection using fluorescent signal and lateral flow readouts.
Design and optimization of the FREM platform
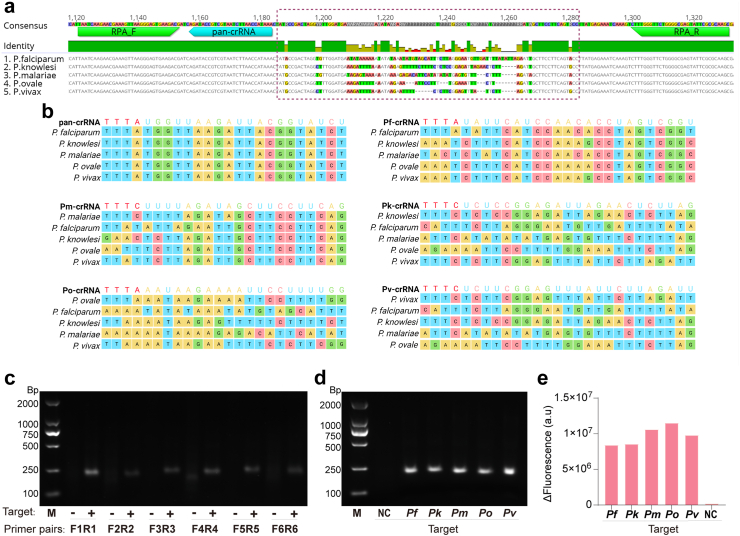
To comprehensively monitor critical malaria infection in an accessible manner, we designed RPA primers and crRNA for a pan-species assay targeting the conserved region of 18S rRNA gene (Fig. 2a). Multiple-sequence alignment demonstrated well-conserved target sites among different Plasmodium species (Fig. 2b). Six pairs of primers were designed based on the highly conserved 18S rRNA sequence in Plasmodium species, and used in the RPA reaction to verify the feasibility of primers design and to screen for the optimal primers. Through extensive optimization, we identified a pair of RPA primers with the highest amplification efficiency (Fig. 2c), which specifically amplified the conserved gene fragment of five species (Fig. 2d). Further, we designed a universal crRNA targeting a specific region between the sequences recognized by the two RPA primers. This pan-crRNA successfully detected all five Plasmodium species, demonstrating its excellent targeting activity (Fig. 2e).
Fig. 2.
Design and evaluation of the RPA primers and crRNAs used in the FREM platform. (a) Sequence alignment for universal RPA primers/and crRNA design in the FREM platform. Nucleotides of RPA primers and crRNA binding regions (highlighted in green and cyan, respectively) of the 18S rRNA gene. The dotted box shows the segment of 18S rRNA gene with the mutable region for the specific-species crRNAs designed for this study. (b) Schematic representation of the Plasmodium species' 18S rRNA gene, showing the region targeted by each crRNA target sequence. (c) Gel analysis of the products for RPA primers screening. The letters F and R represent forward and reverse primer. The numbers indicate the specific primer numbers designed in this study. Therefore, sample names on the x-axis can be understood as follows: F1R1, F2R2, F3R3, F4R4, F5R5and F6R6 represent different primer pairs to screen for the optimal primers. M, represents DL2000 marker. +, represents positive samples. -, represents negative samples. (d) Gel analysis of products for the identification of the five Plasmodium species using RPA universal primers. M, represents DL2000 marker. NC, represents negative control. (e) Quantitative analysis of the reaction of universal crRNA with the five Plasmodium species.
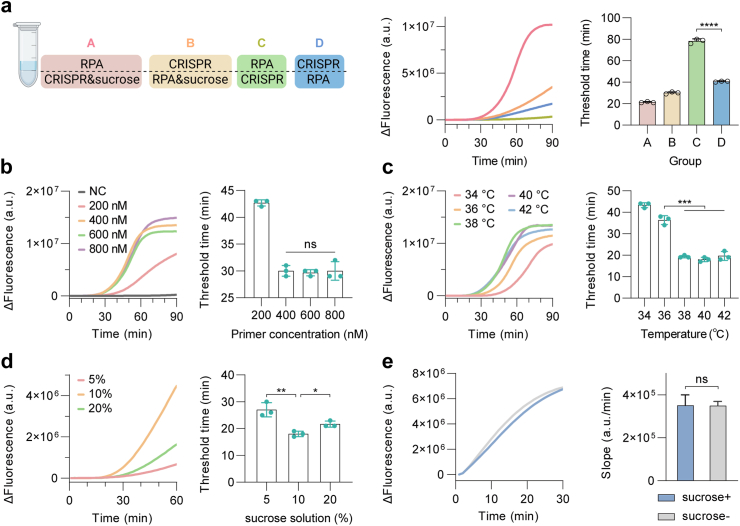
Subsequently, we investigated the possibility of performing RPA and CRISPR detection in a one-pot reaction, to simplify the workflow and reduce carryover contamination. Our previous study has verified that the density difference caused by the sucrose solution helps to establish spatially separated but connected phases in one-pot RPA-CRISPR detection.21 With this in mind, we conducted the one-pot assay with and without sucrose solution, and compared their efficiencies by measuring the end-point and real-time fluorescence signal intensities. Specifically, 10 μL RPA component was added to the CRISPR reaction mixture without or with sucrose solution, and fluorescence was real-time monitored during the reaction. As shown in Fig. 3a, the introduction of sucrose solution facilitated the establishment of spatially separated but connected phases in the one-pot RPA-CRISPR detection, enabling effective one-pot detection without compromising efficiency. We also compared the detection efficiency of the one-pot system with sucrose added to the RPA system or CRISPR solution, and the latter was selected for the following experiments.
Fig. 3.
Establishment and optimization of the sucrose-assisted one-pot RPA-CRISPR/Cas12a reaction. (a) Evaluation of the effect of different addition strategies using sucrose solution on one-pot RPA-CRISPR/Cas12a reaction. In group A, the mixtures of sucrose and CRISPR system were on the bottom of tube and the RPA system was on top; in group B, the mixtures of sucrose and RPA system were on bottom and the CRISPR system was on top; in group C, the RPA component was on bottom and the CRISPR was on top, and the positions were set opposite in group D. (Left panel) Design and comparison of different addition strategies of sucrose solution on one-pot reaction. (Middle panel) Performance of one-pot reaction measured by real-time fluorescence over 90 min. (Right panel) Performance of one-pot reaction determined by threshold time at a given fluorescence; (b) Comparison of the kinetics and detection threshold time of one-pot reaction at different primer concentrations; (c) and (d) Measurement of real-time fluorescence output comparing the detection activity and threshold time of one-pot reaction at five different temperatures (34, 36, 38, 40 and 42 °C) and sucrose solution concentrations (5, 10 and 20%); (e) Assessment of Cas12a activities without and with the sucrose solution on the single CRISPR system. All the error bars represent mean ± s.d., where n = 3 replicates. NC, negative control. Two-tailed Student's t-test was used for each two-group comparison. The asterisks denote values levels of statistical significance: P < 0.0001 (∗∗∗∗); P < 0.001 (∗∗∗); P < 0.01 (∗∗); P < 0.05 (∗); ns, not significant.
To maximize the amplification efficiency of RPA and the signal generation capability of CRISPR/Cas12a, several critical components and reaction concentration were investigated. To optimize RPA primer concentration, we first fixed crRNA and Cas12a concentrations at 1:2 ratio by employing the condition from the previous report22 and investigated the RPA primer concentrations ranging from 200 to 800 nM. As shown in Fig. 3b, the one-pot reaction with primers above 400 nM almost reached plateau at 60 min, and little changes of the threshold time were observed when the concentration of RPA primers was further increased. Next, we explored the effect of working temperature ranging from 34 °C to 42 °C on the one-pot reaction system. Results showed that the one-pot reaction has little difference in detection efficiency at 38–42 °C and 38 °C as chosen as the optimal working temperature for FREM (Fig. 3c). FREM tests on the 3D-printed chip exhibited similar trend on temperature optimization as assays carried in tubes (Figure S2). Since sucrose solved the incompatibility issue of RPA and CRISPR systems by forming different density phases, we then systematically tested the one-pot reaction at various concentrations of sucrose ranging from 5% to 20% to evaluate detection performance. As shown in Fig. 3d, higher concentration of solution was too viscous for two systems to communicate dynamically, while lower concentration characterized lower density, which failed to effectively form different density phases which dynamically separate two systems. The results showed that 10% sucrose solution provided the best detection performance when incorporating into the one-pot reaction (Fig. 3d). It should be noted that our current strategy was different from previous study which added sucrose to RPA system at the bottom, thus we further evaluated whether sucrose hampers Cas12a trans-cleavage efficiency. The result showed that the CRISPR-Cas12a detection was not influenced in addition of sucrose to the system (Fig. 3e). Thus, the introduction of sucrose solution enabled the successful one-pot integration of RPA and CRISPR reactions, increasing the efficiency of the platform and minimizing contamination risks.
Analytical performance of the FREM platform
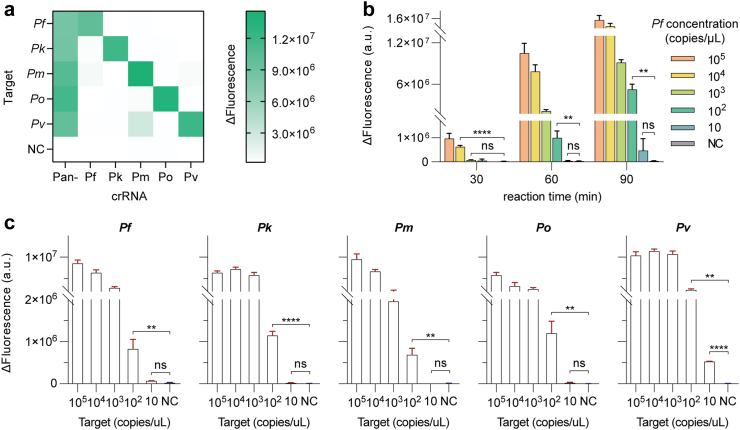
Under the optimized condition, the performance of our developed FREM platform was investigated. First, a two-step strategy was tested by 20 min RPA reaction and the followed CRISPR fluorescence detection for another 10–20 min, provided excellent sensitivity with the limit of detection (LOD) of 10 copies per reaction (Figure S3). The specificity was evaluated through the mutual interference among the five target Plasmodium species. The results verified that the FREM platform could accurately identify different Plasmodium species, demonstrating a high specificity for the crRNAs used in this study (Fig. 4a). The lateral flow readouts were also introduced to enhance accessibility and ease of interpretation, successfully visualizing the presence or absence of target templates (Figure S4).
Fig. 4.
Evaluation of the analytical performance of the FREM platform. (a) Specificity test of the six crRNAs against different Plasmodium species. The fluorescence-based test results are demonstrated as a heatmap; (b) Analytical sensitivity of pan-species assay in a one-pot fashion. Fluorescence values were determined at 30, 60 and 90 min for collateral activities; (c) Analytical sensitivity of the specific-species assays for the five Plasmodium parasites in a one-pot fashion. The end-point fluorescence intensity at 60 min readout for one-pot results with 0.8 μL sample at different concentrations of 105, 104, 103, 102 and 10 copies/μL. All the error bars represent mean ± s.d., where n = 3 replicates. NC, negative control. Two-tailed Student's t-test was used for each two-group comparison. The asterisks denote values levels of statistical significance: P < 0.0001 (∗∗∗∗); P < 0.01 (∗∗); ns, not significant.
As our detection exhibited sufficient specificity and sensitivity levels on separate processes, the one-pot assay was further investigated for applied as a POCT platform. 10-fold serial dilutions of each target synthetic 18S rRNA gene segment (1 to 105 copies/μL) were utilized to investigate the LODs of the one-pot reaction. We first used pan-crRNA and a gradient diluted synthetic P. falciparum 18S rRNA gene segment as target to assess the detection efficiency and best incubation time. As shown in Fig. 4b, the fluorescent values increased accordingly with the increase of target concentrations. By comparing fluorescence intensities at different time point, it was indicated that 60 min incubation time was suitable to exhibit statistical difference and achieve high sensitivity. Next, five specific crRNAs targeting mutable regions were used to evaluate LODs for different species (Fig. 4c), respectively. The result revealed that the detection sensitivity (102 copies/μL) of the one-pot reaction is 10 folds lower than that of the two steps system, in consistent with the previous reports23 (Figures S5 and S6). An exception to the detection of P. vivax may be due to the highly efficient crRNA sequence, as it plays a critical role in targeting analytes and activating Cas effectors.24,25 Our results also demonstrate that the LODs on the chips is comparable to that of the tube-based fluorescent signal readout (Figure S7).
Plasmodium species genotyping using FREM platform
To investigate the potential of FREM detection on a POCT platform (FREM platform), the simultaneous detection of different genomic species with six crRNAs was performed, one targeting a conserved region to confirm malaria infection and five targeting mutable regions to identify five specific Plasmodium species. First, the specific-species crRNAs (Fig. 2b) were designed with a PAM site for Cas12a recognition according to the aforementioned specifications. To simplify operation steps, a microfluidic chip was designed to perform the genotyping assays which can realize the simultaneous detection of different species (Fig. 5a). The developed FREM microfluidic platform demonstrated effective fluid control, enabling the automatic distribution of samples through the fractal branching six-channel microfluidic chip (Fig. 5b; Video S1). Notably, within the reaction chambers, we observed a distinct stratification phenomenon of two liquids.
Fig. 5.
Overview of the microfluidic chip designed for the FREM platform. (a) The image and 3D structures showing the microchannels of the microfluidic chip, with labels; (b) Schematic illustrating the microfluidic chip filled with pink dye to mimic the reaction system loading; (c) Fluorescence grayscale values of real clinical samples tested on microfluidic chip; (d) Fluorescence signal readout of real clinical samples tested on microfluidic chip using blue light. All the error bars represent mean ± s.d., where n = 3 replicates. NC, negative control.
In addition, the FREM platform was tested, which combines RPA and multiplexed CRISPR-based detection with the advantages of the integrated microfluidic chip to screen and precisely identify the Plasmodium species. Our results demonstrated that the FREM platform can be utilized to monitor malaria infection and genotype Plasmodium species (Fig. 5c and d). Notably, there was no signal change before and after the reaction neither in detecting non-target sample, nor in peripheral chambers involving mismatch crRNAs, while each reaction chamber which contained crRNA targeting specific species observed the signal change. Armed with lateral flow readouts the FREM platform would be more accessible for general use and easier to read. After addition of the running buffer, the lateral flow strip was directly inserted into the detection chambers and the positive band at the test line appeared within 5 min specifically in the samples containing the correct amplicons (Figure S8). The visual bands for the presence or absence of target templates from the lateral flow biosensors were easy-to-interpret at all detectable levels. This capability is particularly valuable in regions where multiple Plasmodium species co-exist and mixed infections are common, facilitating comprehensive diagnosis and appropriate treatment.
Clinical sample testing of the FREM platform
To further evaluate the feasibility of FREM Platform for directly analysis of clinical samples, crude DNA extracted from 154 clinical human blood samples, which were confirmed 126 positive and 28 negative by the hourse-used PCR-sequencing assay26 (Fig. 6a). Remarkably, the FREM platform demonstrated superior sensitivity (98.41%) and specificity (92.86%), achieving highly consistent results with the results of PCR-sequencing for malaria detection, which yielded positive predictive agreement of 98.41% and negative predictive agreement of 92.86% (Fig. 6b). The accuracy of the species genotyping of FREM platform was further investigated by comparing with the PCR-sequencing. The consistency between the results obtained from both the FREM and PCR-sequencing methods was observed across 140 samples (Fig. 6c), comprising 42 samples with P. falciparum infections, 68 samples with P. vivax infections, 27 samples where none of the five targeted Plasmodium species, and 3 samples with co-infections. As shown in Fig. 6c, the overall concordance rate of 90.90% between the two methods, where were discrepancies in species genotyping results for 14 samples.
Fig. 6.
Clinical validation of the FREM platform using dry blood spot samples of malaria infection. (a) Clinical characteristics of FREM compared to PCR-sequencing for malaria infection screening; (b) Summary of the clinical validation of FREM platform in terms of sensitivity, specificity, positive and negative predictive agreement; (c) The resulting images and their Plasmodium species genotyping for 154 clinical samples tested by FREM platform; (d) Resolution of discordant results between FREM and PCR-sequencing using arbitration tests. (e) Samples with discordant genotyping results by the FREM method after arbitration tests. FGS, first generation sequencing.
To further investigate samples those exhibited discrepancies between the proposed FREM and PCR-sequencing assays, qPCR analysis27 was performed as an arbitrating test. Despite this, in the context of malaria infection screening, qPCR confirmed concordant results with FREM for three out of four inconsistent samples, while one sample remained unverified even after resolving the discordant outcome (Fig. 6d). The discrepancies in genotyping results of three method could be caused by different primers efficiency of qPCR assay for each species, as well as crRNAs efficiency for of the FREM test. Combining PCR-sequencing and qPCR analyses, the overall concordance of malaria infection screening using the FREM platform reached 99.35% (153/154). In addition, for samples with discrepant Plasmodium species genotyping, additional qPCR analysis revealed that the FREM results were consistent with genotyping results for most samples (Fig. 6e), resulting in an overall accuracy of Plasmodium species genotyping of 94.16% (145/154).
Discussion
Malaria screening is crucial for reducing hotspots of parasite infection and interrupting transmission, especially by identifying asymptomatic infections.28,29 In addition, timely determination and monitoring of malaria infection are essential for understanding the presence and dissemination of diverse Plasmodium species.30 In this study, we presented the development and validation of the FREM platform for malaria screening and Plasmodium species genotyping. Utilizing a sucrose-assisted assay, we successfully achieved spatial separation of RPA and CRISPR reactions, mitigating compatibility concerns and contamination risks. The integration of RPA, CRISPR in microfluidic chip with a single workflow simplifies the detection process and enhances efficiency.
The introduction of CRISPR-based biosensing has revolutionized molecular diagnostics, particularly in point-of-care applications.31 Thanks to their programmability, CRISPR-based detection systems have the potential to transform the landscape of malaria diagnostics.32 Recently, the conventional approach to CRISPR-based biosensing typically involves a two-step process, including target pre-amplification followed by CRISPR-based nucleic acid detection.33 This methodology, while employed for Plasmodium parasite detection,34 potentially increases the risk of carryover contaminations due to multiple liquid-handling steps.33,35 The integration of isothermal amplification with CRISPR-based detection in one-pot is critical for POCT. However, performing both reactions directly in a confined system could drastically compromise detection efficiency due to poor biocompatibility.21 To avoid contamination issues and achieve one-pot detection, we developed a strategy to perform RPA and CRISPR reactions in one-pot by applying a sucrose solution to mitigate biocompatibility issues. Our results confirm that sucrose addition dose not compromise Cas12a effectors' efficacy. We also hypothesize that the sucrose solution serves as a stabilizer rather than an enhancer, restoring Cas12a functionality in seemingly incompatible systems by modulating protein-state dynamics and interaction with the surroundings.36 By combining the amplification and detection steps in a single chamber, the FREM platform reduces processing time and increases sample analysis throughput. The successful implementation of the one-pot reaction demonstrates its potential for optimization and extension to other infectious diseases and molecular diagnostic applications. Particularly, the FREM platform can be adapted to detect and discriminate different viral strains, especially in the context of SARS-CoV-2 variant of concern. Given the global concern of antibiotic resistance, the FREM platform could also be used for rapid identification bacterial infections and antibiotic resistance profiling, aiding in more targeted treatment.
Multiplex capacity is critical not only for immediate diagnostic purposes but also for the prospect of facilitating more comprehensive diagnostic strategies in the future, including differential diagnostics and the detection of mixed infections.37 In this study, microfluidic devices driven by capillary flow have emerged as instrumental tools for enabling multiplex analyses of pathogens and small molecules within diverse samples.16,18 Compared with previously reported CRISPR-based molecular methods for malaria detection,7,34 our methodology focuses on achieving a comprehensive diagnosis of malaria through a unique approach. Specifically, we have integrated a microfluidic device with the capability to simultaneously detect six targets in parallel, controlled by a single inlet. Notably, it streamlines the diagnostic process by exclusively employing a single pair of universal RPA primers tailored to the five Plasmodium species targets. Additionally, the strategic design of six unique crRNAs imparts the capacity to distinguish Plasmodium infections and identify different species. By observing signals in six spatially separated reaction chambers, the Plasmodium infection status and species composition of the detected sample can be identified. This information is effortlessly translated through either fluorescent or lateral flow readouts, allowing users to intuitively interpret the results. Using fluorescent signal output in a reaction volume of 90 μL universal RPA reagent and 60 μL CRISPR reagent in total for simultaneous Plasmodium detection and five-species genotyping, the cost was roughly at 7.4 $ per technical replicate. The resin cost for a 3D-printed microfluidic chip for the FREM platform was ∼0.4 $, leading to a total consumption at ∼7.8 $ per FREM assay. Further, for colorimetric signal readout, the cost for lateral flow readout increased to 8.3 $ due to additional consumption on test strips. Such advances have put forward improvements as a CRISPR-based POCT platform in endemic areas. Subsequently, the accuracy and feasibility of the FREM platform for real-world applications were validated through clinical sample testing. An assay with high sensitivity and specificity is important for identifying individuals in the early stages of infection and limiting the spread of pathogens.38,39 The platform's superior sensitivity and specificity compared to the standard PCR-sequencing assay indicate its potential as a reliable tool for malaria detection.
However, despite the potential benefits of the FREM platform, there are some limitations that should be noted. First, during the evaluation phase, we encountered a notable limitation related to the design of crRNA targeting P. malariae. This crRNA exhibited a degree of cross-reactivity with P. vivax, attributed to significant nucleotide identity within the crRNA target regions of these two Plasmodium species. This limitation highlights the need for further crRNA optimization to enhance specificity and eliminate cross-reactivity risks. In addition, our current microfluidic approach, although effective, involves several sequential steps. The FREM platform involves DNA extraction step, which, while manageable, can still be a bottleneck in resource-limited settings. Simplifying and streamlining sample preparation procedures can enhance the platform's suitability for point-of-care applications. Furthermore, the initial preloading of the species-specific CRISPR components into different reaction chambers, followed by the subsequent introduction of universal RPA system through a central inlet, results in a complex workflow. To streamline this process, we envision a further development in which all required CRISPR and RPA components are loaded into their respective chambers in freeze-dried form. This innovation would allow sequential activation of reactions upon introduction of the test sample through the central inlet, significantly simplifying the workflow. This proposed improvement aims to enhance the feasibility of established platform for real POCT applications.
In conclusion, the FREM platform presents a promising solution for malaria screening and Plasmodium species genotyping. The integration of innovative technologies, such as RPA, CRISPR, and microfluidics, demonstrates its potential to improve diagnostic accuracy and efficiency, particularly in resource-limited settings. To achieve the ambitious goal of malaria eradication by 2040, necessitates targeting all Plasmodium species infecting humans with a comprehensive understanding of origin and dissemination. In this perspective, the FREM platform represents a significant advancement in the field of malaria diagnostics, with its potential to offer rapid, accurate, and field-deployable detection of malaria infection and simultaneous identification of Plasmodium species. Successful validation with clinical samples highlights its practical utility in improving malaria control efforts and potentially bring us closer to the ultimate goal of eradication. Further optimization and expansion of the FREM platform hold promise for broader applications in molecular diagnostics.
Contributors
LX and KY were involved in the concept of the study. LX, JC and KY contributed to the study design and formulating the research question. LX and HL performed the experiments, analyzed the results and wrote the manuscript. YZ revised the manuscript. QH, SC, CW and XZ conducted the experiments and analyzed the results. All authors reviewed and revised the final version of manuscript. LX, HL and KY have accessed and verified the underlying data, LX and KY were responsible for the decision to submit the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Declaration of interests
Leshan Xiu, Kun Yin and Huimin Li have a patent (People's Republic of China) related to this article. All other authors declare no competing interests.
Acknowledgements
This work was financially supported by the International Joint Laboratory on Tropical Diseases Control in Greater Mekong Subregion (21410750200), National Natural Science Foundation of China (22104090), the Natural Science Foundation of Shanghai (22ZR1436200), National Research and Development Plan of China (2020YFE0205700 and 2018YFE0121600), the Health Research in the Public Interest (201202019) and the project from Bill & Melinda Gates foundation (INV-003421).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104898.
Contributor Information
Leshan Xiu, Email: xiuls001@hotmail.com.
Jun-Hu Chen, Email: chenjh@nipd.chinacdc.cn.
Kun Yin, Email: kunyin@sjtu.edu.cn.
Appendix A. Supplementary data
References
- 1.Arora G., Chuang Y.M., Sinnis P., Dimopoulos G., Fikrig E. Malaria: influence of Anopheles mosquito saliva on Plasmodium infection. Trends Immunol. 2023;44(4):256–265. doi: 10.1016/j.it.2023.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giacometti M., Milesi F., Coppadoro P.L., et al. A lab-on-chip tool for rapid, quantitative, and stage-selective diagnosis of malaria. Adv Sci. 2021;8(14) doi: 10.1002/advs.202004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips M.A., Burrows J.N., Manyando C., van Huijsduijnen R.H., Van Voorhis W.C., Wells T.N.C. Malaria. Nat Rev Dis Prim. 2017;3 doi: 10.1038/nrdp.2017.50. [DOI] [PubMed] [Google Scholar]
- 4.Kshirsagar A., Choi G., Santosh V., Harvey T., Bernhards R.C., Guan W. Handheld purification-free nucleic acid testing device for point-of-need detection of malaria from whole blood. ACS Sens. 2023;8(2):673–683. doi: 10.1021/acssensors.2c02169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogeni P., Williams T.N., Omedo I., et al. Detecting malaria hotspots: a comparison of rapid diagnostic test, microscopy, and polymerase chain reaction. J Infect Dis. 2017;216(9):1091–1098. doi: 10.1093/infdis/jix321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi G., Prince T., Miao J., Cui L., Guan W. Sample-to-answer palm-sized nucleic acid testing device towards low-cost malaria mass screening. Biosens Bioelectron. 2018;115:83–90. doi: 10.1016/j.bios.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee R.A., Puig H., Nguyen P.Q., et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc Natl Acad Sci U S A. 2020;117(41):25722–25731. doi: 10.1073/pnas.2010196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong L., Li W., Xu Q., et al. A rapid multiplex assay of human malaria parasites by digital PCR. Clinica chimica acta. Int J Clin Chem. 2023;539:70–78. doi: 10.1016/j.cca.2022.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Zainabadi K. Ultrasensitive diagnostics for low-density asymptomatic Plasmodium falciparum infections in low-transmission settings. J Clin Microbiol. 2021;59(4):e01508–e01520. doi: 10.1128/JCM.01508-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabay G., DeCastro J., Altay A., et al. Emerging biosensing technologies for the diagnostics of viral infectious diseases. Adv Mater. 2022;34(30) doi: 10.1002/adma.202201085. [DOI] [PubMed] [Google Scholar]
- 11.Dai Y., Wu Y., Liu G., Gooding J.J. CRISPR mediated biosensing toward understanding cellular biology and point-of-care diagnosis. Angew Chem. 2020;59(47):20754–20766. doi: 10.1002/anie.202005398. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y., Zong N., Ye F., Mei Y., Qu J., Jiang X. Dual-CRISPR/Cas12a-Assisted RT-RAA for ultrasensitive SARS-CoV-2 detection on automated centrifugal microfluidics. Anal Chem. 2022;94(27):9603–9609. doi: 10.1021/acs.analchem.2c00638. [DOI] [PubMed] [Google Scholar]
- 13.Lin C., Chen F., Huang D., et al. A universal all-in-one RPA-Cas12a strategy with de novo autodesigner and its application in on-site ultrasensitive detection of DNA and RNA viruses. Biosens Bioelectron. 2023;239 doi: 10.1016/j.bios.2023.115609. [DOI] [PubMed] [Google Scholar]
- 14.Hu T., Ke X., Li W., et al. CRISPR/Cas12a-Enabled multiplex biosensing strategy via an affordable and visual nylon membrane readout. Adv Sci. 2023;10(2) doi: 10.1002/advs.202204689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Xie Y., Chen F., et al. Amplification-free CRISPR/Cas detection technology: challenges, strategies, and perspectives. Chem Soc Rev. 2023;52(1):361–382. doi: 10.1039/d2cs00594h. [DOI] [PubMed] [Google Scholar]
- 16.Xie Y., Li H., Chen F., et al. Clustered regularly interspaced short palindromic repeats-based microfluidic system in infectious diseases diagnosis: current status, challenges, and perspectives. Adv Sci. 2022;9(34) doi: 10.1002/advs.202204172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avaro A.S., Santiago J.G. A critical review of microfluidic systems for CRISPR assays. Lab Chip. 2023;23(5):938–963. doi: 10.1039/d2lc00852a. [DOI] [PubMed] [Google Scholar]
- 18.Xu Z., Chen D., Li T., et al. Microfluidic space coding for multiplexed nucleic acid detection via CRISPR-Cas12a and recombinase polymerase amplification. Nat Commun. 2022;13(1):6480. doi: 10.1038/s41467-022-34086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadimisetty K., Song J., Doto A.M., et al. Fully 3D printed integrated reactor array for point-of-care molecular diagnostics. Biosens Bioelectron. 2018;109:156–163. doi: 10.1016/j.bios.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Tholoth M., Bai H., Mauk M.G., Saif L., Bau H.H. A portable, 3D printed, microfluidic device for multiplexed, real time, molecular detection of the porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine deltacoronavirus at the point of need. Lab Chip. 2021;21(6):1118–1130. doi: 10.1039/d0lc01229g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin K., Ding X., Li Z., Zhao H., Cooper K., Liu C. Dynamic aqueous multiphase reaction system for one-pot CRISPR-cas12a-based ultrasensitive and quantitative molecular diagnosis. Anal Chem. 2020;92(12):8561–8568. doi: 10.1021/acs.analchem.0c01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J., Liu Z., Zhang Z., Wu T. Unlocking the full potential of Cas12a: exploring the effects of substrate and reaction conditions on trans-cleavage activity. Anal Chem. 2023;95(28):10664–10669. doi: 10.1021/acs.analchem.3c01307. [DOI] [PubMed] [Google Scholar]
- 23.Lu S., Tong X., Han Y., et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat Biomed Eng. 2022;6(3):286–297. doi: 10.1038/s41551-022-00861-x. [DOI] [PubMed] [Google Scholar]
- 24.Patchsung M., Jantarug K., Pattama A., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4(12):1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 25.Woodside W.T., Vantsev N., Catchpole R.J., et al. Type III-A CRISPR systems as a versatile gene knockdown technology. RNA. 2022;28(8):1074–1088. doi: 10.1261/rna.079206.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B., Han S.S., Cho C., et al. Comparison of microscopy, nested-PCR, and Real-Time-PCR assays using high-throughput screening of pooled samples for diagnosis of malaria in asymptomatic carriers from areas of endemicity in Myanmar. J Clin Microbiol. 2014;52(6):1838–1845. doi: 10.1128/JCM.03615-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefterova M.I., Budvytiene I., Sandlund J., Farnert A., Banaei N. Simple real-time PCR and amplicon sequencing method for identification of Plasmodium species in human whole blood. J Clin Microbiol. 2015;53(7):2251–2257. doi: 10.1128/JCM.00542-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen I., Clarke S.E., Gosling R., et al. "Asymptomatic" malaria: a chronic and debilitating infection that should Be treated. PLoS Med. 2016;13(1) doi: 10.1371/journal.pmed.1001942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galatas B., Bassat Q., Mayor A. Malaria parasites in the asymptomatic: looking for the hay in the haystack. Trends Parasitol. 2016;32(4):296–308. doi: 10.1016/j.pt.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 30.McMorrow M.L., Aidoo M., Kachur S.P. Malaria rapid diagnostic tests in elimination settings--can they find the last parasite? Clin Microbiol Infection. 2011;17(11):1624–1631. doi: 10.1111/j.1469-0691.2011.03639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dongen J.E., Berendsen J.T.W., Steenbergen R.D.M., Wolthuis R.M.F., Eijkel J.C.T., Segerink L.I. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaminski M.M., Abudayyeh O.O., Gootenberg J.S., Zhang F., Collins J.J. CRISPR-based diagnostics. Nat Biomed Eng. 2021;5(7):643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- 33.Joung J., Ladha A., Saito M., et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham C.H., Hennelly C.M., Lin J.T., et al. A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping. eBioMedicine. 2021;68 doi: 10.1016/j.ebiom.2021.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding X., Yin K., Li Z., et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11(1):4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen L.T., Rananaware S.R., Yang L.G., et al. Engineering highly thermostable Cas12b via de novo structural analyses for one-pot detection of nucleic acids. Cell Rep Med. 2023;4(5) doi: 10.1016/j.xcrm.2023.101037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marquez-Costa R., Montagud-Martinez R., Marques M.C., et al. Multiplexable and biocomputational virus detection by CRISPR-cas9-mediated strand displacement. Anal Chem. 2023;95(25):9564–9574. doi: 10.1021/acs.analchem.3c01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tietje K., Hawkins K., Clerk C., et al. The essential role of infection-detection technologies for malaria elimination and eradication. Trends Parasitol. 2014;30(5):259–266. doi: 10.1016/j.pt.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Fozouni P., Son S., Diaz de Leon Derby M., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184(2):323–333.e9. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.