Abstract
Deletion of the nuclear hormone receptor small heterodimer partner (Shp) ameliorates the development of obesity and nonalcoholic steatohepatitis (NASH) in mice. Liver-specific SHP plays a significant role in this amelioration. The gut microbiota has been associated with these metabolic disorders, and the interplay between bile acids (BAs) and gut microbiota contributes to various metabolic disorders. Since hepatic SHP is recognized as a critical regulator in BA synthesis, we assessed the involvement of gut microbiota in the antiobesity and anti-NASH phenotype of Shp−/− mice. Shp deletion significantly altered the levels of a few conjugated BAs. Sequencing the 16S rRNA gene in fecal samples collected from separately housed mice revealed apparent dysbiosis in Shp−/− mice. Cohousing Shp−/− mice with WT mice during a Western diet regimen impaired their metabolic improvement and effectively disrupted their distinctive microbiome structure, which became indistinguishable from that of WT mice. While the Western diet challenge significantly increased lipopolysaccharide and phenylacetic acid (PAA) levels in the blood of WT mice, their levels were not increased in Shp−/− mice. PAA was strongly associated with hepatic peroxisome proliferator-activated receptor gamma isoform 2 (Pparg2) activation in mice, which may represent the basis of the molecular mechanism underlying the association of gut bacteria and hepatic steatosis. Shp deletion reshapes the gut microbiota possibly by altering BAs. While lipopolysaccharide and PAA are the major driving forces derived from gut microbiota for NASH development, Shp deletion decreases these signaling molecules via dysbiosis, thereby partially protecting mice from diet-induced metabolic disorders.
Supplementary key words: bile acids and salts, inflammation, intestine, liver, nuclear receptor/RXR, PPARs, microbiome, lipopolysaccharide, phenylacetic acid, small heterodimer partner
Nuclear hormone receptors play crucial roles in many mammalian physiologies, including metabolic homeostasis, by regulating target gene expression (1). An orphan nuclear hormone receptor, small heterodimer partner (SHP/NR0B2), was identified as an interacting protein of the nuclear hormone receptor constitutive androstane receptor (NRLI3) using a yeast two-hybrid screening strategy (2). Earlier molecular studies found that SHP transcriptionally repressed target gene expression by directly recruiting DNA-bound transcription factors (3, 4, 5). Feedback regulation of cholesterol 7α-hydroxylase (Cyp7a1) gene expression by bile acid (BA) has been identified as one of SHP’s most important and widely studied physiological roles and has been a stepping stone for understanding BA and cholesterol metabolism (6, 7). The generation of Shp−/− mice confirmed the important role of SHP in BA synthesis (8). Subsequent studies using mouse models, including Shp−/− mice, have further identified the potential role of SHP in lipid and glucose metabolism (9, 10, 11). These studies suggested that Shp deletion protects mice from diet-induced obesity and fatty liver but not from diabetes. Suppression of peroxisome proliferator-activated receptor gamma isoform 2 (Pparg2) expression was suggested as one proposed molecular mechanisms for attenuating hepatic lipid accumulation in Shp−/− mice (12). Our accompanying study additionally revealed that Shp deletion ameliorates nonalcoholic steatohepatitis (NASH) development induced by a long-term Western diet (WD) (13).
Obesity and nonalcoholic fatty liver disease (NAFLD) are highly associated metabolic disorders impacting modern society. Recent progress in understanding the underlying mechanisms of these metabolic disorders has strongly implicated gut microbiota in their development (14, 15). Studies on ob/ob mice have provided a mechanistic basis for the association between obesity and gut microbiota, where microbiome enrichment with an increased capacity for dietary energy harvesting was evident (16). Transplantation of this obesity-prone microbiota into germ-free mice resulted in a significantly greater body fat increase than transplantation of microbiota from lean control mice, indicating that the microbiota carries determinant factors for metabolic outcomes in its host, which has also been shown in humans (17). In addition, a high-fat diet (HFD) chronically elevates serum bacterial lipopolysaccharide (LPS) by altering gut flora and intestinal permeability, triggering inflammatory signatures and metabolic dysregulations, eventually leading to the phenotypic presentation of obesity, diabetes, and NAFLD (18).
Dietary fat and BAs play critical roles in modulating the gut microbiota. Taurocholic acid induced by saturated fat consumption is directly involved in an expansion of a low-abundance, sulfite-reducing pathobiont, Bilophila wadswothia, which is associated with colitis development in interleukin 10−/− mice (19). Farnesoid X receptor knockout (Fxr−/−) mice, the primary regulator in BA synthesis, developed many metabolic dysfunctions because of BA-induced alterations in gut microbial composition rather than directly from Fxr deletion (20). The alteration of microbial ecology by BA, diet, or xenobiotics can induce intestinal tauro-β-muricholic acid in mice or glycoursodeoxycholic acid in humans, inhibiting intestinal FXR activity and thereby improving various metabolic dysfunctions in the associated diseases (21, 22, 23).
As discussed previously, the orphan nuclear receptor SHP is directly involved in regulating the expression of enzymes involved in the BA synthesis pathway, with its deletion modifying hepatic BA composition (11). To assess the association of gut microbiota with the phenotypes of Shp−/− mice reported in earlier studies, including an accompanying report (13), we examined the gut microbiota using 16S rRNA gene sequencing of fecal samples from WT and mutant mice and explored phenotypes using mixed-group housing (cohousing) strategies, which enable the sharing of gut microbiota between the two mouse strains (24). Because of BA composition changes, Shp−/− mice formed bacterial operational taxonomy units (OTUs) distinct from WT mice. Cohousing disrupted the OTU distinctiveness and protective phenotypes against diet-induced obesity and NASH development of Shp−/− mice in segregated-group (separated) housing. Shp−/− mice housed in separate cages maintained reduced serum levels of LPS and phenylacetic acid (PAA), a bacterial metabolite highly associated with hepatic fat accumulation in humans, under a WD compared with WT counterparts (25). Moreover, PAA signaling appeared to be associated with hepatic Pparg2 expression, promoting hepatic lipid storage (26).
Materials and Methods
Mouse studies
The Shp−/− mice have been previously described (27). Only male mice were used in the experiments. The cohousing experiment used equal numbers of age-matched WT C57BL/6NHsd and Shp−/− mice placed in a cage immediately after weaning. Each genotype was produced by homozygous (nonlittermate) or heterozygous (littermate) breeding. All cages were housed in a temperature- and light-controlled room with a 12/12 h light-dark cycle (on: 06:30, off: 18:30). Food and water were available ad libitum. Rodent chow was obtained from Lab Diet (MO; catalog no.: 5008). In order to induce obesity and NASH, mice were fed a WD (catalog no.: TD.88137; Envigo, IN; 21.2% [w/w] total lipid [42% kcal], 48.5% [w/w] carbohydrates [42.7% kcal], 17.3% [w/w] proteins [15.2% kcal], and 0.2% cholesterol; total = 4.5 kcal/g) starting at 8 weeks old for 24 weeks. Body weight (BW) was measured every week after WD feeding began. WT and Shp−/− mice in littermate mouse experiments were produced by mating heterozygous Shp+/− mice. In these experiments, cohousing used WT and Shp−/− littermates. They were fed the same diet regimen described above. Tissues were collected after overnight fasting at the end of the diet regimen. Fecal samples were collected twice, immediately before and after 10 weeks of the diet regimen, for 16S rRNA gene sequencing. After 14 weeks of the WD, mouse body composition was determined using an EchoMRI machine (EchoMRI, TX). All animal care and use protocols were approved by the Institutional Animal Care and Use Committee of Northeast Ohio Medical University (Rootstown, OH).
Lipidomics
Lipids were extracted from approximately 20 mg of liver in a chloroform-methanol mixture. The extracted lipids were analyzed using shotgun lipidomics as previously described (28). Protein layers from chloroform-methanol extraction were used to normalize lipid quantities as ion counts based on the obtained protein concentrations.
BA analysis
BAs were extracted from 5 μl of bile from the gallbladder (GB) and of weighed ileum with luminal content as previously described (29). BAs were analyzed in the samples from separately housed mice using LC-MS/MS (6490 Triple Quadrupole LC/MS; Agilent Technologies, Santa Clara, CA) at Baylor College of Medicine (Houston, TX). For GB samples from cohoused mice, 1 μl of bile was diluted with 99 μl of methanol and 10 μl of internal standards containing 2 μg/ml of LCA-d4 and CA-d4 in methanol. The BA composition was analyzed using a Q-Exactive Orbitrap Mass Spectrometer coupled with an ultraperformance liquid chromatography system (Thermo Fisher Scientific, San Jose, CA) at the Northeast Ohio Medical University.
RNA isolation and quantitative real-time PCR
The quantitative PCR analysis was performed as previously described in detail (27). Briefly, complementary DNA was synthesized using PrimeScript RT Master Mix (Clontech, CA) from total RNA extracted from the tissues, and quantitative PCR was performed using a CFX96 Real-Time PCR Machine with iTaq Universal SYBR Supermix (BioRad, CA). Gapdh was used as the internal control, and relative expression was determined from ΔCt values normalized to the expression of separate WT mice fed a chow diet (CD).
Gut microbiome profiling and bacterial pathway analysis
Fecal samples were collected from all cages immediately before initiating the WD and again after 10 weeks of the diet regimen (CD or WD). Mice were placed individually into an autoclave-sterilized cage for 36 h with sterilized water, a matching diet, and a bedding. All feces were collected in a laminar flow hood and stored at −80°C until used. Bacterial DNA was extracted using a Fecal DNA MiniPrep Kit (Zymo Research, CA). PCR was performed using primers targeting the V3 and V4 regions of the 16S rRNA gene (Integrated DNA Technologies, IA). The primers are listed in supplemental Table S1. Pippin Prep cassettes (1.5% agarose) were used to purify and collect the targeted DNA (Sage Science, MA). Purified samples were quantified using Qubit spectroscopy (Qubit Systems, ON, Canada) using a double-stranded DNA High-Sensitivity Assay Kit (Thermo Fisher Scientific, NY). The double-stranded DNA amplicons were sent to the Advanced Genetic Technologies Center (University of Kentucky, Lexington, KY) for Illumina MiSeq sequencing on a dual-indexed, 250-base pair flowcell (Illumina, CA). The output files were demultiplexed using QIIME (version 1.9.1, http://www.qiime.org/ inside of the parenthesis), and OTUs were determined using an open-reference algorithm (30, 31). For samples with a sequence count >104,006, beta diversity principal coordinates were estimated using unweighted UniFrac and Adonis Permanova to measure significance (P value) and effect size (R2) to explain variation, and rarefied alpha diversity was estimated using Faith’s phylogenetic diversity whole tree analysis (32, 33). Unless otherwise specified, a homoscedastic Student’s t-test was used to compare two groups, with a P < 0.05 considered statistically significant. Unless otherwise specified, values are presented as mean ± standard deviation. In order to identify differentially enriched functional pathways from the 16S rRNA gene sequencing data, linear discrimination analysis effect size (LEfSe) was calculated for the Kyoto Encyclopedia of Genes and Genomes orthology functions using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved State (PICRUSt) software package (34, 35). The microbiome analysis of fecal samples from mice obtained via heterozygous mating strategy was performed in EzBiome, Inc (Gaithersburg, MD) as previously described (36).
Serum PAA measurement
Serum PAA levels were measured in blood collected under isoflurane anesthesia from the portal vein of 4-month-old mice on the diet regimen for 2 months after fasting for 5 h. The serum was processed as previously described (37). PAA was quantified using GC-MS (Agilent Technologies) with phenyl-D5-acetic acid (Sigma, St. Louis, MO) as the internal standard.
Serum alanine aminotransferase measurement
Serum alanine aminotransferase concentrations in mice on the diet regimen for 6 months were measured colorimetrically (Infinity Reagent; Thermo Fisher Scientific, Waltham, MA) using Synergy 4 (BioTek, Winooski, VT).
Serum LPS and fecal albumin measurements
Serum LPS concentrations were quantified using an ELISA kit (LSBio, Seattle, WA). Fecal albumin concentrations were quantified in the samples collected from mice at the end of the 6-month WD regimen using an ELISA kit (Immunology Consultation Lab, Inc, Portland, OR). Briefly, about 100 mg of feces were homogenized in 500 μl of phosphate-buffered saline. After centrifugation, supernatants were diluted 400 times before ELISA analysis.
PAA treatment of a mouse hepatoma cell line
Hepa1-6 cells were seeded into 24-well plates and maintained in Dulbecco’s modified Eagle’s medium until 100% confluent. Next, cells were treated with 10 mM PAA as previously described (25) for 0, 1, 3, and 5 h before being harvested for RNA isolation.
Statistical analysis
Unless otherwise specified, values are presented as mean ± standard error of the mean. Student’s t-test was used to compare two groups. A P < 0.05 was considered statistically significant. We also analyzed various datasets using two-way ANOVA. Statistical analyses were performed using GraphPad Prism software (GraphPad Software, Inc) and presented in the associated figures.
Results
Metabolic phenotypes of Shp−/− mice in the cohousing strategy
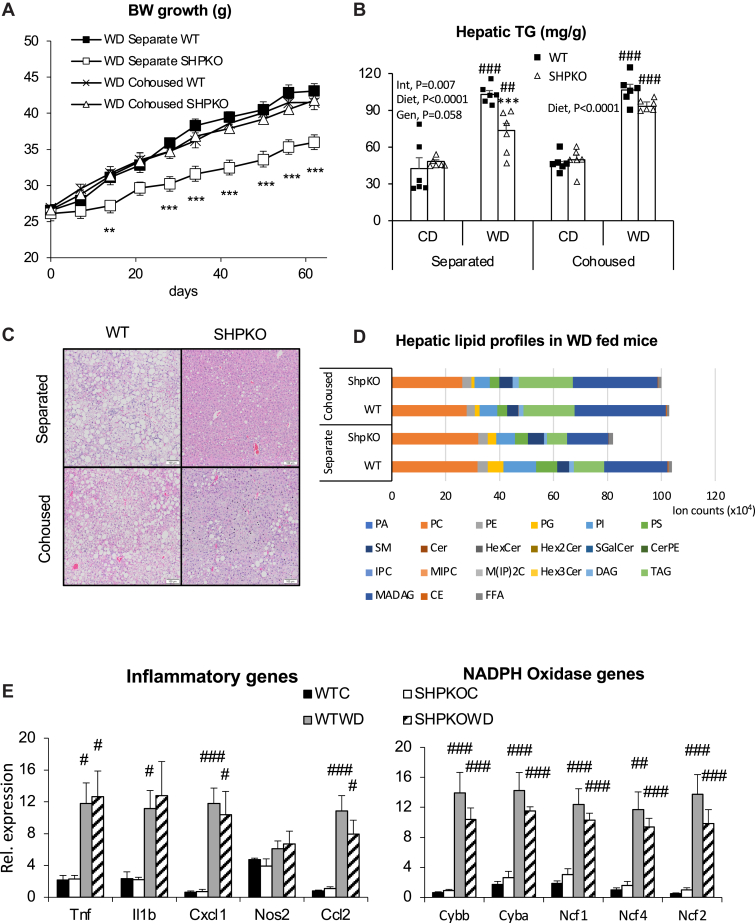
Our earlier studies, including the accompanying report, showed that Shp deletion attenuated obesity and NAFLD or NASH development in mice induced by a WD or methionine/choline-deficient diet. A strong association between BA, gut microbiota, and metabolic phenotypes prompted us to assess the involvement of gut microbiota in the phenotypic manifestations observed in Shp−/− mice. Cohousing strategies have often been used in mouse studies to determine the effect of microbiota on presented metabolic features because of their coprophagic nature (38, 39). Therefore, WT and Shp−/− mice were cohoused at weaning and subjected to a WD feeding regimen starting from 10 weeks old for 6 months for metabolic assessment. Their BW gains were compared with those of WT and Shp−/− mice raised in separate cages (Fig. 1A). While BW gain because of WD feeding was attenuated in Shp−/− mice housed in separate cages, the attenuation was abolished in Shp−/− mice cohoused with WT mice. In addition, Shp−/− mice fed a WD and cohoused with WT mice no longer showed significantly lower liver-to-BW ratios (supplemental Fig. S1A). Similarly, triglyceride (TG) concentrations and hematoxylin and eosin staining showed that reduced hepatic TG accumulation was also disrupted in Shp−/− mice cohoused with WT mice (Fig. 1B, C). Detailed lipid profiles were determined by LC-MS (Fig. 1D), showing a similar pattern to the measured TG levels. Cohousing significantly affected the levels of diacylglycerol, triacylglycerol, monoalkyl diacylglycerol, and cholesteryl ester, which were all significantly lower in separately housed Shp−/− mice (supplemental Fig. S1B). Mysteriously, however, TG values measured using a commercial kit (Fig. 1B) looked different from those measured using a GS-MS (supplemental Fig. S1B). Because the commercial kit quantifies all glycerols released from the extracted lipid after lipase treatment, the obtained values reflected more closely total lipid values measured by the GC-MS (Fig. 1D).
Fig. 1.
The metabolic phenotypes presented by Shp gene deletion are disrupted by cohousing scheme. A: BWs of WT and Shp−/− mice fed WD from separate or cohousing scheme were measured and plotted with standard errors for the first 2-month regimen. (Separate WT, cohoused WT, cohoused Shp−/−; n = 8. Separate Shp−/−; n = 7). B: Hepatic TG concentrations (mg/g of liver weight) were quantified from the WT and Shp−/− mice fed CD or WD in separate or cohoused cages for 6 months, and their averages were plotted with standard errors (n = 6). Two-way ANOVA and Student' t-test were performed to evaluate statistical significance. C: Livers from the experimental mice were processed for H&E staining. Representative H&E sections from mice fed WD were shown. White bars at right lower corner represent 100 μm length. D: Extracted lipids from livers of the WD-fed mice were analyzed using LC-MS. Average values of individual lipid class from each group (n = 4) were plotted in stacked columns. E: mRNA expression of hepatic genes responsible for inflammation were assessed using quantitative PCR analysis in cohoused WT and Shp−/− mice and presented with average ± SEM (n = 4–6). #comparison with CD, ∗comparison with WT counterparts. Three symbols represent P < 0.005, two symbols P < 0.01, and one symbol P < 0.05. CE, cholesteryl ester; Cer, ceramide; CerPE, ceramide phosphatidylethanolamine; DAG, diacylglycerol; FFA, free fatty acid; HexCer, monohexylceramide; Hex2Cer, dihexylceramide; Hex3Cer, trihexosylceramide; IPC, inositolphosphorylceramide; MADAG, monoalkyldiacylglycerol; MIPC, mannosyl-inositol phosphorylceramide; M(IP)2C, mannosyl-di-(inositolphosphoryl) ceramide; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SGalCer, 3-O-sulfogalatisylceramide; SM, sphingomyelin; TAG, triacylglycerol.
In our accompanying report, we also assessed inflammatory gene expression to evaluate the potential impact of gut microbiota on the attenuation of the NASH phenotype in Shp−/− mice (13). Unlike Shp−/− mice housed in separate cages, Shp−/− mice fed a WD showed comparable hepatic expression of inflammatory and nicotinamide adenine dinucleotide phosphate oxidase genes to WT mice when cohoused (Fig. 1E), indicating that cohousing almost entirely abolished the phenotypic attenuation by Shp deletion.
BA profiles of Shp−/− mice
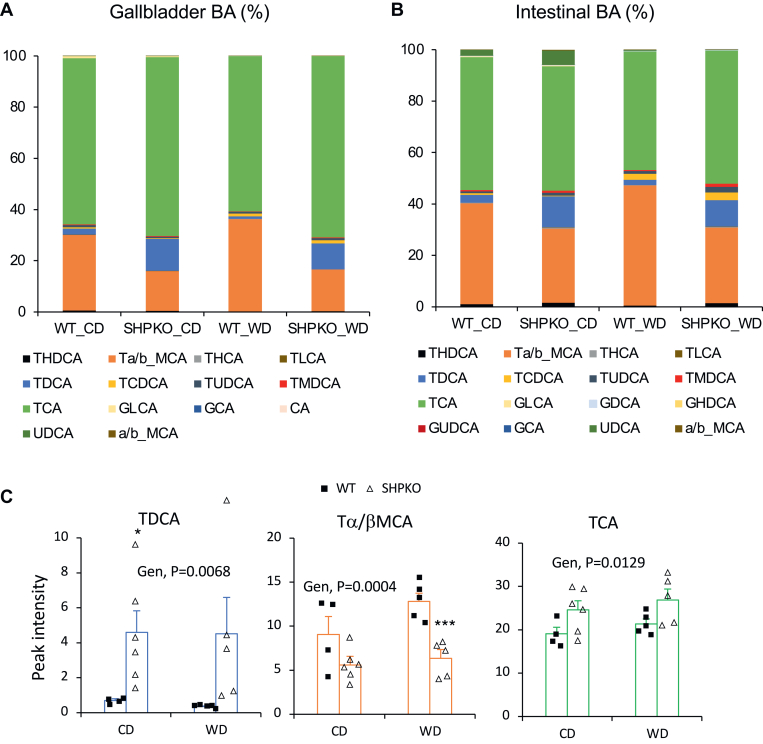
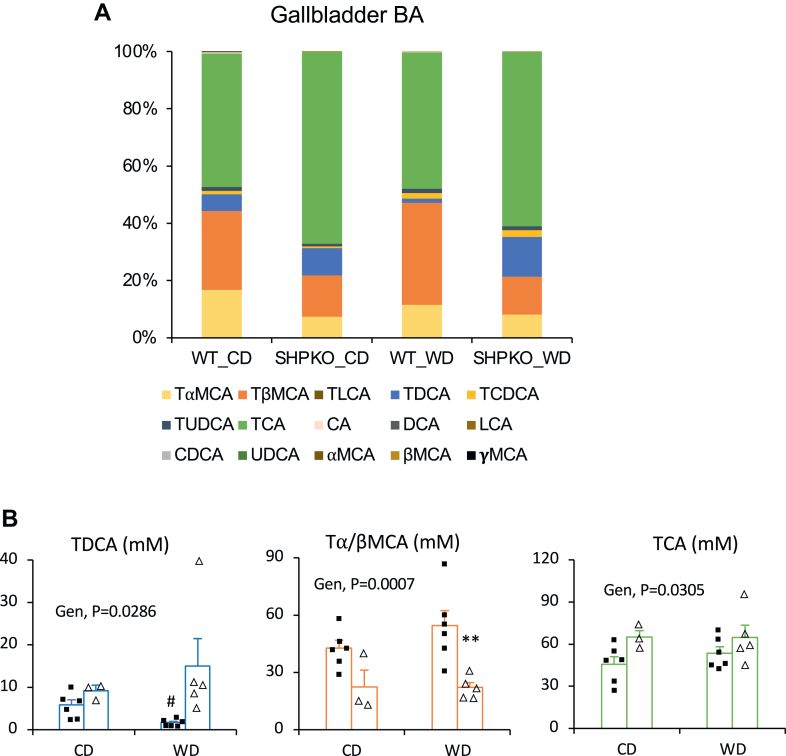
Based on the observations that SHP is important in BA synthesis, we hypothesized that an altered hepatic BA profile may affect the gut microbiome, thereby modulating metabolic and inflammatory responses in Shp−/− mice (8, 40). One of our earlier studies showed that the livers of Shp−/− mice contained higher cholic acid and lower muricholic acid than those of WT mice (11). We extend the earlier study here by reporting their detailed BA profiles, including conjugated BAs (supplemental Fig. S2A). The most abundant hepatic BA in Shp−/− mice was taurocholic acid (TCA; 230.85 nmol/g), higher than in WT mice (147.86 nmol/g). While total taurine-conjugated BAs were more abundant in Shp−/− than in WT mice, likely because of their higher expression of cysteine sulfinic acid decarboxylase, a key enzyme in taurine synthesis (41), total muricholic acids and their taurine-conjugated forms were less abundant in Shp−/− mice probably because of higher Cyp8b1 expression (supplemental Fig. S7). In this study, we analyzed BA profiles in the GB and intestine, which directly affect the structure of gut microbiota, of the mice fed a CD or a WD (Fig. 2). BA concentrations in the GB and liver of Shp−/− mice were similarly higher than those in WT mice (19% for CD and 10% for WD). Because of the strong affinity of two major hepatic canalicular membrane BA transporters (bile salt export pump and multidrug resistance-associated protein 2) for conjugated BAs (42), unconjugated BAs were barely detected in the GB from WT and Shp−/− mice, and the overall conjugated BA profiles were very similar between the liver and GB. However, considering the reported 2.5-fold higher GB volume in Shp−/− mice than in WT mice (11), the absolute amount of each BA in the GB of Shp−/− mice would be >2.5 higher than shown in Fig. 2A compared with BAs in WT mice. Deoxycholic acid (DCA), a secondary BA, and taurodeoxycholic acid, its conjugated form, were most significantly increased by Shp deletion in the liver (3.5-fold) and GB (6.7-fold or 16.75-fold when the increased bile volume is considered), and β-muricholic acid was the most significantly decreased by Shp deletion in the liver and GB (supplemental Fig. S2B, C). Intestinal BA profiles were relatively similar to the liver and GB profiles but with decreased TCA (Fig. 2B), which is supported by an earlier report that TCA is most actively absorbed in the intestine (43). Therefore, the less absorbed tauromuricholic acid appeared to be increased in the intestinal BA profile. In addition, a significant amount of ursodeoxycholic acid, a secondary BA, was detected in the intestine of both WT and Shp−/− mice (from 0.008% ± 0.002% in the GB of both mouse strains to 2.3% ± 1.3% and 5.6% ± 2.9% in the intestine of WT and Shp−/− mice, respectively) fed a CD. However, it was significantly reduced with the WD in both mouse strains. The higher overall DCA concentration indirectly indicates structural changes in the gut microbiota of Shp−/− mice.
Fig. 2.
BA compositions in GBs and small intestines from WT and Shp−/− mice. A: BAs extracted from GBs of experimental animals fed CD or WD were analyzed using an LC–MS. Percent values calculated from their average peak intensities (N; WTC = 4, SHPKOC = 6, WTWD = 5, and SHPKOWD = 5) were plotted into a stacked column graph. B: BAs were extracted from weighed ilea, and their compositions were analyzed. Percent values were calculated from their average peak intensities normalized by weight and were plotted in a stacked column graph (N = 6). C: TDCA, TMC, and TCA values from GBs were plotted with average ± SEM. Two-way ANOVA was performed to obtain statistical values. CA, cholic acid; Gen, genotypes; GCA, glycocholic acid; GLCA, glycolithocholic acid; MCA, muricholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; TDCA, taurodeoxycholic acid; THCA, taurohyocholic acid; THDCA, taurohyodeoxycholic acid; TLCA, taurolithocholic acid; TMCA, tauromurcholic aicd; TMDCA, tauromurideoxycholic acid; TUDCA, tauroursodeoxycholic acid; UDCA, ursodeoxycholic acid.
Alteration of gut microbiota in Shp−/− mice
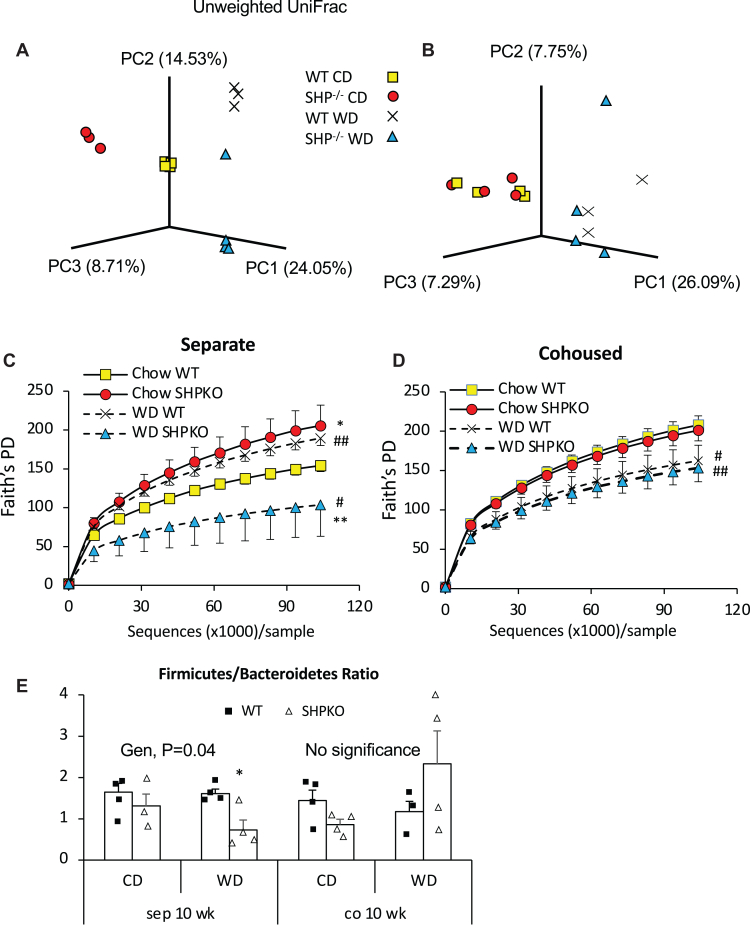
To evaluate the potential impact of Shp deletion on gut microbiota composition, we sequenced the 16S rRNA gene in fecal DNA from WT and Shp−/− mice fed a CD or a WD for 10 weeks. First, we performed principal coordinate analysis using unweighted UniFrac distances between the OTUs obtained from the 16S rRNA gene sequencing to assess β-diversity in each group. Figure 3A shows that each group forms a distinctive cluster depending on their genotype and diet, suggesting that the gut bacterial composition of Shp−/− mice differs significantly from that of WT mice. Faith’s phylogenetic diversity did not differ significantly between mouse strains at a young age before the diet intervention (supplemental Fig. S3A). At 16 weeks of age, phylogenic diversity increased significantly in Shp−/− mice fed a CD. However, 10 weeks of a WD significantly decreased phylogenic diversity in Shp−/− mice compared with WT mice (Fig. 3C). At the phylum level, the WD increased Proteobacteria in both mouse strains. Diet-induced obesity has been reported to increase the abundance of Firmicutes and decrease the abundance of Bacteroidetes (44). The WD decreased Firmicutes and increased Bacteroidetes in Shp−/− mice, significantly decreasing their ratio (Fig. 3E and supplemental Fig. S3B).
Fig. 3.
Gut microbiome structure in the experimental mice fed CD or WD. A and B: Three-dimensional principal coordinate analysis was performed with gut bacterial OUTs obtained from fecal samples of animals in separated housing (A) or cohousing (B) using unweighted UniFrac beta-diversity. C and D: Alpha-diversity was assessed with the obtained OTUs using Faith’s phylogenetic diversity analysis (Faith’s PD) in animals housed in separate cages (C) or cohoused cages (D). E: Ratios of Firmicutes to Bacteriodetes based on 16S rRNA gene sequencing data were plotted with average ± SEM (n = 3–4).
Cohousing disrupted the clear separation of gut bacterial OTUs observed in the separately caged groups and the difference in phylogenic diversity with maintained dietary effect (Fig. 3B, D). The ratio of Firmicutes to Bacteroidetes was significantly increased in cohoused Shp−/− mice fed the WD, which showed an obese phenotype (Fig. 3E). We also assessed the bacterial populations in each group (supplemental Fig. S4). Their abundance did not differ between the two mouse strains, although a stronger bactericidal effect was expected in Shp−/− mice with higher BA pool size (40).
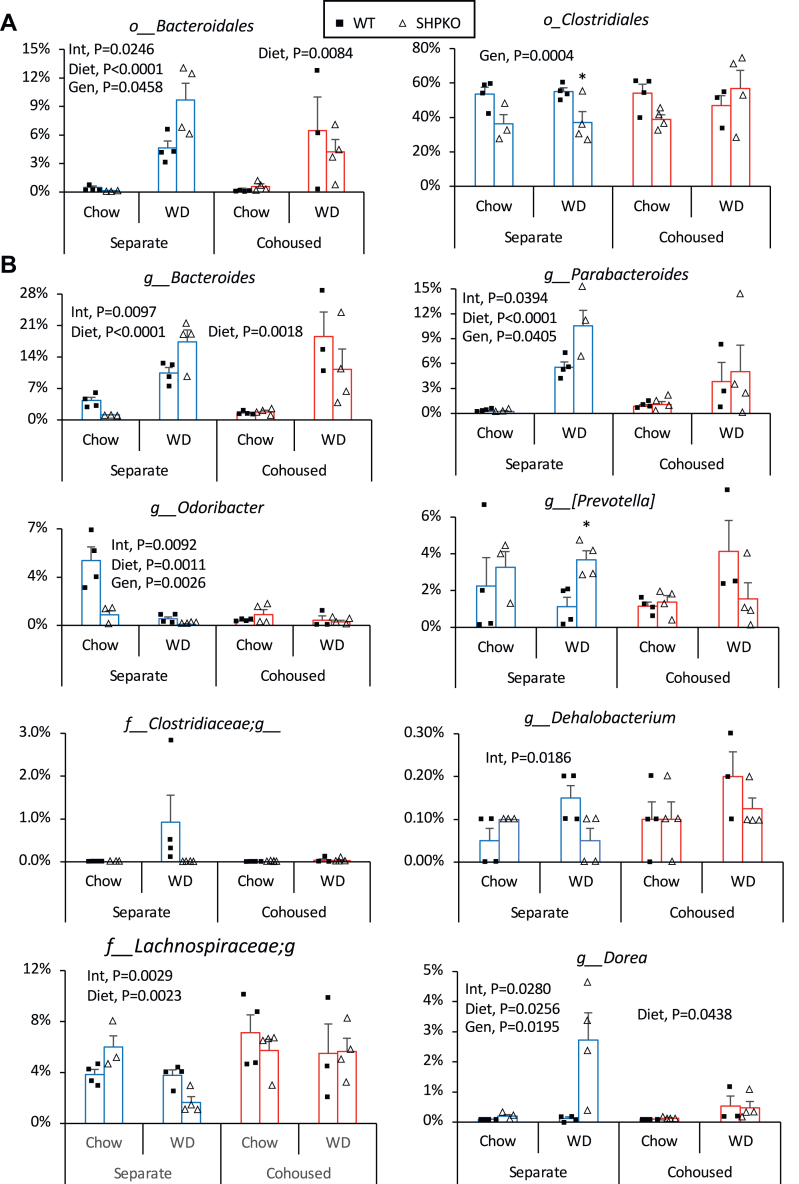
Our 16S rRNA gene sequencing identified gut bacteria up to the genus level. Of the screened 174 genus-level OTUs, 53 had at least two decimal percent values in any mouse. Comparison of these 53 genera between the separate and cohoused groups showed that the two most abundant bacterial orders, Bacteroidales and Clostridiales, were significantly impacted by cohousing in Shp−/− mice (Fig. 4A). Among the genera in the order Bacteroidales, the abundances of Bacteroides, Parabacteroides, Odoribacter, and Prevotella were significantly altered in Shp−/− mice fed a WD compared with WT mice (Fig. 4B). In the order Clostridiales, the abundances of unidentified genera in the family Clostridiaceae and Lachnospiraceae and genera Dehalobacterium and Dorea differed significantly between WT and Shp−/− mice fed a WD. However, these differences were disrupted by cohousing.
Fig. 4.
Gut microbes in Shp−/− mice affected by cohousing. Phylogenetic gut microbes were classified based on fecal 16S rRNA gene sequencing. Their abundances presented as percent values were compared between groups studied in Fig. 3A. Orders of Bacteriodales (left) and Clostridales (right) were presented as percent abundance ± SEM between experimental groups. B: Eight genera including unclassified ones belonging to the two orders were presented as bar graphs based on their average percent abundances with SEM. Two-way ANOVA results were shown with P values. Gen, genotype; Int, interaction.
We also assessed the functional pathways of the microbial communities using PICRUSt and identified differentially enriched pathways using LEfSe analysis (34). The top enriched pathways and their numbers (LEfSe ≥2) in each group were considered in three categories (genotype, diet, and housing; Table 1). The highest number of enriched pathways (61) were found in the separately housed CD-fed genotype category. The number of enriched pathways decreased to 17 when the cohoused group of the same category was analyzed. The smallest number of pathways (3) were enriched when comparing the separate and cohoused WT WD-fed groups. In contrast, the same comparison of Shp−/− WD groups identified 45 enriched pathways. This observation suggests that cohousing greatly affected the gut microbiome of Shp−/− mice compared with WT mice. While only 12 pathways were enriched between cohoused WT and Shp−/− mice fed the WD, these results indicate that cohousing modifies the gut microbiota of Shp−/− mice to become similar to WT mice, consistent with the described phenotypic outcomes. Some of the pathways positively associated with human obesity were enriched in the separately housed WT WD group, and some negatively associated pathways were enriched in the separately housed Shp−/− WD group, confirming the validity of this analysis (supplemental Fig. S5) (16).
Table 1.
Differentially enriched microbial pathways in each group predicted from fecal 16S RNA gene sequencing
| Group |
Number of pathways & top four enriched pathways in each group |
Total |
|||
|---|---|---|---|---|---|
| Genotype | WT | ShpKO | |||
| Separate CD | 18 | Transporters | 43 | Ribosome | 61 |
| Bacterial motility proteins | DNA repair and recombination proteins | ||||
| Flagella assembly | Phosphotransferase system | ||||
| Two-component system | Mismatch repair | ||||
| Separate WD | 12 | Transporters | 22 | Citrate cycle—tricarboxylic acid cycle | 34 |
| Bacterial motility proteins | Carbon fixation pathways in prokaryotes | ||||
| Betalain biosynthesis | Oxidative phosphorylation | ||||
| Flagella assembly | Ala, Asp, and glutamate metabolism | ||||
| Cohoused CD | 6 | Bacterial motility proteins | 11 | Carbon fixation in photosynthetic organisms | 17 |
| Flagella assembly | Nucleotide metabolism | ||||
| Bacterial chemotaxis | Bacterial secretion system | ||||
| Starch and sucrose metabolism | Cell mobility and secretion | ||||
| Cohoused WD | 12 | Pyrimidine metabolism | 0 | 12 | |
| Peptidases | |||||
| Purine metabolism | |||||
| Ribosome | |||||
| Diet | CD | WD | |||
|---|---|---|---|---|---|
| Separate WT | 5 | Peptidases | 16 | Other ion-coupled transporters | 21 |
| Streptomycin biosynthesis | Cysteine and methionine metabolism | ||||
| Other glycan degradation | Replication recombination repair proteins | ||||
| Drug metabolism other enzymes | Restriction enzyme | ||||
| Separate ShpKO | 3 | Phosphotransferase system | 12 | Other ion-coupled transporters | 15 |
| Aminoacyl tRNA biosynthesis | Pyruvate metabolism | ||||
| Staphylococcus aureus infection | Carbon fixation in photosynthetic organisms | ||||
| Inorganic ion transport and metabolism | |||||
| Cohoused WT | 4 | Methane metabolism | 5 | General function prediction only | 9 |
| Lysine biosynthesis | Other ion-coupled transporters | ||||
| C5 branched dibasic acid metabolism | Amino sugar and nucleotide sugar metabolism | ||||
| DDT degradation | Inorganic ion transport and metabolism | ||||
| Cohoused ShpKO | 35 | Ribosome | 11 | Two-component system | 46 |
| DNA repair and recombination proteins | Others | ||||
| Amino acid–related enzymes | Function unknown | ||||
| Pyrimidine metabolism | Secretion system |
| Housing | Separate | Cohoused | |||
|---|---|---|---|---|---|
| WT CD | 7 | Protein folding and associated process | 10 | Phe, Tyr, and tryptophan biosynthesis | 17 |
| Function unknown | Cysteine and methionine metabolism | ||||
| Other glycan degradation | Methane metabolism | ||||
| Amino sugar and nucleotide sugar metabolism | Nitrogen metabolism | ||||
| ShpKO CD | 0 | 5 | Val, Leu, and isoleucine biosynthesis | 5 | |
| Cell motility and secretion | |||||
| Phenylalanine metabolism | |||||
| Geraniol degradation | |||||
| WT WD | 1 | Methane metabolism | 2 | Photosynthesis proteins | 3 |
| Starch and sucrose metabolism | |||||
| ShpKO WD | 34 | Ribosome | 11 | Two-component system | 45 |
| Energy metabolism | Function unknown | ||||
| Ala, Asp, and glutamate metabolism | Secretion system | ||||
| DNA replication proteins | Others |
PICRUSt and LEfSe analyses were performed to predict enriched microbial pathways in each experimental group from fecal 16S RNA sequencing results described in Fig. 3.
Serum LPS and phenylacetate responsible for NASH development are reduced in Shp−/− mice fed the WD
Gut dysbiosis and disease phenotypes manifested in earlier Nlrp inflammasome gene knockout mouse studies (45, 46), which used nonlittermate control WT mice, were recently challenged by studies using littermate mice (47, 48). To determine whether cage or maternal effects contributed to the observed phenotypes in Shp−/− mice, biochemical and physiological features were reassessed in Shp−/− mice generated from heterozygous breeding and compared with their WT littermates. Figure 5 shows that all the metabolic phenotypes and gene expression patterns in littermate WT and Shp−/− mice were almost identical to those in nonlittermate studies. These observations suggest that phenotypes manifested by separately housed and cohoused Shp−/− mice were not caused by housing or maternal inheritance but by Shp gene deletion, which changes the BA profile and microbial ecosystem in the gastrointestinal tract. In addition, structures of gut microbes from these two genotypes generated by heterozygous breeding were clearly separated in principal coordinate analysis based on 16S rRNA gene sequencing data regardless of their diet (supplemental Fig. S6).
Fig. 5.
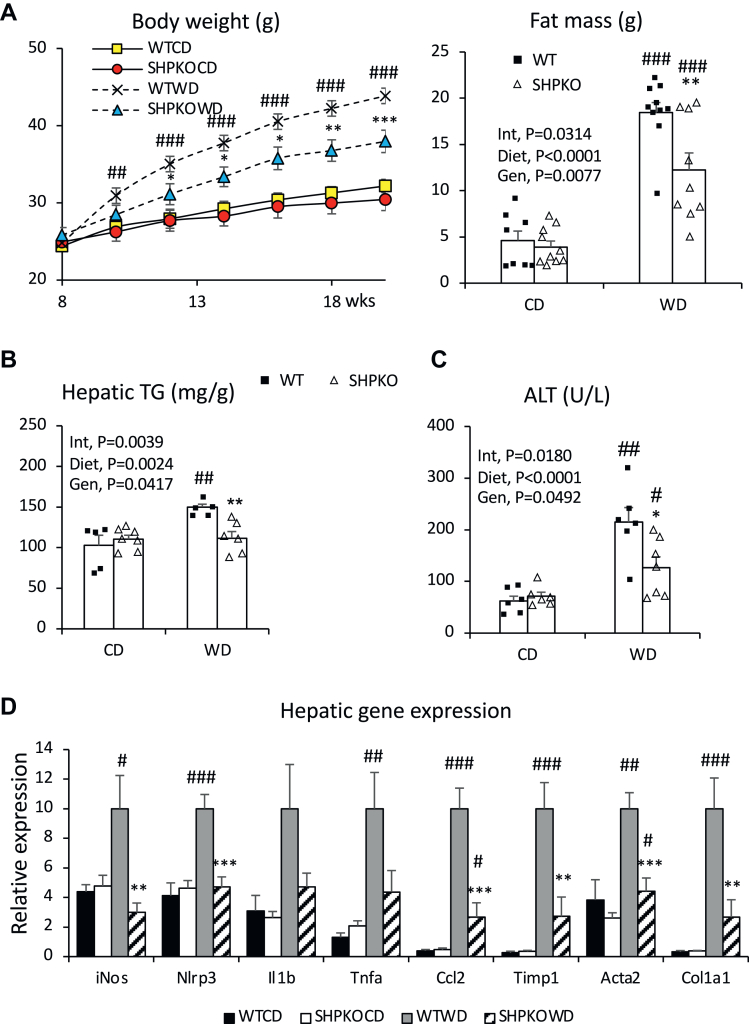
Metabolic features in Shp−/− mice generated from heterozygous breeding scheme. A: Their body weights upon WD regimen were monitored, and the average values were plotted with SEM (left). Their fat masses were assessed using EchoMRI one day before euthanasia for tissue collection (right). The average values were presented with SEM. n = 8–10. B: Hepatic TG levels were presented as means ± SEM. n = 5–6. C: Serum ALT levels were assessed, and their average values were presented with SEM. n = 6–7. D: mRNA levels of indicated hepatic inflammatory and fibrogenic genes were quantified, and their mean values were plotted with SEM (n = 6–8). Two-way ANOVA and Student's t-test were performed to evaluate statistical significance. #comparison with CD, ∗comparison with WT counterparts. Three symbols represent P < 0.005, two symbols P < 0.01, and one symbol P < 0.05.
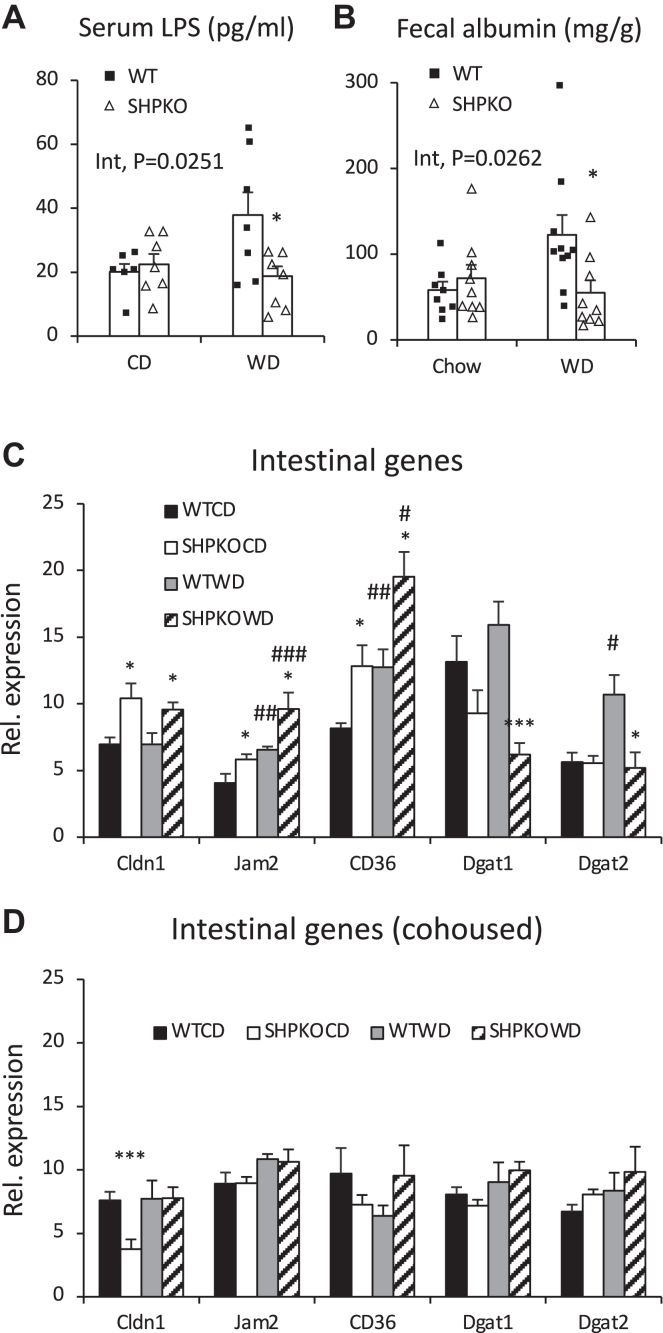
Endotoxemia has been considered one of the major contributors causing diet-associated metabolic diseases such as obesity and fatty liver (18). In order to test the involvement of LPS in phenotypic manifestations, serum LPS levels were measured in the littermates of WT and Shp−/− mice fed a CD or a WD. Figure 6A shows that LPS levels increased approximately 2-fold after the WD challenge in WT mice but not in Shp−/− mice, causing LPS levels to differ significantly between WT and Shp−/− on the WD regimen. Since higher LPS penetration into the circulation can result from increased intestinal permeability, mRNA levels of genes involved in intestinal permeability were examined. The mRNA levels of claudin 1 (Cldn1), junction adhesion molecule 2 (Jam2), and cluster of differentiation 36 (Cd36) were significantly higher in Shp−/− mice than in WT mice fed a WD (Fig. 6C). Since the deletion of Cd36 in endothelial cells affected gut permeability via extracellular matrix remodeling (49), we assessed fecal albumin levels, an indicator of intestinal barrier disruption, in our mice (50). Like serum LPS levels, fecal albumin concentrations were significantly lower in WD-fed Shp−/− mice than in WT mice but were similar in CD-fed WT and Shp−/− mice (around 60 ng/mg; Fig. 6B). While increased endotoxemia was attributed to increased serum alanine aminotransferase levels in WT mice fed a WD, these diet-induced insults were attenuated in Shp−/− mice (Fig. 5C).
Fig. 6.
Reduced endotoxemia and gut permeability in Shp−/− mice fed WD. A: LPS levels were measured in serum collected from WT and Shp−/− mice generated by heterozygous mating. Their average values were presented with SEM (n = 6–7). B: Fecal albumin levels in the experimental animals were quantified and plotted as means ± SEM (n = 8–10). C and D: quantitative PCR analysis was performed to quantify mRNA levels of indicated genes involved gut permeability and fat absorption in their small intestines from separately housed (C) and cohoused mice (D). n = 5–8. Two-way ANOVA and Student's t-test were performed to evaluate statistical significance. #comparison with CD, ∗comparison with WT counterparts. Three symbols represent P < 0.005, two symbols P < 0.01, and one symbol P < 0.05.
In addition to the genes directly involved in gut barrier function, a few genes involved in fatty acid esterification were also examined. Among those genes, diacylglycerol O-acyltransferase 1 (Dgat1) and 2 (Dgat2) expression was significantly downregulated in WD-fed Shp−/− mice compared with WT mice (Fig. 6C). The expression of these two genes was reported to be associated with specific intestinal microbes in response to an HFD for fat absorption (51). Since many genus-level bacteria belonging to the family Clostridiaceae were reported to be responsible for the overexpression of these genes and dietary lipid absorption, this observation indicated an altered gut microbiome (especially the data in Fig. 4B) and impaired intestinal fat absorption in Shp−/− mice (52).
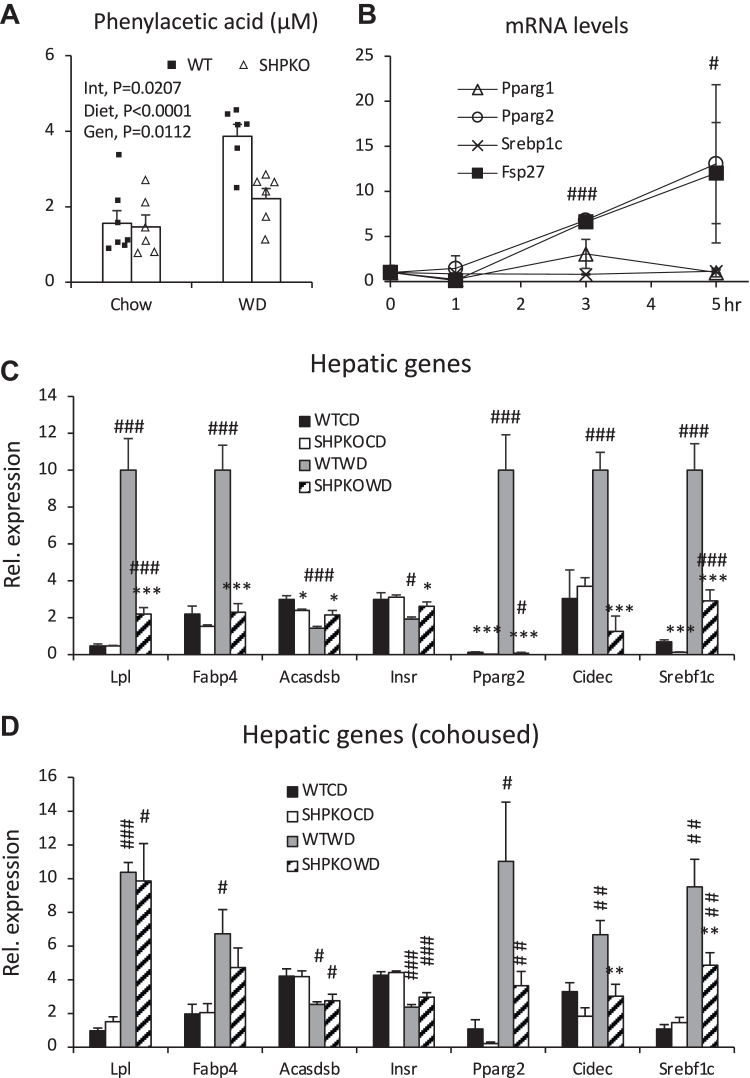
A recent integrative multiomics study on the molecular mechanisms responsible for the integrated interactions between signals from gut bacteria and host hepatic steatosis identified serum PAA as the strongest positive marker of hepatic steatosis among microbial-mammalian cometabolites in women with morbid obesity (25). Therefore, we quantified PAA levels in the blood collected from the portal vein to assess their impact on the hepatic steatosis phenomes. The PAA levels were significantly lower in WD-fed Shp−/− than in WT mice, consistent with their fatty liver phenotypes (Fig. 7A). Interestingly, PAA was reported to function as a PPARG agonist via direct binding and to activate its gene expression (53). Since low hepatic Pparg2 mRNA levels are believed to contribute to the attenuation of the fatty liver phenotype in Shp−/− mice (12), PAA involvement in Pparg2 expression was assessed using a mouse hepatoma cell line (Fig. 7B). PAA treatment strongly induced the expression of Pparg2 and its downstream target fat-specific protein 27 (Fsp27) within 5 h but not peroxisome proliferator-activated receptor gamma isoform 1 (Pparg1) or sterol regulatory element-binding protein 1 (Srebp1). To confirm these results in vivo, we assessed the mRNA levels of many other genes associated with PAA signaling in WT and Shp−/− mice fed a WD. Figure 7C shows that the expression of positively associated genes (lipoprotein lipase [Lpl] and fatty acid binding protein 4 [Fabp4]) and Pparg2 and its target genes was significantly increased in WT mice upon WD challenge. However, their increases were significantly attenuated in Shp−/− mice. In addition, negatively associated genes were downregulated only in WT but not in Shp−/− mice fed a WD. Altogether, these results suggest that the regulatory role of orphan nuclear receptor SHP in BA synthesis affects metabolic phenomes, at least in part, by altering the gut microbiota, thereby decreasing fat absorption and serum levels of LPS and PAA, the two main gut bacterial products contributing to metabolic syndrome, mainly by enhancing gut barrier function and/or their production.
Fig. 7.
Levels of PAA in portal veins and its association with hepatic PPARG2 activation. A: PAA levels were determined in the blood collected from portal veins using a GC-MS. Their averages were presented with SEM (n = 6–7). B: Hepa1-6, a mouse hepatoma cell line, cells were treated with PAA for indicated time points. Levels of mRNA of the indicated genes were quantified by quantitative PCR analysis from triplicates. Statistical results are from comparison of both Pparg2 and Fsp27 with 0 time points (n = 3). C and D: Expression of hepatic genes associated with microbial PAA was assessed in WT and Shp−/− mice fed CD or WD from separately grouped (C) and cohoused strategies (D) using quantitative PCR analysis (n = 5–8). Two-way ANOVA or Student's t-test was performed to evaluate statistical significance. #comparison with CD, ∗comparison with WT counterparts. Three symbols represent P < 0.005, two symbols P < 0.01, and one symbol P < 0.05.
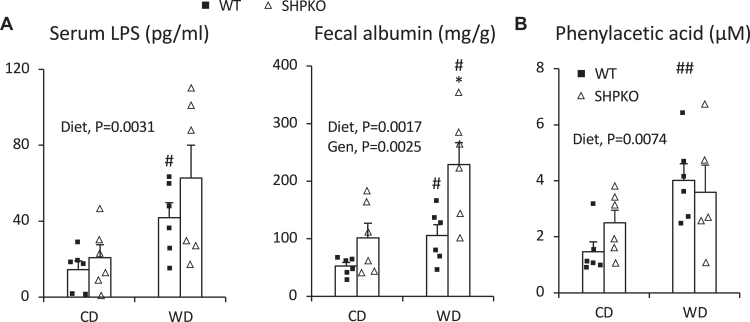
Cohousing disrupts many metabolic parameters, including serum LPS and PAA levels observed in separately housed Shp−/− mice
Serum PAA and LPS levels were also determined in cohoused mice to confirm their association with the phenotypes observed in the separately housed mice. Fig. 8A, B shows that LPS and PAA levels did not differ significantly between WT and Shp−/− mice, especially those fed a WD. WD-fed Shp−/− mice showed higher fecal albumin levels than the WT mice (Fig. 8A). The diet effect in two-way ANOVA was significantly higher in all WD-fed groups for all three parameters. The expression levels of intestinal and hepatic genes differentially regulated by Shp deletion were also assessed in cohoused mice. Intestinal genes involved in barrier function and TG absorption were similarly expressed in cohoused WT and Shp−/− mice (Fig. 6D vs. C). Expression differences in hepatic genes associated with serum PAA between WT and Shp−/− mice were partially or entirely blunted by cohousing (Fig. 7D vs. C), consistent with the disruption of partially protective phenotypes in cohoused Shp−/− mice.
Fig. 8.
Serum LPS, fecal albumin, and serum PAA levels in cohoused littermate WT and Shp−/− mice. A: Concentrations of serum LPS and fecal albumin were quantified in cohoused WT and Shp−/− mice generated by heterozygous mating using commercial ELISA kits. The average values were plotted with SEM (n = 6). B: PAA levels were measured in the blood collected from portal veins of 4-month-old cohoused mice on 2 month diet regimen after 5 h fasting using a GC-MS. Average mean values were presented with SEM (n = 5–6). Two-way ANOVA was used to evaluate statistical significances. Student's t-test was also performed to evaluate statistical significance. #comparison with CD, ∗comparison with WT counterparts. Two symbols represent P < 0.01, and one symbol represents P < 0.05.
Since BA sharing between the cohoused WT and Shp−/− mice would be expected to influence their BA composition, we analyzed the BA compositions in the GB bile from the cohoused WT and Shp−/− mice (Fig. 9A). Each group’s BA composition was highly similar to that of corresponding separately housed mice (Fig. 2A), including the three major BAs, taurodeoxycholic acid, tauromuricholic acid, and TCA (Fig. 9B). We also analyzed the mRNA levels of major genes associated with BA synthesis and transport. supplemental Fig. S7 shows that the expression patterns of the major genes were almost indistinguishable between separately housed and cohoused mice. The coprophagic behavior of the mice appeared to have a minimal impact on BA composition change. Similar to hepatic TG, cohousing effectively disrupted the decrease in hepatic cholesterol level induced by Shp deletion in mice fed the WD, suggesting that amelioration of hepatic cholesterol in WD-fed Shp−/− mice was also linked to changes in the gut microbiome but not changes in BA metabolism or profile. However, the mechanism linking the Shp−/−altered microbiome, LPS, PAA, and fecal albumin to changes in hepatic cholesterol metabolism remain to be determined (supplemental Fig. S8).
Fig. 9.
BA compositions in GBs of cohoused littermate animals. A: BA composition was determined in GBs of cohoused WT and Shp−/− mice fed CD or WD. Their absolute concentrations were converted to percent values to be compared with values obtained from separately grouped animals in Fig. 2A. The average percent values of each BA species were presented as stacked bar graphs (n = WTCD [6], SHPKOCD [3], WTWD [6], and SHPKOWD [5]). B: Average concentrations of three major tauro-conjugated BAs were plotted with SEM values. Two-way ANOVA was performed to measure statistical significance. Student's t-test was also performed to compare two corresponding groups. #P < 0.05 compared with CD counterpart, ∗∗P < 0.01 compared with WT counterpart.
Discussion
The orphan nuclear hormone receptor SHP has been identified as a transcriptional repressor that directly binds to numerous transcriptional activators, including other nuclear hormone receptors (4, 54). The repression is achieved by recruiting transcriptional repressors (55, 56). The suppression of Cyp7a1 gene expression has been considered the major and well-studied SHP-driven transcriptional regulation. This critical regulation in BA metabolism has been confirmed in several mouse models, including Shp−/− mice. Many mouse model studies have found that Shp deletion attenuates diet-induced obesity and NAFLD in the mice, whereas Shp overexpression has the opposite effect. While a few transcriptional targets responsible for these phenotypic manifestations have been proposed, the association of its BA metabolism with the observed metabolic phenotypes has not been studied or suggested. A recent study demonstrated that FXR (also called nuclear receptor subfamily 1, group H, member 4), a BA receptor, regulates obesity and fatty liver by altering the gut microbiota (20).
This study focused on assessing change in gut microbiota through the BA profile change and its impact on the partially protective phenotype against NASH development in Shp−/− mice. Since DCA has been recognized as a major BA contributing to the inflammatory response and hepatocellular carcinoma (57), increased DCA levels in the serum and liver of Shp−/− mice were unexpected and could not explain their phenotype. Therefore, altered gut microbiota composition because of Shp deletion was the major contributing factor to the phenotypic manifestations, which was proved by the cohousing scheme and gut microbiome structure determined by 16S rRNA gene sequencing analysis of fecal samples. The cohousing had little effect on BA composition in these mouse models. While phylum-level bacterial composition may agree with their metabolic phenotypes, many genus-level bacterial groups showed positive and negative correlations with phenotypes in previous studies. For example, one species in the genus Bacteroides, Bacteroides vulgatus, was overrepresented in mice resistant to diet-induced NAFLD (58). The genus Bacteroides was more abundant in Shp−/− mice fed a WD than in WT mice, and its abundance was disrupted by cohousing. In contrast, while the genus Dorea was elevated in HFD-fed mice in an earlier study (59), this genus was not elevated by the WD in WT mice but was in Shp−/− mice, resulting in a higher abundance in Shp−/− mice than in WT mice fed a WD. In addition, the abundance of Akkermansia muciniphila was reported to inversely correlate with BW and diabetes, and treatment of mice with diet-induced obesity with this bacteria improved their adverse metabolic profile (60). Our study showed a higher abundance of the genus Akkermansia in Shp−/− mice fed a WD than in WT mice housed in separate cages, consistent with the previous study. However, the difference in abundance was not reversed by cohousing, indicating that Akkermansia muciniphila may not contribute to the observed metabolic phenotypes in Shp−/− mice, although our sequencing depth was limited to the genus level. Since similar discrepancies in bacterial association with metabolic disorders have been frequently documented in various studies (61), it is challenging to pinpoint specific bacteria responsible for the SHP-driven metabolic phenotypes with current approaches. Nevertheless, a few genera were found to differ significantly between WD-fed WT and Shp−/− mice housed in separate cages but not when cohoused. Among these genera, Dehalobacterium in the phylum Firmicutes showed the strongest positive association with the phenotypic manifestations in the Shp−/− mice, consistent with earlier studies (62, 63).
While BA composition and gut microbiota structure underlie the phenotype manifestations in Shp−/− mice, the responsible pathways underlying their potential impacts remain unknown. The PICURSt assessment of 16S rRNA sequencing data showed that some enriched pathways in WT and Shp−/− mice fed the WD correlated with the previously identified positive and negative pathways, respectively (16). These enriched pathways include flagellar assembly and bacterial motility proteins for WT mice and aromatic amino acid biosynthesis, β-alanine metabolism, pyruvate metabolism, and citrate cycle for Shp−/− mice. The associations between these pathways and metabolic outcomes must be explored extensively to understand the underlying mechanisms and establish concrete connections.
Our study’s significant findings were the decreased serum LPS and PAA levels in Shp−/− mice fed a WD compared with WT mice. In addition, hepatic Pparg2 expression was shown to be a direct positive target of PAA. Shp deletion provides better circulatory environments to cope with WD-induced obesity and NASH by modifying the BA profile and gut microbiota. Chronic endotoxemia, represented by high bacterial LPS levels in the blood, is a well-known trigger for the onset of metabolic syndromes, including obesity, diabetes, and NASH (18). Notably, the Gram-negative phylum Proteobacteria was greatly increased in WT mice fed the WD compared with Shp−/− mice. PAA is a recently identified phenylalanine metabolite in blood circulation strongly associated with steatosis among the microbial-mammalian cometabolites in patients with hepatic steatosis (25). In their studies, LPL and FABP4, two major targets of PPARγ2 (64), were among the strongly induced hepatic genes in patients with steatosis. While not normally expressed in the liver, these two genes were highly induced in the livers of human patients with NAFLD or mice fed a WD, along with PPARG2 (65, 66, 67). The proteins encoded by these genes are essential components for the adipogenic program in adipose tissues. Given their specific roles in fat storage, surplus fats delivered to the liver by an HFD would be stored rather than oxidized or packaged into very low-density lipoprotein by activating these proteins. The direct link between bacterial metabolite PAA and hepatic PPARγ2 activation would be the first step for NASH development under the two-hit theory (53, 68).
Higher serum LPS induces inflammation in the steatotic liver caused concomitantly by PAA, eventually developing into NASH when persistently fed a WD. The bacterial gene responsible for PAA production from phenylalanine is mapped to several species belonging to the genus Clostridium (69). Unfortunately, the contribution of this genus to phenotypes in our experimental mice remains unknown because of variations in the abundance of various Clostridium species in our 16S rRNA gene sequencing analysis. An earlier study reported that PAA functions as a ligand for PPARγ and transcriptionally activates Pparg expression simultaneously (53). Therefore, we hypothesize that PAA-bound PPARγ1 (the abundant isoform in the liver) can activate the expression of another isoform (Pparg2). PPARγ binds direct repeat 1 elements in the nuclear receptor binding site with the retinoid X receptor. Our previous study identified a possible direct repeat 1 element in the Pparg2 promoter that is bound by the hepatocyte nuclear factor 4 alpha-hairy and enhancer of split 6 to repress expression (70). We hypothesize that under normal conditions, the hepatocyte nuclear factor 4 alpha-hairy and enhancer of split 6 complex represses Pparg2 expression, and increased PAA under an HFD may relieve this repression via activated PPARγ1 binding to induce an adipogenic program in the liver, inducing the expression of Pparg2 and its downstream target genes. Hepatic TG synthesis rate was not measured in the present study to corroborate this hypothesis.
In summary, Shp deletion reshapes gut microbiome especially in WD-fed condition, in such a way as to (1) enhance gut barrier function and/or (2) reduce LPS and PAA production in the gut. This study also demonstrated that hepatic cholesterol is positively associated with NAFLD/NASH development. All these molecular responses to dietary-modified gut microbiota contribute to the development of obesity and NASH. While a germ-free mouse approach is necessary to provide a definitive conclusion, our cohousing data suggest that Shp−/− mice show reduced molecular responses possibly because of BA-mediated dysbiosis, which remains to be determined.
Data Availability
All data are contained within this article.
Supplemental data
This article contains supplemental data (11, 16).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Byunghun So at Inha University, Incheon, Korea for graph revision.
Author contributions
Y.-K. L. conceptualization; R. M., J. E. P., M. L., Y.-J. Y. W., H. A. B., T. K., X. W., F. L., R. N., and L. P. S. methodology; R. M., J. E. P., and M. L. validation; R. M., J. E. P., M. L., P. K. J., M. A., L. L., R. N., and H. H. formal analysis; R. M., J. E. P., M. L., P. K. J., M. A., L. L., and H. H. investigation; Y.-J. Y. W., H. A. B., T. K., X. W., F. L., and L. P. S. resources; R. M., J. E. P., and M. L. data curation; R. M. writing–original draft; J. E. P., M. L., P. K. J., T. K., L. P. S., and Y.-K. L. writing–review & editing; R. M., J. E. P., and M. L. visualization; Y.-K. L. supervision; Y.-K. L. project administration; Y.-K. L. funding acquisition.
Funding and additional information
This study was supported by the National Institutes of Health R01DK093774, R01AA013623, a bridge fund from NEOMED (to Y. K. L.). The content is solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Current address for Prasant Kumar Jena: Department of Pediatrics, Cedars Sinai Medical Center, Los Angeles, CA, USA.
Supplementary Data
References
- 1.Aranda A., Pascual A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 2.Seol W., Choi H.S., Moore D.D. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 3.Johansson L., Thomsen J.S., Damdimopoulos A.E., Spyrou G., Gustafsson J., Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J. Biol. Chem. 1999;274:345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y.K., Dell H., Dowhan D.H., Hadzopoulou-Cladaras M., Moore D.D. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol. Cell Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y.K., Moore D.D. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. J. Biol. Chem. 2002;277:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- 6.Lu T.T., Makishima M., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J., et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin B., Jones S.A., Price R.R., Watson M.A., McKee D.D., Moore L.B., et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 8.Wang L., Lee Y.-K., Bundman D., Han Y., Thevananther S., Kim C.-S., et al. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 9.Boulias K., Katrakili N., Bamberg K., Underhill P., Greenfield A., Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman H.B., Lai K., Evans M.J. Loss of small heterodimer partner expression in the liver protects against dyslipidemia. J. Lipid Res. 2009;50:193–203. doi: 10.1194/jlr.M800323-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Park Y.J., Qatanani M., Chua S.S., LaRey J.L., Johnson S.A., Watanabe M., et al. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008;47:1578–1586. doi: 10.1002/hep.22196. [DOI] [PubMed] [Google Scholar]
- 12.Kim S.C., Kim C.K., Axe D., Cook A., Lee M., Li T., et al. All-trans-retinoic acid ameliorates hepatic steatosis in mice by a novel transcriptional cascade. Hepatology. 2014;59:1750–1760. doi: 10.1002/hep.26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y.K., Park J.E., Lee M., Mifflin R., Xu Y., Novak R., et al. Deletion of hepatic small heterodimer partner ameliorates development of nonalcoholic steatohepatitis in mice. J. Lipid Res. 2023;64 doi: 10.1016/j.jlr.2023.100454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Backhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Backhed F., Manchester J.K., Semenkovich C.F., Gordon J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U. S. A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 17.Vrieze A., Van Nood E., Holleman F., Salojarvi J., Kootte R.S., Bartelsman J.F., et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916.e917. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 18.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 19.Devkota S., Wang Y., Musch M.W., Leone V., Fehlner-Peach H., Nadimpalli A., et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parseus A., Sommer N., Sommer F., Caesar R., Molinaro A., Stahlman M., et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66:429–437. doi: 10.1136/gutjnl-2015-310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K., et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Jiang C., Xie C., Li F., Zhang L., Nichols R.G., Krausz K.W., et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest. 2015;125:386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Xie C., Wang G., Wu Y., Wu Q., Wang X., et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018;24:1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elinav E., Thaiss C.A., Flavell R.A. Analysis of microbiota alterations in inflammasome-deficient mice. Methods Mol. Biol. 2013;1040:185–194. doi: 10.1007/978-1-62703-523-1_14. [DOI] [PubMed] [Google Scholar]
- 25.Hoyles L., Fernandez-Real J.M., Federici M., Serino M., Abbott J., Charpentier J., et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018;24:1070–1080. doi: 10.1038/s41591-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y.K., Park J.E., Lee M., Hardwick J.P. Hepatic lipid homeostasis by peroxisome proliferator-activated receptor gamma 2. Liver Res. 2018;2:209–215. doi: 10.1016/j.livres.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J.E., Lee M., Mifflin R., Lee Y.K. Enhanced ethanol catabolism in orphan nuclear receptor SHP-null mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;310:G799–807. doi: 10.1152/ajpgi.00343.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H., Taraboletti A., Shriver L.P. Dimethyl fumarate modulates antioxidant and lipid metabolism in oligodendrocytes. Redox Biol. 2015;5:169–175. doi: 10.1016/j.redox.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K.H., Choi J.M., Li F., Arizpe A., Wooton-Kee C.R., Anakk S., et al. Xenobiotic nuclear receptor signaling determines molecular pathogenesis of progressive familial intrahepatic cholestasis. Endocrinology. 2018;159:2435–2446. doi: 10.1210/en.2018-00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rideout J.R., He Y., Navas-Molina J.A., Walters W.A., Ursell L.K., Gibbons S.M., et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faith D.P., Baker A.M. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol. Bioinform. Online. 2006;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng L., Jena P.K., Liu H.X., Kalanetra K.M., Gonzalez F.J., French S.W., et al. Krishnan V.V., Mills D.A., Wan Y.Y. Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci. Rep. 2017;7:1748. doi: 10.1038/s41598-017-01576-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alrehaili B.D., Lee M., Takahashi S., Novak R., Rimal B., Boehme S., et al. Bile acid conjugation deficiency causes hypercholanemia, hyperphagia, islet dysfunction, and gut dysbiosis in mice. Hepatol. Commun. 2022;6:2765–2780. doi: 10.1002/hep4.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasumov T., Brunengraber L.L., Comte B., Puchowicz M.A., Jobbins K., Thomas K., et al. New secondary metabolites of phenylbutyrate in humans and rats. Drug Metab. Dispos. 2004;32:10–19. doi: 10.1124/dmd.32.1.10. [DOI] [PubMed] [Google Scholar]
- 38.Garrett W.S., Gallini C.A., Yatsunenko T., Michaud M., DuBois A., Delaney M.L., et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerr T.A., Saeki S., Schneider M., Schaefer K., Berdy S., Redder T., et al. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerr T.A., Matsumoto Y., Matsumoto H., Xie Y., Hirschberger L.L., Stipanuk M.H., et al. Cysteine sulfinic acid decarboxylase regulation: a role for farnesoid X receptor and small heterodimer partner in murine hepatic taurine metabolism. Hepatol. Res. 2014;44:E218–228. doi: 10.1111/hepr.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kullak-Ublick G.A., Stieger B., Meier P.J. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. 2004;126:322–342. doi: 10.1053/j.gastro.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Aldini R., Roda A., Montagnani M., Cerre C., Pellicciari R., Roda E. Relationship between structure and intestinal absorption of bile acids with a steroid or side-chain modification. Steroids. 1996;61:590–597. doi: 10.1016/s0039-128x(96)00119-5. [DOI] [PubMed] [Google Scholar]
- 44.Turnbaugh P.J., Backhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W.Z., Strowig T., et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mamantopoulos M., Ronchi F., Van Hauwermeiren F., Vieira-Silva S., Yilmaz B., Martens L., et al. Nlrp6- and ASC-dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity. 2017;47:339–348.e334. doi: 10.1016/j.immuni.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 48.Lemire P., Robertson S.J., Maughan H., Tattoli I., Streutker C.J., Platnich J.M., et al. The NLR protein NLRP6 does not impact gut microbiota composition. Cell Rep. 2017;21:3653–3661. doi: 10.1016/j.celrep.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 49.Cifarelli V., Ivanov S., Xie Y., Son N.H., Saunders B.T., Pietka T.A., et al. CD36 deficiency impairs the small intestinal barrier and induces subclinical inflammation in mice. Cell Mol. Gastroenterol. Hepatol. 2017;3:82–98. doi: 10.1016/j.jcmgh.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., Llorente C., Hartmann P., Yang A.M., Chen P., Schnabl B. Methods to determine intestinal permeability and bacterial translocation during liver disease. J. Immunol. Methods. 2015;421:44–53. doi: 10.1016/j.jim.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Guryn K., Hubert N., Frazier K., Urlass S., Musch M.W., Ojeda P., et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23:458–469.e455. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park Y.J., Kim S.C., Kim J., Anakk S., Lee J.M., Tseng H.T., et al. Yechoor V., Park J., Choi J.S., Jang H.C., Lee K.U., Novak C.M., Moore D.D., Lee Y.K. Dissociation of diabetes and obesity in mice lacking orphan nuclear receptor small heterodimer partner. J. Lipid Res. 2011;52:2234–2244. doi: 10.1194/jlr.M016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samid D., Wells M., Greene M.E., Shen W., Palmer C.N., Thibault A. Peroxisome proliferator-activated receptor gamma as a novel target in cancer therapy: binding and activation by an aromatic fatty acid with clinical antitumor activity. Clin. Cancer Res. 2000;6:933–941. [PubMed] [Google Scholar]
- 54.Seol W., Choi H.-S., Moore D.D. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol. Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 55.Kemper J.K., Kim H., Miao J., Bhalla S., Bae Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol. Cell Biol. 2004;24:7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boulias K., Talianidis I. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 2004;32:6096–6103. doi: 10.1093/nar/gkh947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshimoto S., Loo T.M., Atarashi K., Kanda H., Sato S., Oyadomari S., et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 58.Le Roy T., Llopis M., Lepage P., Bruneau A., Rabot S., Bevilacqua C., et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 59.Velazquez K.T., Enos R.T., Bader J.E., Sougiannis A.T., Carson M.S., Chatzistamou I., et al. Prolonged high-fat-diet feeding promotes non-alcoholic fatty liver disease and alters gut microbiota in mice. World J. Hepatol. 2019;11:619–637. doi: 10.4254/wjh.v11.i8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tilg H., Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Del Chierico F., Abbatini F., Russo A., Quagliariello A., Reddel S., Capoccia D., et al. Gut microbiota markers in obese adolescent and adult patients: age-dependent differential patterns. Front Microbiol. 2018;9:1210. doi: 10.3389/fmicb.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zietak M., Kovatcheva-Datchary P., Markiewicz L.H., Stahlman M., Kozak L.P., Backhed F. Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab. 2016;23:1216–1223. doi: 10.1016/j.cmet.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tontonoz P., Hu E., Spiegelman B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid- activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 65.Westerbacka J., Kolak M., Kiviluoto T., Arkkila P., Siren J., Hamsten A., et al. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56:2759–2765. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 66.Yamazaki T., Shiraishi S., Kishimoto K., Miura S., Ezaki O. An increase in liver PPARgamma2 is an initial event to induce fatty liver in response to a diet high in butter: PPARgamma2 knockdown improves fatty liver induced by high-saturated fat. J. Nutr. Biochem. 2011;22:543–553. doi: 10.1016/j.jnutbio.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 67.Zhang L., Zhang Z., Li Y., Liao S., Wu X., Chang Q., et al. Cholesterol induces lipoprotein lipase expression in a tree shrew (Tupaia belangeri chinensis) model of non-alcoholic fatty liver disease. Sci. Rep. 2015;5 doi: 10.1038/srep15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Day C.P., James O.F. Hepatic steatosis: innocent bystander or guilty party? Hepatology. 1998;27:1463–1466. doi: 10.1002/hep.510270601. [DOI] [PubMed] [Google Scholar]
- 69.Dodd D., Spitzer M.H., Van Treuren W., Merrill B.D., Hryckowian A.J., Higginbottom S.K., et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park J.E., Lee M., Kim S.C., Zhang Y., Hardwick J.P., Lee Y.K. Hairy and enhancer of split 6 prevents hepatic lipid accumulation through inhibition of Pparg2 expression. Hepatol. Commun. 2017;1:1085–1098. doi: 10.1002/hep4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this article.