Abstract
Covalent chemistry represents an attractive strategy for expanding the ligandability of the proteome, and chemical proteomics has revealed numerous electrophile-reactive cysteines on diverse human proteins. Determining which of these covalent binding events impact protein function, however, remains challenging. Here, we describe a base-editing strategy to infer the functionality of cysteines by quantifying the impact of their missense mutation on cancer cell proliferation. The resulting atlas, which covers >13,800 cysteines on >1,750 cancer dependency proteins, confirms the essentiality of cysteines targeted by covalent drugs and, when integrated with chemical proteomic data, identifies essential, ligandable cysteines in >160 cancer dependency proteins. We further show that a stereoselective and site-specific ligand targeting an essential cysteine in TOE1 inhibits the nuclease activity of this protein through an apparent allosteric mechanism. Our findings thus describe a versatile method and valuable resource to prioritize the pursuit of small-molecule probes with high function-perturbing potential.
Introduction
Small molecules are powerful tools for studying the functions of proteins in biological systems and can serve as starting points for therapeutics1. Advanced chemical probes and drugs frequently target established small-molecule binding pockets in proteins, such as the active sites of enzymes or ligand-binding pockets of receptors. Many protein types, however, are not known to bind endogenous small molecules and are consequently more difficult to gauge in terms of their potential for targeting by chemical probes. The timeliness and importance of this problem are accentuated by the output of modern large-scale sequencing and genetic screening efforts, which are identifying a broad array of human disease-relevant genes that code for protein types such as DNA/RNA-binding or adaptor/scaffolding proteins2,3 that have historically been challenging to target with small molecules.
Several platforms for the discovery of small-molecule binders of proteins have recently been introduced as a means to expand the ligandability of the human proteome4. These ‘binding-first’ technologies, which include fragment-based screening5, DNA-encoded libraries6, and chemical proteomic methods such as activity-based protein profiling (ABPP)7,8, have led to the discovery of small-molecule ligands for diverse arrays of proteins. Nonetheless, whether such ligands affect the functions of protein targets often remains unclear and can be challenging to determine experimentally, especially for proteins with poorly characterized biochemical or cellular activities. In cases where newly discovered small molecule-protein interactions have been shown to be functional, it is notable that they frequently act by allosteric mechanisms9,10, underscoring the diverse and unanticipated ways that chemical probes can modulate the activity of proteins.
Among binding-first approaches, ABPP of electrophilic small molecules has demonstrated considerable potential for ligand discovery7,8,11–15. Advantages of this approach include the deployment of covalent chemistry14,16,17, which can address shallow and dynamic pockets on protein surfaces that are less amenable to binding reversibly to small molecules, as well as provide a selectivity filter in the form of targeting isotype-restricted nucleophilic residues (e.g., cysteines) within paralogous proteins, as have been demonstrated by several recent cancer therapeutics (e.g., osimertinib for EGFR_C79718, sotorasib for KRAS_G12C19,20). Additionally, by evaluating small molecules directly in native biological systems, ABPP can identify cryptic ligandable pockets regulating aspects of protein function that are difficult to discern with purified proteins or protein domains12,21,22.
The human genome encodes over 200,000 cysteines distributed across virtually all proteins. A growing, but still modest proportion of these cysteines has been shown to interact with electrophilic small molecules in ABPP experiments7,11–14,16, and, even in these cases, the impact of electrophile-cysteine interactions on protein function remains mostly unknown. Here, we integrate base editing with ABPP to globally assess the essentiality and ligandability of cysteines in the context of cancer dependency proteins as defined by the Cancer Dependency Map (DepMap) 2. Recognizing that gene editing of certain structurally buried cysteines may create an essentiality outcome due to perturbations in protein folding, we also present a general strategy to categorize essential, but as-of-yet unliganded cysteines based on their chemical reactivity in native versus denatured proteomes. The approach reported herein offers residue-level functional annotation and small-molecule reactivity maps to guide the ongoing and future pursuit of covalent chemical probes and drugs.
Results
Base editing identifies cysteines targeted by cancer drugs
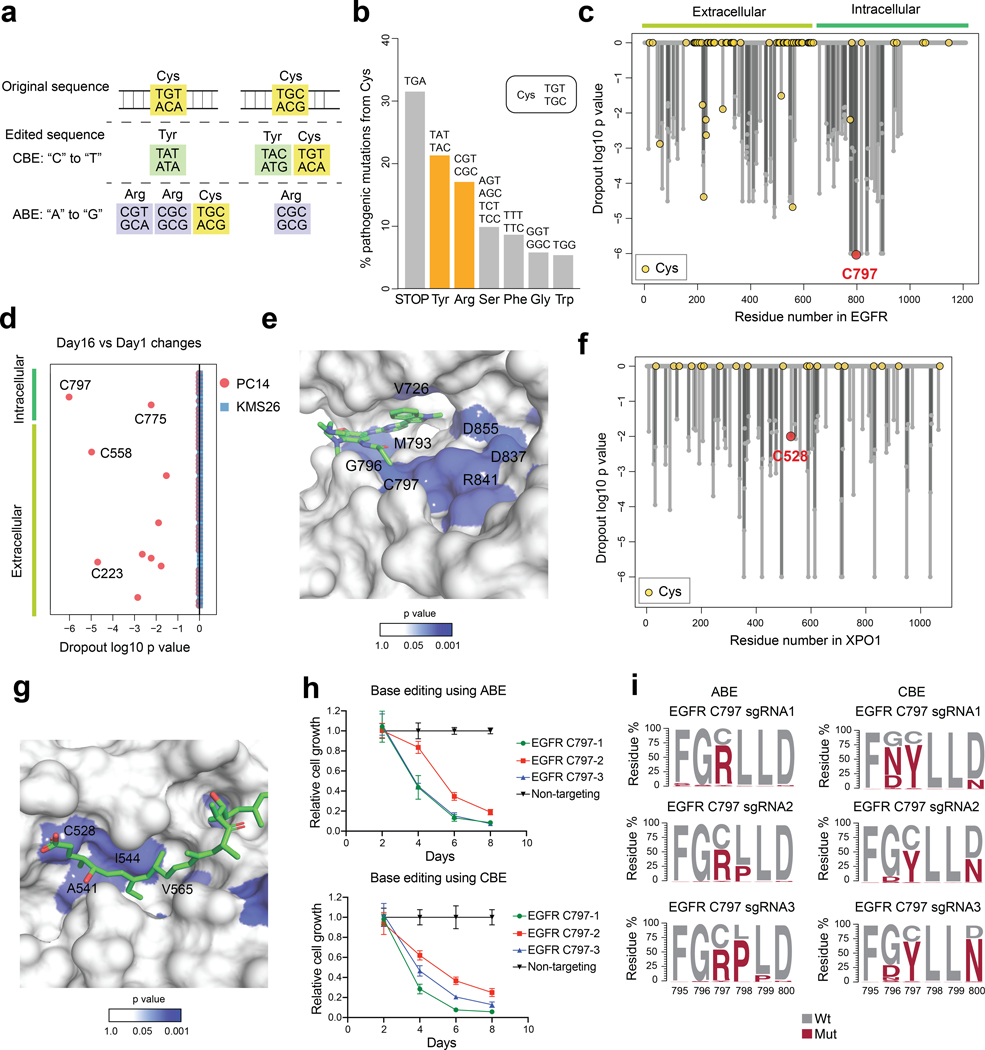
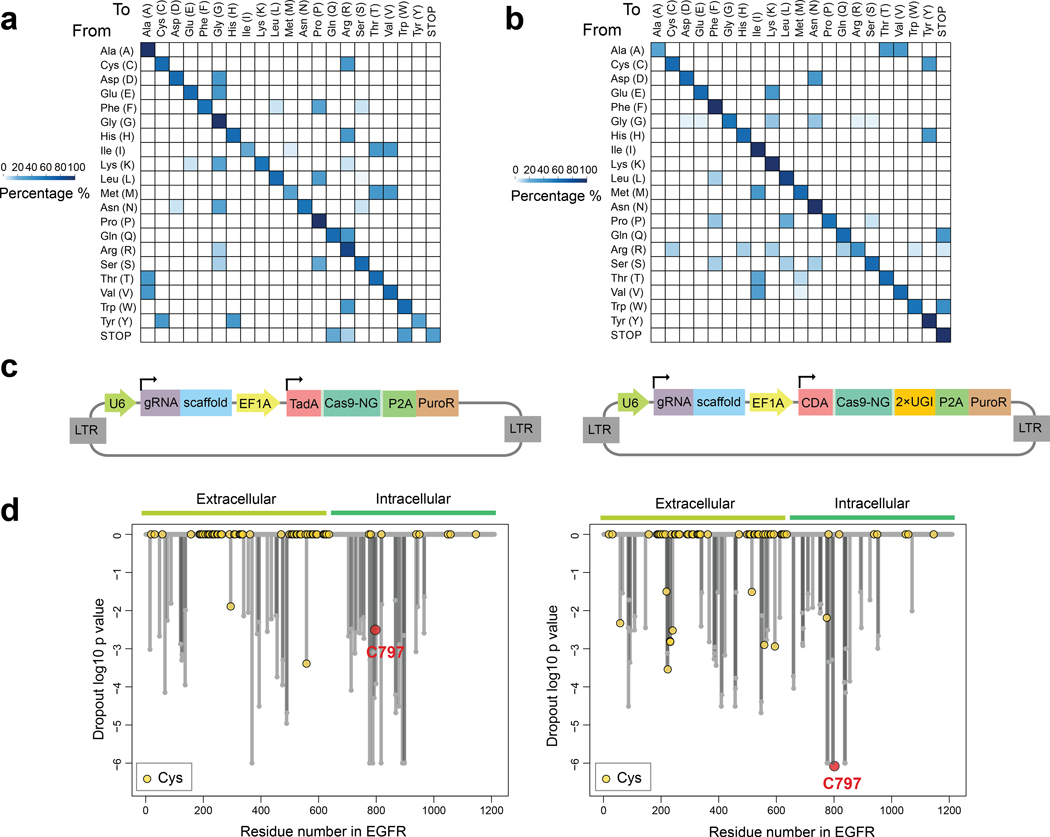
Programmable base editors comprising Cas9 nickases fused to cytidine deaminases or laboratory-evolved deoxyadenosine deaminases23,24 can introduce nucleotide substitutions at targeted genomic DNA sites. Within an “editing window”, deoxyadenosine deaminases in adenine base editors (ABEs) convert an A•T base pair into a G•C base pair23 and cytidine deaminases in cytosine base editors (CBEs) convert a C•G base pair into a T•A base pair24. Consistent with the mutagenic paths for cysteine afforded by ABEs (cysteine-to-arginine) and CBEs (cysteine-to-tyrosine) (Fig. 1a and Extended Data Fig. 1a, b) having a good probability of altering protein function, we found that, among >5,100 pathogenic cysteine mutations in the ClinVar database25 (version 2023–04), 17.1% and 21.4% represented conversions to arginine and tyrosine, respectively (Fig. 1b). These frequencies were much larger than the pathogenic missense mutations that convert cysteine to serine, phenylalanine, or glycine, despite equivalent or greater codon availability for these mutational paths (Fig. 1b).
Fig. 1 |. Base editing assigns essentiality to cysteines targeted by anti-cancer drugs.
a, Codon analysis of cysteine mutagenesis patterns generated by ABE or CBE. b, Barplot showing the distribution of pathogenic missense mutations of cysteine residues (n=5,184) from the ClinVar database. c, Waterfall plot showing the dropout significance calculated per EGFR residue between day 16 vs day 1 using PC14 cells (cutoff: Log2 Fold Change, LFC < −0.5; p < 0.05). Data represent average values of two independent experiments. One-sided p values were calculated using randomization tests. d, Dot plot comparing the day 16 vs day 1 dropout significance of EGFR cysteine residues in PC14 and in KMS26 cells (cutoff: LFC < −0.5; p < 0.05, one-sided randomization test). Data represent average values of two independent experiments. e, Structure of the EGFR kinase domain bound to osimertinib (PDB: 6JXT). The p values are derived from c. f, Waterfall plot showing the dropout significance calculated per XPO1 residue between day 16 vs day 1 using KMS26 cells (cutoff: LFC < −0.5; p < 0.05). Data represent average values of two independent experiments. One-sided p values were calculated using randomization tests. g, Structure of the XPO1 cargo-binding cleft in association with the natural product Leptomycin B (PDB: 6TVO). The p values are derived from f. h, Proliferation graphs for PC14 cells with EGFR_C797 editing in an arrayed format measured by CellTiterGlo. The relative growth was normalized using the group treated with a non-targeting sgRNA as 100%. Data represent average values ± SEM for 6 independent experiments. i, EGFR amino acid mutagenesis generated by different sgRNAs and the indicated base editors in PC14 cells. Results are representative of two independent experiments.
We selected ABE8e (ABE) and evoCDA (CBE) in conjunction with SpCas9-NG to recognize an “NG” protospacer adjacent motif (PAM)26–28 as base editors because they provided a combination of high editing efficiency and broad genome-wide targeting. The 5–10 nt editing windows allowed for missense mutation of a greater number of cysteines within reach of a PAM site, but also introduced the potential to edit neighboring residues. We considered this approach to offer a reasonable balance of specificity and generality and anticipated that edits at nearby residues could provide useful information on the essentiality of the local environment surrounding a ligandable cysteine.
We first evaluated the performance of our cysteine editing protocol with two cancer dependency proteins – EGFR and XPO1 – that are targets of cysteine-directed covalent drugs. Specifically, C797 of EGFR is engaged by covalent inhibitors such as osimertinib to treat non-small cell lung cancer (NSCLC)18, and C528 of XPO1 is engaged by selinexor to treat multiple myeloma29. We designed a pooled base-editing library containing >5,800 sgRNAs representing a mix of ABE and CBE sites that covered most residues in EGFR and XPO1 and delivered this library to the EGFR-dependent NSCLC cell line PC14 and the multiple myeloma cell line KMS26 using lentiviral vectors (Extended Data Fig. 1c). We then allowed the cells to grow for 16 days and performed targeted amplicon sequencing of the sgRNA cassette region on days 1 and 16 to obtain relative sgRNA frequency changes associated with cancer cell proliferation. The dropout values for all sgRNAs predicted to edit the same residue were averaged to give a significance value of residue-level essentiality (Supplementary Dataset 1). Encouragingly, this analysis identified C797 among the most essential residues and the top dropout among all intracellular cysteines in EGFR in PC14 cells (Fig. 1c, d and Extended Data Fig. 1d). Most other essential cysteines in EGFR were extracellular residues involved in structural disulfide bonds (Fig. 1c, d). We did not observe essentiality for EGFR_C797 or, in general, other EGFR residues in KMS26 cells (Fig. 1d and Supplementary Dataset 1), consistent with this cell line being independent of EGFR for growth. We also found evidence of essentiality for other residues located near C797 in and around the kinase catalytic pocket (Fig. 1e). Similar findings emerged for XPO1, an essential protein involved in nuclear export, where C528 and additional residues in the XPO1 substrate/selinexor-binding cleft displayed substantial dropout in KMS26 cells (Fig. 1f, g, Supplementary Dataset 1). Finally, we validated the pooled screening results by individual cloning and gene editing of six selected EGFR_C797-targeting sgRNAs in an arrayed format. We observed that all six sgRNAs caused prominent decreases in PC14 proliferation (Fig. 1h), and targeted genomic sequencing revealed editing of EGFR_C797 and the region flanking this residue (Fig. 1i).
These proof-of-principle experiments with established cancer dependency proteins supported that our strategy combining base editing with pooled cell proliferation assays can create sufficient single or composite mutations at and around cysteines targeted by covalent small molecules to reveal the functionality of these residues and the druggable pockets where they are located, thus complementing previously described indel-based genome editing approaches to identify functional protein domains30.
Global analysis of cysteines in cancer dependency proteins
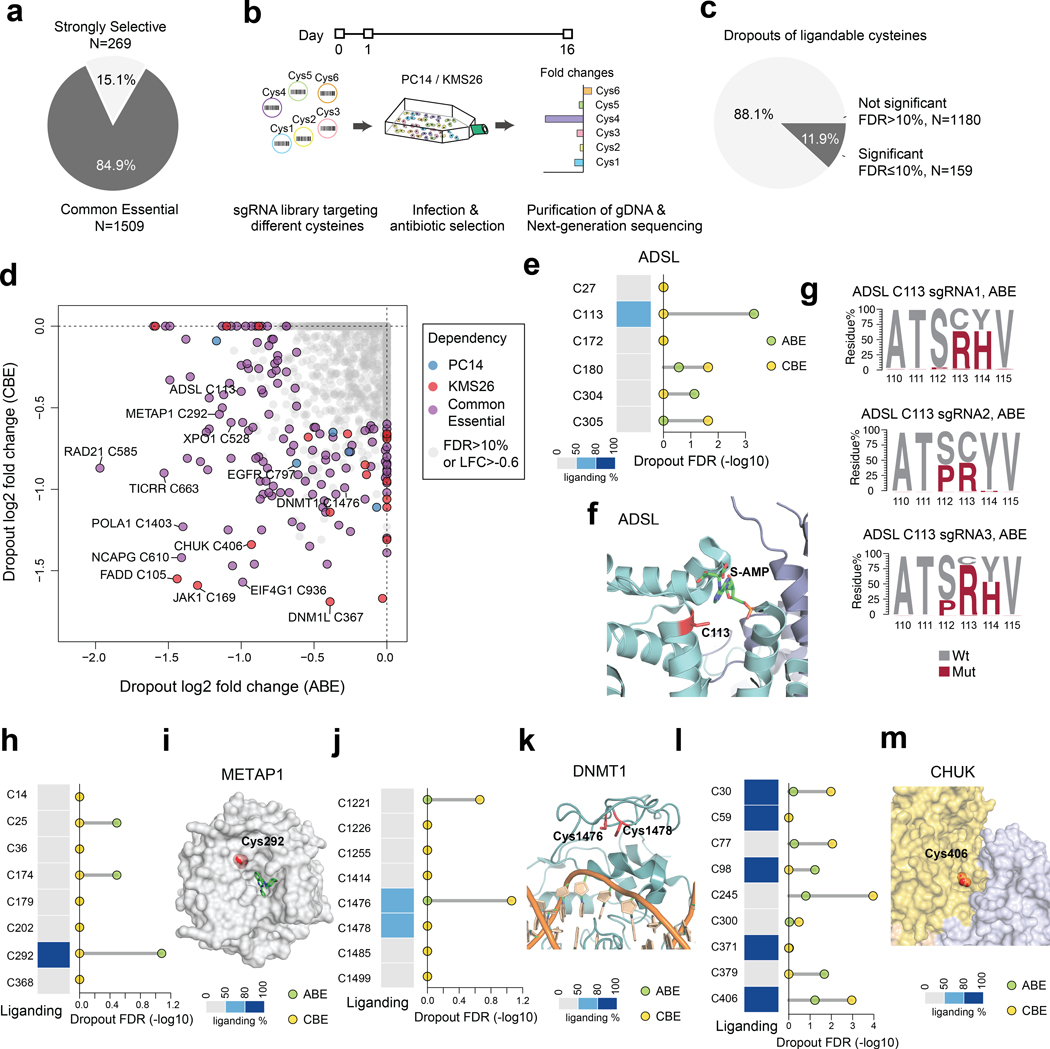
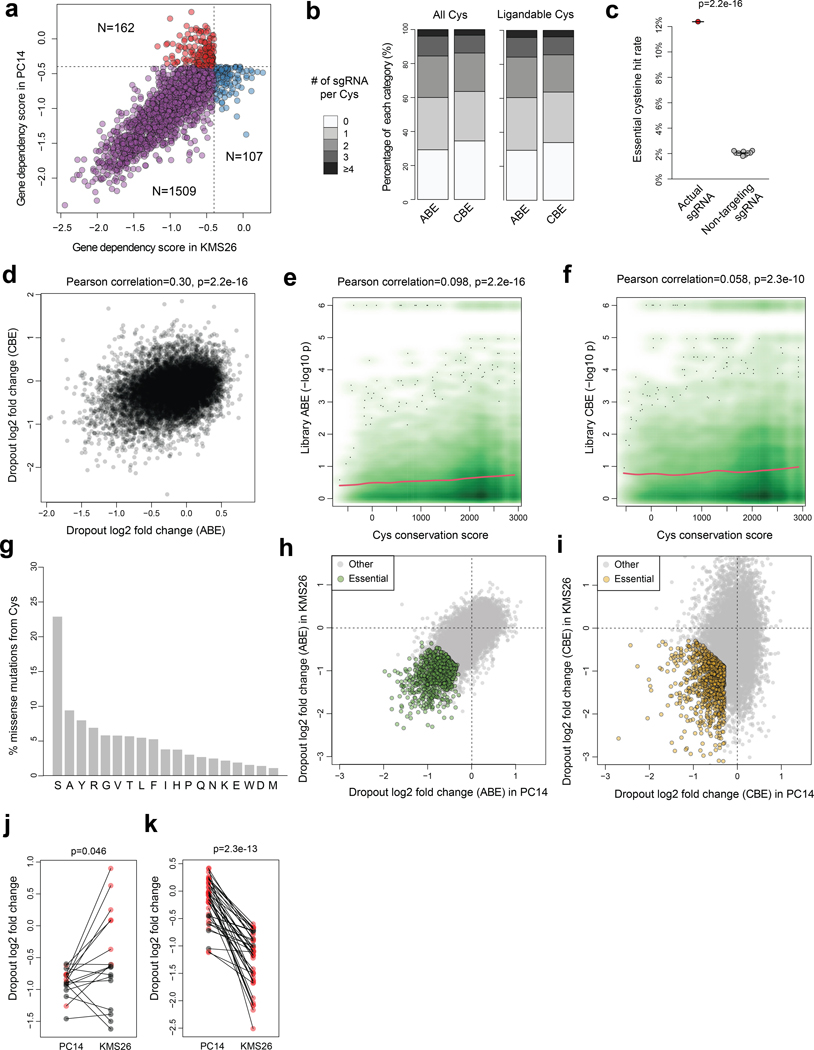
We next sought to globally assess the essentiality of cysteines in a broad set of cancer dependency proteins. We designed ~50,000 sgRNAs using both ABE and CBE that covered >13,800 cysteines from >1,750 cancer dependency proteins. Of these proteins, ~270 were defined as Strongly Selective in the DepMap, reflecting a restricted dependency relationship with a subset of cancer cell lines, and another ~1500 were defined as Common Essential to indicate their general requirement for the growth (Fig. 2a, Extended Data Fig. 2a, Supplementary Dataset 2). The sgRNA library contained at least one sgRNA that satisfied the NG PAM requirements of the ABE or CBE for about 70% of all cysteines in the cancer dependency proteins (Extended Data Fig. 2b). These sgRNAs were screened as a pooled library, along with ~1,200 non-targeted control sgRNAs, in separate experiments performed in PC14 and KMS26 cells (Fig. 2b).
Fig. 2 |. Global discovery of essential cysteines in cancer dependency proteins.
a, Pie chart showing the proportion of Strongly Selective and Common Essential dependency proteins evaluated in the screen. b, Schematic showing the workflow for determining cysteine essentiality by base editing dropout screens. c, Pie chart showing the proportion of ligandable cysteines with or without significant dropouts in gene editing screens (FDR cutoff: 10%, LFC cutoff: −0.6). d, Plot of essential, ligandable cysteines identified in gene editing dropout screens. The LFC of each cysteine is calculated by comparing the frequency of corresponding sgRNAs between day 16 vs day 1. For cysteines from Common Essential proteins, the average dropout values of both model cell lines were used in the calculations, and, for cysteines from Strongly Selective proteins, either PC14 (blue points) or KMS26 (red points) dropout data were used. e, The fragment electrophile ligandability profile (left) and significance of dropout (right) for cysteines in ADSL. f, Structure of ADSL in complex with its substrate adenylosuccinate (PDB: 2VD6). g, ADSL amino acid mutagenesis generated by individual sgRNAs and the indicated base editor in PC14 cells. Results are representative of two independent experiments. h, The fragment electrophile ligandability profile (left) and significance of dropout (right) for cysteines in METAP1. i, Structure of METAP1 in complex with the non-covalent inhibitor FZ1 (PDB: 4IU6). j, The fragment electrophile ligandability profile (left) and significance of dropout (right) for cysteines in DNMT1. C1476 and C1478 are on the same tryptic peptide, so their ligandability cannot be distinguished by cysteine-directed MS-ABPP. k, Structure of DNMT1 in complex with DNA (PDB: 7SFG). l, The fragment electrophile ligandability profile (left) and significance of dropout (right) for cysteines in CHUK. m, Structure of CHUK showing the homo-oligomerization surface (PDB: 5EBZ). For d, e, h, j, l, the one-sided p values were first estimated by randomization tests of dropout data from two independent base-editing experiments and then adjusted by Benjamini-Hochberg procedure to calculate the FDR.
Using parameters of gene-level effect2 (CERES score < −0.4) and residue-level dropout (log2 fold changes, LFC < −0.6; false discovery rate, FDR < 10%), we identified a total of 1,718 essential cysteines across 846 proteins (Supplementary Dataset 2). This % hit rate for essential cysteines (12.4% of 13,872 total screened cysteines) was much higher than the % hit rate observed with non-targeted sgRNAs (2.0%, Extended Data Fig. 2c). We found similar hit rates using either editor (ABE: 6.6%; CBE: 7.4%) and the dropout fold-changes for each library were overall correlated (Extended Data Fig. 2d), suggesting a similar potential for functional impact of Cys-to-Arg and Cys-to-Tyr mutations. By calculating cysteine conservation scores, we observed a moderate but significant correlation between increased evolutionary conservation of cysteines and greater dropout potential (Extended Data Fig. 2e, f, Supplementary Dataset 2). Additionally, conservative mutations (e.g., Ser (22.9%), Ala (9.5%)) were most frequently observed for essential cysteines across orthologous proteins compared to conversions to Tyr (8.0%) and Arg (7.0%) (Extended Data Fig. 2g).
We next cross-referenced the base editing results with chemical proteomic maps of cysteine engagement by broadly reactive fragment electrophiles (derived from past work7,11,12 supplemented by additional cysteine-directed ABPP experiments performed herein, Supplementary Dataset 2). This analysis yielded 159 cysteines showing combined features of essentiality and ligandability (Fig. 2c, d and Supplementary Dataset 2). A much larger set of ligandable cysteines (1,180 in total) in cancer dependency proteins were assigned as nonessential from the base-editing screens (Fig. 2c and Supplementary Dataset 2), suggesting only a limited relationship between cysteine ligandability and functionality, at least as determined by base editing-mediated mutagenesis analyzed in cell proliferation assays. Essential cysteines were distributed into sub-groups showing selective effects in PC14 or KMS26 cells, or in both cell lines (Fig. 2d and Supplementary Dataset 2), and this distribution generally matched that predicted from gene-level disruption2 (Extended Data Fig. 2h-k). The pooled screening strategy, for instance, rediscovered the selective essentiality of EGFR_C797 in PC14 cells and the common essentiality of XPO1_C528 in both PC14 and KMS26 cells (Fig. 2d). Additionally, nine of the ligandable cysteines in cancer dependency proteins have disease associations in the ClinVar database, and four (FADD_C105, STAT3_C712, DNM1L_C367, CREBBP_C1775) were assigned as essential in our base editing screens (Supplementary Dataset 2).
Among the essential, ligandable cysteines were residues in enzyme active sites, including C113 located in the substrate binding pocket of adenylosuccinate lyase (ADSL) (Fig. 2e, f). The ABE created the expected C113R mutation along with S112P or Y114H mutations in ADSL (Fig. 2g). An essential and ligandable active-site cysteine (C292) was also identified in methionine aminopeptidase 1 (METAP1) (Fig. 2h), and this residue is near the binding site for a reversible METAP1 inhibitor31 (Fig. 2i). Other essential, ligandable cysteines were found at protein-DNA and protein-protein interfaces, such as C1476 of the DNA methyltransferase DNMT1 (Fig. 2j, k) and C406 of the kinase CHUK (Fig. 2l, m), respectively.
These findings show how the integration of gene editing and ABPP data can identify essential, ligandable cysteines required for cancer cell growth.
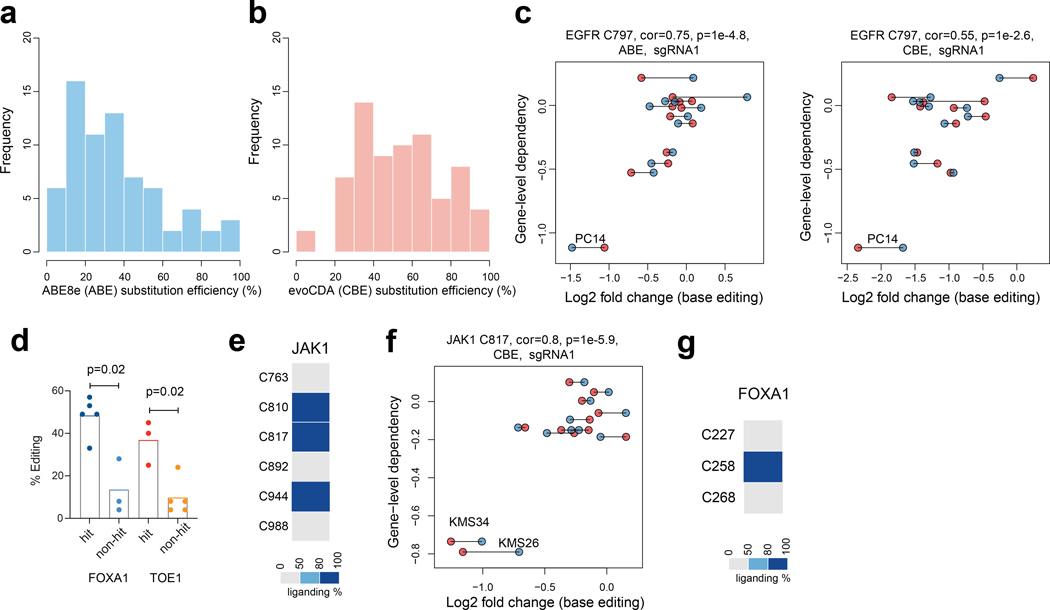
Saturated local base editing of ligandable cysteines
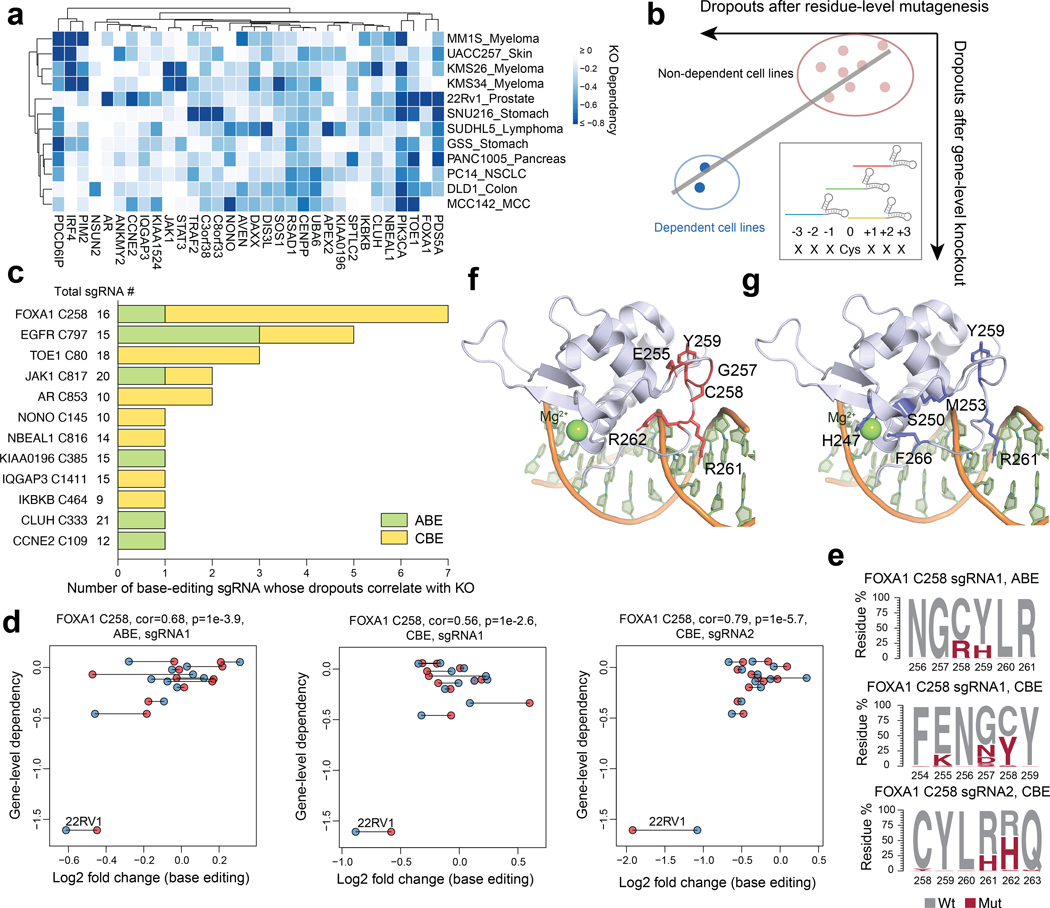
The PC14 and KMS26 cell lines displayed dependency on a substantial portion of Common Essential proteins (1,509 of 2,664), but, as expected, required far fewer Strongly Selective proteins for growth (269 of 3,082)2. Considering that many of the most compelling therapeutic targets in cancer fall into the Strongly Selective category, we devised an alternative strategy to expand our analysis of essential cysteines for this group of proteins. We specifically adopted a pooled dropout screening approach to enable parallel analysis of essential cysteines across twelve human cancer lines from nine different tissues that together captured ~700 Strongly Selective proteins, of which ~30 possessed one or more ligandable cysteines (Fig. 3a and Supplementary Dataset 3). We focused our pooled dropout screen on these ligandable cysteines and expanded the targeting editing window to include three residues on either side of each cysteine, which we anticipated would increase our probability of obtaining high-efficiency editing events for assessing the essentially of the local cysteine region. This adaptation was made in response to finding that the editing efficiency for a substantial proportion of cysteines targeted in our initial screen (13% and 47% for CBE- and ABE-editing events, respectively) was < 30% (Extended Data Fig. 3a, b), which pointed to the possibility of overlooking the functional impact of ligandable cysteines due to inefficient editing, especially for sites targetable by very few sgRNAs. Finally, an additional advantage of this pooled dropout screen is that individual cell lines should serve as cross-validating internal controls, where essential cysteines in Strongly Selective proteins would be expected to only impair the growth of the subset of cell lines that are dependent on that protein (Fig. 3b).
Fig. 3 |. Base editing reveals essentiality of ligandable cysteine regions in Strongly Selective cancer dependency proteins.
a, Heatmap showing Strongly Selective dependency proteins with ligandable cysteines in the twelve-cancer cell line panel. b, Schematic showing how base-editing can assign essentiality to ligandable cysteine regions in Strongly Selective proteins. sgRNAs were designed to edit ligandable cysteines +/− 3 surrounding residues. An sgRNA is considered a hit if the resulting missense mutation leads to preferential dropouts of cancer cell lines that also show selective dependency on the corresponding protein containing the ligandable cysteine based on gene-level disruptions in the DepMap. c, Essential, ligandable cysteines identified in the twelve-cell line panel dropout screen, and the number of sgRNAs assigning essentiality to each cysteine. The total number of sgRNAs included in the analysis for each cysteine region is also provided. d, Scatter plots for the FOXA1_C258 region showing the correlation between base editing dropout data acquired herein and gene disruption dropout data (DepMap). The two independent experiments are shown in blue and red. Pearson correlations and associated two-sided p values are shown. e, FOXA1 amino acid mutagenesis generated by individual sgRNAs and the indicated base editors in the FOXA1-dependent cell line 22Rv1. Results are representative of two independent experiments. f, g, FOXA1 DNA-binding domain homology model based on the crystal structure of FOXA3 in complex with DNA (PDB: 1VTN). The side chains of essential residues identified in the base-editing dropout screen and residues representing hotspot pro-cancerous mutations33,34 are highlighted in red and blue in f and g, respectively.
We compared the magnitude of base editing effects to reference dropout values for the corresponding gene disruptions in the DepMap. We considered a cysteine to be essential when the base editor targeting that cysteine’s region produced: i) LFC < −0.5 for the cell line(s) showing selective dependency on the corresponding protein; and ii) the magnitude of base-editing dropout was greatest for the cell line(s) showing selective dependency on the corresponding protein compared to other cell lines in the panel (correlation > 0.5 and FDR < 10%). Based on these criteria, we identified 12 ligandable, essential cysteines (Fig. 3c and Supplementary Dataset 3), including EGFR_C797, which showed the expected preferential dropout in PC14 cells (Extended Data Fig. 3c).
We noted that only a subset of the tested sgRNAs registered as hits for each essential, ligandable cysteine, and the total number of hit sgRNAs also varied widely for these residues (Fig. 3c). Consistent with differences in editing efficiency being a potential source for such variability, we found that, among 16 representative sgRNAs targeting the same cysteine regions that were individually cloned and arrayed, hit sgRNAs produced an average of 44% editing whereas non-hit sgRNAs produced only 11% editing (Extended Data Fig. 3d). These data indicated that the total number of hit sgRNAs per cysteine region may not correlate with the degree of essentiality, but rather with the extent of editing efficiency achieved by the sgRNAs targeting this region, and, conversely, even a single hit sgRNA may be sufficient to assign essentiality to a cysteine region. Also supportive of this conclusion, we observed that JAK1_C817 was designated as essential by a single sgRNA (Fig. 3c and Extended Data Fig. 3e, f), and covalent ligands targeting this cysteine have recently been found to allosterically inhibit JAK1-dependent signaling in human cells21.
Among the other essential, ligandable cysteines was C258 of the pioneer transcription factor FOXA1 (Fig. 3c, Extended Data Fig. 3g). FOXA1 is a Strongly Selective dependency in subsets of prostate and breast cancer cell lines, and several base-editing sgRNAs for FOXA1 showed preferential dropout in the 22Rv1 prostate cancer line (Fig. 3c, d). We arrayed and quantified the genome-editing outcomes for three representative sgRNAs and found that they possessed distinct sets of missense mutations at C258 and/or neighboring residues (C258R+Y259H, E255K+G257N+C258Y, and R261H+R262H) (Fig. 3e). Structural analysis using a FOXA3 homology model32 indicated that C258, as well as the additional residues altered by base editing, are part of the Wing2 region of the forkhead (FKHD) domain and reside in proximity to the DNA-binding site (Fig. 3f). Interestingly, the region surrounding C258 is also enriched in oncogenic mutations that have been shown to remodel chromatin accessibility and gene expression, as well as affect the differentiation state, of breast and prostate cancer cells33–35 (Fig. 3g). Taken together, these data indicate that genetic alteration of the C258 region can modulate the activity of FOXA1, and the covalent ligandability of C258 further points to the possibility for small molecules to produce similar functional outcomes.
Functional effects of covalent ligands targeting TOE1
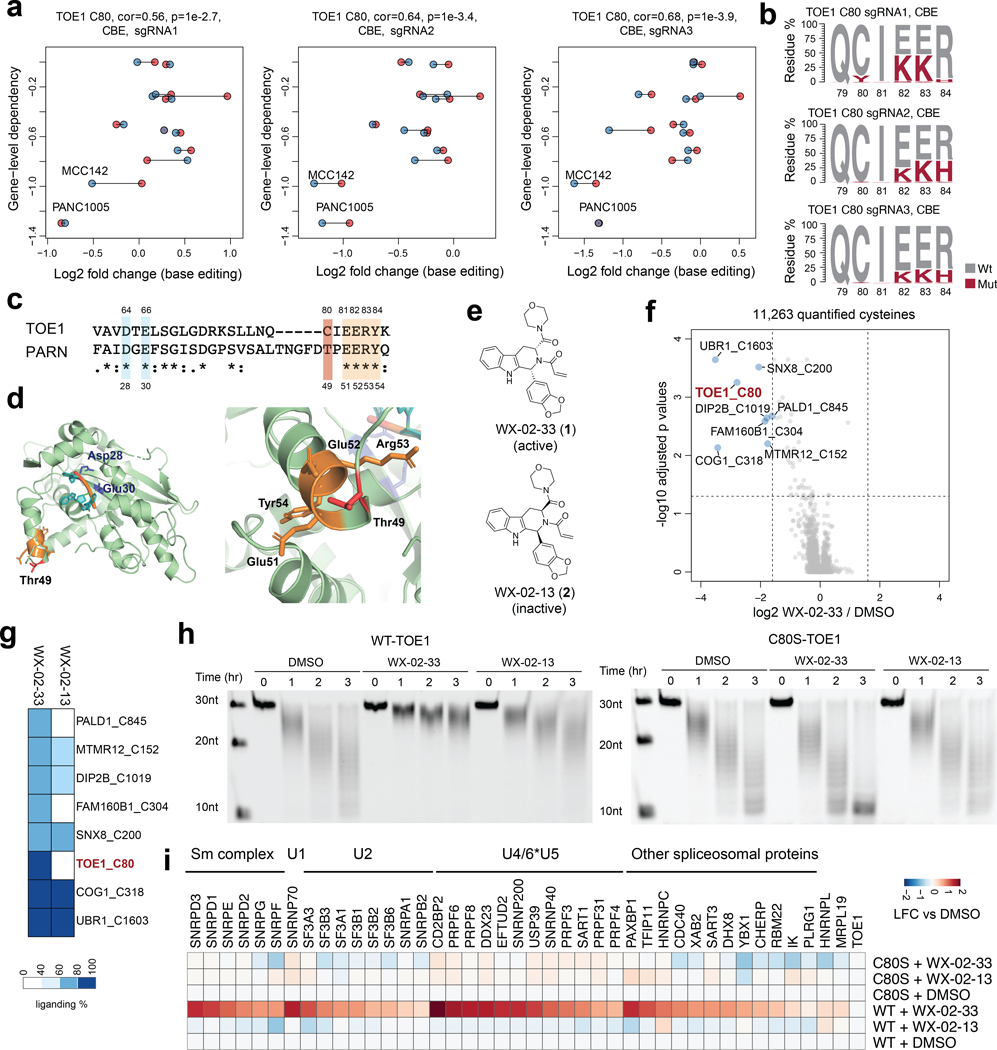
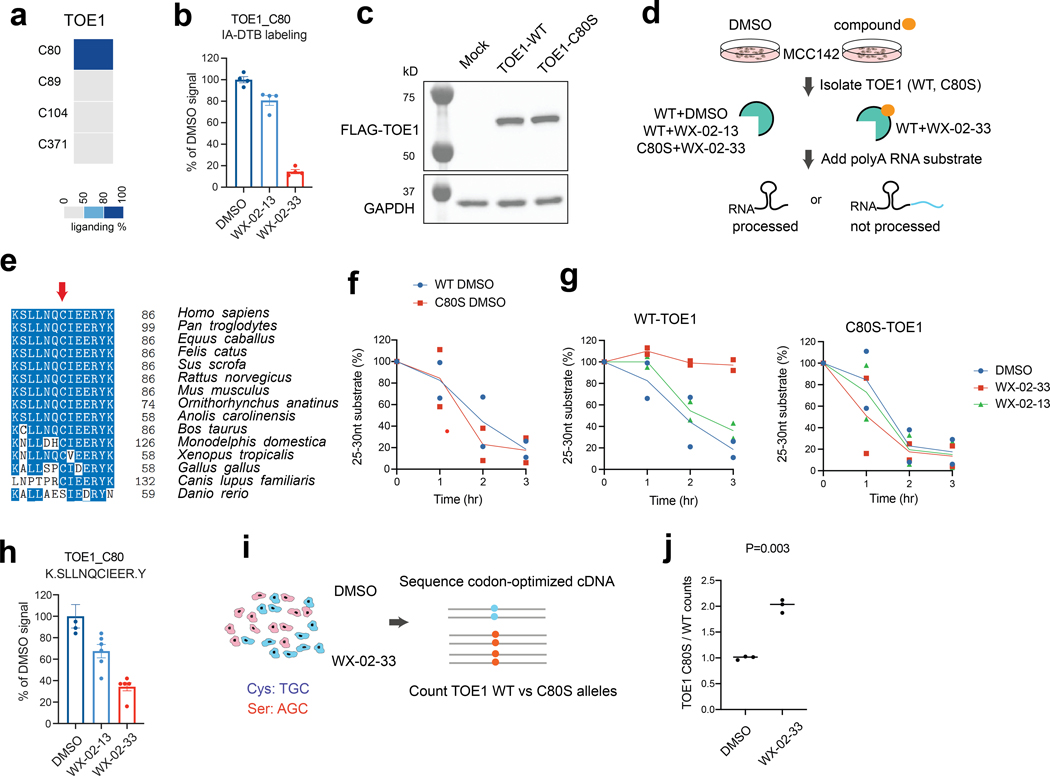
Essential, ligandable cysteines were also discovered in Strongly Selective proteins that have less well-understood roles in cancer. One example was C80 in TOE1 (Extended Data Fig. 4a), a 3’ RNA exonuclease that trims the tails of non-coding RNAs (e.g., small nuclear RNAs (snRNAs)) as part of their maturation36–38. In the twelve-cell line panel, MCC142 (Merkel cell carcinoma) and PANC1005 (pancreatic adenocarcinoma) cells showed the greatest dependency on TOE1 (Fig. 4a), with hit sgRNAs targeting the C80 region causing missense mutations that included C80Y and, to an even greater extent, E82K, E83K, and/or R84H (Fig. 4b). The E82-E83-R84 sequence, but not C80, is conserved in the homologous nuclease PARN, despite this protein and TOE1 only sharing 30% overall identity (Fig. 4c), and the crystal structure of PARN indicates these conserved residues are located far from the nuclease active site (Fig. 4d). The proximity of C80 to a stretch of conserved and essential residues in the TOE1/PARN nuclease family suggested that covalent compounds targeting this residue may have the potential to impact the function of TOE1.
Fig. 4 |. Functional characterization of covalent ligands targeting an essential cysteine C80 in TOE1.
a, Scatter plots for the TOE1_C80 region showing the correlation between base editing dropout data acquired herein and gene disruption dropout data (DepMap). The two independent experiments are shown in blue and red. Pearson correlations and associated two-sided p values are shown. b, TOE1 amino acid mutagenesis generated by individual sgRNAs and the indicated base editors. Results are representative of two independent experiments. c, Alignment of TOE1 and PARN protein sequences. The conserved catalytic residues are highlighted in blue and the EERY sequence is highlighted in orange. d, TOE1 homology model based on the crystal structure of PARN in complex with RNA (PDB: 2A1R). PARN_T49 (corresponding to TOE1_C80) and the EERY sequence are highlighted in red and orange respectively. e, Structures of the TOE1_C80 ligand tryptoline acrylamide WX-02–33 and the less active enantiomer WX-02–13. f, Volcano plot of ABPP data showing cysteines substantially and significantly (P adjusted by Benjamini-Hochberg procedure < 0.01, two-sided Student’s t test, n = 2 independent experiments) engaged by WX-02–33 (20 μM, 3 h) in Ramos cells. g, Heatmap comparing WX-02–33 and WX-02–13 engagement for the cysteines substantially and significantly engaged by WX-02–33 identified in f. Data are average values from n = 2 independent experiments. h, Time-dependent TOE1 deadenylation activity from MCC142 cells stably expressing recombinant WT-TOE1 or a C80S-TOE1 mutant treated with WX-02–33 or WX-02–13 (20 μM, 6 h) or DMSO. The images are representative of 2 independent experiments. i, Heatmap showing proteins that were enriched in FLAG-WT- and C80S-TOE1 IP-MS experiments from stably expressing MCC142 cells treated with WX-02–33 or WX-02–13 (20 μM, 6 h) or DMSO. Displayed proteins exhibited at least 2-fold enrichment in FLAG-WT-TOE1-expressing cells compared to mock MCC142 cells. Results are average values from 2 independent experiments. Data are normalized to TOE1 signals within each experiment.
As part of our ongoing efforts to discover covalent ligands by cysteine-directed ABPP12,22,39, we identified a tryptoline acrylamide WX-02–33 (1) that engaged TOE1_C80 with good potency (> 80% engagement), stereoselectivity (compared to the enantiomer WX-02–13 (2)), and proteome-wide selectivity in cancer cells (20 μM compound, 3 h) (Fig. 4e-g, Extended Data Fig. 4b, and Supplementary Dataset 4). Only seven other cysteines among >11,000 quantified cysteines were engaged >67% by WX-02–33, and among these additional targets, only two cysteines (FAM160B1_C304; PALD1_C845) were preferentially engaged by WX-02–33 compared to WX-02–13 (Fig. 4g). We next treated MCC142 cells stably expressing recombinant FLAG-tagged WT-TOE1 protein or a C80S-TOE1 mutant (Extended Data Fig. 4c) with WX-02–33 or WX-02–13 (20 μM, 6 h) and measured TOE1 activity with a polyadenylated RNA substrate following anti-FLAG immunoprecipitation (Extended Data Fig. 4d). We selected a C80S-TOE1 mutant for these studies because certain vertebrate orthologues of TOE1 have C80 replaced with serine (Extended Data Fig. 4e), suggesting that a C80S mutation should be tolerated while also conferring resistance to potential functional effects of WX-02–33. Consistent with this hypothesis, we found that WX-02–33 inhibited the activity of WT-TOE1, but not C80S-TOE1, which otherwise displayed similar nuclease activity compared to WT-TOE1 (Fig. 4h and Extended Data Fig. 4f). The enantiomeric compound WX-02–13 showed much less inhibitory activity on WT-TOE1 (Fig. 4h), consistent with exhibiting weaker engagement of TOE1_C80 (Extended Data Fig. 4b).
The distal location of TOE1_C80 relative to the nuclease active site suggested that covalent ligands targeting this residue inhibited TOE1 activity by an allosteric mechanism. Immunoprecipitation-mass-spectrometry (IP-MS) studies of FLAG-WT-TOE1 and FLAG-C80S-TOE1 from cells treated with WX-02–33 or WX-02–13 (20 μM, 6 h) revealed that WX-02–33 site-specifically and stereoselectively promoted TOE1 binding to various spliceosome-related protein complexes involved in pre-mRNA splicing (Fig. 4i, Extended Data Fig. 4h, and Supplementary Dataset 4). While we do not yet understand if or how this WX-02–33-induced stabilization of TOE1-spliceosome interactions relates to the inhibition of TOE1 nuclease activity, we speculate, given the role of TOE1 in snRNA tail processing36–38 and that snRNAs are core components of the spliceosome, that allosteric blockade of TOE1 activity by WX-02–33 may result in incompletely processed snRNAs remaining bound to TOE1, which in turn leads to increased TOE1-spliceosome interactions. Finally, to evaluate how WX-02–33 engagement of TOE1 affects the growth rate of the TOE1-dependent cell line MCC142, we devised a competition assay where the relative number of cells carrying WT- and C80S-TOE1 alleles were quantified after several days of pooled culture in the presence of WX-02–33 (Extended Data Fig. 4i). This experiment revealed that the C80S-TOE1 mutant-expressing cells grew significantly better than WT-TOE1-expressing cells in the presence of WX-02–33 (Extended Data Fig. 4j). Taken together, these data support that covalent ligands targeting TOE1_C80 – a cysteine discovered to be essential in base-editing screens – act as allosteric inhibitors of TOE1 nuclease activity and promote TOE1 binding to spliceosome complexes in cancer cells.
Identifying essential cysteines with ligandability potential
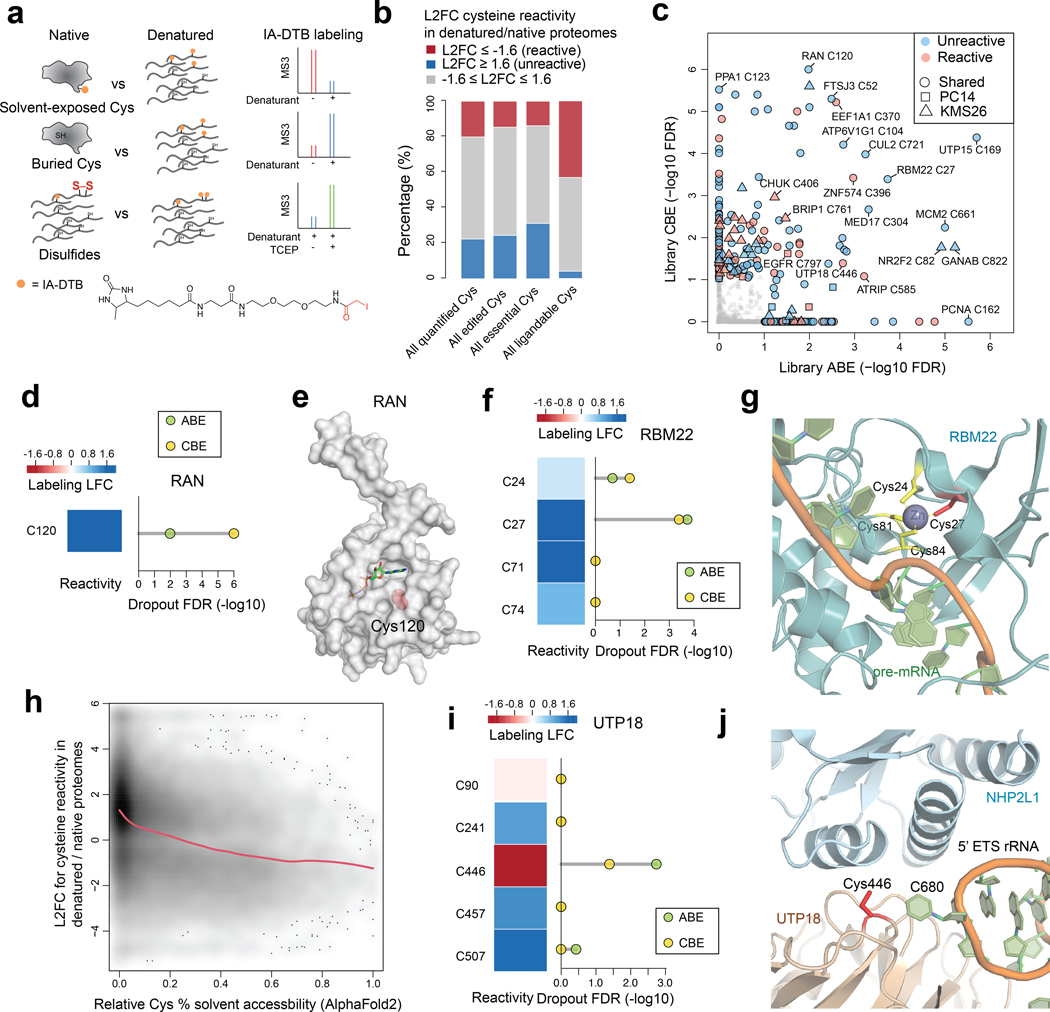
In considering the scope and limitations of our approach, we grappled with the small fraction of essential cysteines mapped by base editing that had evidence of covalent ligandability in chemical proteomic studies performed to date (159 of 1,718 total essential cysteines in the global pooled screen). This can be explained, at least in part, by the limited diversity of chemistry evaluated in original ABPP experiments, which have so far have only screened a handful of electrophilic fragments7,11,12 and therefore likely underestimate the complete small-molecule interaction potential of cysteines in the human proteome. We also wondered, however, if other factors may be at play. For instance, some essential cysteines may have an intrinsically low likelihood of interacting with small molecules if, for instance, they serve structural roles at buried locations in proteins with limited or no access to solvent. Distinguishing such buried, essential cysteines from those that have greater potential to interact with small molecules would provide a way to prioritize cysteines for the future pursuit of chemical probes. With this goal in mind, we adapted established ABPP protocols7,8 to enable quantitative comparisons of cysteine reactivity with an iodoacetamide-desthiobiotin (IA-DTB) probe in denatured versus native proteomes (Fig. 5a). We hypothesized that this comparison may reveal bidirectional changes in cysteine reactivity that were informative for classifying different types of functional residues: i) essential cysteines showing increased reactivity following protein denaturation may serve structural roles, but be inaccessible to solvent in the native protein state; and ii) essential cysteines showing decreased reactivity following protein denaturation may be solvent-accessible residues proximal to pockets that promote enhanced IA-DTB reactivity.
Fig. 5 |. Prioritizing essential cysteines with ligandability potential by quantitative cysteine reactivity profiling in native and denatured proteomes.
a, Schematic showing an ABPP workflow to distinguish solvent-exposed and buried cysteines based on quantitative differences in IA-DTB reactivity in native and denatured proteomes. b, Stack barplots summarizing the distribution of reactivity changes for the indicated categories of cysteines, including 1) all cysteines quantified in the ABPP experiments of native versus denatured proteomes; 2) all cysteines that were base-edited; 3) all essential cysteines (i.e., cysteines with editing-induced dropouts LFC<−0.6, FDR<10%); and 4) all ligandable cysteines using electrophilic fragments. c, Summary of editing-induced dropout values for cysteines showing substantially greater or lesser reactivity in native proteomes (designated as reactive and unreactive, respectively). Substantial reactivity changes were defined as cysteines showing denaturing/native LFC scores < −1.6 (reactive) or > 1.6 (unreactive). d, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for the edited C120 in RAN. e, Structure of RAN in complex with GNP (PDB: 5JLJ). The side chain of essential, unreactive (buried) cysteine C120 is highlighted in red. f, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in RBM22. g, Structure of RBM22 in complex with pre-mRNA as part of the human spliceosome (PDB: 5XJC). h, Density plot showing the association between predicted cysteine solvent accessibility based on AlphaFold2 protein structures and quantified cysteine reactivity values. A fitted smoothing spline is shown in red. Pearson correlation = −0.36 with two-sided p = 2.2e-16. i, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in UTP18. j, Structure of UTP18 as part of the human small subunit processome (PDB: 7MQ8). For b, c, d, f, h, i, the ABPP data of native versus denatured proteomes represent average values from 6 independent experiments. For c, d, f, i, the one-sided p values were first estimated by randomization tests of dropout data from two independent base-editing experiments and then adjusted by Benjamini-Hochberg procedure to calculate the FDR.
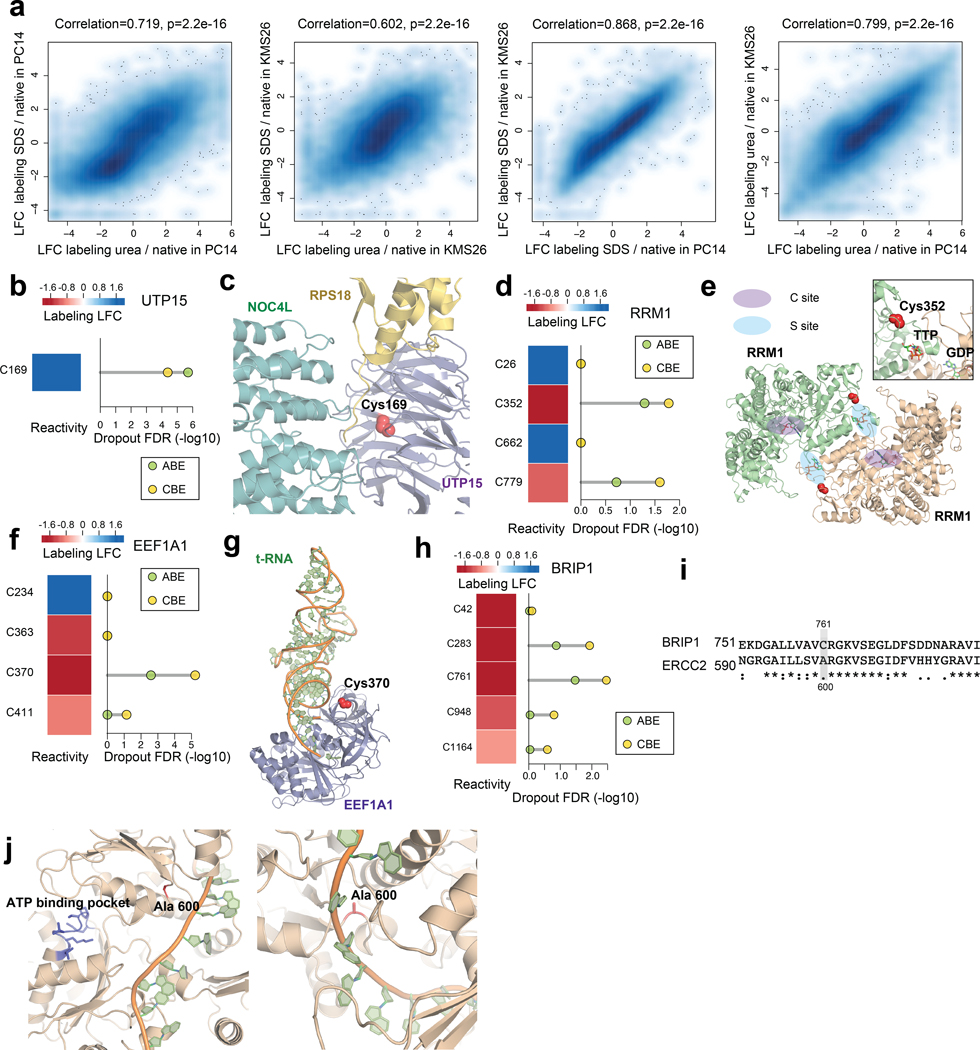
An initial evaluation of SDS- and urea-treated proteomes revealed that each denaturant induced similar cysteine reactivity changes in KMS26 and PC14 proteomes (Extended Data Fig. 5a), and we therefore combined data from both conditions for a more detailed comparison to native proteomes (Supplementary Dataset 5). Across ~29,000 quantified cysteines, ~20% and 22% showed substantial (LFC > 1.6) decreases or increases in IA-DTB reactivity in denatured proteomes, respectively (referred to hereafter as reactive and unreactive cysteines, respectively; Fig. 5b and Supplementary Dataset 5). A similar distribution of cysteine reactivity changes was observed for the ~5,000 quantified cysteines that had been edited in our dropout screens (Fig. 5b and Supplementary Dataset 5). In contrast, we found that a greater relative fraction of the ~600 essential cysteines quantified by ABPP were unreactive (Fig. 5b and Supplementary Dataset 5), indicating an enrichment in cysteines that were preferentially accessible to the IA-DTB probe in unfolded protein states. Consistent with previous studies7, a striking converse relationship was observed for the ~5,700 ligandable cysteines quantified by ABPP, which were strongly enriched in reactive cysteines with virtually no representation of unreactive cysteines (Fig. 5b and Supplementary Dataset 5).
Structural analysis of representative proteins with essential, unreactive cysteines (blue points, Fig. 5c) supported the buried location of these residues. For instance, an essential, unreactive cysteine C120 in the GTP-binding nucleocytoplasmic transport protein RAN has a solvent-accessible surface area in the folded protein structure of 0 Å2 (Fig. 5d, e). Likewise, an essential, unreactive cysteine C169 in the ribosome biogenesis factor UTP15 has its accessibility to solvent restricted by interactions with the associated proteins NOC4L and RPS18 (Extended Data Fig. 5b, c). Additional essential, unreactive cysteines were found to chelate metals, such as C27 of the splicing factor RBM22 (Fig. 5f, g). Finally, using AlphaFold2-generated structural models40, we predicted the solvent accessibility for ~ 20,000 cysteines quantified in our ABPP experiments (Supplementary Dataset 5), which revealed that unreactive cysteines were generally less accessible to solvent (Fig. 5h). These data, taken together, indicate that a substantial number of essential cysteines may lack evidence of ligandability because they serve structural roles at buried sites within proteins that have limited accessibility to small molecules.
Our analysis conversely revealed a set of 88 essential, reactive cysteines (red points, Fig. 5c; also see Supplementary Dataset 5). Considering that heightened reactivity in native proteomes was a feature strongly enriched in ligandable cysteines (Fig. 5b), we interpret essential cysteines showing this property as prioritized targets for the future pursuit of covalent probes. An initial analysis of these essential, reactive cysteines revealed several located at macromolecular interfaces, including, for instance, C352 in ribonucleotide reductase 1 (RRM1), which resides at the dimerization interface of this enzyme in proximity to an allosteric regulatory site where specificity effector nucleoside triphosphates bind to regulate RRM1 activity41 (Extended Data Fig. 5d, e). Other essential, reactive cysteines were located at protein-RNA/DNA interfaces, including C446 in processome protein UTP18 (Fig. 5i, j) and C370 in the elongation factor EEF1A1 (Extended Data Fig. 5f, g). Essential, reactive cysteines were also identified in Strongly Selective cancer dependencies, such as C761 in BRIP1 (Extended Data Fig. 5h), a DNA helicase that is required for DNA double-stranded break repair and the maintenance of chromosomal stability42. Based on a homology model generated from the related protein ERCC2, we localized C761 to the helicase domain at a site predicted to be proximal to BRIP1-DNA substrate interactions (Extended Data Fig. 5i, j).
Together, these data show how the integration of base editing data with global maps of cysteine reactivity in native and denatured proteomes, can illuminate solvent-accessible, essential cysteines as prioritized targets for future chemical probe development.
Discussion
Key to the implementation of a base editing strategy for assessing the functionality of ligandable cysteines was the knowledge afforded by – i) ABPP of site-level resolution of covalent compound-cysteine interactions across the human proteome7,11–13,43,44; and ii) the DepMap of protein-encoding genes required for cancer cell proliferation2 – which, together, enabled focused, base editing-mediated mutational screens of ligandable cysteines and surrounding regions to assess their essentiality for cell growth. Recently, an alternative homology-directed repair (HDR)-based oligo recombineering approach was described to assess the functionality of cysteines in Toxoplasma gondii45. This approach is well-suited for focused, array-based screens, particularly in haploid systems, but may lack the throughput and efficiency to perform large-scale analyses of cysteines in diploid mammalian cells. We chose base editing to install mutations because this technology can introduce nucleotide changes for residue-level functional assessment, while minimizing DNA-double-strand breaks and uncontrolled mixture of indels 23,24 that are observed with HDR CRISPR-Cas9 nuclease systems. Base editing has also been used to evaluate cancer-associated missense mutations emerging from clinical genomics data46,47, as well as to perform focused saturated scans to explore missense mutations that cause resistance to targeted therapeutics48 or alter enzymatic activities using a reporter readout49.
The discovery of essential, ligandable cysteines in proteins with Strongly Selective designations in the DepMap, such as CHUK, FOXA1, and TOE1, suggests that the development of more advanced covalent probes for these proteins might serve as starting points for targeted cancer therapies. Considering further that the ligandable cysteine in FOXA1 (C258) is proximal to cancer hotspot mutations33,34, we wonder whether it might be possible to create covalent probes that specifically target such pro-tumorigenic mutant forms of proteins, as has been done for EGFR18 and KRAS19,20. Our identification of a covalent inhibitor targeting C80 in TOE1 that strengthens binding of this protein to spliceosome subcomplexes36 highlights how integrated base-editing and cysteine-directed ABPP data can illuminate essential, ligandable sites for allosteric regulation of cancer dependency proteins.
The base editing of more than 1,100 ligandable cysteines in cancer dependency proteins did not affect cancer cell growth in our screens. One interpretation of these results is that ligands targeting these cysteines may not substantially affect protein function, which points to the potential of using the ligands as components of heterobifunctional small molecules to promote, for instance, protein degradation50. However, multiple technical caveats should also be considered. First, some covalent ligand-cysteine interactions may exert functional effects that qualitatively or quantitatively differ from those produced by genetic mutation of proteins. We attempted to account for this difference, at least in part, in our focused screen of ~35 ligandable cysteines in Strongly Selective proteins by using base editors with expanded targeting windows, allowing for mutation of additional residues that neighbor ligandable cysteines, which we anticipated would more fully assess the functionality of the local cysteine region (as we found to be the case for EGFR_C797 and XPO1_C528; Fig. 1e, g). Additionally, some ligandable cysteines that were designated as non-essential by base-editing screens may lack interpretability if insufficient gene editing of that cysteine region was technically achieved in our screens. Future genetic interrogation of cysteine essentiality could benefit from editors with improved efficiency that use, for instance, PAM-less Cas9 and methods that provide direct monitoring of editing efficiencies in parallel46,47.
Our base editing screens also identified a large number of essential cysteines for which ligands have not yet been discovered. Tracking the evolving landscape of cysteine ligandability is an important objective, and community-curated repositories of cysteine-directed ABPP data are under development43,44. Determining which essential, but not yet liganded, cysteines have the highest potential for targeting by covalent chemistry could also enable more focused screens compatible with larger compound libraries. We discovered that cysteines with established ligandability were much more likely to show decreased reactivity after denaturation, suggesting that essential cysteines displaying this property may have greater potential for targeting by covalent chemistry. We also speculate that the opposite profile – denaturation-induced increases in cysteine reactivity – may flag a category of residues for which essentiality reflects mutation-induced disruption of protein folding and/or expression.
In summary, by integrating base editing and chemical proteomic technologies, we have generated a rich resource of cysteines and cysteine regions that are essential for the growth of cancer cells alongside a status report on the current and future potential for targeting these cysteines with covalent small molecules. In the future, we envision performing integrated base-editing and chemical proteomic screens coupled to high-throughput readouts of other features of cell biology beyond proliferation and, through doing so, further enriching our understanding of functional cysteines that can be targeted by covalent chemistry.
Methods
Cell culture
Cancer cell lines used in this study include MM1S (ATCC, CRL-2974), UACC257 (NCI), KMS26 (JCRB, JCRB1187), KMS34 (JCRB, JCRB1195), 22Rv1 (ATCC, CRL-2505), SNU216 (KCLB, 00216), SUDHL5 (ATCC, CRL-2958), GSS (RIKEN, RCB2277), PANC1005 (ATCC, CRL-2547), PC9/14 (ECACC, 90071810), DLD1 (ATCC, CCL-221), MCC142 (ECACC, 10092303). Human cell lines were verified based on Short Tandem Repeat (STR) profiles by the providers. All cancer cell lines were grown in RPMI-1640 (Gibco) media using standard cell culture conditions (37°C, 5% CO2) and were free of microbial contamination including mycoplasma. All media were supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco), 10% FBS (Omega Scientific), and 2 mM GlutaMAX (Gibco).
Lentivirus particle production
The plasmids including all-in-one base editor vector, lentiviral packaging vector (pCMV-dR8.91) and envelope vector (VSV-G) were mixed at 9:6:1 ratio in OPTI-MEM media. 3ul of 1mg/ml PEI (Polysciences) was added per ug of total plasmids. After 20 min of incubation at room temperature, the mixture was dripped gently on 293T cells (ATCC) at 50% confluence. After 8 hrs, the media was changed to fresh DMEM (Corning) with 30% FBS plus pen-strep and 2mM GlutaMAX. The virus was collected both 2 days and 3 days later and 0.45 μm syringe filters (Millipore) were used to eliminate cells.
Arrayed base editing
On day 0, cells were seeded in a 96-well plate at a density of 0.5–1e4/well and were mixed with virus supernatant with 8 μg/ml polybrene (Millipore). Spin infection was performed (900g, 1 hr at 30C). On day 1, the virus containing media was removed, and fresh media was added. On day 2, puromycin was added to start the selection. The transduced cells were collected on day 5 after PBS wash once.
Targeted genomic sequencing
Genomic sites with base editing were amplified and indexed using a two-step PCR method as previously described with minor modifications51. Briefly, 100ul lysis buffer (10 mM Tris pH 7.5, 0.5% Tween 20, 0.02% SDS plus 20 ug/ml freshly added proteinase K) was added per 1e5 cells and the samples were incubated at 55°C for 2 hr before heat inactivation at 95°C for 30 min. Primers with illumina adapters were used for PCR1. Specifically, in each reaction, 5ul of genomic DNA extract, 12.5 ul Phusion PCR master mix (ThermoFisher), 1.25ul of 10uM forward and 1.25ul of 10uM reverse primers were added to a total volume of 25ul. The PCR1 reactions were carried out as following: 95°C for 3 min, 30 cycles of (20s at 95°C, 20s at 60°C, 25s at 72°C), followed by final extension at 72°C for 2 min. PCR1 products were cleaned using Ampure beads (Beckman) according to manufacturer’s instructions and were eluted in 50ul of 10mM Tris (pH=7.5). For each PCR2 reaction, 5 ul of PCR1 product, 12.5 ul Phusion PCR master mix (Thermo Scientific), 1.25ul of 10uM forward and 1.25ul of 10uM reverse index primers were added to a total volume of 25ul. The PCR2 reactions were carried out as following: 98°C for 3 min, then 12 cycles of (20s at 95°C, 20s at 60°C, 25s at 72°C), followed by final extension at 72°C for 2 min. The PCR2 products were then pooled and cleaned using Ampure beads. The library was quantified using PicoGreen dsDNA assay kits (ThermoFisher) and sequenced on an Illumina Miniseq instrument (GenerateFASTQ - 2.0.1) with 10% PhiX spike-in. Paired-end reads were demultiplexed based on combinatorial dual indexes (Supplementary Dataset 6). The genome editing quantification was performed using CRISPResso252. The parameters were set as following: CRISPResso --fastq_r1 {r1_file} --fastq_r2 {r2_file} --amplicon_seq {amplicon_sequence} -g {guide} -wc −14 -w 20 -q 30 --base_editor_output
Design of pooled base editing libraries
The dependency “CERES” scores were downloaded from the DepMap (21Q3, https://depmap.org/portal/). To design the libraries in this study, we focused on protein-coding transcripts annotated by the GENCODE database. For each protein, we selected the principal isoform based on the APPRIS database53. We further defined the approximate editing window center to be 15 nucleotides upstream of the NG PAM with 5nt width for ABE8e and 10nt width for evoCDA. The sgRNA sequences were selected if the editing window were predicted to create missense mutations to the target position or nearby residues if relevant. We then appended BsmBI sites to allow restriction digestion and PCR primer binding sites to allow oligo pool amplification. The final oligo structure is: 5′-(primer forward)CGTCTCACACCG(sgRNA, 20 nt)GTTTCGAGACG (primer reverse).
Cloning of pooled base editing libraries
All-in-one lenti-ABE8e-NG and lenti-evoCDA-NG vectors (Addgene #200447 and #200449) were designed based on the ABE8e/evoCDA editors and the lentiCRISPR v2 backbone (gifts from David Liu and Feng Zhang, Addgene #138491, #125613, and #52961) and were assembled using golden gate cloning (NEB). To clone sgRNA inserts, the plasmids were digested with BsmBI (NEB) following manufacturer’s recommendations and the backbone DNA with sticky ends were then separated in 1% agarose gel and purified for later use. The sgRNA oligo pool with restriction sites and flanking primer sequences was synthesized by Twist Bioscience and was amplified using Q5 polymerase (NEB). The PCR reaction was carried out as following: 98°C for 3 min, then 10 cycles of (20s at 95°C, 20s at 53°C, 20s at 72°C), followed by final extension at 72°C for 2 min. The PCR products were cleaned using DNA Clean & Concentrator (Zymo). The ligation product was assembled as following: 5 ng insert, 1 ug digested backbone, 1x Tango buffer, 1mM DTT, 1mM ATP, 1ul Esp3I (ThermoFisher), 1ul T7 ligase (Qiagen Beverly) in a total volume of 50ul. The ligation products were cleaned using isopropanol precipitation and were electroporated into electrocompetent cells (Thermo). After 1 hr incubation in recover media at 37°C, the bacteria cells were spread onto 145mm * 145mm plates for overnight culture at 30°C. For each sgRNA, at least 1,000 colonies were obtained to get sufficient library representation. After 20hr, the bacteria cells were scraped and centrifuged at 3,000g for 10 min and the plasmids were then extracted (Qiagen). For validation, selected individual sgRNA base editor constructs were cloned with reduced reagent input.
Cysteine reactivity profiling of native versus denatured proteomes evaluated by ABPP
10 million cells (PC14 or KMS26) were collected for each condition or replicate after three PBS washes. Each frozen cell pellet was mixed with 300 ul PBS and was sonicated with 3*8 pulses on ice. The protein was then normalized to 1 mg in a total of 500 ul volume with different treatments (8M urea at 65°C for 15 min, 1% SDS at 95 °C for 5 min, or 1% SDS+2 mM TCEP at 95°C for 5min). The native groups were mock treated with PBS and were kept on ice before further use. After equilibrating to room temperature, the samples were then treated with 5 ul 10 mM stock of IA-DTB (SCBT) and were incubated at room temperature for 1 hr. To precipitate proteins, 500ul cold methanol and 200ul cold chloroform was added to each tube. The samples were vortexed and then centrifuged at 16,000g for 30 min at 4°C. After removing the liquid phase, the protein disk was washed with cold methanol and was centrifuged again at 16,000g for 30 min at 4°C. The liquid phase was then aspirated, and the pellet is frozen at −80°C for later use. The subsequent sample processing and LC-MS instrumentation is the same as the previously described protocol for cysteine ligandability profiling12,22,39. Briefly, the protein pellets were reduced by DTT, alkylated by iodoacetamide (Sigma), and digested by trypsin (Promega) overnight. The peptides were then enriched using streptavidin beads (ThermoFisher), labeled with TMT tags (ThermoFisher), followed by desalting using Sep-Pak C18 cartridges (Waters). The peptides were fractionated using high-pH HPLC methods and were then analyzed in Orbitrap Fusion™ Mass Spectrometer with Xcalibur v4.3 (ThermoFisher).
Analysis of cysteine reactivity profiling data
The raw MS data files were uploaded to and converted by Integrated Proteomics Pipeline (IP2, v 6.7.1). The data files were then processed using the ProLuCID program based on a reverse concatenated, non-redundant version of the Human UniProt database (release 2016–07). Cysteine residues were searched with a static modification for carboxyamidomethylation (+57.02146 Da). N-termini and lysine residues were searched with a static modification corresponding to the TMT tag (+229.16293 Da). To search for the cysteine IA-DTB labeling, a dynamic modification (+398.25292 Da) was used. The census output files from IP2 were further processed by aggregating TMT reporter ion intensities to obtain signals based on unique peptides that are further annotated with protein-cysteine residue numbers. The resulting data were then median normalized per TMT channel and log2 fold changes between the native versus denatured conditions were calculated for each cysteine.
Analysis of cysteine solvent accessibility
The cysteine solvent accessibility data was extracted from the AlphaFold Protein Structure Database. The residue-level solvent accessibility scores were calculated using the DSSP function from BioPython (https://biopython.org/). Only cysteine residues with confidence scores pLDDT > 70 were used. A smoothing spline was fitted using the smooth.spline function in R with cross-validations (lambda=0.01).
Cysteine ligandability with electrophilic fragments evaluated by ABPP
The list of ligandable cysteines derived from previous reactive fragment electrophile profiling studies7,11,12 was used without change of analysis workflow or threshold. The additional cysteine-directed ABPP experiments performed in this study were based on the previously described protocol12. These data were generated by treating three different model cancer cell lines (Ramos, 22Rv1 and MCF7) with covalent ligands including 200uM KB02 and KB057 in situ for 1 hour. The unliganded cysteine were then labeled by IA-DTB, following by streptavidin enrichment, trypsin digestion and multiplexed proteomic quantification. We define a site to be ligandable if the fragment can engage > 50% of the IA-DTB-reactive cysteine in at least one of the model cell lines.
Screens of base-editing libraries
On day 0, cells were seeded in 12-well plates at a density of 0.5–1e6/well and were infected in virus supernatant containing 8 μg/ml polybrene. Spin infection was performed by centrifugation at 900g for 1 hour. One day after infection, 20% of cells were frozen as pellets for library normalization and the rest of cells were split to 15cm plates or T175 flasks. On day 2, puromycin was added and was maintained throughout the entire screen. About 30–50% infection rate was achieved in the screen to get an optimal multiplicity of infection (MOI). Each sgRNA was screened in at least 1000 cells and the cells were cultured for additional 14 days. Genomic DNA from the cell pellets was purified using the NucleoSpin blood L kits (Macherey-Nagel) according to the manufacturer’s instructions and was quantified using PicoGreen dsDNA assay kits (ThermoFisher).
The number of total PCR reactions performed for each cell line is calculated by sgRNA number in the library * 1000 cells /1e6 and was rounded to the next integer. The one-step PCR appended Illumina adapters and indexes using the P5 primer and the P7 primer described previously48 (Supplementary Dataset 6). For each targeted sgRNA cassette amplification PCR, 5ug of genomic DNA, 50ul Phusion PCR master mix (ThermoFisher), 5ul of 10uM forward and 5ul of 10uM reverse primers with illumina adaptors and indexes were added to a total volume of 100ul. The PCR reactions were carried out as following: 98°C for 3 min, repeated cycles of (20s at 98°C, 20s at 53°C, 20s at 72°C), followed by final extension at 72°C for 10 min. The PCR products were then pooled and cleaned using Ampure beads (Beckman) and were sequenced in an Illumina sequencer (HiSeq, NextSeq or NovaSeq) with 20% PhiX spike-in.
Analysis of pooled screen data
The frequency of sgRNA on day 1 and day 16 were counted using PoolQ (Broad Institute) and were normalized by total reads per sample to get counts per million (CPM). The significance of cysteine dropouts was calculated by comparing day 16 vs day 1 average log2 fold CPM changes of targeting sgRNA versus non-targeting sgRNA (null distribution). Here, the targeting sgRNAs include those that are predicted to mutate the cysteine of interest or nearby residues. Additionally, the sgRNAs that are predicted to introduce stop codons or make mutations near splicing sites were excluded from further analysis. The log2 fold CPM changes of non-targeting sgRNA were randomly resampled to get a null distribution based on the number of targeting sgRNA. The p value for each site was then estimated as the percentage of null observations that have greater average dropouts than the observed average dropout. The false discovery rate (FDR) was calculated using the Benjamini-Hochberg procedure for all edited cysteines in each protein.
For the cell panel screens, the sgRNA data was similarly counted and normalized. The day 16 vs day 1 dropouts were calculated for each sgRNA in each cell line. To select hit sgRNAs for each dependency, the base editing-induced dropout log2 fold CPM changes in the evaluated cell lines should correlate with the corresponding gene knockout-induced dropout scores (CERES 21Q3 release, https://depmap.org/portal/) with Pearson correlation greater than 0.5 and FDR < 10%.
Synthesis of WX-02–13 and WX-02–33
The synthesis and characterization of WX-02–13 and WX-02–33 have been reported previously22.
Immunoblot analysis
MCC142 cells stably expressing recombinant FLAG-WT or C80S-TOE1 were scraped from 6 well plates and were collected by centrifugation (400g, 5 min). After PBS washes, protein was extracted using RIPA lysis buffer before quantification using a BCA assay kit (Thermo). After mixing with 4X LDS sample buffer (Invitrogen), proteins were then resolved by SDS-PAGE, transferred to nitrocellulose membrane using a power blotter semi-dry transfer system (Thermo). The membrane was blocked with 5% milk in TBST buffer (20 mM Tris-HCl pH=7.5, 150 mM NaCl, 0.1% tween 20) for 1 hour at room temperature. The blot was then incubated with anti-FLAG HRP antibody (Sigma Cat# A8592, Clone# M2, 1:10,000 dilution) or antiGAPDH HRP antibody (Proteintech Cat# HRP-60004, Clone# 1E6D9, 1:10,000 dilution) at room temperature for 1hr. After three TBST washes, the blot was incubated with chemiluminescent HRP substrates (Thermo) and images were acquired on a Bio-Rad ChemiDoc imaging system.
TOE1 deadenylation assay
MCC142 cell line with stable expression of N-FLAG-tagged wild type TOE1 or C80S mutant was generated by spin-infection with packaged lentiviral particles. 10 million cells were treated with 20uM probes for 6 hours in situ and were frozen at −80°C after PBS washes. The collected cell pellets were lysed with 500 μl of isotonic lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μM aprotinin, 1 μM leupeptin) for 10 minutes on ice. Cell debris was removed by centrifugation at 20,000 g for 15 min at 4°C. The supernatant was subsequently supplemented with 125 μg/ml RNase A and incubated with 25 μl anti-Flag M2 agarose beads (Sigma) at 4°C, rotating overnight. Beads were washed eight times with Net-2 buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100) and Flag-tagged TOE1 protein was eluted with 150 μg/ml Flag peptide (ApexBio) in 100 μl Net-2 containing 10% glycerol for 30 minutes, rotating at 4°C. Eluates were analyzed by immunoblotting with anti-TOE1 antibody54 and protein concentration was measured by BCA assay following the manufacturer’s protocol (Thermo Scientific). Eluates were stored at −80 °C until further use.
In-vitro deadenylation assays were performed by incubating 200 nM fluorescein-labelled poly-A20 RNA substrate (Fl.C.U.U.U.C.C.C.C.U.G.A.A.A.A.A.A.A.A.A.A.A.A.A.A.A.A.A.A.A.A; Horizon Discovery Biosciences, PAGE purification) with 100 nM Flag-tagged TOE1-WT or TOE1-C80S in 10 μl of deadenylation buffer (20 mM HEPES-KOH pH 7.4, 2 mM MgCl2, 0.1 mg/ml bovine serum albumin, 1 mM spermidine, 0.1% NP-40, 0.5 U/μl RNaseOut, 5 μg/μl yeast total RNA) at 37°C for 0, 1, 2, or 3 hours. Deadenylation reactions were stopped by addition of 10 μl 2× loading buffer (95% formamide, 10 mM EDTA, 0.01% bromophenol blue). Reaction mixtures were heated at 80°C for 10 minutes and then separated in 20% polyacrylamide-TBE gels containing 6M urea. Fluorescein-labelled RNAs, 10, 20, and 30 nucleotides in length, were used as markers. Gels were imaged using a ChemiDoc Imaging System (Bio-Rad) and band intensities were quantified using GelAnalyzer 19.1.
TOE1 IP-MS studies
MCC142 cells with or without stable expression of N-FLAG-tagged wild type TOE1 or C80S mutant were treated in situ with probes or vehicle control for 6hrs. Cells were collected and washed with ice cold PBS. Frozen cell pellets were lysed in IP lysis buffer (50mM EPPS pH 7.5, 150mM NaCl, 1% Triton X100, 10% glycerol, 1 mM MgCl2) with cOmplete Protease Inhibitor Cocktail (Roche) and benzonase (40U/ml), and were sonicated with a probe sonicator (2*15 pulses). After incubation at room temperature for 20 min, the insoluble debris was removed by centrifuging at 16,000 g for 5 min. The protein concentration in the supernatant was quantified using BCA assay (Pierce). 2mg protein was taken from each sample and was mixed with 40ul anti-Flag M2 agarose slurry (Sigma) pre-washed using the lysis buffer, followed by rotation at 4°C for 3hrs. After incubation, the beads were then spun down, washed with IP wash buffer (50mM EPPS pH 7.5, 150mM NaCl, 0.2% Triton X100) three times (2000g, 1min) and then twice with 50mM EPPS pH 7.5 (2000g, 1min). The proteins were eluted using 50ul 8M urea in EPPS at 65 °C for 10 min and then reduced with 200 mM DTT at 65 °C for 15 min, alkylated with 400 mM iodoacetamide at 37 °C for 30 min, and diluted to 2 M urea by addition of 50mM EPPS pH 7.5. The proteins were then digested using MS-grade trypsin (Promega) at 37 °C overnight, followed by TMT labeling (Thermo Scientific) and desalting using Sep-Pak C18 cartridges (Waters) as described previously12,22,39. The labeled peptides were analyzed in Orbitrap Fusion™ Mass Spectrometer (ThermoFisher).
Conservation score analysis
The available protein ortholog sequences from a panel species were downloaded from Ensembl using REST API (https://rest.ensembl.org/, 2023–04). The multiple sequence alignment algorithm ClustalOmega in the msa package (1.24.0) was used to align the obtained protein sequences and conservation scores were then calculated using the BLOSUM62 matrix. The panel of species include Anolis carolinensis, Bos taurus, Caenorhabditis elegans, Canis lupus familiaris, Danio rerio, Drosophila melanogaster, Equus caballus, Felis catus, Gallus gallus, Macaca mulatta, Monodelphis domestica, Mus musculus, Ornithorhynchus anatinus, Pan troglodytes, Rattus norvegicus, Sus scrofa, Xenopus tropicalis.
Statistics and Reproducibility
Statistical analyses in this paper were performed using R (v4.1.1) and Python (v3.7.4). Data visualization was done in R and Prism 9 (GraphPad). To compare the means between two groups of data points, two-sided Student’s t test was used to calculate the p values. Two-sided statistical tests were performed unless stated otherwise. The Benjamini–Hochberg procedure was used to adjust multiple hypothesis testing when applicable.
Extended Data
Extended Data Fig. 1 |. Base editing assigns essentiality to cysteines targeted by anti-cancer drugs.
a, b, Heatmap showing the mutagenesis effects of Adenine Base Editors (ABE) (a) and Cytosine Base Editors (CBE) (b) on different amino acid residues. The total percentage is normalized to 100% per row. c, Schematic showing the design of lentivirus vectors delivering base editor libraries used in this study. CDA: cytidine deaminase; TadA: deoxyadenosine deaminase; UGI: uracil glycosylase inhibitor; PuroR: puromycin resistance gene. d, Waterfall plots for ABE (left) and CBE (right) libraries showing the dropout significance calculated per EGFR residue between day 16 vs day 1 from saturated scanning experiments using PC14 cells (cutoff: LFC < −0.5; p < 0.05). Data represent average values of two independent experiments. One-sided p values were calculated using randomization tests (no multiple comparison adjustments, see source code in Github).
Extended Data Fig. 2 |. Global discovery of essential cysteines in cancer dependency proteins.
a, Scatter plot showing the dependencies included in the base-editing screens. b, Barplots summarizing the number of designed sgRNAs per cysteine. (Left) all or (right) ligandable cysteines in the dependency proteins. c, Plot comparing the essential cysteine hit rate using sgRNA targeting cysteines in dependency proteins or non-targeting sgRNA data randomly resampled from the library 10 times. The p value was calculated using two-sided Student’s t test. d, Scatter plot showing the correlation between cysteine dropouts observed with the ABE or the CBE library for Common Essential proteins in PC14 and KMS26. The Pearson correlation and two-sided p value is shown. e, f, Density scatter plots comparing the evolutionary conservation and significance of dropout of cysteines using the ABE (e) or CBE (f). Pearson correlations and two-sided p values are shown. g, Barplot showing the distribution of missense mutations for essential cysteines in orthologous proteins. h, i, Scatter plots comparing the day 16 vs day 1 LFC values of cysteines in Common Essential dependencies in KMS26 versus PC14 cells. The essential cysteines are highlighted for (h) ABE and (i) CBE libraries. j, k, Dot plots comparing the dropouts of cysteines from Strongly Selective dependencies using (j) PC14 or (k) KMS26 cells. Each pair of points compares the same cysteine. The cysteines were included based on the first essentiality filter (FDR < 10%, LFC < −0.6) only using data from one cell line without considering the dependency difference between the two cell lines. A second selectivity filter (greater dropout in the more dependent cell line, LFC difference > 0.3) was applied, and cysteines passing this filter are shown in red. Only Strongly Selective proteins with gene-level dependency scores that are sufficiently different (CERES difference > 0.8) between PC14 and KMS26 were included. The p values were calculated using two-sided paired Student’s t test based on all cysteines that pass the first filter. For c, d, e, f, h, i, j, k, the base-editing dropout screen data represent two independent experiments in PC14 and KMS26.
Extended Data Fig. 3 |. Base editing reveals essentiality of ligandable cysteine regions in Strongly Selective cancer dependency proteins.
a, b, Histogram showing the nucleotide substitution efficiencies for 70 sgRNAs using (a) ABE8e-NG or (b) evoCDA-NG in PC14 cells. The relative residue mutation frequency five days after lentivirus infection was quantified using targeted genomic PCR and amplicon sequencing. The percentage of gene editing was estimated by the ratio of reads that contain nucleotide substitutions over total aligned reads. Data represent average values of two independent experiments. c, Scatter plots for the EGFR_C797 region showing the correlation between base editing dropout data acquired herein and gene disruption dropout data (DepMap). The two independent experiments are shown in blue and red. Pearson correlations and associated two-sided p values are shown. d, Bar graph comparing the editing efficiencies among the sgRNAs that did or did not create significant base editing-knockout correlations. Each point represents a different sgRNA designed for FOXA1 or TOE1. Data represent average values of two independent experiments. The p value was calculated using two-sided Student’s t test. e, The fragment electrophile ligandability profile for cysteines in JAK1, showing ligandability of C817. Note that C810 and C817 on the same tryptic peptide, so their ligandability cannot be distinguished by cysteine-directed MS-ABPP. However, other studies have verified that C817 is the liganded cysteine in JAK121. f, Scatter plots for the JAK1_C817 region showing the correlation between base editing dropout data acquired herein and gene disruption dropout data (DepMap). The two independent experiments are shown in blue and red. Pearson correlations and associated two-sided p values are shown. g, The fragment electrophile ligandability profile for cysteines in FOXA1, showing ligandability of C258.
Extended Data Fig. 4 |. Functional characterization of covalent ligands targeting an essential cysteine C80 in TOE1.
a, The fragment electrophile ligandability profile for cysteines in TOE1, showing ligandability of C80. b, Quantification of TOE1_C80 engagement by the indicated compounds as measured by cysteine-directed ABPP performed in Ramos cells (20 μM, 3 h). Data are average values ± SEM normalized to DMSO from n = 2 independent experiments. c, Western blot showing recombinantly expressed FLAG-tagged TOE1 in MCC142 cells. GAPDH was used as a loading control. This was repeated in 3 independent experiments with similar results. d, Schematic summarizing the TOE1 deadenylation assay workflow. e, Alignment of TOE1 orthologous proteins in a panel of selected vertebrate species. Only the aligned sequences near human TOE1_C80 are shown. f, Comparison of deadenylase activity of recombinant FLAG-WT- and C80S-TOE1 proteins immunoprecipitated from MCC142 cells. Data represent two independent experiments. g, Quantification of time-dependent TOE1 deadenylation activity as shown in Fig. 4h. Data represent 2 independent experiments. h, Quantification of TOE1_C80 engagement by the indicated compounds (20 μM, 6 h) as measured by monitoring the C80-containing tryptic peptide in IP-MS experiments from FLAG-WT-TOE1-expressing MCC142 cells. Data are average values ± SEM normalized to DMSO from n = 2 independent experiments. i, Schematic for growth competition assay. Briefly, the TOE1-dependent cell line MCC142 was spin-infected with lentiviral vectors expressing codon-optimized FLAG-WT- or C80S-TOE1. After antibiotic (G418) selection, cells were mixed at a 1:1 ratio and treated with WX-02–33 (10 μM) or DMSO and grown for 8 days, after which genomic DNA was extracted and targeted amplicon sequencing was used to quantify the relative frequency of C80S vs WT. j, Relative allele frequency of MCC142 cells expressing WT- or C80S-TOE1 treated with 10 μM WX-02–33 or DMSO. The p value was calculated using two-sided Student’s t test based on n = 3 independent experiments.
Extended Data Fig. 5 |. Prioritizing essential cysteines with ligandability potential by quantitative cysteine reactivity profiling in native and denatured proteomes.
a, (from left to right) Density scatter plots comparing the effect of i) denaturants urea vs SDS on IA-DTB labeling in PC14 cells (left) and KMS26 cells (left middle); and ii) SDS (right middle) or urea (right) on IA-DTB labeling in PC14 vs KMS26 cells. Each point represents a quantified IA-DTB labeled cysteine. Pearson correlations and associated two-sided P values were calculated based on data from 2 biologically independent samples. b, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in UTP15. c, Structure of UTP15 in complex with NOC4L and RPS18 as part of the human small subunit processome (PDB: 7MQ8). The side chain of essential, unreactive (buried) C169 is highlighted in red. d, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in RRM1. e, Structure of RRM1 bound to effector metabolite TTP at the specificity site (S site) and substrate GDP at the catalytic site (C site) (PDB: 3HND). f, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in EEF1A1. g, Structure of EEF1A1 in complex with tRNA as part of the human ribosome complex (PDB: 6ZMO). h, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in BRIP1. i, Alignment of BRIP1 and ERCC2 protein sequences. j, BRIP1 homology model based on the crystal structure of ERCC2 in complex with DNA (PDB: 6RO4). The BRIP1_C761 corresponding residue ERCC2_A600 and the ATP-binding pocket is highlighted. For b, d, f, h, the ABPP data of native versus denatured proteomes represent average values from 6 independent experiments. For b, d, f, h, the one-sided p values were first estimated by randomization tests of dropout data from two independent base-editing experiments and then adjusted by Benjamini-Hochberg procedure to calculate the FDR.
Supplementary Material
Acknowledgements
We thank J. Doench (Broad institute) and J. Luo (NIH) for helpful discussions regarding CRISPR library cloning. This work was supported by the NIH (R35 CA CA231991 awarded to B.F.C., U01 AI142756 awarded to D.R.L., RM1 HG009490 awarded to D.R.L., R35 GM118062 awarded to D.R.L., R35 GM118069 awarded to J.L.), the Damon Runyon Cancer Research Foundation (DRG: 2406-20 awarded to H.L.), Jane Coffin Childs Fund (awarded to K.E.D)., the Mark Foundation for Cancer Research (H.L.), and the Howard Hughes Medical Institute (D.R.L.).
Footnotes
Competing Interest Statement
B.F.C. is a founder and scientific advisor to Vividion Therapeutics. D.R.L. is a consultant and/or equity owner for Prime Medicine, Beam Therapeutics, Pairwise Plants, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome editing or epigenome engineering agents. The other authors declare no competing interests.
Code availability
Custom code used in the analysis is available on github (https://github.com/cravattlab/Cys_editing).
Data availability
Proteomics data have been deposited to the ProteomeXchange Consortium (PXD038232, PXD038239 and PXD041314). Sequencing data have been deposited in the NCBI Sequence Read Archive (PRJNA905477). Processed screen data and proteomics data are provided as Supplementary Dataset. Source data are provided with this paper. Databases used in this study include the DepMap (https://depmap.org/portal/, 21Q3), UniProt (https://www.uniprot.org/, release 2016-07), AlphaFold (https://alphafold.ebi.ac.uk/, 2022), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, version 2023-04), GENCODE (http://www.gencodegenes.org/, 2020), Ensembl (https://rest.ensembl.org/, 2023-04), and APPRIS (https://appris.bioinfo.cnio.es/#/, 2020).
References
- 1.Schreiber SL et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell 161, 1252–1265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsherniak A. et al. Defining a cancer dependency map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghandi M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schreiber SL A chemical biology view of bioactive small molecules and a binder-based approach to connect biology to precision medicines. Isr J Chem 59, 52–59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott DE, Coyne AG, Hudson SA & Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry 51, 4990–5003 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Brenner S. & Lerner RA Encoded combinatorial chemistry. Proceedings of the National Academy of Sciences 89, 5381–5383 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backus KM et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weerapana E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arkin MR & Wells JA Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat Rev Drug Discov 3, 301–317 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Wakefield AE, Kozakov D. & Vajda S. Mapping the binding sites of challenging drug targets. Curr Opin Struct Biol 75, 102396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-Peled L. et al. Chemical proteomics identifies druggable vulnerabilities in a genetically defined cancer. Cell 171, 696–709.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinogradova EV et al. An activity-guided map of electrophile-cysteine interactions in primary human T cells. Cell 182, 1009–1026.e29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuljanin M. et al. Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat Biotechnol 39, 630–641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurais AJ & Weerapana E. Reactive-cysteine profiling for drug discovery. Curr Opin Chem Biol 50, 29–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbasov ME et al. A proteome-wide atlas of lysine-reactive chemistry. Nat Chem 13, 1081–1092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spradlin JN, Zhang E. & Nomura DK Reimagining druggability using chemoproteomic platforms. Acc Chem Res 54, 1801–1813 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Lu W. et al. Fragment-based covalent ligand discovery. RSC Chem Biol 2, 354–367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross DAE et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 4, 1046–1061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrem JM, Peters U, Sos ML, Wells JA & Shokat KM K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanman BA et al. Discovery of a covalent inhibitor of KRAS G12C (AMG 510) for the treatment of solid tumors. J Med Chem 63, 52–65 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Kavanagh ME et al. Selective inhibitors of JAK1 targeting an isoform-restricted allosteric cysteine. Nat Chem Biol 18, 1388–1398 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldman HC et al. Selective inhibitors of SARM1 targeting an allosteric cysteine in the autoregulatory ARM domain. Proceedings of the National Academy of Sciences 119, e2208457119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landrum MJ et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42, D980–D985 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimasu H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science (1979) 361, 1259–1262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter MF et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol 38, 883–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thuronyi BW et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol 37, 1070–1079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogl DT et al. Selective inhibition of nuclear export with oral Selinexor for treatment of relapsed or refractory multiple myeloma. Journal of Clinical Oncology 36, 859–866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang F. et al. Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells. J Med Chem 56, 3996–4016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark KL, Halay ED, Lai E. & Burley SK Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Parolia A. et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 571, 413–418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams EJ et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 571, 408–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arruabarrena-Aristorena A. et al. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell 38, 534–550.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lardelli RM et al. Biallelic mutations in the 3′ exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat Genet 49, 457–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lardelli RM & Lykke-Andersen J. Competition between maturation and degradation drives human snRNA 3′ end quality control. Genes Dev 34, 989–1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Son A, Park J-E & Kim VN PARN and TOE1 constitute a 3′ end maturation module for nuclear non-coding RNAs. Cell Rep 23, 888–898 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Lazear MR et al. Proteomic discovery of chemical probes that perturb protein complexes in human cells. Mol Cell 83, 1725–1742.e12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairman JW et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol 18, 316–322 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Litman R. et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8, 255–265 (2005). [DOI] [PubMed] [Google Scholar]
- 43.White MEH, Gil J. & Tate EW Proteome-wide structural analysis identifies warhead- and coverage-specific biases in cysteine-focused chemoproteomics. Cell Chem Biol 30, 828–838.e4 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Boatner LM, Palafox MF, Schweppe DK & Backus KM CysDB: a human cysteine database based on experimental quantitative chemoproteomics. Cell Chem Biol 30, 683–698.e3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benns HJ et al. CRISPR-based oligo recombineering prioritizes apicomplexan cysteines for drug discovery. Nat Microbiol 7, 1891–1905 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sánchez-Rivera FJ et al. Base editing sensor libraries for high-throughput engineering and functional analysis of cancer-associated single nucleotide variants. Nat Biotechnol 40, 862–873 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y. et al. High-throughput functional evaluation of human cancer-associated mutations using base editors. Nat Biotechnol 40, 874–884 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanna RE et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e20 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Lue NZ et al. Base editor scanning charts the DNMT3A activity landscape. Nat Chem Biol 19, 176–186 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Békés M, Langley DR & Crews CM PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov 21, 181–200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang TP, Newby GA & Liu DR Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc 16, 1089–1128 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Clement K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez JM et al. APPRIS 2017: principal isoforms for multiple gene sets. Nucleic Acids Res 46, D213–D217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner E, Clement SL & Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear Cajal bodies. Mol Cell Biol 27, 1686–1695 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited to the ProteomeXchange Consortium (PXD038232, PXD038239 and PXD041314). Sequencing data have been deposited in the NCBI Sequence Read Archive (PRJNA905477). Processed screen data and proteomics data are provided as Supplementary Dataset. Source data are provided with this paper. Databases used in this study include the DepMap (https://depmap.org/portal/, 21Q3), UniProt (https://www.uniprot.org/, release 2016-07), AlphaFold (https://alphafold.ebi.ac.uk/, 2022), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, version 2023-04), GENCODE (http://www.gencodegenes.org/, 2020), Ensembl (https://rest.ensembl.org/, 2023-04), and APPRIS (https://appris.bioinfo.cnio.es/#/, 2020).