Abstract
In vitro, the methyl-directed mismatch repair system of Escherichia coli requires the single-strand exonuclease activity of either ExoI, ExoVII, or RecJ and possibly a fourth, unknown single-strand exonuclease. We have created the first precise null mutations in genes encoding ExoI and ExoVII and find that cells lacking these nucleases and RecJ perform mismatch repair in vivo normally such that triple-null mutants display normal mutation rates. ExoI, ExoVII, and RecJ are either redundant with another function(s) or are unnecessary for mismatch repair in vivo.
The methyl-directed mismatch repair (MMR) system of Escherichia coli is a key enforcer of genetic stability. The MMR system corrects DNA polymerase errors (reviewed in reference 22) and prevents the recombination of partially diverged DNA sequences (18, 19, 26, 39). E. coli strains lacking any essential component of this system display a mutator phenotype in which mutation rates are 100- to 1,000-fold above normal (22, 30) and are better able to recombine partially diverged DNA sequences (18, 26, 39). Both the elevated mutation rate and the relaxed sequence stringency of recombination of mutator strains may contribute to the pathogenicity of E. coli (14, 17). Homologs of the E. coli MutS and MutL MMR proteins have been identified in yeasts, mice, and humans and, as predicted from studies with E. coli, their absence results in increased mutation, genome instability, and, in mammals, cancer (reviewed in references 12, 23, and 24).
The molecular mechanism of methyl-directed MMR in E. coli, as defined biochemically, includes the following steps (reviewed in references 15, 22, 23, and 28). Repair is initiated by the binding of MutS to the mismatch, of MutL to MutS, and of MutH to a nearby d(GATC) sequence. An incision is made by MutH 5′ to the d(GATC) sequence on an unmethylated DNA strand. The nicked DNA strand is displaced by the coordinated activities of MutS, MutL, and MutU (helicase II) and is degraded by exonucleases specific for single-strand DNA. The exonuclease required depends on the position of the incision relative to the mismatch: if the incision is located 3′ of the mismatch, repair requires the 3′ to 5′ exonucleolytic activity of exonuclease I (ExoI) in a purified system and/or that of an unidentified component in crude extracts (7); if the incision is located 5′ of the mismatch, repair requires the 5′ to 3′ exonucleolytic activity of either RecJ or exonuclease VII (ExoVII). The final steps in MMR require the activities of a single-strand DNA-binding protein, DNA polymerase III, and DNA ligase for filling the single-strand gap left by excision.
Of the components required biochemically, it is clear that MutS, MutL, MutH, and MutU are also needed to perform their respective functions in MMR in vivo. Null mutations in the genes encoding any one of these disable MMR, resulting in cells displaying a mutator phenotype (30–32) and decreased recombination sequence stringency (18, 26, 39). For the single-strand exonucleases, their roles in vivo have been less obvious, in part because of the absence of precise null alleles of the genes encoding them. Razavy et al. (27) constructed the first known precise null allele of the gene encoding ExoI in E. coli, a deletion-insertion allele called ΔxonA300::cat. Previous E. coli alleles either are altered-function and dominant alleles (e.g., sbcB15) (27), have not been demonstrated to be null alleles (e.g., xonA2 and xonA6) (25), or remove a large segment of the E. coli chromosome such that phenotypes cannot be attributed unambiguously to the lack of the ExoI-encoding gene [Δ(sbcB-his); cited in reference 7]. Similarly, for ExoVII, the only previously known null alleles remove not only the xseA gene encoding the enzyme’s large subunit but also a neighboring guanosine biosynthesis gene causing a guanosine requirement (37). This could alter normal DNA metabolism, which could be relevant to DNA MMR. A useful recJ-null allele has been described (16). Here we report the construction of the first precise null allele of ExoVII, ΔxseA18::amp (Table 1), and the first strains carrying precise null mutations in genes encoding all three known E. coli single-strand-dependent exonucleases. These strains are described and used to examine the role of the exonucleases in MMR in vivo.
TABLE 1.
E. coli K-12 strains and plasmids
| Strain or plasmid | Relevant genotype or characteristics | Reference, source, or construction |
|---|---|---|
| E. coli strains | ||
| AB1157 | F−thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 | 1 |
| JC11450 | AB1157, spontaneous Su− | A. J. Clark |
| FC40 | ara Δ(lac-proAB)XIIIthi Rifr [F′ proAB+ lacI33ΩlacZ] | 3 |
| CAG12176 | zef-3189::Tn10kan | 33 |
| CAG18604 | zgf-3156::Tn10kan | 33 |
| STL160 | AB1157 Δ(xseA-guaB) zfh-3139::Tn10kan | S. T. Lovett |
| SMR91 | mutL211::Tn5 | Laboratory collection |
| SMR423 | recD1903::Tn10 | Laboratory collection |
| SMR690 | FC40 recJ284::Tn10 | 10 |
| SMR838 | JC11450 ΔxonA300::cat his+ | Laboratory collection |
| SMR839 | JC11450 ΔxonA300::cat | 27 |
| SMR1403 | JC11450 ΔxonA300::cat recJ284::Tn10 | 27 |
| SMR2597 | FC40 Δ(xseA-guaB) zfh-3139::Tn10kan | Laboratory collection |
| SMR3070 | FC40 ΔxonA300::cat | FC40 × P1(SMR839) |
| SMR3404 | FC40 mutL211::Tn5 | FC40 × P1(SMR91) |
| SMR3465 | recD1903::Tn10 ΔxseA18::amp | This studya |
| SMR3472 | FC40 ΔxseA18::amp | SMR2597 × P1(SMR3465)b |
| SMR3481 | FC40 ΔxonA300::cat recJ284::Tn10 ΔxseA18::amp | FC40 × P1(SMR838), P1(SMR690), P1(SMR3472)c |
| SMR3488 | JC11450 ΔxonA300::cat recJ284::Tn10 ΔxseA18::amp | SMR1403 × P1(SMR3472)c |
| SMR3524 | JC11450 mutL211::Tn5 | JC11450 × P1 (SMR3404) |
| SMR4035 | FC40 ΔxonA300::cat zef-3189::Tn10kan | SMR3070 × P1(CAG12176) |
| SMR4036 | FC40 recJ284::Tn10 zgf-3156::Tn10kan | SMR690 × P1(CAG18604) |
| SMR4037 | FC40 ΔxseA18::amp zfh-3139::Tn10kan | SMR3472 × P1(STL160) |
| Plasmids | ||
| pMJ3 | pACYC184 derivative containing xseA and guaBA | This studyd |
| pMJ6 | pMJ3 derivative containing ΔxseA18::amp | This studye |
Constructed by transforming SMR423 with the 5-kb AvaI-BamHI fragment of pMJ6 which contains ΔxseA18::amp and selecting ampicillin-resistant transformants (29).
This transduction confirmed the chromosomal location of ΔxseA18::amp, as all Ampr transductants were Gua+ and the expected linkage to zfh-3139::Tn10kan (4) was observed.
The presence of the null alleles in the triple mutants was confirmed by P1 transduction of each mutation into genetic backgrounds in which the following characteristic phenotypes were observed: recJ284::Tn10 makes recB21 recC22 sbcB15 sbcC201 strains extremely UV sensitive (16); xonA-null mutations decrease transductional recombination via the RecF pathway (2); and xseA mutations enhance sensitivity to low concentrations of nalidixic acid (4). The triple mutants also display UV light sensitivity (discussed in the text), as expected from their reduced-recombination phenotype (27).
A 5-kb BglI-BamHI fragment from Kohara phage λ427 (λ8E3) (11) containing xseA and guaBA was ligated into BglI-BamHI-digested pACYC184 (see, e.g., reference 21). BglI 3′ overhangs were removed with T4 DNA polymerase (exonuclease activity) prior to ligation.
The 690-bp EagI-AflII fragment of xseA (6) (GenBank accession no., J02599) was replaced with an EagI-AflII-digested 1,035-bp PCR fragment containing the bla gene of pBR322 (see, e.g., reference 21). bla was amplified with primers that create EagI and AflII sites (EagI primer: 5′GTACGGCCGAGTAAACTTGGTCTGACA; AflII primer: 5′ATGCTTAAGTAGACGTCAGGTGGCACT).
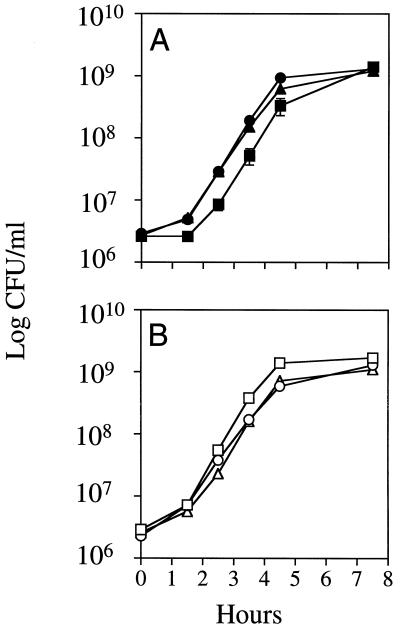
The viabilities of strains of two different genetic backgrounds carrying the null alleles of xonA, xseA, and recJ (Table 1) are normal (Table 2), and their growth curves are not markedly different from those of their Exo+ parents (Fig. 1). It was shown previously that strains defective for all three exonucleases display decreased homologous recombination and Chi activity when Chi stimulates recombination opposite heterologous DNA (27). The latter phenotype was observed only in the triple mutant and not in any of the double-exonuclease mutants. As predicted on the basis of their recombination-depressed phenotypes, our strains carrying the null alleles of xonA, xseA, and recJ also display elevated UV light sensitivity. In both genetic backgrounds, their sensitivities are greater than that of any of the single mutants and less than those of recA recombination-deficient strains (data not shown).
TABLE 2.
Viabilities of single-strand exonuclease-deficient strains
| Strain | Relevant genotype | CFU/ml (109)a | No. of observed cells/ml (109)b | Viability (CFU/cell counted) |
|---|---|---|---|---|
| JC11450 | xonA+ xseA+ recJ+ F− | 2.6 ± 0.13 | 2.9 ± 0.26 | 0.9 |
| SMR3488 | JC11450 ΔxonA ΔxseA recJ | 2.7 ± 0.14 | 2.1 ± 0.28 | 1.3 |
| SMR3524 | JC11450 mutL | 2.9 ± 0.11 | 2.4 ± 0.75 | 1.2 |
| FC40 | xonA+ xseA+ recJ+ F′ | 2.9 ± 0.09 | 2.8 ± 0.31 | 1.0 |
| SMR3481 | FC40 ΔxonA ΔxseA recJ | 2.6 ± 0.18 | 2.3 ± 0.23 | 1.1 |
| SMR3404 | FC40 mutL | 2.3 ± 0.07 | 1.2 ± 0.17 | 1.9 |
Mean number of CFU per milliliter ± one standard error of the mean for five independent cultures grown to saturation in Luria-Bertani-Herskowitz (LBH) broth (see, e.g., reference 36).
Mean number of cells per milliliter ± one standard error of the mean in the same five saturated cultures determined with a Petroff-Hausser counter.
FIG. 1.
Growth curves of strains carrying null mutations in xonA, xseA, and recJ. Each point represents the mean of the viable cell counts from three independent cultures. Error bars, one standard error of the mean. (A) JC11450 derivatives; (B) FC40 derivatives. Squares, Exo+ strains; triangles, ΔxonA300::cat ΔxseA18::amp recJ284::Tn10 strains; circles, mutL strains.
Studies of MMR in vitro were inconclusive as to whether the absence of the three exonucleases should be sufficient to block MMR in cells. On the one hand, in a purified system, the presence of ExoVII or RecJ was required for MMR with a d(GATC) sequence 5′ to the mismatch and ExoI was required for repair with the d(GATC) sequence 3′ to the mismatch (7). ExoVII could not substitute for ExoI on 3′ substrates in the purified system (7), despite the fact that ExoVII has been found to possess both 3′ and 5′ single-strand nuclease activity in other in vitro assays (5). On the other hand, in a crude system, MMR of 3′ substrates occurred in extracts of cells lacking ExoI, leading the authors to suggest the possible existence of a fourth single-strand exonuclease in E. coli which substitutes for ExoI in their crude system (7). However, ExoVII was present in the crude extract lacking ExoI. Because the ability of ExoVII to digest 3′ ends in the crude system is unknown, it remains possible that ExoVII was supporting the repair of 3′ substrates.
In vivo studies in which recombination was assayed suggested links between the single-strand exonucleases and MMR (8, 9). The authors described MMR protein-dependent recombination of UV-irradiated DNA. They found that single- and double-exonuclease mutants display small decreases in the frequency of such recombination, indicating roles for these nucleases in either MMR, recombination, or both (8). Because single- and double-exonuclease mutants have since been shown to have similarly small decreases in their frequencies of normal (MMR-independent) recombination (20, 27), it now seems likely that recombination rather than MMR was inhibited by exonuclease deficiency in the previous study (8). The observation of single-strand DNA formation in nuclease-proficient cells, but not in cells deficient for one of the single-strand exonucleases (8), similarly cannot distinguish whether such single-strand DNA was an intermediate in MMR or recombination or in both processes or neither process.
To test whether single-strand exonucleases are required for MMR in vivo, we asked whether cells lacking ExoI, ExoVII, and RecJ display the mutator phenotype characteristic of MMR-deficient cells. For two separate E. coli K-12 strain backgrounds, we found that cells lacking ExoI, ExoVII, and RecJ displayed mutation rates similar to those of their xonA+ xseA+ recJ+ parents (Table 3). In contrast, isogenic strains lacking MutL, an essential component of MMR in vivo, showed greatly elevated mutation rates. Thus, the activities of ExoI, ExoVII, and RecJ appear not to be essential for MMR in vivo.
TABLE 3.
Mutation ratesa
| Strain | Genetic background | Relevant genotype | Expt no. | Rate of mutationb (10−10) phenotype indicated
|
||
|---|---|---|---|---|---|---|
| Nalr | Strr | Arg+ | ||||
| JC11450 | JC11450 | xonA+ xseA+ recJ+ mutL+ | 1 | 1.5 | NAc | 12 |
| 2 | 1.2 | NA | 11 | |||
| 3 | 3.7 | NA | 45 | |||
| SMR3488 | JC11450 | ΔxonA300::cat recJ284::Tn10 ΔxseA18::amp | 1 | <1.6d | NA | 44 |
| 2 | <1.2d | NA | 36 | |||
| 3 | 3.0 | NA | 59 | |||
| SMR3524 | JC11450 | mutL211::Tn5 | 1 | 580 | NA | 740 |
| 2 | 700 | NA | 430 | |||
| 3 | 700 | NA | 220 | |||
| FC40 | FC40 | xonA+ xseA+ recJ+ mutL+ | 3 | 5.0 | 1.3 | NA |
| 4 | 4.7 | 0.83 | NA | |||
| 5 | 3.6 | 0.82 | NA | |||
| SMR3481 | FC40 | ΔxonA300::cat recJ284::Tn10 ΔxseA18::amp | 3 | 2.4 | 0.74 | NA |
| 4 | <2.9d | <1.0d | NA | |||
| 5 | <3.8d | <1.1d | NA | |||
| SMR3404 | FC40 | mutL211::Tn5 | 3 | 1,100 | 91 | NA |
| 4 | 1,000 | 40 | NA | |||
| 5 | 1,200 | 53 | NA | |||
For each strain and experiment, the number of cultures was 25.
Number of mutations per cell per generation. Nalidixic acid-resistant (Nalr) and streptomycin-resistant (Strr) colonies were selected on Luria-Bertani-Herskowitz (LBH) plates (see, e.g., reference 36) supplemented with 40 μg of nalidixic acid or 100 μg of streptomycin per ml. Arginine prototrophs (Arg+) were selected on minimal M9 plates (21) with the addition of 0.1% glycerol and 5 μg of the appropriate amino acids per ml. Mutants were scored after ca. 24 (LBH) or 72 h (M9) of incubation at 37°C. Mutation rates were calculated by the method of the median as modified by von Borstel (38).
Not applicable (JC11450 is Strr and FC40 is Arg+).
In these cases more than half of the cultures produced no mutant colonies and the mutation rate was calculated using a median of <1; the rates are thus overestimates and are preceded by “<.”
Could the triple-exonuclease mutants be MMR deficient but fail to display the mutator phenotype? For example, it might be that ExoI, ExoVII, and RecJ are necessary and sufficient exonuclease activities for MMR in vivo but that triple-mutant cells initiating MMR die from accumulation of the nicked but not displaced DNA intermediate. This would kill those cells that experienced polymerase errors and so prevent a mutator phenotype. This possibility seems unlikely for two reasons. First, the viabilities of strains lacking ExoI, ExoVII, and RecJ are normal (Table 2), providing no evidence that those attempting MMR die. Second, mutU helicase mutants would be expected to accumulate a similar DNA intermediate (a nicked but not displaced strand) but these are viable and display a mutator phenotype (31, 32). This argues that failure to complete MMR after nicking is not a lethal event.
We will discuss three possible explanations for the results presented. First, another as yet uncharacterized exonuclease(s) may be sufficient for MMR. Cooper et al. (7) postulated that another exonuclease must contribute after they found repair of 3′ but not 5′ substrates in cells lacking ExoI and RecJ. If such an activity catalyzes MMR in vivo in our assay, then it is interesting that apparently normal levels of MMR can be accomplished with only this 3′ nuclease.
Regardless of the polarity of a putative substituting nuclease(s), the existence of one or more is suggested by the discoveries of MMR-associated single-strand exonucleases in the yeasts Schizosaccharomyces pombe (34) and Saccharomyces cerevisiae (35). In these systems a single 5′ single-strand-dependent exonuclease associates with (35) and/or is required for proper function of (34) the MMR apparatus.
There are two uncharacterized open reading frames in the E. coli genome sequence that contain conserved exonuclease motifs, although neither gene’s product has been tested yet for nuclease function (13). Either of these or an as yet unidentified gene might supply a function that substitutes for that of ExoI, ExoVII, and RecJ exonucleases in MMR in vivo.
Second, it is formally possible that the three single-strand exonucleases are normally required for MMR in vivo and that cells lacking them do not show an in vivo mutator phenotype because their absence induces the expression of a new substituting activity. This idea could be tested by in vitro analysis of MMR in crude extracts of our triple-exonuclease-defective strains, as both the crude and purified in vitro MMR assays require exonuclease, at least when 5′ substrates are used (7). If a new exonuclease-bypassing (or substituting) activity were expressed only in the triple mutant, one might expect 5′ substrates to be repaired in crude extracts of the triple-exonuclease mutants, but not in double-mutant extracts, as was reported previously for 5′ substrates (7). Also, any 3′ substrate repair detected would be unambiguously independent of ExoVII.
One specific version of this general idea is addressed by data shown in Table 4. If the triple-nuclease-defective strain grew slowly or were inviable, then cells already harboring a secondary (suppressor) mutation might usually be selected when constructing triple-exonuclease mutants. When this kind of problem occurs, most cells receiving the deleterious mutation during the strain construction (the third nuclease allele in our constructions) are lost, and the rare suppressor-carrying mutants predominate among progeny that carry the deleterious mutation. This possibility was tested by determining the efficiency of recovering the third nuclease mutation in P1 transduction experiments in which the third nuclease mutation is introduced by selection for a nearby, linked marker rather than by direct selection. We find that there is no bias against recovering the third exonuclease mutation in double-exonuclease mutants, as compared with exonuclease-proficient strains (Table 4). This argues strongly against the presence of secondary suppressor mutations in triple-exonuclease-deficient strains.
TABLE 4.
Ability to construct triple-exonuclease mutants without direct selectiona
| Genetic background | Relevant genotype of:
|
No. of col.c | Frequency of cotransducing exo mutation with linked Tn10kan (mean ± SD)b | |
|---|---|---|---|---|
| Donor | Recipient | |||
| JC11450 | ΔxseA18::amp zfh-3139::Tn10kan | xonA+ xseA+ recJ+ | 148 | 0.19 ± 0.03 |
| ΔxonA300::cat recJ284::Tn10 | 573 | 0.19 ± 0.03 | ||
| ΔxonA300::cat zef-3189::Tn10kan | xonA+ xseA+ recJ+ | 161 | 0.24 ± 0.05 | |
| recJ284::Tn10 ΔxseA18::amp | 170 | 0.27 ± 0.05 | ||
| recJ284::Tn10 zgf-3156::Tn10kan | xonA+ xseA+ recJ+ | 123 | 0.13 ± 0.04 | |
| ΔxonA300::cat ΔxseA18::amp | 244 | 0.20 ± 0.05 | ||
| FC40 | ΔxseA18::amp zfh-3139::Tn10kan | xonA+ xseA+ recJ+ | 886 | 0.19 ± 0.01 |
| ΔxonA300::cat recJ284::Tn10 | 1,420 | 0.18 ± 0.01 | ||
| ΔxonA300::cat zef-3189::Tn10kan | xonA+ xseA+ recJ+ | 1,010 | 0.27 ± 0.01 | |
| recJ284::Tn10 ΔxseA18::amp | 902 | 0.21 ± 0.01 | ||
| recJ284::Tn10 zgf-3156::Tn10kan | xonA+ xseA+ recJ+ | 540 | 0.17 ± 0.01 | |
| ΔxonA300::cat ΔxseA18::amp | 576 | 0.15 ± 0.02 | ||
If secondary suppressor mutations are necessarily selected when constructing triple-exonuclease null mutants, then most transductants of a double-exonuclease mutant with the final nuclease allele would be absent from the transduction progeny—only the rare double-nuclease mutant that already carries a secondary suppressor mutation would form a detectable transductant colony. The frequency of cotransduction of the third nuclease allele with a nearby selectable marker would then appear to be much lower when transducing into double-nuclease mutant recipient strains than into exonuclease-proficient recipients. These frequencies were compared for all of the possible orders of construction of triple-nuclease mutants in both strain backgrounds used in this study. In each case, a nearby selectable kanamycin resistance-encoding transposon, Tn10kan, was selected, and the number of colonies also carrying the linked nuclease allele was determined by assaying the (other) drug resistance encoded by the cassette disrupting each nuclease gene.
Three separate P1 transductional crosses were performed. Standard transduction methods were used (21).
The total number of transductant colonies screened.
Finally, it could be that single-strand exonuclease activity is not required for MMR in vivo, even though it is required in vitro. Complete displacement of the unmethylated DNA strand by MutU helicase may be unfavorable in vitro, perhaps because the displaced single-strand DNA can reanneal. Exonuclease activity would then be required to degrade the displaced DNA strand. This requirement might be bypassed in vivo if MutU, MutS, and MutL could remove the unmethylated DNA strand completely. This might occur in vivo but not in vitro for any of several possible reasons including the following.
(i) In vivo, the displaced strand might not reanneal because it competes for reannealing with the (perfectly complementary) parental strand at the replication fork. A competitor strand is not provided in vitro.
(ii) A single nick was provided to direct MutU helicase in the experiments demonstrating nuclease requirements in vitro. Perhaps the normal in vivo substrate is a mismatch flanked by two nicks, making complete removal of the displaced strand possible without exonucleolytic digestion.
(iii) The displaced strand might be stabilized and prevented from reannealing in vivo by single-strand binding proteins or other activities or conditions that might not have been optimized in the in vitro systems.
With any of these possibilities, the single-strand exonucleases might still degrade the displaced single strand, but this would not be an obligate step in MMR.
Acknowledgments
We thank D. Berg for Kohara phage; H. J. Bull, S. Gottesman, P. J. Hastings, P. Modrich, H. Razavy, R. Sidhu, and an anonymous reviewer for comments on the manuscript; H. Razavy and S. Szigety for strain construction; C. Thulin for excellent technical assistance; and the E. coli Genetic Stock Center for strains.
This work was supported by grants from the National Cancer Institute of Canada, funded by the Canadian Cancer Society, from the Medical Research Council of Canada (MRC), and grant GM53158 from the National Institute of General Medical Sciences (United States). A graduate studentship (R.S.H.) and a postdoctoral fellowship (M.-J.L.) were provided by the Alberta Heritage Foundation for Medical Research, and R.S.H. held an Honorary Izaak Walton Killam Memorial Scholarship. S.M.R. was supported by MRC Scientist and Alberta Heritage Senior Medical Scholar awards.
REFERENCES
- 1.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 2.Benson N R, Roth J. Suppressors of recB mutations in Salmonella typhimurium. Genetics. 1994;138:11–29. doi: 10.1093/genetics/138.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns J, Foster P L. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chase J W, Richardson C C. Escherichia coli mutants deficient in exonuclease VII. J Bacteriol. 1977;129:934–947. doi: 10.1128/jb.129.2.934-947.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chase J W, Richardson C C. Exonuclease VII of Escherichia coli. J Biol Chem. 1974;249:4553–4561. [PubMed] [Google Scholar]
- 6.Chase J W, Rubin B A, Murphy J B, Stone K L, Williams K R. Escherichia coli exonuclease VII: cloning and sequencing of the gene encoding the large subunit (xseA) J Biol Chem. 1986;261:14929–14935. [PubMed] [Google Scholar]
- 7.Cooper D L, Lahue R S, Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 8.Feng W-Y, Hays J B. DNA structures generated during recombination initiated by mismatch repair of UV-irradiated phage DNA in Escherichia coli: requirements for helicases, exonucleases, and RecF and RecBCD functions. Genetics. 1995;140:1175–1186. doi: 10.1093/genetics/140.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng W-Y, Lee E, Hays J B. Recombinagenic processing of UV-light photoproducts in nonreplicating phage DNA by the methyl-directed mismatch repair system. Genetics. 1991;129:1007–1020. doi: 10.1093/genetics/129.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris R S, Longerich S, Rosenberg S M. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 11.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 12.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 13.Koonin E V. A conserved ancient domain joins the growing superfamily of 3′ to 5′ exonucleases. Curr Biol. 1997;7:604–606. doi: 10.1016/s0960-9822(06)00311-3. [DOI] [PubMed] [Google Scholar]
- 14.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 15.Linn S M, Lloyd R G, Roberts R J. The nucleases. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 16.Lovett S T, Clark A J. Genetic analysis of the recJ gene of Escherichia coli K-12. J Bacteriol. 1984;157:190–196. doi: 10.1128/jb.157.1.190-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 18.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 19.Matic I, Taddei F, Radman M. Genetic barriers among bacteria. Trends Microbiol. 1996;4:69–73. doi: 10.1016/0966-842X(96)81514-9. [DOI] [PubMed] [Google Scholar]
- 20.Miesel L, Roth J R. Evidence that functions of the “RecF pathway” contribute to RecBCD-dependent transductional recombination. J Bacteriol. 1996;178:3146–3155. doi: 10.1128/jb.178.11.3146-3155.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 22.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 23.Modrich P. Mismatch repair, genetic stability and tumour avoidance. Philos Trans R Soc Lond B. 1995;347:89–95. doi: 10.1098/rstb.1995.0014. [DOI] [PubMed] [Google Scholar]
- 24.Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 25.Philips G J, Prasher D C, Kushner S R. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli. J Bacteriol. 1988;170:2089–2094. doi: 10.1128/jb.170.5.2089-2094.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 27.Razavy H, Szigety S K, Rosenberg S M. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in Chi stimulated recombination in vivo. Genetics. 1996;142:333–339. doi: 10.1093/genetics/142.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rupp W D. DNA repair mechanisms. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2277–2294. [Google Scholar]
- 29.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaaper R M. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 31.Siegel E C. Mutator mutations in Escherichia coli induced by the insertion of phage Mu and the transposable elements Tn5 and Tn10. Mutat Res. 1982;93:25–33. doi: 10.1016/0027-5107(82)90122-1. [DOI] [PubMed] [Google Scholar]
- 32.Siegel E C, Kamel F. Reversion of frameshift mutations by mutator genes in Escherichia coli. J Bacteriol. 1974;117:994–1001. doi: 10.1128/jb.117.3.994-1001.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szankasi P, Smith G R. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 35.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torkelson J, Harris R S, Lombardo M-J, Nagendran J, Thulin C, Rosenberg S M. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vales L D, Chase J W, Murphy J B. Orientation of the guanine operon of Escherichia coli by using strains containing guaB-xse and guaB-upp deletions. J Bacteriol. 1979;139:320–322. doi: 10.1128/jb.139.1.320-322.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Borstel R C. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- 39.Zahrt T C, Maloy S. Barriers to recombination between closely related bacteria: MutS and RecBCD inhibit recombination between Salmonella typhimurium and Salmonella typhi. Proc Natl Acad Sci USA. 1997;94:9786–9791. doi: 10.1073/pnas.94.18.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]