Abstract
COL2A1 gene encodes the alpha-1 chain of type-II procollagen. Heterozygous pathogenic variants are associated with the broad clinical spectrum of genetic diseases known as type-II collagenopathies. We aimed to characterize the NM_001844.5:c.1330G>A;p.Gly444Ser variant detected in the COL2A1 gene through trio-based prenatal exome sequencing in a fetus presenting a severe skeletal phenotype at 31 Gestational Weeks and in his previously undisclosed mild-affected father. Functional studies on father's cutaneous fibroblasts, along with in silico protein modeling and in vitro chondrocytes differentiation, showed intracellular accumulation of collagen-II, its localization in external Golgi vesicles and nuclear morphological alterations. Extracellular matrix showed a disorganized fibronectin network. These results showed that p.Gly444Ser variant alters procollagen molecules processing and the assembly of mature type-II collagen fibrils, according to COL2A1-chain disorganization, displayed by protein modeling. Clinical assessment at 38 y.o., through a reverse-phenotyping approach, revealed limp gait, short and stocky appearance. X-Ray and MRI showed pelvis asymmetry with severe morpho-structural alterations of the femoral heads bilaterally, consistent with a mild form of type-II collagenopathy. This study shows how the fusion of genomics and clinical expertise can drive a diagnosis supported by cellular and bioinformatics studies to effectively establish variants pathogenicity.
Keywords: COL2A1, Type-II collagenopathies, Reverse-phenotyping, Exome sequencing, Functional characterization
1. Introduction
COL2A1 (OMIM*120140) gene encodes for the alpha-1 chain of type-II procollagen. Mature type-II collagen provides the structural function of the cartilage matrix forming a covalently cross-linked fibrillar network. It is synthesized by proliferating chondrocytes and formed after procollagen type-II secretion into the extracellular matrix and processing.
More than 600 pathogenic variants have been described in the COL2A1 gene; most of them are missense variants, localized in the triple helix domain of the protein, consisting in 330 repeats of the triplet Gly-X-Y. Missense variants replacing the Glycine residue, with a bulkier amino acid in the Gly-X-Y triplet, lead to disruption of the helical structure and function of type-II collagen. Pathogenic COL2A1 variants are associated with type-II collagenopathies, a large and clinically heterogeneous group of conditions inherited with a dominant pattern. Main known features are skeletal dysplasia, ocular and otorhinolaryngological anomalies, and hearing loss. According to the clinical severity they can be divided in three main groups: lethal, severe non-lethal, and intermediate/mild non-lethal forms.
Lethal type-II collagenopathies include Achondrogenesis type-2 and Hypochondrogenesis (OMIM #200610), and Platyspondylic lethal dysplasia Torrance-type (OMIM#151210). The typical onset is in the prenatal period with two hallmarks: lack of spinal and ilio-pubic branches ossification (Handa et al., 2021). Severe non-lethal forms include Spondyloepiphyseal dysplasia (SED) congenita (OMIM#183900), Spondyloepimetaphyseal dysplasia, Strudwick-type (OMIM#184250), Kniest Dysplasia (OMIM#156550) and Spondyloperipheral dysplasia (OMIM#271700). All three of the above conditions may be associated with myopia, deafness, cleft palate and/or Pierre Robin sequence (Terhal et al., 2015).
The subgroup of type-II collagenopathies associated with an intermediate/mild phenotype is broad, ranging from Stickler syndrome type 1 (OMIM#108300), Osteoartritis with mild chondrodysplasia (OMIM#604864), Czech dysplasia (OMIM#609162) to Vitreoretinopathy with phalangeal epiphyseal dysplasia (OMIM#619248) (Kozlowski et al., 2004; Richards et al., 2002). All these conditions are marked by epiphyseal involvement of variable degree, modest radiological lesions poorly detectable in early childhood, joint pain, evolution to osteoarthritis or osteochondritis and inconstant but suggestive associated signs such as myopia, retinal detachment, and deafness (Terhal et al., 2015). The mildest phenotypic expression in the spectrum of COL2A1-related conditions is represented by a form of skeletal dysplasia known as Avascular Necrosis of the Femoral Head (ANFH, OMIM#608805), characterized by the collapse of the femoral head and subsequent loss of coxo-femoral joint function. This condition is manifested by progressive pain, pain on exertion, limp, and leg length discrepancy. Other clinical manifestations include early hip osteoarthritis and short stature compared to the family target (Liu et al., 2005).
The underlying pathogenesis of these diseases remains largely unclear.
The aim of this work was to characterize clinically and functionally a postnatal undisclosed mild phenotype, unraveled through trio-prenatal exome sequencing required for a severe prenatal skeletal phenotype presentation.
2. Materials and methods
2.1. Subjects and trio-based prenatal exome sequencing (pES)
A non-consanguineous couple was referred for genetic counseling after pregnancy interruption of a fetus presenting with polydramnios, biometry <2/3 SD of long bones, cephalic biometry between 90 and 97° pc and subcutaneous tissue oedema spreading at the retronuchal space, prefrontally and at the body at 31 Gestational Weeks (GW).
The pregnancy was uneventful until 30 GW, with a regular I trimester scan (NT 1.40 mm) and a regular II trimester morphologic scan. After uninformative standard chromosomal and microarray analysis, trio-based prenatal exome sequencing (pES) was performed on genomic DNA extracted from amniocytes and parental leukocytes, collected after written informed consent and in accordance with the Principles of the Declaration of Helsinki (supplementary materials and methods). Library preparation and clinical exome capture were performed by using the Twist Human Core Exome Kit (Twist Bioscience) according to the manufacturer's protocol and sequenced on the Illumina NovaSeq 6000 platform. The BaseSpace pipeline and the Geneyx software LifeMap Sciences were respectively used for the variant calling and annotating variants. Sequencing data were aligned to the hg19 human reference genome. The variants were filtered analyzed in silico by using Combined Annotation Dependent Depletion (CADD), Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping v2 (PolyPhen-2) and Mutation Taster for the prediction of deleterious non-synonymous SNVs for human diseases. Global minor allele frequency (MAF) for analyzed variants was calculated according to Genome Aggregation Database (gnomAD). Based on the guidelines of the American College of Medical Genetics and Genomics (ACMG), a minimum depth coverage of 30× was considered suitable for analysis. Variants were also examined for Qscore and visualized by the Integrative Genome Viewer (IGV).
Deep clinical reverse-phenotyping and skin biopsy was performed on the father and compared to an age-matched control (CTR) subject, after fibroblasts isolation and culture.
To further characterize the pathogenicity of the variant, in silico and in vitro studies were performed in patient's cells and compared to control's cells.
2.2. In silico modeling
In silico protein modeling was performed using AlphaFold Protein Structure Database (Jumper et al., 2021) along with a phosphorylation prediction analysis with GPS6.0 (Chen et al., 2023).
2.2.1. Phosphorylation prediction
The COL2A1 protein sequence was retrieved from UniProt (UniProt Consortium, 2023) selecting the Human P02458 reviewed entry. A peptide of 15 amino acids centered on the Gly444 was selected and mutated to allow for the introduction of the Ser444 (WT: RGPPGPQGATGPLGP; MT: RGPPGPQSATGPLGP).
Subsequently two more peptides centered on the Thr446 containing respectively Gly444 and Ser444 were generated (WT: PPGPQGATGPLGPKG; MT: PPGPQSATGPLGPKG) and were used as input in the GPS6.0 web server (Chen et al., 2023) for the prediction of the phosphorylation using the species-specific modality.
2.2.2. 3D model reconstruction
A collagen 3D model using the SWISS-MODEL web server (Waterhouse et al., 2018) and the 2D3H PDB structure as template for both Gly444 and Ser444 was built. The chosen template corresponds to the collagen-like peptides containing the Pro-Pro-Gly motifs (Okuyama et al., 2009). The 3D model of each single collagen molecule was superposed on the proline residues of the triple helix. The phosphate group was added on the threonine 446 using CHARMM-GUI (Park et al., 2023). The two collagen triple helices have been shown using the Chimera v1.16 software (Pettersen et al., 2004).
2.3. Chondrogenic differentiation
Chondrogenic differentiation was induced on confluent cell monolayer seeded in 24-well plate of both control and patient's cells using a basal chondrogenic medium DMEM (Life Technologies, Bleiswijk, The Nedherlands) supplemented with 50 mg/mL ITS+premix, 10−7 M dexamethasone, 50 μL ascorbate-2phosphate, 1 mM sodium pyruvate, and 100 U/mL-100 μg/mL penicillin-streptomycin, (Life Technologies) and the chondrogenic factors TGFβ and BMP6 (10 ng/mL, Miltenyi Biotech, Auburn,CA, US).The medium was changed two times a week and the differentiation process was checked at Day 0, Day 21 and Day 28 by Safranin-O staining.
The chondrogenic monolayers at Day 0, Day 21 and Day 28 were washed twice with PBS and fixed with PFA4% solution for 20 min. at room temperature. After fixation the samples were washed three times with PBS and stained with 0.1 % Safranin-O solution for 30 min at room temperature. The samples were washed 3 time with PBS and the plate were dried at room temperature. The specimens staining was quantified at the absorbance of 540 nm by the spectrophotometer (Infinite® M200 Nanoquant plate reader-TECAN, Switzerland).
2.4. Immunofluorescence analysis and antibodies
Cells grown on coverslips were fixed with hot absolute methanol at room temperature for 10 min or 4 % paraformaldehyde 15 min at RT and permeabilized with 0,05 % triton for 5 min at RT. After saturation of non-specific binding with 4 % BSA in PBS 1× solution for 20 min, coverslips were incubated with primary antibodies overnight at 4 °C O·N. or 1 h at RT, and revealed with FITC or TRIC-conjugated secondary antibodies diluted 1:200 (incubated for 1 h at RT). DAPI (4,6-diamidino-2-phenylindole) blue staining was used to counterstained cell nuclei. Samples were mounted with an anti-fade reagent (Molecular Probes Life Technologies) with DAPI and observed with Nikon Eclipse Ni epifluorescence microscope with a 40×, 60× and 100× objectives. The images captured with NIS- Elements 4.3 AR software, were elaborated using Photoshop CS. Antibodies employed were: anti-collagen 2 (Chemicon, MAB8887) at 1:100 dilution O·N; anti-golgin 97 (Cell Signaling, D8P2K) 1:100 O.N; anti-fibronectin (Proteintech, 66,042) 1:100 O.N.
3. Results
3.1. Molecular results
Trio-pES disclosed the heterozygous NM_001844.5:c.1330G>A;p.Gly444Ser paternally inherited variant in COL2A1 gene, classified as Likely Pathogenic (LP) and three maternally heterozygous inherited variants: the NM_006031.6:c.8583_8584insGA;p.Glu2866GlyfsTer11 LP variant in PCNT and two variants of uncertain significance (VUS) NM_001135.3:c.551C>T;p.Thr184Met in ACAN and NM_012224.3:c.3708G>T;p.Gln1236His in NEK1 genes.
According to current prenatal International Society for Prenatal Diagnosis and American College Medical Genetics and Genomics recommendations (Van den Veyver et al., 2022; Monaghan et al., 2020), all the detected variants were deeply multidisciplinary analyzed and the maternally inherited variants were not considered causative, either because did not fit the fetal phenotype or the expected disease inheritance pattern. On the contrary, the predicted LP COL2A1 variant needed further clinical and biological evaluation. As a matter of fact, the never described before COL2A1 p.Gly444Ser variant, had a CADD score of 28.6, was present in ClinVar (variation ID:423267) classified as Pathogenic without clinical details, and was not present in control population databases: gnomAD (v.2.1.1 and v.3.1), 1000 Genomes Consortium and Exome Variant Server. Two pathogenic alternative changes at the same amino acid position were reported in the Human Gene Mutation Database and literature.
3.2. Clinical and radiological characterization
The couple had an unremarkable family history; however the father reported a personal history of femoral head anomaly in infancy (at three years old) without anamnestic predisposing conditions.
A clinical reassessment was required, along with a total body X-Ray and MRI examinations.
The father at 38 y.o., showed limp gait, asymmetry of the legs, proportionate short and stocky appearance, not associated with Intellectual Disability/Developmental Delay. He did not report any visual problem or hearing impairment. Height was 1.60, resulting below familial target (mother 1.65 and father 1.70). Unfortunately, biological samples from the parents of the father were not available to perform segregation analysis.
The radiograph of the thoracolumbar spine showed mild left-convex scoliosis, with reduced dorsal kyphosis and lumbar lordosis. Moderate flattening of some vertebral bodies, particularly the posterior somatic walls, were observed in the dorsal segment, a finding most appreciable at the level of the D10 and D11 soma (Fig. 1 A and B). An anteroposterior radiograph documented an asymmetry of the pelvis with an elevation to the left of the bicresto-iliac line (27 mm), the bi-cephalo-femoral line (26 mm) and the bi-cotyloid line (25 mm) (Fig. 1 D). Severe morpho-structural alteration of the meta- and epiphyseal region of the femoral heads was also observed bilaterally, characterized by moderate fragmentation, reduction of physiological convexity, and irregularity of cortical bone profiles. Reduced development of the femoral necks bilaterally and extra-rotation of the coxo-femoral joint on the left was also noted (Fig. 1 C and D).
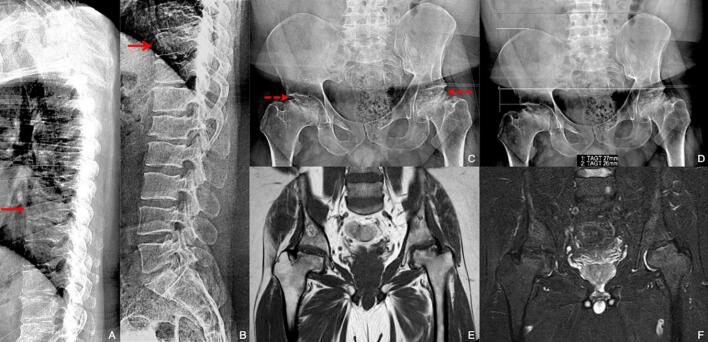
Fig. 1.
Thoracic (A) and lumbar (B) spine lateral x-ray views demonstrated a «pear-shaped» appearance of T10 (red open arrow) and T8 (red closed arrow) vertebrae. Antero-posterior (C and D) hip x-ray view; to note the severe remodeling of femoral heads with fragmentation and flattened profile (red dotted arrows). In (D) quantification of sopraelevation of the right iliac crest and ipsilateral upper profile of acetabulum. Coronal T1-TSE (E) and T2-STIR (F) MRI of hip: to note small amount of trabecular oedema of femoral heads.
A concomitant moderate narrowing of the joint space was observed, resulting in acetabular flattening, more evident on the left (Fig. 1 C and D).
MRI of the pelvis confirmed the morphostructural alterations and showed modest residual trabecular oedema at the level of the bilateral femoral head and neck (Fig. 1 E and F). The above findings were suggestive of residual alterations of previous aseptic osteonecrosis on a vascular basis, a clinical picture consistent with a mild form of a type-II collagenopathy, i.e. ANFH.
Concerning the fetus, unfortunately a babygram was not available, neither autopsy results, refused from the couple.
3.3. In silico modeling and phosphorylation prediction
AlphaFold COL2A1 modeling prediction showed that the p.Gly444Ser variant falls in the non-structured part of triple-helical domain, critical for the stability of collagen (Supplementary Fig. 1).
To investigate if the p.Gly444Ser could influence the phosphorylation of the COL2A1 protein, the prediction analysis with GPS6.0 (Chen et al., 2023) on the wild-type and on the mutant peptides (supplementary Fig. 2 A,B) were compared. The serine residue (Ser444) in COL2A1 was predicted not to be a new phosphorylation site, but this variant can affect the phosphorylation of a neighboring threonine (Thr446). Thr446 was predicted to be phosphorylated by Protein Kinase-Like (PKL) and Receptor Guanylate Cyclases (RGC) kinases with a significant score only in the mutant peptide (supplementary Fig. 2, Table 1).
3.4. In vitro characterization
To determine the functional role of Ser444-COL2A1 on chondrogenic differentiation, Safranin O staining was performed to detect proteoglycan deposition, the major component of the animal extracellular matrix. The combination of proteoglycans and collagen form cartilage. In vitro, Ser444 variant showed to affect chondrocytes differentiation at day-21 and day-28 compared to control cells (Fig. 2 panel A). Under basal conditions, patient's cells tended to overlap, leaving empty spaces (Fig. 2 panel B).
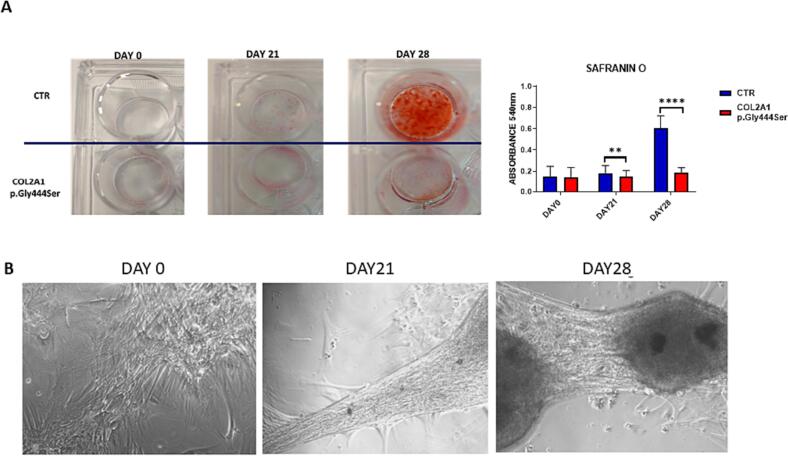
Fig. 2.
A) Representative images of Safranin O staining of chondrogenic differentiated cells at days 0, 21 and 28. Graph showing the quantification of Safranin O positive cells. Two-way ANOVA with Sidak's multiple comparison test, ** p < 0.002, **** p < 0.0001 B) Phase contrast microscope images (20× objective) of patient's cells at days 0, 21 and 28 of chondrogenic differentiation.
After differentiation with a chondrogenesis mix, the overlapping cells formed small clusters and left large empty spaces (Fig. 2 panel B).
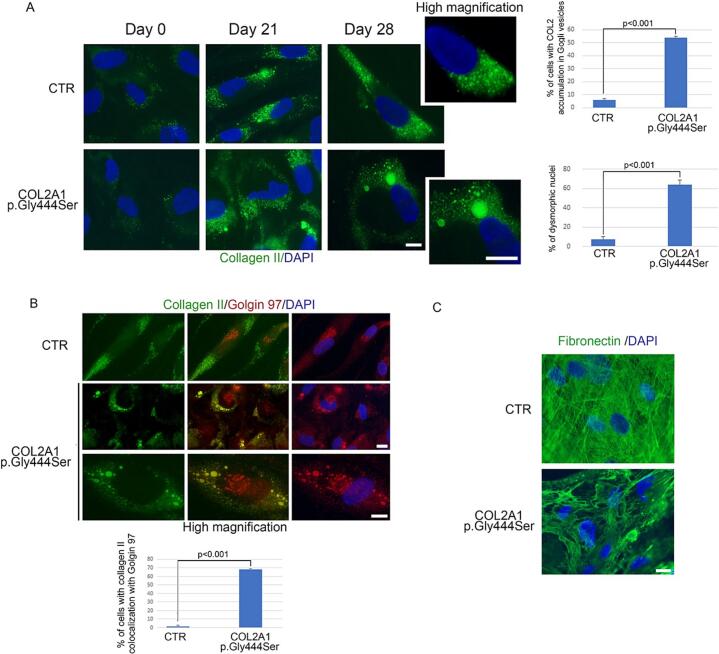
In addition, Collagen-II staining in patient's cells indicated excessive intracellular accumulation of mutant protein, observed at day-21 and day-28 (Fig. 3 panel A). No collagen-II was observed in the extracellular matrix and alterations at the nuclear level were also visible (Fig. 3 panel A). A co-localization of Collagen-II with golgina97 at the level of external Golgi vesicles only, was also observed (Fig. 3 panel B). These should be vesicles intended for extracellular delivery.
Fig. 3.
Immunofluorescence analysis of human chondrocytes in control (CTR) and in a patient carrying COL2A1 (p.Gly444Ser) variant. (A) Immunostaining of collagen II (green) in cells at days 0, 21 and 28 of chondrogenic differentiation and high magnification on the right (high magnification). Statistical analysis is reported in the graphs on the right. (B) Double immunofluorescence analysis of collagen II (green), and golgin 97 (red) in human chondrocytes at day 28 of differentiation. High magnification in the panels below (high magnification). Statistical analysis is reported in the graph below. (C) Immunostaining of fibronectin (green) in human chondrocytes at day 28 of differentiation. 4,6-diamidino-2-phenylindole (DAPI, blue) was used to counterstained cell nuclei. Scale bars, 10 μm. Three biological replicates were used in each experiment and statistically significant differences (p < 0.001) between values are indicated.
By localization of fibronectin, it has been observed that in the extracellular matrix, fibronectin failed to form a well-structured network in the patient's cells compared to control's cells (Fig. 3 panel C).
4. Discussion
We identified a glycine substitution, p.Gly444Ser, falling in the Gly-X-Y repeat of COL2A1 protein. In patient's cells, the observation of impaired chondrogenic differentiation (Fig. 2 panel A and B), intracellular accumulation of mutant protein at the level of external Golgi vesicles (Fig. 3 panel B) and extracellular matrix fibronectin network disorganization (Fig. 3 panel C) showed that p.Gly444Ser variant impairs transport of procollagen molecules, as well as the assembly of mature type-II collagen fibrils.
Literature data suggest that missense variants involving the substitution of glycine residues with amino acids with higher steric hindrance exert a dominant negative effect, which involves an abnormal assembly of alpha-1 chains in collagen molecules (Körkkö et al., 2000; Liberfarb et al., 2003).
Glycine-serine substitutions have been reported with severe or milder skeletal phenotypes in the SED spectrum (G504S, G513S, G1197S) associated or not with otorhinolaryngological manifestations (G513S, G1197S) (Nishimura et al., 2005) and characterized by marked inter- and intra-familial expression variability.
Chen et al. (2017) described a family, in which the proband presented with a spondyloepimetaphyseal dysplasia with severe metaphyseal lesions, while her mother and her sister, carriers of the same COL2A1 variant c.1636G>A, showed spondyloepiphyseal dysplasia features without metaphyseal involvement (Chen et al., 2017).
Clinical reassessment of the father reported here was compatible with a previous undisclosed COL2A1-related disorder in the milder spectrum, such as ANFH, highlighting a risk of recurrence for a COL2A1-related disorder.
To date, only six different variants in COL2A1 have been reported in literature in association with ANFH cases and 5 out 6 are glycine-serine substitutions in the Gly-X-Y sequence of the triple helix (Zhang et al., 2021). Concerning the fetus, we cannot rule out that the severity of the antenatal phenotype could be linked to the additional effect of the maternally inherited variants or other undetected causes by the extensive molecular analyses performed. In particular, the heterozygous VUS NM_001135.3:c.551C>T;p.Thr184Met in ACAN gene (MIM *155760) resulted the more interesting one, as it encodes for aggregan, a major component together with collagen type-2 of the cartilage matrix in cartilage tissue. The mechanical properties of the matrix depend on both major components and their stabilization by intermolecular interactions. Moreover, pathogenic variants in ACAN gene belong to the spectrum of molecular causes associated to SED.
Recently another COL2A1 variant (p.Arg904Cys), with father-fetus transmission detected through trio-pES and associated to Stickler Syndrome type-1, has been reported with a more complex phenotype in the fetus, presenting additional and more severe findings in respect to the previously undiagnosed father (Janicki et al., 2023).
These cases also illustrate how using inheritance filtering in the prenatal setting, based on the assumption that both parents are unaffected, and the lack of a pre- or peri-conceptional genetic counseling can miss diagnoses. Conversely, examination of parents and ascertaining the family history in clinical genetics is crucial for a complete analysis. Many autosomal dominant conditions have variable expressivity and age of onset and parents may often be unaware of carrier status.
In literature, some extremely variable familial cases have been also explained by gonado-somatic mosaicism in one of the parents, such as families in which the offsprings presented with lethal skeletal dysplasia and the affected parent showed only a mild phenotype (Désir et al., 2012).
This significant extra- and intrafamilial different presentation in patients carrying the same variant (Terhal et al., 2015) demonstrates that the position of the variant in the alpha-1 chain of procollagen type-II and within the triple helix region cannot totally account for the effect of the genetic alteration on the affected individual. These results suggest that there may be genetic, epigenetic, and environmental acting modifiers, underlying the difficulties that can arise in genetic counseling.
It has been suggested that phosphorylation/dephosphorylation mechanisms may play a regulatory role in collagen processing, stability, assembly, degradation, or binding (Acevedo-Jake et al., 2017; Qiu et al., 2018).
In collagen type-1, the phosphorylation of Ser and Thr residues in the collagen Gly-X-Y sequence have been shown by phospho-proteomics studies. It has been hypothesized that the Gly-X-Y repeat of collagen and its triple-helical structure could influence its ability to serve as a substrate for a kinase (Qiu et al., 2018). However, the kinases involved in collagen phosphorylation, the location of the phosphorylation process, and the physiological mechanisms remain unknown.
In silico studies predicted that the serine residue (Ser444) in COL2A1 is not a new phosphorylation site, but this variant can affect the phosphorylation of a neighboring threonine (Thr446) (Supplementary Fig. 2 A, B). Thr446 was predicted to be phosphorylated by PKL and RGC kinases with a significant score in the mutant peptide only (Supplementary Fig. 2, Table 1).
Moreover, previous studies proposed that phosphorylation may be a mechanism of controlling collagen triple-helix stability (Acevedo-Jake et al., 2017; Qiu et al., 2018). Interestingly, collagen has been proposed as a potential substrate of Fam20c (a PKL kinase), secreted in the Golgi apparatus and representing the major secretory pathway protein kinase (Xu et al., 2021). Further characterization of the structural basis of collagen phosphorylation and the effects of such phosphorylation on triple-helix properties will help clarify the biological role of this post-translational modification and its possible role in the complex scenario of type-II collagenopathies.
5. Conclusions
To date, in type-II collagenopathies there is no clear phenotype–genotype correlation, and the impact of COL2A1 variants is unpredictable. In routine settings, the clinical significance of a new variant is often speculative and based on software prediction because they generally do not benefit from functional validation (Barat-Houari et al., 2016). Genomic ascertainment and reverse phenotyping have been recently proposed as a model of predictive genomic medicine and genomic screening (Wilczewski et al., 2023).
We demonstrated the pathogenicity of the variant in the father, clinically through a targeted reverse phenotyping approach and functionally, through in silico and in vitro studies. During genetic counseling we gave a risk of 50 % of variant transmission, underlying the fact that the severity of a COL2A1-related phenotype cannot be predicted.
A diagnosis of a type-II collagenopathy is critical in providing accurate genetic risk counseling, offering antenatal or preimplantation genetic testing for future pregnancies and appropriate clinical management for probands.
A combination of clinical expertise, genomics, functional and modeling studies is essential to establish effective variants pathogenicity.
Funding
This work was supported by Piano Sviluppo e Coesione Salute - FSC 2014-2020 GENERA Project to GN (cod. T3-AN-04) and AIRC Project to MHC (IG 23539).
CRediT authorship contribution statement
Enrica Marchionni: Conceptualization, Data curation, Resources, Writing – original draft, Writing – review & editing. Maria Rosaria D'Apice: Conceptualization, Data curation, Formal analysis, Methodology, Writing – review & editing. Viviana Lupo: Data curation, Writing – original draft. Giovanna Lattanzi: Conceptualization, Formal analysis, Methodology, Writing – review & editing. Elisabetta Mattioli: Formal analysis, Methodology, Writing – review & editing. Gina Lisignoli: Formal analysis, Methodology, Writing – review & editing. Elena Gabusi: Formal analysis, Methodology. Gerardo Pepe: Formal analysis, Investigation, Methodology. Manuela Helmer Citterich: Formal analysis, Funding acquisition, Methodology, Writing – review & editing. Elena Campione: Investigation, Methodology. Anna Maria Nardone: Formal analysis, Investigation. Paola Spitalieri: Formal analysis, Investigation. Noemi Pucci: Data curation, Writing – original draft. Dario Cocciadiferro: Data curation, Methodology, Writing – review & editing. Eliseo Picchi: Data curation, Investigation, Writing – original draft. Francesco Garaci: Data curation, Investigation, Writing – review & editing. Antonio Novelli: Data curation, Formal analysis, Methodology, Writing – review & editing. Giuseppe Novelli: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision.
Ethics
The study was conducted in accordance with the Declaration of Helsinki and patients involved in the study signed an informed consent accordingly to ethical guidelines. Patients were collected in a path that belongs to common clinical practice.
Declaration of competing interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bonr.2023.101728.
Appendix A. Supplementary data
Supplementary material
References
- Acevedo-Jake A.M., Ngo D.H., Hartgerink J.D. Control of collagen triple Helix stability by phosphorylation. Biomacromolecules. 2017;18(4):1157–1161. doi: 10.1021/acs.biomac.6b01814. [DOI] [PubMed] [Google Scholar]
- Barat-Houari M., Dumont B., Fabre A., et al. The expanding spectrum of COL2A1 gene variants IN 136 patients with a skeletal dysplasia phenotype. Eur. J. Hum. Genet. 2016;24(7):992–1000. doi: 10.1038/ejhg.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ma X., Zhou Y., et al. Recurrent c.G1636A (p.G546S) mutation of COL2A1 in a Chinese family with skeletal dysplasia and different metaphyseal changes: a case report. BMC Pediatr. 2017;17(1):175. doi: 10.1186/s12887-017-0930-9. (Published 2017 Jul 24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhang W., Gou Y., et al. GPS 6.0: an updated server for prediction of kinase-specific phosphorylation sites in proteins. Nucleic Acids Res. 2023;51(W1):W243–W250. doi: 10.1093/nar/gkad383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Désir J., Cassart M., Donner C., et al. Spondyloperipheral dysplasia as the mosaic form of platyspondylic lethal skeletal dyplasia torrance type in mother and fetus with the same COL2A1 mutation. Am. J. Med. Genet. A. 2012;158A(8):1948–1952. doi: 10.1002/ajmg.a.35301. [DOI] [PubMed] [Google Scholar]
- Handa A., Grigelioniene G., Nishimura G. Radiologic features of type II and type XI collagenopathies. Radiographics. 2021;41(1):192–209. doi: 10.1148/rg.2021200075. [DOI] [PubMed] [Google Scholar]
- Janicki E., De Rademaeker M., Meunier C., et al. Implementation of exome sequencing in prenatal diagnostics: chances and challenges. Diagnostics (Basel) 2023;13(5):860. doi: 10.3390/diagnostics13050860. Published 2023 Feb 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körkkö J., Cohn D.H., Ala-Kokko L., et al. Widely distributed mutations in the COL2A1 gene produce achondrogenesis type II/hypochondrogenesis. Am. J. Med. Genet. 2000;92(2):95–100. doi: 10.1002/(sici)1096-8628(20000515)92:2<95::aid-ajmg3>3.0.co;2-9. (PMID: 10797431) [DOI] [PubMed] [Google Scholar]
- Kozlowski K., Marik I., Marikova O., et al. Czech dysplasia metatarsal type. Am. J. Med. Genet. A. 2004;129A(1):87–91. doi: 10.1002/ajmg.a.30132. [DOI] [PubMed] [Google Scholar]
- Liberfarb R.M., Levy H.P., Rose P.S., et al. The Stickler syndrome: genotype/phenotype correlation in 10 families with Stickler syndrome resulting from seven mutations in the type II collagen gene locus COL2A1 [published correction appears in Genet Med. 2003 Nov-Dec;5(6):478] Genet. Med. 2003;5(1):21–27. doi: 10.1097/00125817-200301000-00004. [DOI] [PubMed] [Google Scholar]
- Liu Y.F., Chen W.M., Lin Y.F., et al. Type II collagen gene variants and inherited osteonecrosis of the femoral head. N. Engl. J. Med. 2005;352(22):2294–2301. doi: 10.1056/NEJMoa042480. [DOI] [PubMed] [Google Scholar]
- Monaghan K.G., Leach N.T., Pekarek D., et al. The use of fetal exome sequencing in prenatal diagnosis: a points to consider document of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2020;22(4):675–680. doi: 10.1038/s41436-019-0731-7. [DOI] [PubMed] [Google Scholar]
- Nishimura G., Haga N., Kitoh H., et al. The phenotypic spectrum of COL2A1 mutations. Hum. Mutat. 2005;26(1):36–43. doi: 10.1002/humu.20179. [DOI] [PubMed] [Google Scholar]
- Okuyama K., Hongo C., Wu G., et al. High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)4-Xaa-Yaa-Gly-(Pro-Pro-Gly)4]: implications for triple-helix hydration and Hyp(X) puckering. Biopolymers. 2009;91(5):361–372. doi: 10.1002/bip.21138. [DOI] [PubMed] [Google Scholar]
- Park S.J., Kern N., Brown T., et al. CHARMM-GUI PDB manipulator: various PDB structural modifications for biomolecular modeling and simulation. J. Mol. Biol. 2023;435(14) doi: 10.1016/j.jmb.2023.167995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Poppleton E., Mekkat A., et al. Enzymatic phosphorylation of Ser in a Type I collagen peptide. Biophys. J. 2018;115(12):2327–2335. doi: 10.1016/j.bpj.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A.J., Morgan J., Bearcroft P.W., et al. Vitreoretinopathy with phalangeal epiphyseal dysplasia, a type II collagenopathy resulting from a novel mutation in the C-propeptide region of the molecule. J. Med. Genet. 2002;39(9):661–665. doi: 10.1136/jmg.39.9.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhal P.A., Nievelstein R.J., Verver E.J., et al. A study of the clinical and radiological features in a cohort of 93 patients with a COL2A1 mutation causing spondyloepiphyseal dysplasia congenita or a related phenotype. Am. J. Med. Genet. A. 2015;167A(3):461–475. doi: 10.1002/ajmg.a.36922. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 2023;51(D1):D523–D531. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Veyver I.B., Chandler N., Wilkins-Haug L.E., et al. International Society for Prenatal Diagnosis Updated Position Statement on the use of genome-wide sequencing for prenatal diagnosis. Prenat. Diagn. 2022;42(6):796–803. doi: 10.1002/pd.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczewski C.M., Obasohan J., Paschall J.E., et al. Genotype first: clinical genomics research through a reverse phenotyping approach. Am. J. Hum. Genet. 2023;110(1):3–12. doi: 10.1016/j.ajhg.2022.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Tan H., Zhang J., et al. Fam20C in human diseases: emerging biological functions and therapeutic implications. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.790172. (Published 2021 Dec 20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhu K., Dai H., et al. A novel mutation of COL2A1 in a large Chinese family with avascular necrosis of the femoral head. BMC Med. Genet. 2021;14(1):147 doi: 10.1186/s12920-021-00995-y. Published 2021 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sitography (URLs)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material