Abstract
The mechanism of repression of the β-glucoside utilization (bgl) operon of Escherichia coli by a carboxy-terminally truncated derivative of the nucleoid-associated protein H-NS which is defective in DNA binding was investigated. The DNA-binding function of the H-NS-like protein StpA was found to be necessary for repression, which is consistent with a role for StpA as a DNA-binding adapter for mutant derivatives of H-NS.
The H-NS protein of Escherichia coli is a major component of the bacterial nucleoid and has been identified as a pleiotropic regulator of gene expression, recombination, and genome stability (reviewed in reference 22). Among other functions, H-NS was identified as a negative regulator of expression of the cryptic bgl operon (bglY [1]) and of the osmoregulated proU operon (osmZ [8]). The bgl operon encodes the functions required by the cell for the uptake and utilization of β-glucoside sugars, such as salicin, and can be activated by insertion sequence element insertions upstream of its promoter, point mutations in its upstream cyclic AMP receptor protein binding site, or mutations in DNA gyrase as well as by H-NS inactivation (12, 15). H-NS is necessary but not sufficient for in vitro repression of the bgl operon, which involves DNA elements located both upstream and downstream of the promoter (14, 15). Heterologous promoters placed into the bgl context are also repressed (14), and the repression of this operon is probably the eubacterial example closest to the classic eukaryotic process of gene silencing.
It was observed earlier that of hns alleles selected on the basis of proU derepression at low osmolarity, only a subset led to high-level expression of the bgl operon (2, 8). More recently, the nature of this effect was clarified as part of a systematic structure-function analysis of the H-NS protein (21). In that study it was found that while proU-derepressing point mutations near the N terminus of H-NS also caused bgl derepression, mutations near the C terminus of the protein, and also a C-terminal truncation of H-NS at amino acid 92, caused no bgl derepression despite derepressing proU significantly. Intriguingly, an in vitro analysis of the latter class of proteins showed that they appeared to have lost their ability to bind DNA, which is consistent with an earlier study suggesting that the DNA-binding function of H-NS may be localized in the C terminus (17). In contrast, the N-terminal mutants which did derepress bgl retained the ability to bind DNA, implying that they were specifically impaired in repression function. These observations led Ueguchi et al. (21) to conclude that bgl silencing employed a (possibly unique) mechanism which merely required the targeting of an N-terminal repression domain of H-NS to the bgl promoter via an interaction with a heterologous DNA-binding protein.
The E. coli stpA gene encodes a 15.3-kDa protein which is 58% identical to H-NS at the amino acid level. StpA and H-NS have similar in vitro DNA-binding properties, including a preference for intrinsically curved DNA, and can repress similar spectra of genes in vivo (16, 24). Although normally expressed at a low level in rich medium, stpA is strongly induced in hns mutants (5, 19, 24) and StpA may take over some of the functions of H-NS in these strains. Moreover, StpA has been shown both genetically in vivo and by protein-protein cross-linking in vitro to interact with H-NS, and these interactions seem to require the respective N termini of the proteins (23). Thus, StpA is a likely candidate for a molecular adapter which could target the N-terminal repression domain of a mutant or truncated H-NS derivative to the bgl promoter region. We have investigated this possibility by studying the derepression of bgl in the presence of various combinations of hns and stpA mutations.
hns mutations with differing effects on bgl expression.
In order to assess a possible adapter role for StpA in mediating bgl repression by truncated derivatives of H-NS, we used two different mutant alleles of hns. The first, hns-205::Tn10, is a well-characterized allele which harbors a Tn10 insertion in codon 93 of hns (9) and produces both a truncated hns mRNA species (6) and a truncated N-terminal protein fragment (2, 3) consistent with the termination of hns transcription and translation close to this insertion point. In contrast, the hns2 allele was isolated as a spontaneous proU-derepressing mutant in our laboratory and carries an ≈750-bp insertion (probably IS1) within the first 22 codons of the hns open reading frame. This allele produces no detectable RNA transcript or protein product (3, 6). Duplicate cultures of the E. coli strain GM37 (8) and its hns-205::Tn10 and hns2 derivatives GM230 and CJD829 were grown overnight at 37°C in M9 minimal medium supplemented with 0.4% (wt/vol) succinate, 0.2% (wt/vol) Casamino Acids, and 5 mM β-methyl-d-glucoside, and phospho-β-glucosidase activity was assayed in growing cells (13). The results (Table 1) show that the hns-205::Tn10 allele causes no derepression of bgl expression, while the hns2 allele allows a sigificant level of bgl expression. This is consistent with the model proposed by Ueguchi et al. (21) in which N-terminal mutations in H-NS lead to bgl derepression whereas the H-NS C terminus is dispensable for its repression function at this promoter.
TABLE 1.
Effects of hns and stpA mutations and plasmids on expression of the bgl operon as assayed by phospho-β-glucosidase activity
| Strain | Relevant genotype | Phospho-β-glucosidase activitya |
|---|---|---|
| GM37 | Wild type | 0.78 (0.05) |
| CJD1124 | ΔstpA::Tcr | 0.51 (0.03) |
| CJD898 | hnrG::Apr | 0.59 (0.05) |
| GM230 | hns-205::Tn10 | 0.59 (0.04) |
| CJD899 | hns-205::Tn10 hnrG::Apr | 1.03 (0.02) |
| CJD1126 | hns-205::Tn10 ΔstpA::TcrhnrG::Apr | 57.3 (0.42) |
| CJD1126(pYCStpA) | hns-205::Tn10 ΔstpA::TcrhnrG::Apr (StpA+) | 0.54 (0.03) |
| CJD1126(pYCStpAΔ65C) | hns-205::Tn10 ΔstpA::TcrhnrG::Apr (StpA−) | 64.5 (1.37) |
| CJD829 | hns2 | 83.0 (3.17) |
| CJD1125 | hns2 ΔstpA::Tcr | 105 (5.66) |
Duplicate cultures were assayed; mean values, in relative units calculated as for β-galactosidase, are shown. Standard deviations are in parentheses.
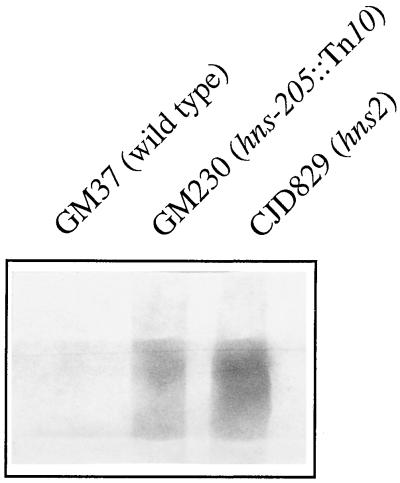
We also examined the effect of these two hns mutations on expression of the stpA gene. This was to rule out the possibility that the above-mentioned effects on bgl expression could be explained by a differential induction of StpA by the two alleles, which might substitute for H-NS in repression of bgl. Strains GM37, GM230, and CJD829 were grown in L broth at 37°C to an optical density at 600 nm of ≈0.6, and total cellular RNA was isolated as described previously (4). Five-microgram aliquots of these RNA samples were electrophoresed on a 1.5% 3′-(N-morpholino)propanesulfonic acid (MOPS)-formaldehyde-agarose gel, transferred to a Hybond-N membrane, and probed for the stpA transcript, using a specific RNA probe by previously published methods (4, 5). The blot (Fig. 1) shows significant induction of the stpA transcript in both hns mutants; the level of induction is slightly higher in the hns2 mutant CJD829 than in the hns-205::Tn10 strain GM230. Western blotting with a polyclonal antiserum that recognizes both H-NS and StpA suggested that levels of StpA protein are also similar in the two hns mutant strains (data not shown). This is inconsistent with the continued repression of bgl in GM230 being due to an enhanced induction of StpA in this strain compared to that in CJD829.
FIG. 1.
Northern blot of the stpA transcript from wild-type strain GM37 and its hns mutant derivatives GM230 and CJD829.
StpA is necessary for repression of bgl by truncated H-NS.
We then used P1cml transduction (18) to introduce the ΔstpA::Tcr allele (24) into GM37 and also into the hns mutant CJD829, creating strains CJD1124 and CJD1125. Because both the ΔstpA::Tcr allele and the hns-205::Tn10 allele encode tetracycline resistance, we used a derivative of GM230 containing the linked hnrG::Apr allele (CJD899 [3]) as a P1cml donor to transfer the hns-205::Tn10 allele into the ΔstpA::Tcr strain CJD1124, generating CJD1126. The hns-205::Tn10 and hnrG::Apr alleles are ≈95% linked, and cotransduction of hns-205::Tn10 into CJD1126 was verified by Southern blotting (data not shown). Duplicate cultures of these strains were then grown overnight in M9 medium supplemented with 0.4% (wt/vol) succinate, 0.2% (wt/vol) Casamino Acids, and 5 mM β-methyl-d-glucoside, and phospho-β-glucosidase was assayed as before (Table 1). It can be seen that the stpA mutation on its own causes no derepression of bgl expression, which is consistent with the dominant effect of H-NS over StpA in many systems in wild-type strains (24) (see below). Likewise, the combination of the ΔstpA::Tcr and hns2 mutations leads to little extra derepression of bgl over that seen in the hns2 single mutant strain despite the fact that stpA expression is strongly induced in this strain (Fig. 1), suggesting that StpA by itself is a poor repressor of bgl. However, when the stpA mutation is combined with the hns-205::Tn10 allele, strong derepression of bgl expression to a level similar to that in the hns2 mutant is observed. Thus, the continued repression of bgl by the truncated H-NS protein produced by hns-205::Tn10 strains is absolutely dependent upon the presence of StpA. The most likely explanation for this observation is that StpA provides the DNA-binding function allowing this fragment of H-NS to be targeted to and repress the bgl promoter. However, it remains possible that StpA is merely required for the synthesis of a second protein which functions as a corepressor at the bgl promoter. As a control, we confirmed that the hnrG::Apr allele used as a transductional marker has no effect on bgl expression (Table 1).
To investigate further the role of StpA in the presence of the hns-205::Tn10 mutation, we transformed CJD1126 with plasmid pYCStpA, a pACYC184 derivative carrying the wild-type stpA gene (23). pYCStpA was able to restore full repression of phospho-β-glucosidase production, as predicted if the plasmid-encoded StpA protein can act as a DNA-binding adapter for the truncated H-NS derivative produced in this strain. Moreover, when plasmid pYCStpAΔ65C, which encodes a truncated StpA derivative lacking the presumptive DNA-binding domain (23), was transformed into CJD1126, repression was not restored (Table 1), suggesting that DNA binding by StpA is essential for this repression mechanism to operate.
The combined effect of H-NS and StpA on bgl expression contrasts with that on proU expression.
We sought to compare the effects of these combinations of hns and stpA alleles on bgl repression with their abilities to repress the proU operon at low osmolarity. The wild-type and mutant strains previously assayed for bgl expression were grown overnight in L broth; under these low-osmolarity conditions the proU-lacZ fusion in GM37 remains repressed (8) (Table 2). When β-galactosidase activity in the mutant strains was assayed (as described by Miller [11]), both the hns-205::Tn10 and hns2 alleles were seen to allow significant derepression of proU, although this was greater in the latter case (Table 2). The ΔstpA::Tcr mutation had no effect on proU expression when H-NS was present but led to further enhanced transcription of the fusion when combined with either of the hns alleles. This demonstrates that H-NS is the dominant repressor of proU in a wild-type cell, as is the case for bgl, but that StpA exerts a measurable negative effect on proU expression when it is induced in an hns mutant strain. The significant derepression of proU in the hns-205::Tn10 strain GM230, in contrast to the continued repression of bgl in this strain, implies that StpA cannot provide a DNA-binding function to allow the truncated H-NS molecule to function as a repressor at this promoter, presumably because the H-NS′-StpA-DNA complex thus formed is not conformationally suited to repression (see, however, the discussion below). Interestingly, it has recently been shown that merely targeting H-NS to the vicinity of the proU promoter is insufficient to repress transcription (10), suggesting that the architecture of the repressive nucleoprotein complex formed by H-NS at this promoter is more subtle than that at bgl.
TABLE 2.
Effects of hns and stpA mutations on expression of the proU-lacZ fusion in GM37 and its derivatives as assayed by β-glucosidase activity
| Strain | Relevant genotype | β-Galactosidase activitya |
|---|---|---|
| GM37 | Wild type | 8.57 (0.07) |
| CJD1124 | ΔstpA::Tcr | 10.6 (0.29) |
| CJD898 | hnrG::Apr | 6.87 (0.07) |
| GM230 | hns-205::Tn10 | 117 (2.83) |
| CJD899 | hns-205::Tn10 hnrG::Apr | 101 (0) |
| CJD1126 | hns-205::Tn10 ΔstpA::TcrhnrG::Apr | 457 (8.49) |
| CJD829 | hns2 | 355 (0.71) |
| CJD1125 | hns2 ΔstpA::Tcr | 409 (12.0) |
Assays were carried out in duplicate; mean values, in Miller units, are shown. Standard deviations are in parentheses.
Conclusions.
By combining two different hns mutations with a knockout allele of stpA, we have shown that the presence of the StpA protein is necessary for the ability of a truncated N-terminal fragment of H-NS to repress the bgl promoter. Derepression of bgl in this hns-205::Tn10 ΔstpA::Tcr double mutant can be complemented by wild-type StpA but not truncated StpA lacking the presumptive DNA-binding domain, expressed from a multicopy plasmid. This suggests that StpA acts as an adapter molecule which provides a DNA-binding function to target this truncated H-NS′ protein to the bgl regulatory region, although it remains possible that the effect of StpA is indirect. In contrast, in a strain containing a mutation in hns which leads to the production of no detectable H-NS protein fragment, the stpA mutation has only a limited derepressive effect on β-glucosidase production, suggesting that StpA by itself is a rather poor repressor of the bgl operon. This is despite the significant induction of the chromosomal stpA gene in this strain and implies that differences in the N-terminal regions of H-NS and StpA affect their respective repression abilities. These results contrast with those obtained when the same strains are assayed for derepression of the H-NS-regulated proU operon at low osmolarity. In this case the operon is derepressed no matter which hns mutation is present, although again StpA may partially substitute as a low-efficiency repressor in an H-NS-independent fashion at this promoter.
Although the situation in which StpA is coexpressed in the cell with a truncated derivative of H-NS is an artificial one, the implications of these results for wild-type cells are twofold. Firstly, they provide further evidence for the different mechanisms of H-NS-mediated repression in operation at the bgl and proU promoters. The conclusion of Ueguchi et al. (21) that an H-NS DNA-binding function is not necessary for repression of bgl is taken further by the demonstration that what is necessary is the DNA-binding function of a related protein (StpA) to substitute. Therefore, it seems that the bgl promoter is quite tolerant of substantial changes in the nature of the repressor-DNA complex in its regulatory region in a way that the proU promoter is not. In this context, it is interesting that a heterologous DNA curve-binding protein, Btcd from mouse cells, is able to substitute for H-NS in bgl regulation but not in proU regulation (20), confirming that the bgl regulatory system is much more amenable to manipulation. This may in itself aid mechanistic studies on the role of H-NS in repressing this promoter. Intriguingly, Btcd can also restore flagellar expression to an hns mutant strain (20), as can an H-NS-related protein from Bordetella pertussis (7), suggesting that the bgl system is not unique in possessing this flexibility.
The second implication of these results may be for that class of H-NS-regulated promoters which, like proU, are stringent in their requirements for H-NS-mediated repression. In these systems, any subtle changes in the nature of the H-NS-DNA complex could be sufficient to affect the regulation of the promoter and may be important for transcription under inducing conditions. It is noteworthy, though, that while proU is significantly (>10-fold) activated by the hns-205::Tn10 allele, this activation is still 3- to 4-fold less than that caused by hns2 or by combining hns and stpA mutations. Therefore, there may be a component of proU repression which can be mediated by StpA in conjunction with truncated H-NS, on top of which the more stringent H-NS-dependent repression works. Elucidation of the molecular mechanisms of proU repression and induction will be required in order to evaluate these hypotheses.
Acknowledgments
This work was financed by Wellcome Trust grant 044711/Z/95/Z. A.F. was supported by a Wellcome Trust Prize Research Fellowship.
We thank Marlene Belfort for supplying the ΔstpA::Tcr mutant, Henri Buc and Sylvie Rimsky for sending the plasmids pYCStpA and pYCStpAΔ65C and for helpful discussions, and Megan Porter for constructing strain CJD1126. We also acknowledge useful discussions with members of the Dorman laboratory.
REFERENCES
- 1.Defez R, De Felice M. Cryptic operon for β-glucoside metabolism in Escherichia coli K-12: genetic evidence for a regulatory protein. Genetics. 1981;97:11–25. doi: 10.1093/genetics/97.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dersch P, Kneip S, Bremer E. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol Gen Genet. 1994;245:875–889. doi: 10.1007/BF00283274. [DOI] [PubMed] [Google Scholar]
- 3.Free A. H-NS and the regulation of transcription in Escherichia coli. Ph.D. thesis. Dublin, Ireland: Trinity College, University of Dublin; 1995. [Google Scholar]
- 4.Free A, Dorman C J. Escherichia coli tyrT gene transcription is sensitive to DNA supercoiling in its native chromosomal context: effect of DNA topoisomerase IV overexpression on tyrT promoter function. Mol Microbiol. 1994;14:151–161. doi: 10.1111/j.1365-2958.1994.tb01275.x. [DOI] [PubMed] [Google Scholar]
- 5.Free A, Dorman C J. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J Bacteriol. 1997;179:909–918. doi: 10.1128/jb.179.3.909-918.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Free, A., and C. J. Dorman. Unpublished data.
- 7.Goyard S, Bertin P. Characterization of BpH3, an H-NS-like protein in Bordetella pertussis. Mol Microbiol. 1997;24:815–823. doi: 10.1046/j.1365-2958.1997.3891753.x. [DOI] [PubMed] [Google Scholar]
- 8.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 9.Hulton C S J, Seirafi A, Hinton J C D, Sidebotham J M, Waddell L, Pavitt G D, Owen-Hughes T, Spassky A, Buc H, Higgins C F. Histone-like protein H1 (H-NS), DNA supercoiling and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 10.Jordi B J A M, Fielder A E, Burns C M, Hinton J C D, Dover N, Ussery D W, Higgins C F. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J Biol Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- 11.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 12.Mukerji M, Mahadevan S. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol Microbiol. 1997;24:617–627. doi: 10.1046/j.1365-2958.1997.3621725.x. [DOI] [PubMed] [Google Scholar]
- 13.Prasad I, Schaefler S. Regulation of the β-glucoside system in Escherichia coli K-12. J Bacteriol. 1974;120:638–650. doi: 10.1128/jb.120.2.638-650.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnetz K. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 1995;14:2545–2550. doi: 10.1002/j.1460-2075.1995.tb07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnetz K, Wang J C. Silencing of the Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 1996;24:2422–2428. doi: 10.1093/nar/24.12.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Bennett G N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shindo H, Iwaki T, Ieda R, Kurumizaka H, Ueguchi C, Mizuno T, Morikawa S, Nakamura H, Kuboniwa H. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS Lett. 1995;360:125–131. doi: 10.1016/0014-5793(95)00079-o. [DOI] [PubMed] [Google Scholar]
- 18.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 19.Sondén B, Uhlin B E. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 1996;15:4970–4980. [PMC free article] [PubMed] [Google Scholar]
- 20.Timchenko T, Bailone A, Devoret R. Btcd, a mouse protein that binds to curved DNA, can substitute in Escherichia coli for H-NS, a bacterial nucleoid protein. EMBO J. 1996;15:3986–3992. [PMC free article] [PubMed] [Google Scholar]
- 21.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 22.Ussery D W, Hinton J C D, Jordi B J A M, Granum P E, Seirafi A, Stephen R J, Tupper A E, Berridge G, Sidebotham J M, Higgins C F. The chromatin-associated protein H-NS. Biochimie. 1994;76:968–980. doi: 10.1016/0300-9084(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 23.Williams R M, Rimsky S, Buc H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]