Abstract
GDP–l-fucose, the substrate for fucosyltransferases for addition of fucose to polysaccharides or glycoproteins in both procaryotes and eucaryotes, is made from GDP–d-mannose. l-Fucose is a component of bacterial surface antigens, including the extracellular polysaccharide colanic acid produced by most Escherichia coli strains. We previously sequenced the E. coli colanic acid gene cluster and identified one of the GDP–l-fucose biosynthetic pathway genes, gmd. We report here the identification of the gene (fcl), located downstream of gmd, encoding the fucose synthetase.
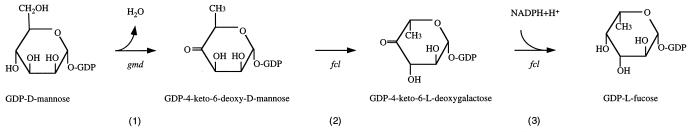
Fucose is quite widely present in bacterial polysaccharides and eucaryote glycoproteins, and it is known that GDP–l-fucose is the precursor of such fucose residues in both bacterial and eucaryote systems. A three-step pathway for converting GDP–d-mannose to GDP–l-fucose has been described (Fig. 1) (3). GDP-mannose dehydratase (GMD) converts GDP-mannose into GDP–4-keto-6-deoxymannose, the first step of the three-step pathway. This is followed by epimerase and reductase reaction steps to give GDP–l-fucose.
FIG. 1.
Pathway for the synthesis of GDP–l-fucose from GDP–d-mannose.
Colanic acid (CA) is an extracellular polysaccharide produced by most Escherichia coli strains, as well as by other species of the family Enterobacteriaceae (4). It contains l-fucose, d-glucuronic acid, d-glucose, d-galactose, and pyruvate (1, 2, 4, 5). We previously sequenced the E. coli K-12 CA gene cluster, which contains 17 genes, and identified some of these genes (9). GDP–d-mannose is synthesized from d-mannose-6-phosphate by two enzymes encoded by manB and manC, which are both found in the K-12 CA gene cluster, and we identified a gene, also located in the K-12 CA gene cluster, which encodes GMD and named it gmd (9). In this study; we identified a gene which is located in the K-12 CA gene cluster and encodes a protein that carries out both the epimerase and reductase reaction steps for GDP–l-fucose synthesis.
Cloning of the potential gene for the last two steps of GDP–l-fucose synthesis.
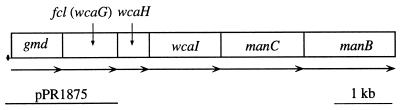
The genes for synthesis of a nucleotide sugar precursor are generally clustered together within the gene cluster for the particular bacterial polysaccharide (8). In the CA gene cluster of E. coli K-12, manC is followed by manB, as is common for this pair of genes, while gmd and manC are separated by three open reading frames (ORFs) (Fig. 2), and we therefore looked at these ORFs for the gene(s) encoding the remaining steps in the synthesis of GDP–l-fucose. One of them, wcaG, encodes a protein which shares 53% similarity with the human FX protein (11), which has been shown to carry out the last two steps to convert GDP–4-keto-6-deoxy-d-mannose to GDP–l-fucose (Fig. 3).
FIG. 2.
Organization of the GDP–l-fucose biosynthetic pathway genes of the K-12 CA gene cluster. The potential Shine-Dalgarno sequence upstream of gmd is shown as a black dot, and the insert of plasmid pPR1875 is also indicated. The arrows indicate the direction of transcription.
FIG. 3.
Alignment of FX protein with amino acid sequences of Fcl proteins from the E. coli K-12 CA gene cluster (Ec_CA [WcaG]), the E. coli O157 O-antigen gene cluster (Ec_O157 [Orf8]), and the O-antigen cluster of Y. enterocolitica O8 (Ye_O8 [WbcJ]). Positions where more than 50% of the sequences have identical amino acids are shaded.
K-12 CA DNA from positions 8621 to 10722, carrying both gmd and wcaG, was PCR amplified by using oligonucleotides 861 (5′ GAGGAATAATCCATGGCAAAAGTCGC) and 1057 (5′ TAAAGGTACCTTACCCCCGAAAGCGGTC). gmd starts at position 8633 with the potential Shine-Dalgarno sequence located from positions 8622 to 8625.
The PCR product was directly cloned into pGEM-T to make plasmid pPR1857, in which gmd and wcaG are under the control of the T7 phage promoter of the vector. Plasmids pPR1857 and pGP1-2 (encoding the T7 RNA polymerase gene [10]) were cotransformed into K-12 strain SØ874 (Table 1), which lacks both O-antigen and CA gene clusters, to make strain P5470. The expression of both the gmd and wcaG genes was confirmed by inducing the expression of the T7 RNA polymerase gene and [35S]methionine labelling of GMD and WcaG (data not shown).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Laboratory stock no. | Relevant characteristics | Reference or source |
|---|---|---|---|
| E. coli K-12 strain SØ874 | P4052 | Δ(sbcB-dcd) | 7 |
| P5470 | P4052 carrying plasmids pPR1857 and pGP1-2 | This study | |
| P5282 | P4052 carrying plasmids pGEM-T and pGP1-2 | This study | |
| Plasmids | |||
| pGEM-T | Cloning vector; Apr; replicon, pMB1 | Promega | |
| pGP1-2 | Bacteriophage T7 RNA polymerase gene cloned into pACYC177; replicon, p15A | 10 | |
| pPR1857 | K-12 CA DNA from positions 8621 to 10722 cloned into pGEM-T | This study |
wcaG (fcl) encodes the fucose synthetase.
We assayed the synthesis of GDP–l-fucose from GDP–d-mannose by the cell lysate of P5470 and detected the conversion of GDP–d-mannose to GDP–l-fucose by high-pressure liquid chromatography (HPLC) and thin-layer chromatography (TLC). Cell lysates were prepared as previously described (9). The reactions were carried out at 37°C for 90 min as described by Tonetti et al. (11), and the incubation mixture contained 50 μM Tris-HCl (pH 8.0), 5 μM MgCl2, 0.2 μM NAD+, 0.2 μM NADPH, 5 μM ATP, and 10 μM nicotinamide in a total volume of 1 ml with 400 μl of cell lysate (7.4 mg of protein per ml) and 0.025 μCi of GDP–[U-14C]mannose as the substrate. The reaction was stopped by heating for only 20 s at 100°C to reduce the amount of degradation (6).
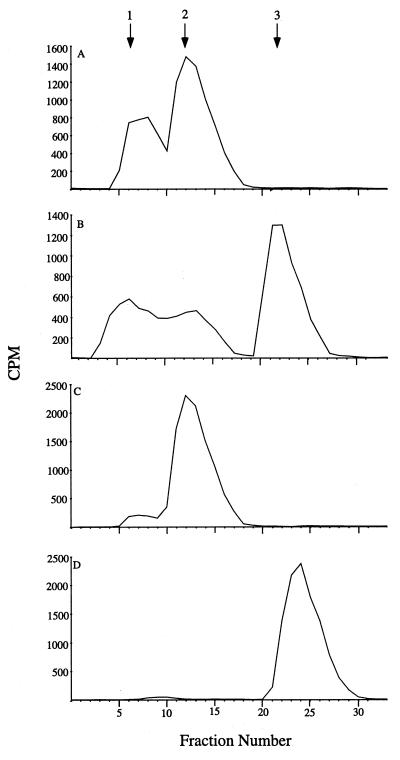
HPLC was carried out as described by Martin et al. (6). A 0.1-ml volume of 1 M sodium perchlorate (pH 4.1) was added to 0.2 ml of the reaction mixture, which was incubated for 15 min before the addition of 0.2 ml of 1 M potassium acetate. The samples were then subjected to a freeze-thaw cycle and centrifuged to achieve complete precipitation of potassium perchlorate. A 200-μl volume of the supernatant was filtered and injected onto a reverse-phase C18 octyldecyl silane-224 Spheri-5 (220 by 4.6 mm) column (Brownlee Labs) with 0.5 M KH4PO4 (pH 4.6) as the mobile phase at a 0.8-ml/min flow rate and 25°C. Fractions (0.4 ml) of the eluate were collected, and the radioactivity was measured in a MINAXI liquid scintillation counter. The results are shown in Fig. 4. A reaction mixture without the cell lysate but subjected to incubation and 100°C treatment is shown in Fig. 4C, and a reaction mixture with GDP–l-[14C]fucose substituted for GDP–[U-14C]mannose is shown in Fig. 4D. GDP–d-[U-14C]mannose and GDP–l-[U-14C]fucose were used as standards for HPLC (data not shown) and gave the same major peaks as in Fig. 4C and D, respectively.
FIG. 4.
Identification of 14C-labelled GDP–l-fucose (peak 3) and GDP–d-mannose (peak 2) by HPLC on a C18 column. A, reaction product obtained by using the cell lysate of P5282. B, reaction product obtained by using the cell lysate of P5470. C and D, 14C-labelled GDP–d-mannose and GDP–l-fucose treated the same way as in A and B but without addition of the cell lysate. The data presented is from a representative experiment.
The conversion of GDP–[U-14C]mannose (peak 2) to GDP–l-[14C]fucose (peak 3) by the cell lysate of P5470 is shown in Fig. 4B. Peak 1 represents the chemically and enzymatically degraded product of GDP–d-mannose: GDP–d-mannose was degraded to produce a small amount of material in peak 1 when no cell lysate was present in the incubation mixture (Fig. 4C) or the reaction was stopped immediately after addition of the cell lysate (data not shown), while after incubation with the cell lysate for 90 min, about twice that amount of material was produced (Fig. 4A [note the different scales for counts per minute]). A reaction mixture containing a cell lysate of strain P5282 (SO/874 carrying pGEM-T and pGP1-2) was used as a control and was not able to convert GDP–[U-14C]mannose to GDP–l-[14C]fucose (Fig. 4A).
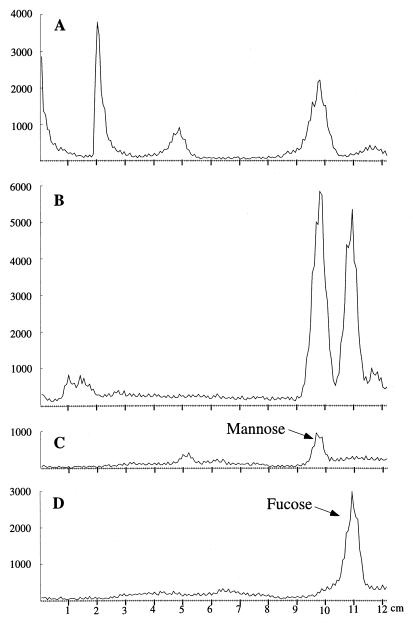
To further confirm the identity of the reaction product, TLC analysis of the monosaccharides derived by acid hydrolysis of the nucleoside diphosphate sugars was carried out. Aliquots (1 ml) of the reaction mixture or nucleoside diphosphate sugar standards were hydrolyzed by addition of 100 μl of 2 N HCl and incubation at 100°C for 20 min, followed by neutralization with 100 μl of 2 N NaOH and desalting with an Amberlite MB20 column before being freeze-dried. Samples were then resuspended in 30 μl of water and spotted on silica gel 60 TLC plates which had been pretreated with 0.1 M Na2S2O5 and 0.009 M sodium citrate, pH 4.8, and dried at 120°C for 30 min. As shown in Fig. 5, the sample from P5470 had both fucose and mannose while the sample from P5282 had only mannose.
FIG. 5.
TLC analysis of the 14C-labelled monosaccharides obtained by acid hydrolysis of the nucleoside diphosphate sugars in the reaction products of P5282 (A) and P5470 (B). 14C-labelled GDP–d-mannose (C) and GDP–l-fucose (D) were hydrolyzed and used as standards. Authentic unlabelled fucose and mannose were also used as standards (data not shown). TLC was carried out on a Merck silica plate (20 by 20 cm) with a solvent comprising 50 ml of n-butanol, 30 ml of pyridine, and 20 ml of 0.1 M HCl. The plate was then dried, the distribution of radioactivity was quantified by using a PhosphorImager 400B (Molecular Dynamics), and the data was analyzed by using the Molecular Dynamic ImageQuant software package. The plots show relative pixel values for radioactively labelled sugars obtained from the PhosphorImager.
The results show that a clone carrying gmd and wcaG confers the ability to synthesize GDP-fucose from GDP-mannose, and we conclude that wcaG encodes an enzyme which resembles its human homolog, FX, in being able to catalyze the two steps required to convert GDP–4-keto-6-deoxymannose to GDP–l-fucose. We therefore renamed the gene fcl.
We are aware that we have not demonstrated directly that Fcl carries out each of the two reactions (reactions 2 and 3 of Fig. 1) and that it is formally possible that it carries out only one of the reactions while the other is carried out by a protein present in E. coli K-12 strain SØ874. However, strain SØ874 has a deletion which extends to dcd (7) and so will lack both O-antigen and CA clusters, including the gmd and fcl genes of the CA gene cluster and the manB and manC genes of both the CA and O-antigen gene clusters. It would be remarkable if this strain carried a gene for just one step of the five-step pathway for synthesis of GDP-fucose from d-mannose-6-phosphate. We feel confident that all four steps are encoded in the CA gene cluster and that Fcl must have both functions.
We know of three sequenced gene clusters for bacterial polysaccharides which contain fucose, i.e., those for CA, the E. coli O157 cluster (11a), and the O-antigen cluster of Yersinia enterocolitica O8 (12). All three clusters contain manB, manC, gmd, and fcl genes, as judged by sequence similarities (Fig. 3), but no other genes are common to all three. This provides strong supporting evidence for the assignment of fcl, and we suggest that wbcJ of Y. enterocolitica O8 should therefore be known as fcl. The function(s) of the two remaining genes between fcl and manC is unknown, but it is of interest that one of them, wbdQ, is a homolog of wcaH in the E. coli O157 cluster (11a), where it is again immediately after fcl, but in this case there is no gene between it and manC.
Acknowledgments
We thank Darryl Nelson and Trevor Duxbury for discussions and help with HPLC.
This work was supported by a grant from the Australian Research Council.
REFERENCES
- 1.Anderson E S, Rogers A H. Slime polysaccharide of the Enterobacteriaceae. Nature. 1963;198:714–715. [Google Scholar]
- 2.Garegg P J, Lindberg B, Onn T, Sutherland I W. Comparative structural studies on the M-antigen from Salmonella typhimurium, Escherichia coli and Aerobacter cloacae. Acta Chem Scand. 1971;25:2103–2108. doi: 10.3891/acta.chem.scand.25-2103. [DOI] [PubMed] [Google Scholar]
- 3.Ginsburg V. Studies of the biosynthesis of guanosine diphosphate L-fucose. J Biol Chem. 1961;236:2389–2393. [PubMed] [Google Scholar]
- 4.Grant W D, Sutherland I W, Wilkinson J F. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J Bacteriol. 1969;100:1187–1193. doi: 10.1128/jb.100.3.1187-1193.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markovitz A. Genetics and regulation of bacterial capsular polysaccharide synthesis and radiation sensitivity. In: Sutherland I W, editor. Surface carbohydrates of the prokaryotic cell. I. New York, N.Y: Academic Press, Inc.; 1977. pp. 415–462. [Google Scholar]
- 6.Martin A, Ruggiero-Lopez D, Broquet P, Richard M, Louisot P. High-performance liquid chromatographic study of GDP-mannose and GDP-fucose metabolism. J Chromatogr. 1989;497:319–325. doi: 10.1016/0378-4347(89)80036-2. [DOI] [PubMed] [Google Scholar]
- 7.Neuhard J, Thomassen E. Altered deoxyribonucleotide pools in P2 eductants of Escherichia coli K-12 due to deletion of the dcd gene. J Bacteriol. 1976;126:999–1001. doi: 10.1128/jb.126.2.999-1001.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeves P R. Biosynthesis and assembly of lipopolysaccharide. New Compr Biochem. 1994;27:281–314. [Google Scholar]
- 9.Stevenson G, Andrianopoulos K, Hobbs H, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tonetti M, Sturla L, Bisso A, Benatti U, De Flora A. Synthesis of GDP-L-fucose by the human FX protein. J Biol Chem. 1996;271:27274–27279. doi: 10.1074/jbc.271.44.27274. [DOI] [PubMed] [Google Scholar]
- 11a.Wang, L., and P. R. Reeves. Unpublished data.
- 12.Zhang L, Radziejewska-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O8. Mol Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]