Abstract
The O antigen is an important cell wall antigen of gram-negative bacteria, and the genes responsible for its biosynthesis are located in a gene cluster. We have cloned and sequenced the DNA segment unique to the O-antigen gene cluster of Salmonella enterica group D3. This segment includes a novel O-antigen polymerase gene (wzyD3). The polymerase gives α(1→6) linkages but has no detectable sequence similarity to that of group D2, which confers the same linkage. We find the remnant of a D3-like wzy gene in the O-antigen gene clusters of groups D1 and B and suggest that this is the original wzy gene of these O-antigen gene clusters.
Lipopolysaccharide (LPS), an important component of the outer membrane of Gram-negative bacteria, usually consists of three distinct regions: lipid A, core oligosaccharide, and O-specific polysaccharide (O antigen). O antigens consist of repeats of an O unit of generally two to six sugars. In Salmonella enterica, there are 46 types of O antigens (32), which differ in sugar composition and in linkages between sugars and between O units. The O units of S. enterica groups B and D1 have the same backbone sugar residues, the only difference being the presence of different dideoxyhexose (DDH) branch sugars, with abequose in group B (12) and tyvelose in D1 (11) (Fig. 1). This difference determines distinctive epitopes for these two groups: O4 for group B and O9 for group D1.
FIG. 1.
(A) Structures of the O antigens of S. enterica groups B, D1, D1 carrying phage P27, D2, D3, and E1. Abbreviations: Abe, abequose; Gal, galactose; Man, mannose; Rha, rhamnose; Tyv, tyvelose. O-antigen epitopes are also shown. (B) Proposed epitopes of O antigen of S. enterica serovar Zuerich group D3. The bold outline around the oligosaccharide unit symbolizes the importance of the sugars in the structure of the epitopes; the thicker the line, the greater the participation of the sugars in the combining site. Dotted lines indicate that the additional sugars may weakly participate in the expression of the epitopes (modified after Nghiem et al. [28]).
The O unit of S. enterica group D2 (O9,46) differs from that of group D1 (O9,12) in the anomeric configuration of the mannosyl residue (reviewed by Kauffmann [15]), with α-mannose in group D1 (11) and β-mannose in group D2 (7, 10, 27), and in the galactosyl-mannose linkage formed on O-antigen polymerization, which is α(1→2) in group D1 and α(1→6) in group D2 (10, 11) (Fig. 1). The O unit of S. enterica group E1 (O3,10) has the same backbone as that of group D2, with the tyvelose side branch absent (34) (Fig. 1).
S. enterica group D3 (O9,12,27,46) has the α(1→6) galactosyl-mannose linkage of group D2 but two kinds of O unit with either α- or β-mannosyl residues (28–30) (Fig. 1); these different O units can coexist on the same O-polysaccharide chain (28, 30).
The biosynthesis of the O antigen begins with synthesis of nucleotide sugars and is followed by assembly of O units by sequential transfer of sugars onto undecaprenyl phosphate by transferases. The polymerization of O units is catalyzed by O-antigen polymerase, Wzy (Rfc). Finally, the O polysaccharides are ligated to lipid A-core to form LPS. The genes coding for the enzymes responsible for synthesis of the nucleotide sugars and the transferases necessary for O-unit assembly are located in the O-antigen gene cluster. The O-antigen gene clusters of S. enterica groups B, D1, D2, and E1 have been sequenced, and most genes in these clusters have been identified (13, 19–21, 36–38, 40). The transferase which catalyzes the mannosyl α(1→4) linkage, named WbaU (RfbU), is found in the O-antigen gene clusters of groups B and D1 (19), while the transferase which catalyzes the mannosyl β(1→4) linkage was named WbaO (RfbO) and is found in the O-antigen gene clusters of groups D2 and E1 (19) (Fig. 2). For most O antigens, the wzy gene is located in the O-antigen gene cluster (3, 17, 23, 25), and this is true for groups D2 and E1 (40). However, the wzy gene for the α(1→2) linkage in groups B and D1 is elsewhere on the chromosome (26). We have shown previously (40) that the group D2 O-antigen gene cluster comprises 5′ and 3′ components homologous to those of the D1 and E1 gene clusters, respectively, separated by an H-repeat (H-rpt)-like element (Fig. 2) which we suggested could have mediated a recombination event between D1 and E1 gene clusters to give the D2 cluster (40).
FIG. 2.
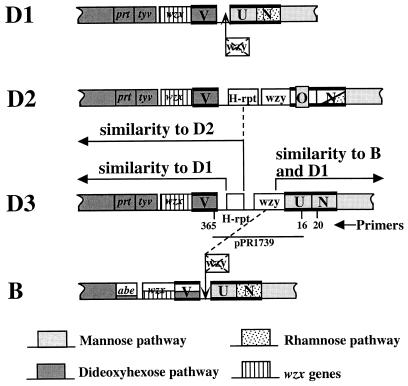
Central regions of the O-antigen gene clusters of S. enterica groups D1, D2, D3, and B. N, O, U, and V indicate genes wbaN, -O, -U and -V, respectively. Mannose, rhamnose (transferase only shown), DDH, and wzx genes are indicated by shading. Heavy bars above and below indicate transferase genes. Level of shading indicates degree of amino acid identity to homologous gene of group D1. Note that wbaO has barely detectable amino acid identity to wbaU. The wbaV-wbaU intergenic region of groups B and D1, shown to be remnant wzy genes, are indicated by a cross-out on the wzy gene. The similarities of the D3 cluster are indicated. The insert of plasmid pPR1739 is indicated, as are the binding sites for primers 365, 16, and 20, used for PCR of D3 DNA; primer 365 (TAGAATTCAAAGGGCTGGCTAGCTACA) is based on group D2 sequence (positions 314 to 332) (40), while primers 16 (GCGGTAGGCTTTAGAATA) and 20 (CTCTTGGAATCCAGAACG) are based on sequences of group B (positions 16160 to 16143 and 17238 to 17221, respectively) (13). The 5′ end of all four clusters (not shown) comprises rhamnose genes (rmlABCD) and DDH genes (ddhABCD), and the 3′ end comprises mannose genes (manBC) and the wbaP gene (not shown).
We have investigated the O-antigen gene cluster of S. enterica group D3 strain M840 (Table 1) which was used in a previous study (39). It had been proposed that group D3 strains may have two O-antigen gene clusters of the D1 and D2 types, which would provide a very simple explanation of the phenotype (24). However, Southern blotting with several probes for genes common to both provided no support for this possibility (39), and one would therefore expect to find an O-antigen gene cluster similar to that of D2, with the addition of a wbaU gene. Southern hybridization of chromosomal DNA of M840 with probes from the wbaOE1, wbaUB, and wzyE1 genes was positive only with the wbaUB probe (39) (subscripts indicate the groups from which the genes were derived). Further Southern hybridization (39) showed that the D3 cluster resembled that of group D1 upstream of wbaV and downstream of wbaU, with a region of difference in the center of the cluster. This finding suggests that strain M840 has novel β-mannosyl transferase and wzy genes in the central region. In this study, we examined the central region of the O-antigen gene cluster of D3 to determine the basis of the specific characteristics of the D3 O antigen.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Lab no. | Characteristics | Reference or source |
|---|---|---|---|
| E. coli K-12 DH10b | P5135 | araD139 Δ(ara-leu7697) galU galK mcrA Δ(mrr-hsdRMS-mcrBC) rpsL deoR, φ80dlacZDM15 endA1 nupG recA1 | 9 |
| S. enterica LT2 group B | |||
| LV386 | P9350 | wzy::aph (kanamycin, spectinomycin, and streptomycin resistant) | 4 |
| SL1654 | P9003 | hsdL6 trpC2 fliB flaA66 rpsL120 xylT404 ilvE452 metE551 metA22 hsdSA29 (streptomycin resistant) | 31 |
| P9487a | P9487 | SL1654 carrying wzy::aph (kanamycin and streptomycin resistant) | This study |
| S. enterica group D3 | |||
| 877 K | M840 | S. II sv. 1,9,12,46,27:C:Z39 (formerly called serovar Zuerich) | Pasteur Institute |
| 2764/79 | M1168 | S. II sv. 1,9,12,46,27:Z10:e,n,x | Pasteur Institute |
| 8316/94 | M1169 | S. II sv. 1,9,12,46,27:a:Z6 | Pasteur Institute |
| 149/66 | M1170 | S. II sv. 1,9,12,46,27:y:Z39 | Pasteur Institute |
| 3334/81 | M1171 | S. II sv. 1,9,12,46,27:Z10:1,5 | Pasteur Institute |
| Plasmids | |||
| pPR1739 | 3.2-kb PCR product from group D3 carrying wzy gene cloned into pGEM-T vector (ampicillin resistance) | This study | |
| pPR1820 | wzyD3 and wbaOE1 genes cloned into vector pPR637 (13) (spectinomycin and streptomycin resistance) | This study | |
| pPR618 | tyv and prt genes cloned into pUC18 (ampicillin resistance) | 37 |
Made as follows: chromosomal DNA of LV386 was sheared by vortexing and transformed into strain SL1654 by electroporation; a kanamycin-resistant recombinant was selected.
Sequencing the region between wbaV and wbaU of group D3.
PCR was used to amplify the region between wbaV and wbaU of strain M840, using primers (365 and 16) indicated in Fig. 2; a 3.2-kb fragment was obtained. This fragment was then cloned into pGEM-T (Promega), and Escherichia coli DH10b (Table 1) was used as the host strain for the resulting plasmid, pPR1739. The region between the end of wbaV and the beginning of wbaU of M840 was completely sequenced in both directions, using dye-primer sequencing kits (ABI), a Perkin-Elmer Cetus DNA Thermal Cycler, and an ABI model 377 Sequencer. The sequencing of this region was done by using primer walking with each internal region amplified and cloned. A 2,497-bp segment between the end of wbaV and the start of wbaU was sequenced.
The sequence was analyzed by using ANGIS (Australian National Genomic Information Service) (33) and the following programs: BESTFIT (5) and SEQH (14) for identification, similarity of DNA, or deduced amino acid sequences; ALOM (programs developed by M. Kanehisa, National Institutes of Health) for predication of potential transmembrane segments by the method of Klein et al. (16); and BLAST (1, 8) for database similarity searches. The first 877 bp are very similar to the corresponding region of group D2, including the last 50 bp of wbaV, 376 bp of noncoding DNA, followed by 451 bp of the H-rpt (insertion sequence-like element) (40). The average level of DNA sequence identity in this region is 78.7%. After the H-rpt sequence, there is a 418-bp noncoding region which has no detectable homology with any sequence in the database (GenBank), and then an open reading frame (ORF) of 1,176 bp (Fig. 2), with its stop codon overlapped by the start codon of the wbaU gene (ATGA).
Identification of the wzyD3 gene.
We were expecting an α(1→6) O-antigen polymerase gene and a β(1→4) mannose transferase gene. Analysis of the deduced amino acid sequence of the protein product of the only ORF (392 amino acids) showed no detectable sequence similarity to any protein in GenBank; however it has a hydropathy profile very similar to those of Wzy proteins, with 10 potential transmembrane segments and a large periplasmic loop (data not shown), making it a candidate for the predicted α(1→6) O-antigen polymerase gene.
Plasmid pPR1739 was transferred into S. enterica serovar Typhimurium strain C5 wzy mutant LV386 (Table 1), and LPS of the resulting strain was analyzed by electrophoresis using a sodium dodecyl sulfate–12.5% polyacrylamide gel (22) (Fig. 3). The results showed that LV386/pPR1739 produced long-chain O antigen, indicating that the wzy mutation could be complemented by the only ORF in pPR1739, which is thereby shown to be an O-antigen polymerase gene and named wzy. It is identified as wzyD3 in this report. Strain LV386/pPR1739 agglutinated with O27 antiserum in addition to O4 antiserum (Table 2), indicating that this polymerase ligates O units with an α(1→6) linkage. However, our O27 antiserum, which agglutinated LV386/pPR1739, did not agglutinate strain M840, which should be O1,9,27,46 (see below for explanation).
FIG. 3.
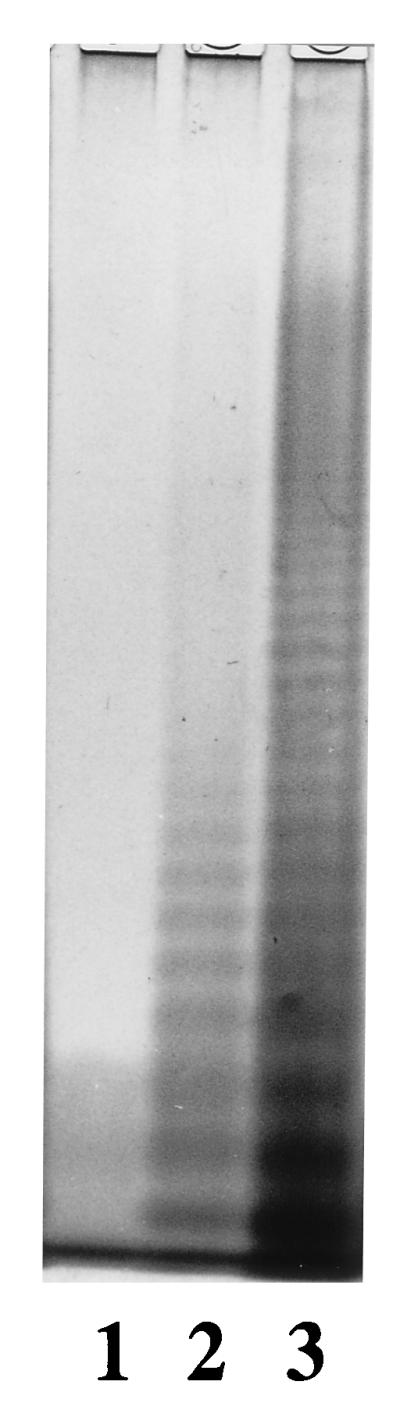
Identification of the wzyD3 gene by analysis of the LPS on a sodium dodecyl sulfate–12.5% polyacrylamide gel and silver staining. Lane 1, LV386 (S. enterica LT2 wzy mutant); lane 2, LV386/pPR1739; lane 3, SL1854 (LT2 wild type).
TABLE 2.
Identification of wzyD3 gene
| Strainb | Additional gene(s) | Expected O-antigen structure(s) | Presence of antigena
|
||
|---|---|---|---|---|---|
| O4 | O27 | O46 | |||
| LV386/pPR1739 | wzyD3 | -6-(αAbe-1,3-)-αMan-1,4-αRha-1,3-αGal]-1- | + | + | − |
| P9487/pPR1820 | wzyD3, wbaOE1 | -6-(αAbe-1,3-)-α(β)Man-1,4-αRha-1,3-αGal]-1- | + | + | − |
| P9487/pPR1820/pPR618 | wzyD3, wbaOE1, tyv, prt | -6-(αAbe-1,3-)-α(β)Man-1,4-αRha-1,3-αGal]-1- and | + | + | + |
| -6-(αTyv-1,3-)-α(β)Man-1,4-αRha-1,3-αGal]-1- | |||||
Detected by agglutination.
Strains LV386 and P9487 have all of the genes for group B O-antigen biosynthesis and are wzy mutants.
Group D3 has O units with both mannosyl α(1→4) and mannosyl β(1→4) linkages, and we investigated if both are polymerized by WzyD3 by combining the wzyD3 and β(1→4) mannose transferase genes in a wzy mutant. We made plasmid pPR1820, which carries the wbaO gene from group E1 and the wzyD3 in low-copy-number vector pPR637 (13). We cloned the 2.1-kb HincII-PstI fragment from pPR966 (positions 10045 to 12217 of the group E1 O-antigen gene cluster [38]) into HindIII (end-filled) and PstI sites on the polylinker of pPR637, followed by cloning of the 2.1-kb insert as a BglII-SacI (vector) fragment from pPR1739 into the resulting plasmid, using BamHI and SacI sites. Our only wzy mutant, LV386, was unexpectedly found to be spectinomycin and streptomycin resistant and therefore not suitable for transfer of pPR1820; therefore, we made P9487, a wzy mutant of LT2 (Table 1). Strain P9487/pPR1820 has α(1→6) polymerase, α-mannosyl, and β-mannosyl transferase genes and should produce two kinds of O antigen: -6-[(αAbe-1,3-)-αMan-1,4-αRha-1,3-αGal]-1-, and -6-[(αAbe-1,3-)-βMan-1,4-αRha-1,3-αGal]-1- (see the legend to Fig. 1 for abbreviations). Strain P9487/pPR1820 was agglutinated with O27 antiserum but not O46 antiserum (Table 2). Factors 27 and 46 have been related to the oligosaccharide -6-[(αDDH-1,3-)-α(β)Man-1,4-αRha-1,3-αGal]-1- with α-mannose for O27 and β-mannose for O46. Agglutination with O27 serum indicates that the wzyD3 gene in this construct is active. The failure of the O46 serum to agglutinate suggested that the β-mannosyl (1→4) linkage is absent in the polymer, but see below.
Plasmid pPR618, which carries prt (rfbS) and tyv (rfbE) genes of group D1, allowing synthesis of CDP-tyvelose in a group B strain (37), was transferred into P9487/pPR1820. Strain P9487/pPR1820/pPR618 could potentially produce two additional O antigens: -6-[(αTyv-1,3-)-αMan-1,4-αRha-1,3-αGal]-1- and -6-[(αTyv-1,3-)-βMan-1,4-αRha-1,3-αGal]-1-; this strain was agglutinated with O4, O9, O27, and O46 antisera (Table 2), the latter indicating the presence of the mannosyl β(1→4) linkage, showing that WzyD3 could polymerize O units containing β-mannose in addition to those containing α-mannose. It is interesting that the polymerase making the galactosyl-mannose linkage is not specific for the anomeric state of the mannosyl residue, although the situation is not known for the E1/D2 and group B polymerases, which have not been tested with the alternate form of the O unit.
Specificity of O27 and O46 sera.
In group B and D1 strains lysogenized by phage P27, a phage-encoded polymerase forms α(1→6) linkages between O units, conferring the new epitope, O27 (2, 18). The failure of the O27 serum to agglutinate the parental group D3 strain in our hands (see above) probably reflects the specificity of the O27 sera for the DDH. Distinct epitopes O27A, O27B, and O27D were distinguished in the early literature (35) as cross-reacting specificities for the O antigens of group A, B, and D1 strains, respectively, when lysogenized by phage P27. However, the current protocol for O27 serum (6) uses a lysogenized group B strain, and the Difco O27 antiserum used in this study had been raised by using S. enterica serovar Schleissheim (group B, phage 27-lysogenized strain [O1,4,12,27], absorbed with S. enterica serovars Paratyphi B [O1,4,5,12] and Essen [O4,12]) (4a). Given that three related but distinct specificities were recognized earlier, it probably has much lower specificity for tyvelose-containing O antigens than for those with abequose and hence at the dilution provided failed to agglutinate our group D3 strain.
We suggest that the current nomenclature, treating O27 as a single specificity, can be misleading as the standard O27 serum, made with a group B lysogen (6), does not appear to agglutinate all strains carrying the O27 specificity at the dilution present in commercial sera, the activity being dependent on the DDH present. This may well have led to underreporting of group D3 strains, which would appear as group D2 strains if the O27 serum was not effective at the dilution used. It is of interest that the O27 serum used in a specific study of group D3 (28) was made not by the standard protocol but by using a lysogenized group D1 strain, presumably to gain better reaction with the tyvelose-containing D3 O antigen.
The O46 epitope was identified in group D2 and is also present in group D3; agglutination by O46 serum requires the α(1→6) linkage between galactose and β-mannose (not α-mannose). The O antigen of group E1, which has the structure described above (Fig. 1), does not react to O46 serum (32), showing that DDH is also required. We suggest that the β-mannosyl linkage is present in both P9487/pPR1820/pPR618 and P9487/pPR1820, and the failure of the O46 serum to react with the O antigen of P9487/pPR1820 while reacting with P9487/pPR1820/pPR618 reflects a requirement for tyvelose (not abequose) as DDH. The O antigen of P9487/pPR1820 has a unique structure with α(1→6) linkage between galactose and β-mannose and with abequose as DDH. It is possible that as for O27, there are three forms of O46 with preference for abequose, paratose, and tyvelose, but this has not been tested by titration of sera against the three O-antigen forms.
Lack of the β-mannosyl transferase gene.
We did not find the expected β-mannosyl transferase gene in the O-antigen gene cluster of group D3. We therefore looked at regions between wbaV and wbaN of four additional group D3 strains (Table 1) together with M840 to determine whether M840 was typical. PCR using primers 365 and 20 (Fig. 2) showed that a 4.3-kb DNA fragment could be amplified from all five strains, indicating that the gene organization in this region is the same in all of them. The wbaO gene of group D3 has not been located but must have little or no sequence similarity to the known wbaO gene in group E1 and D2, and it might be present on a phage on elsewhere of the chromosome.
Ancestral form of group B and D1 O antigens.
The 318-bp remnant wzy gene of group B between wbaV and wbaU (bp 15061 to 15,378 [13]) has a high level of identity to the wzyD3 gene at the nucleic acid level (Fig. 4). The corresponding region of group D1 is more complex: there are 577 bp (positions 2303 to 2879 [21]), of which the first 344 show low-level similarity to wzyD2/E1 at the amino acid level (40), while the following 233-bp sequence has a high level of identity to wzyD3 (Fig. 4).
FIG. 4.
DNA sequence alignment of the second half of the wzyD3 gene and the intergenic regions of groups D1 and B. Numbering of the group D3 gene starts from the beginning of the gene; numberings of group D1 and group B sequences are from references 21 and 13, respectively. Asterisks indicate the overlap of the stop codon of the wzy gene and the start codon of the wbaU gene. Strike-through and italic sections of the hypothetical wzyB gene are alternate alignments of one sequence displayed twice to indicate similarity to two segments of wzyD3 which may represent the basis for deletion by homologous recombination.
Alignment of the functional wzyD3 gene with the remnants of such a gene in the wbaV-wbaU intergenic spaces of groups B and D1 (Fig. 4) gives a strong indication that groups B and D1 once shared a common wzy gene for an α(1→6) linkage in addition to their many other similarities: they now have an α(1→2) linkage wzy gene of unknown origin elsewhere on the chromosome (the original rfc gene of strain LT2). Presumably the ancestral α(1→6) wzy gene suffered inactivation and substantial deletion after the introduction of the new α(1→2) polymerase gene. There are possible parallels with E2 strains which have the E1 antigen gene cluster but in addition a bacteriophage (ɛ15) which encodes a β(1→6) linkage polymerase which functionally replaces the α(1→6) linkage polymerase of the E1 O-antigen cluster (24).
It is interesting that the presumed ancestral α(1→6) linkage was regained later in many group B strains by lysogenization with the P27 bacteriophage which carries an α(1→6) linkage polymerase (24), but in this case nothing is known of the wzy gene. O27 strains thought to be of this type comprise 74 of 144 group B serovars (32). The α(1→6) linkage is also present in group D2; in this case the gene involved is derived from group E1 (40).
Origins of the D3 O-antigen gene cluster.
The location of the H-rpt suggests that it was involved in transfer of the wzy gene to create the D3 gene cluster, just as we proposed for wzyE1 of D2. In the case of D2, both wzy and wbaO are almost identical to genes of group E1 whereas the rest of the gene cluster is from group D1, making an origin by recombination very convincing. However, in the case of D3, the one additional gene not present in group D1, wzyD3, is the wzy gene that we now also believe to have been in the ancestral O-antigen D1 and B gene clusters. It is not yet clear how the D3 gene cluster arose, as there are alternative possibilities. In some ways, the simplest hypothesis is that it derives from the ancestral D1 cluster by addition of an H-rpt, which subsequently suffered deletion of its downstream end. However, in that case we have no explanation for the presence of the H-rpt in the D3 cluster.
The alternative hypothesis is that the D3 cluster arose by transfer of the wzy gene mediated by the H-rpt. The insertion site of the H-rpt is the same in D2 and D3 (only the left-hand end survives in D3), suggesting that the same H-rpt insertion was involved in the formation of both D2 and D3. However, the sequence of events is not clear, as two different wzy genes are involved. The recipient would have been a D1 strain or B strain which had lost its α(1→6)-linkage wzy gene and regained it by this recombination. It is too early to speculate in detail, but the alignments are shown in Fig. 2.
Nucleotide sequence accession number.
The GenBank accession number of the sequence shown in Fig. 2 is AF017148.
Acknowledgments
This work was supported by a grant from the Australian Research Council.
We thank M. Hobbs for his useful discussion, and we thank C. Murray and staff at the Institute of Medical and Veterinary Science, Adelaide, Australia, and M. Y. Popoff at the Pasteur Institute, Paris, France, for helpful discussion on serology.
REFERENCES
- 1.Altschul A F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bagdian G, Luderitz O, Staub A M. Immunochemical studies on Salmonella. XI. Chemical modification correlated with conversion of group B by bacteriophage 27. Ann N Y Acad Sci. 1966;133:405–424. doi: 10.1111/j.1749-6632.1966.tb52380.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown P K, Romana L K, Reeves P R. Molecular analysis of the rfb gene cluster of Salmonella serovar Muenchen (strain M67): genetic basis of the polymorphism between groups C2 and B. Mol Microbiol. 1992;6:1385–1394. doi: 10.1111/j.1365-2958.1992.tb00859.x. [DOI] [PubMed] [Google Scholar]
- 4.Collins L V, Attridge S, Hackett J. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect Immun. 1991;59:1079–1085. doi: 10.1128/iai.59.3.1079-1085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Crossingham, C. (Difco Laboratories). Personal communication.
- 5.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewing W H. Edwards and Ewing’s identification of the Enterobacteriaceae. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. [Google Scholar]
- 7.Fukuda M, Egami F. A reinvestigation of the anomeric configuration of mannose in the antigens of Salmonella groups B, D and E. Eur J Biochem. 1971;20:438–441. doi: 10.1111/j.1432-1033.1971.tb01411.x. [DOI] [PubMed] [Google Scholar]
- 8.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 9.Grant S G, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellerqvist C G, Lindberg B, Pilotti A, Lindberg A A. Structural studies on the O-specific side-chains of the cell-wall lipopolysaccharide from Salmonella strasbourg. Acta Chem Scand. 1970;24:1168–1174. doi: 10.3891/acta.chem.scand.24-1168. [DOI] [PubMed] [Google Scholar]
- 11.Hellerqvist C G, Lindberg B, Svensson S, Holme T, Lindberg A A. Structural studies on the O-specific side chain of the cell wall lipopolysaccharide from Salmonella typhi and Salmonella enteritidis. Acta Chem Scand. 1969;23:1588–1596. doi: 10.3891/acta.chem.scand.23-1588. [DOI] [PubMed] [Google Scholar]
- 12.Hellerqvist C G, Lindberg B, Svensson S, Holme T, Lindberg A A. Structural studies on the O-specific side-chains of the cell-wall lipopolysaccharide from Salmonella typhimurium LT2. Carbohydr Res. 1969;9:237–241. [Google Scholar]
- 13.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 14.Kanehisha M J. Los alamos sequence analysis package for nucleic acids and proteins. Nucleic Acids Res. 1982;10:183–196. doi: 10.1093/nar/10.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauffmann F. Classification of bacteria. Copenhagen, Denmark: Munksgaard; 1975. [Google Scholar]
- 16.Klein P, Kanehisa M, Delisi C. The detection and classification of membrane spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 17.Klena J D, Schnaitman C A. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 18.Lindberg A A, Helleqvist C G, Bagdian-Motta G, Mäkelä P H. Lipopolysaccharide modification accompanying antigenic conversion by phage P27. J Gen Microbiol. 1978;107:279–287. [Google Scholar]
- 19.Liu D, Haase A M, Lindqvist L, Lindberg A A, Reeves P R. Glycosyl transferases of O-antigen biosynthesis in Salmonella enterica: identification and characterization of transferase genes of groups B, C2, and E1. J Bacteriol. 1993;175:3408–3413. doi: 10.1128/jb.175.11.3408-3413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Lindquist L, Reeves P R. Transferases of O-antigen biosynthesis in Salmonella enterica: dideoxhexosyl transferases of groups B and C2 and acetyltransferase of group C2. J Bacteriol. 1995;177:4084–4088. doi: 10.1128/jb.177.14.4084-4088.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, Verma N K, Romana L K, Reeves P R. Relationships among the rfb regions of Salmonella serovars A, B, and D. J Bacteriol. 1991;173:4814–4819. doi: 10.1128/jb.173.15.4814-4819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lugtenberg B, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the major outer membrane proteins of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 23.Lukomski S, Hull R A, Hull A I. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J Bacteriol. 1996;178:240–247. doi: 10.1128/jb.178.1.240-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mäkelä P H, Stocker B A D. Genetics of lipopolysaccharide. In: Rietschel E T, editor. Handbook of endotoxin. I. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. pp. 59–137. [Google Scholar]
- 25.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naide Y, Nikaido H, Mäkelä P H, Wilkinson R G, Stocker B A D. Semirough strains of Salmonella. Proc Natl Acad Sci USA. 1965;53:147–153. doi: 10.1073/pnas.53.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nghiem H O, Bagdian G, Staub A M. Études immunochimiques sur les Salmonella. 13. Détermination de la structure du polyoside spécifique d’une Salmonella du group D2 (S. strasbourg) Eur J Biochem. 1967;2:392–398. doi: 10.1111/j.1432-1033.1967.tb00151.x. [DOI] [PubMed] [Google Scholar]
- 28.Nghiem H O, Himmelspach K, Mayer H. Immunochemical and structural analysis of the O polysaccharides of Salmonella zuerich [1,9,27,(46)] J Bacteriol. 1992;174:1904–1910. doi: 10.1128/jb.174.6.1904-1910.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nghiem H O, Staub A M. Molecular immunological heterogeneity of the Salmonella zuerich [1, 9, 12, (46), 27] cell-wall polysaccharides. Carbohydr Res. 1974;40:153–169. doi: 10.1016/s0008-6215(00)82678-6. [DOI] [PubMed] [Google Scholar]
- 30.Nghiem H O, Staub A M, Galanos C, Luderitz O. Distribution and antigen properties of the O-determinants of Salmonella zeurich (1, 9, 27, 46) Eur J Biochem. 1982;125:431–436. doi: 10.1111/j.1432-1033.1982.tb06701.x. [DOI] [PubMed] [Google Scholar]
- 31.Ornellas E P, Stocker B A D. Relation of lipopolysaccharide character to P1 sensitivity in Salmonella typhimurium. Virology. 1974;60:491–502. doi: 10.1016/0042-6822(74)90343-2. [DOI] [PubMed] [Google Scholar]
- 32.Popoff M Y, Minor L L. Antigenic formulas of the Salmonella serovars, 7th revision. WHO Collaborating Centre for Reference and Research on Salmonella. Paris, France: Institut Pasteur; 1997. [Google Scholar]
- 33.Reisner A H, Bucholtz C A, Smelt J, McNeil S. Proceedings of the Twenty-Sixth Annual Hawaii International Conference on Systems Science. Vol. 1. 1993. Australia’s National Genomic Information Service; pp. 595–602. [Google Scholar]
- 34.Robbins P W, Uchida T. Chemical and macromolecular structure of O-antigens from Salmonella anatum strains carrying mutants of bacteriophage ɛ15. J Biol Chem. 1965;240:375–383. [Google Scholar]
- 35.Staub A M, Bagdian G. Etudes immunochimique sur les Salmonella. XII. Analyse immunologique des facteures 27A, 27B en 27D. Ann Inst Pasteur (Paris) 1966;110:849–852. [PubMed] [Google Scholar]
- 36.Verma N K, Quigley N B, Reeves P R. O-antigen variation in Salmonella spp.: rfb gene clusters of three strains. J Bacteriol. 1988;170:103–107. doi: 10.1128/jb.170.1.103-107.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma V, Reeves P R. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D Salmonella. J Bacteriol. 1989;171:5694–5701. doi: 10.1128/jb.171.10.5694-5701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Romana L K, Reeves P R. Molecular analysis of a Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics. 1992;130:429–443. doi: 10.1093/genetics/130.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang S-H. Variation in rfb gene clusters of Salmonella enterica and origin of group D2. Ph.D. thesis. Sydney, New South Wales, Australia: University of Sydney; 1995. [Google Scholar]
- 40.Xiang S H, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]