Abstract
The plant cell wall structure can be altered by pathogen-secreted polygalacturonases (PGs) that cleave the α-(1→4) linkages occurring between D-galacturonic acid residues in homogalacturonan. The activity of the PGs leads to cell wall maceration, facilitating infection. Plant PG inhibiting proteins (PGIPs) impede pathogen PGs, impairing infection and leading to the ability of the plant to resist infection. Analyses show the Glycine max PGIP11 (GmPGIP11) is expressed within a root cell that is parasitized by the pathogenic nematode Heterodera glycines, the soybean cyst nematode (SCN), but while undergoing a defence response that leads to its demise. Transgenic experiments show GmPGIP11 overexpression leads to a successful defence response, while the overexpression of a related G. max PGIP, GmPGIP1 does not, indicating a level of specificity. The analyses presented here have identified PGIPs from 51 additional studied proteomes, many of agricultural importance. The analyses include the computational identification of signal peptides and their cleavage sites, O-, and N-glycosylation. Artificial intelligence analyses determine the location where the processed protein localize. The identified PGIPs are presented as a tool base from which functional transgenics can be performed to determine whether they may have a role in plant-pathogen interactions.
Keywords: Plant interactions, Polygalacturonase inhibiting protein (PGIP), Soybean, Heterodera glycines, Beta vulgaris, Sugar beet
Specifications Table
| Subject | Biological sciences |
| Specific subject area | Omics: Genomics |
| Data format | Raw, Analysed, Filtered |
| Type of data | Table, Figure |
| Data collection |
Analysed proteomes The 11 G. max PGIP protein sequences are used in Basic Local Alignment Search Tool program (BLAST) searches of the proteomes (BLASTP) using the default parameters at Phytozome (http://www.phytozome.net/). The identified PGIP proteins are compiled using a Bitscore of 140 as a cutoff. To identify the PGIP proteins, each of the 11 G. max PGIP protein sequences are queried into the studied proteomes. The individual queries for GmPGIP1 through GmPGIP11 are stored in individual tabs in Excel. Then, the PGIPs that have Bitscores of 140 or higher are compiled for all of the queries for the individual GmPGIPs. The duplicate PGIPs then are removed in Excel. The analysis results in a list of PGIP proteins that include the products of alternate splicing so the numbers in some cases are higher than the numbers of genes in some genomes. Signal peptide prediction Signal peptide prediction is done using SignalP 6.0. The default parameters are used. O-glycosylation determination O-glycosylation is determined using NetOGlyc - 4.0. The parameters are set on default. N-glycosylation determination N-glycosylation is determined using NetNGlyc - 1.0. The parameters are on set default. Protein alignment Protein alignment is performed using CLUSTAL Omega, CLUSTAL O(1.2.4) multiple sequence alignment. The analysis is performed using default parameters. Artificial intelligence Prediction of eukaryotic protein subcellular localization using deep learning is done using DeepLoc-1.0 in default settings. |
| Data source location | Data obtained from Phytozome (http://www.phytozome.net/) |
| Data accessibility | Direct URL to data: https://data.mendeley.com/datasets/66r9pkckjz/1 |
1. Value of the Data
-
•
Why are these data valuable?
Plants have a 2-tiered defense platform allowing them to defend themselves from pathogens [1]. The plant recognizes epitopes produced directly or indirectly as a consequence of the plant-pathogen interaction [1]. The epitopes are collectively called pathogen activated molecular patterns (PAMPs) acting within a 2-tiered defense system involving PAMP (pattern) triggered immunity (PTI) and effector triggered immunity (ETI) [1].
Plant cell walls are an important barrier to pathogen infection. Up to 60% of the cell wall pectic moieties of dicot and nongraminaceous monocot primary cell walls are homogalacturonans (HGs), the major component of the middle lamella [2]. Pathogen polygalacturonases (PGs) are effective in facilitating pathogenicity because they break down cell wall polymers, permitting infection [3]. The study presented here is valuable to those interested in understanding plant defence, the evolutionary processes behind defence processes, cell signalling, and an understanding of basic cellular processes.
-
•
Who can benefit from these data?
In order to impede pathogen PGs, plants secrete polygalacturonase inhibiting proteins (PGIPs). PGIPs have a bimodal function. Firstly, PGIPs directly inhibit PGs. Secondly, PG activity leads to oligogalacturonide (OG) accumulation, eliciting a defence response [4]. Therefore, PGIPs deactivate the pathogen effector while also leading to the production and amplification of a signalling cascade. This signal cascade further impairs the pathogen, leading to their demise. For example, a Beta vulgaris (sugar beet) PGIP, when expressed in Nicotiana benthamiana, limits the pathogenicity of Rhizoctonia solani, Fusarium solani, and Botrytis cinerea whose pathogenicity is normally driven by their PGs [5]. Previous work on G. max PGIPs (GmPGIPs) have functionally examined them [6], benefitting stakeholders interested in the development of pathogen-resistant crops, including Beta vulgaris ssp. vulgaris (sugar beet). Novel signalling events can also be determined through the study presented here.
-
•
How can these data be reused by other researchers?
Using 11 G. max PGIP protein sequences, the analysis presented here extracts the PGIPs that exist in 51 additional genomes of other important crops and other flowering plants. Analyses determine whether the 469 proteins have signal sequences, compatible with them being secreted proteins, a cleavage site, and whether they are O- and/or N-glycosylated. Artificial Intelligence analyses show which cellular locale the proteins can be expected to exist, complementing recent transgenic studies of the GmPGIP11. The provided analysis and accompanying data can be re-used to basic aspects of plant cell biology and generate pathogen-resistance in a wide spectrum of agriculturally-important crops. The evolution of defence and signalling processes can also be examined.
2. Data Description
A total of 469 proteins obtained from Phytozome, not including G. max, are analysed, spanning 51 proteomes (Table 1, Supplemental Data File 1) [7]. The proteins annotated as probable PGIPs pass a cutoff between 300 and 399 AAs, within the range of known PGIPs. Among them, 394 putative PGIPs are between 300 and 399 AAs (84%). Among the 51 proteomes, 45 (9.6%) are shorter than 300 AAs. Furthermore, 30 (6.4%) proteins annotated as PGIPs are identified as being 400 AAs or larger. LRRS have a low overall homology based on the LRR composition. For example, BvPGIP6 (EL10Ac4g07809.1) is annotated as being 1,383 AAs. When a BLASTP analysis is run, it is shown to be homologous to the 1,249 AA GASSHO1 (GSO1) (OAO97463.1) as well as the 332 AA polygalacturonase inhibiting protein 1 (AAM65836.1). A re-annotation of the PGIPs is beyond the scope of the study.
Table 1.
The proteomes under study.
| Genome number | Species | genome | Order | Family |
|---|---|---|---|---|
| 1 | Amborella trichopoda | Amborella trichopoda v1.0 | Amborellales | Amborellaceae |
| 2 | Amaranthus hypochondriacus | Amaranthus hypochondriacus v2.1 | Caryophyllales | Amaranthaceae |
| 3 | Beta vulgaris | Beta vulgaris EL10_1.0 | Caryophyllales | Amaranthaceae |
| 4 | Chenopodium quinoa | Chenopodium quinoa v1.0 | Caryophyllales | Amaranthaceae |
| 5 | Spinacia oleracea | Spinacia oleracea Spov3 | Caryophyllales | Amaranthaceae |
| 6 | Coffea arabica | Coffea arabica v0.5 | Gentianales | Rubiaceae |
| 7 | Daucus carota | Daucus carota v2.0 | Apiales | Apiaceae |
| 8 | Helianthus annuus | Helianthus annuus r1.2 | Asterales | Asteraceae |
| 9 | Lactuca sativa | Lactuca sativa V8 | Asterales | Asteraceae |
| 10 | Mimulus guttatus | M.guttatus_TOL v5.0 | Lamiales | Phrymaceae |
| 11 | Olea europaea | Olea europaea v1.0 | Lamiales | Oleaceae |
| 12 | Solanum lycopersicum | Solanum lycopersicum ITAG4.0 | Solanales | Solanaceae |
| 13 | Solanum tuberosum | Solanum tuberosum v6.1 | Solanales | Solanaceae |
| 14 | Vaccinium darrowii | V.darrowii v1.2 | Ericales | Ericaceae |
| 15 | Eucalyptus grandis | Eucalyptus grandis v2.0 | Myrtales | Myrtaceae |
| 16 | Vitis vinifera | Vitis vinifera v2.1 | Vitales | Vitaceae |
| 17 | Arachis hypogaea | Arachis hypogaea v1.0 | Fabales | Fabaceae |
| 18 | Castanea dentata | Castanea dentata v1.1 | Fagales | Fagaceae |
| 19 | Cicer arietinum | Cicer arietinum v1.0 | Fabales | Fabaceae |
| 20 | Cucumis sativus | Cucumis sativus v1.0 | Cucurbitales | Cucurbitaceae |
| 21 | Fragaria vesca | Fragaria vesca v4.0.a2 | Rosales | Rosaceae |
| 22 | Malus domestica | Malus domestica v1.1 | Rosales | Rosaceae |
| 23 | Medicago truncatula | Medicago truncatula Mt4.0v1 | Fabales | Fabaceae |
| 24 | Phaseolus vulgaris | Phaseolus vulgaris v2.1 | Fabales | Fabaceae |
| 25 | Prunus persica | Prunus persica v2.1 | Rosales | Rosaceae |
| 26 | Quercus rubra | Quercus rubra v2.1 | Fagales | Fagaceae |
| 27 | Trifolium pratense | Trifolium pratense v2 | Fabales | Fabaceae |
| 28 | Carya illinoinensis | Carya illinoinensis v1.1 | Fabales | Juglandaceae |
| 29 | Vigna unguiculata | Vigna unguiculata v1.2 | Fabales | Fabaceae |
| 30 | Linum usitatissimum | Linum usitatissimum v1.0 | Malpighiales | Linaceae |
| 31 | Manihot esculenta | Manihot esculenta v8.1 | Malpighiales | Euphorbiaceae |
| 32 | Carica papaya | Carica papaya ASGPBv0.4 | Brassicales | Caricaceae |
| 33 | Theobroma cacao | Theobroma cacao v2.1 | Malvales | Malvaceae |
| 34 | Arabidopsis thaliana | Arabidopsis thaliana TAIR10 | Brassicales | Brassicaceae |
| 35 | Schrenkiella parvula | Schrenkiella parvula v2.2 | Brassicales | Brassicaceae |
| 36 | Brassica oleracea capitata | Brassica oleracea capitata v1.0 | Brassicales | Brassicaceae |
| 37 | Brassica rapa | Brassica rapa FPsc v1.3 | Brassicales | Brassicaceae |
| 38 | Sinapis alba | Sinapis alba v3.1 | Brassicales | Brassicaceae |
| 39 | Gossypium hirsutum | Gossypium hirsutum v2.1 | Malvales | Malvaceae |
| 40 | Citrus sinensis | Citrus sinensis v1.1 | Sapindales | Rutaceae |
| 41 | Ananas comosus | Ananas comosus v3 | Poales | Bromeliaceae |
| 42 | Dioscorea alata | Dioscorea alata v2.1 | Dioscoreales | Dioscoreaceae |
| 43 | Musa acuminata | Musa acuminata v1 | Zingiberales | Musaceae |
| 44 | Hordeum vulgare | Hordeum vulgare r1 | Poales | Poaceae |
| 45 | Oryza sativa | Oryza sativa v7.0 | Poales | Poaceae |
| 46 | Triticum aestivum | Triticum aestivum v2.2 | Poales | Poaceae |
| 47 | Brachypodium distachyon | Brachypodium distachyon v3.2 | Poales | Poaceae |
| 48 | Miscanthus sinensis | Miscanthus sinensis v7.1 | Poales | Poaceae |
| 49 | Sorghum bicolor | Sorghum bicolor v3.1.1 | Poales | Poaceae |
| 50 | Zea mays | Zea mays RefGen_V4 | Poales | Poaceae |
| 51 | Panicum hallii | Panicum hallii v3.2 | Poales | Poaceae |
| 52 | Glycine max | G.max Wm82.a2.v1 | Fabales | Fabaceae |
2.1. Signal peptide prediction
Signal peptide prediction is performed to determine whether the identified 469 proteins exhibiting homology to PGIP have characteristics of secreted proteins (Supplemental Data File 2). The protein sequences are imported into SignalP 6.0 [8,9]. The number of putative PGIPs with predicted signal peptides are identified (Table 2; Supplemental Data File 3).
Table 2.
The identified PGIPs.
| Genome number | Species | genome | PGIP proteins |
|---|---|---|---|
| 1 | Amborella trichopoda | Amborella trichopoda v1.0 | 2 |
| 2 | Amaranthus hypochondriacus | Amaranthus hypochondriacus v2.1 | 6 |
| 3 | Beta vulgaris | Beta vulgaris EL10_1.0 | 9 |
| 4 | Chenopodium quinoa | Chenopodium quinoa v1.0 | 22 |
| 5 | Spinacia oleracea | Spinacia oleracea Spov3 | 6 |
| 6 | Coffea arabica | Coffea arabica v0.5 | 15 |
| 7 | Daucus carota | Daucus carota v2.0 | 9 |
| 8 | Helianthus annuus | Helianthus annuus r1.2 | 8 |
| 9 | Lactuca sativa | Lactuca sativa V8 | 13 |
| 10 | Mimulus guttatus | M.guttatus_TOL v5.0 | 10 |
| 11 | Olea europaea | Olea europaea v1.0 | 5 |
| 12 | Solanum lycopersicum | Solanum lycopersicum ITAG4.0 | 5 |
| 13 | Solanum tuberosum | Solanum tuberosum v6.1 | 3 |
| 14 | Eucalyptus grandis | Eucalyptus grandis v2.0 | 9 |
| 15 | Vitis vinifera | Vitis vinifera v2.1 | 4 |
| 16 | Arachis hypogaea | Arachis hypogaea v1.0 | 17 |
| 17 | Castanea dentata | Castanea dentata v1.1 | 5 |
| 18 | Cicer arietinum | Cicer arietinum v1.0 | 8 |
| 19 | Cucumis sativus | Cucumis sativus v1.0 | 3 |
| 20 | Fragaria vesca | Fragaria vesca v4.0.a2 | 4 |
| 21 | Malus domestica | Malus domestica v1.1 | 8 |
| 22 | Medicago truncatula | Medicago truncatula Mt4.0v1 | 23 |
| 23 | Phaseolus vulgaris | Phaseolus vulgaris v2.1 | 9 |
| 24 | Prunus persica | Prunus persica v2.1 | 5 |
| 25 | Quercus rubra | Quercus rubra v2.1 | 7 |
| 26 | Trifolium pratense | Trifolium pratense v2 | 16 |
| 27 | Carya illinoinensis | Carya illinoinensis v1.1 | 3 |
| 28 | Vigna unguiculata | Vigna unguiculata v1.2 | 10 |
| 29 | Linum usitatissimum | Linum usitatissimum v1.0 | 9 |
| 30 | Manihot esculenta | Manihot esculenta v8.1 | 6 |
| 31 | Carica papaya | Carica papaya ASGPBv0.4 | 3 |
| 32 | Theobroma cacao | Theobroma cacao v2.1 | 4 |
| 33 | Arabidopsis thaliana | Arabidopsis thaliana TAIR10 | 6 |
| 34 | Schrenkiella parvula | Schrenkiella parvula v2.2 | 6 |
| 35 | Brassica oleracea capitata | Brassica oleracea capitata v1.0 | 19 |
| 36 | Brassica rapa | Brassica rapa FPsc v1.3 | 19 |
| 37 | Sinapis alba | Sinapis alba v3.1 | 26 |
| 38 | Gossypium hirsutum | Gossypium hirsutum v2.1 | 6 |
| 39 | Citrus sinensis | Citrus sinensis v1.1 | 4 |
| 40 | Ananas comosus | Ananas comosus v3 | 5 |
| 41 | Dioscorea alata | Dioscorea alata v2.1 | 9 |
| 42 | Musa acuminata | Musa acuminata v1 | 5 |
| 43 | Hordeum vulgare | Hordeum vulgare r1 | 13 |
| 44 | Oryza sativa | Oryza sativa v7.0 | 9 |
| 45 | Triticum aestivum | Triticum aestivum v2.2 | 19 |
| 46 | Brachypodium distachyon | Brachypodium distachyon v3.2 | 7 |
| 47 | Miscanthus sinensis | Miscanthus sinensis v7.1 | 13 |
| 48 | Sorghum bicolor | Sorghum bicolor v3.1.1 | 8 |
| 49 | Zea mays | Zea mays RefGen_V4 | 9 |
| 50 | Panicum hallii | Panicum hallii v3.2 | 7 |
| 51 | Vaccinium darrowii | V.darrowii v1.2 | 13 |
| TOTAL | 469 |
2.2. Comparison of O- and N-glycosylation of GmPGIPs
A companion analysis demonstrates that GmPGIP11 but not GmPGIP1 functions in the defence response that G. max has toward H. glycines parasitism. A comparative analysis of G. max PGIPs is undertaken to determine whether O- and/or N-glycosylation could be correlated to these differences. The O-glycosylation analysis demonstrates that while GmPGIP1 is O-glycosylated, GmPGIP11 is not (Table 3; Supplemental Data File 4).
Table 3.
The signal peptide prediction, O-, N-glycosylation prediction, cellular location prediction.
| Species/PGIP protein | SP | O glyc | N-glyc | Location | Species/PGIP protein | SP | O glyc | N-glyc | Location |
|---|---|---|---|---|---|---|---|---|---|
| Amborella trichopoda | Vigna unguiculata | ||||||||
| AmtPGIP1 | y | Y | y | Extracellular | VuPGIP1 | y | y | y | Extracellular |
| AmtPGIP2 | y | Y | y | Extracellular | VuPGIP2 | y | n | y | Extracellular |
| Amaranthus hypochondriacus | VuPGIP3 | y | n | y | Extracellular | ||||
| AhyPGIP1 | y | N | y | Extracellular | VuPGIP4 | y | n | y | Extracellular |
| AhyPGIP2 | y | N | y | Extracellular | VuPGIP5 | y | n | y | Extracellular |
| AhyPGIP3 | y | N | y | Extracellular | VuPGIP6 | n | y | y | Extracellular |
| AhyPGIP4 | n | N | y | Cytoplasm | VuPGIP7 | y | n | y | Extracellular |
| AhyPGIP5 | y | Y | y | Extracellular | VuPGIP8 | y | n | y | Extracellular |
| AhyPGIP6 | n | N | y | Extracellular | VuPGIP9 | y | n | y | Extracellular |
| Beta vulgaris | VuPGIP10 | y | n | y | Extracellular | ||||
| BvPGIP1 | y | Y | y | Extracellular | Linum usitatissimum | ||||
| BvPGIP2 | y | Y | y | Extracellular | LuPGIP1 | y | y | y | Extracellular |
| BvPGIP3 | y | Y | y | Extracellular | LuPGIP2 | y | y | y | Extracellular |
| BvPGIP4 | y | Y | y | Extracellular | LuPGIP3 | y | y | y | Extracellular |
| BvPGIP5 | y | N | y | Extracellular | LuPGIP4 | y | n | y | Extracellular |
| BvPGIP6 | y | Y | y | Extracellular | LuPGIP5 | y | y | y | Extracellular |
| BvPGIP7 | y | N | y | Extracellular | LuPGIP6 | y | n | y | Extracellular |
| BvPGIP8 | n | Y | y | Extracellular | LuPGIP7 | n | n | y | Cytoplasm |
| BvPGIP9 | y | Y | y | Extracellular | LuPGIP8 | y | y | y | Extracellular |
| Chenopodium quinoa | LuPGIP9 | y | y | y | Extracellular | ||||
| CqPGIP1 | n | Y | y | Extracellular | Manihot esculenta | ||||
| CqPGIP2 | y | Y | y | Extracellular | MePGIP1 | y | n | y | Extracellular |
| CqPGIP3 | y | Y | y | Extracellular | MePGIP2 | y | n | y | Extracellular |
| CqPGIP4 | y | Y | y | Extracellular | MePGIP3 | y | n | y | Extracellular |
| CqPGIP5 | y | N | y | Extracellular | MePGIP4 | y | n | y | Extracellular |
| CqPGIP6 | y | Y | y | Extracellular | MePGIP_5 | n | y | y | Extracellular |
| CqPGIP7 | y | N | y | Extracellular | MePGIP6 | y | y | y | Celll membrane |
| CqPGIP8 | n | N | y | Extracellular | Carica papaya | ||||
| CqPGIP9 | n | N | y | Extracellular | CpPGIP1 | y | n | y | Extracellular |
| CqPGIP10 | y | N | y | Extracellular | CpPGIP2 | y | n | y | Extracellular |
| CqPGIP11 | n | Y | y | Nucleus | CpPGIP3 | y | n | y | Extracellular |
| CqPGIP12 | n | N | y | Cytoplasm | Theobroma cacao | ||||
| CqPGIP13 | n | N | y | Cytoplasm | TcPGIP1 | y | n | y | Extracellular |
| CqPGIP14 | y | N | y | Extracellular | TcPGIP2 | y | n | y | Extracellular |
| CqPGIP15 | n | N | y | Extracellular | TcPGIP3 | y | y | y | Extracellular |
| CqPGIP16 | n | N | y | Nucleus | TcPGIP4 | y | n | y | Extracellular |
| CqPGIP17 | y | N | y | Extracellular | Arabidopsis thaliana | ||||
| CqPGIP18 | y | N | y | Extracellular | AtPGIP1 | y | n | y | Extracellular |
| CqPGIP19 | y | Y | y | Lysosome | AtPGIP2 | y | y | y | Extracellular |
| CqPGIP20 | n | N | y | Extracellular | AtPGIP3 | y | y | y | Extracellular |
| CqPGIP21 | n | N | y | Extracellular | AtPGIP4 | y | y | n | Extracellular |
| CqPGIP22 | y | Y | y | Extracellular | AtPGIP5 | y | n | y | Extracellular |
| Spinacia oleracea | AtPGIP6 | y | n | y | Extracellular | ||||
| SoPGIP1 | y | Y | y | Extracellular | Schrenkiella parvula | ||||
| SoPGIP2 | y | Y | y | Extracellular | SpPGIP1 | y | n | y | Extracellular |
| SoPGIP3 | n | Y | y | Cytoplasm | SpPGIP2 | y | n | y | Extracellular |
| SoPGIP4 | y | Y | y | Extracellular | SpPGIP3 | y | y | y | Extracellular |
| SoPGIP5 | y | N | y | Extracellular | SpPGIP4 | y | n | y | Extracellular |
| SoPGIP6 | y | N | n | Extracellular | SpPGIP5 | y | n | y | Extracellular |
| Coffea Arabica | SpPGIP6 | y | y | y | Extracellular | ||||
| CaPGIP1 | y | Y | y | Extracellular | Brassica oleracea capitata | ||||
| CaPGIP2 | y | N | y | Extracellular | BoPGIP1 | y | n | y | Extracellular |
| CaPGIP3 | y | Y | y | Extracellular | BoPGIP2 | n | y | y | Extracellular |
| CaPGIP4 | y | Y | y | Extracellular | BoPGIP3 | y | n | y | Extracellular |
| CaPGIP5 | y | Y | y | Extracellular | BoPGIP4 | y | y | y | Extracellular |
| CaPGIP6 | n | N | y | Cytoplasm | BoPGIP5 | y | y | y | Extracellular |
| CaPGIP7 | n | Y | y | Cytoplasm | BoPGIP6 | n | n | y | Extracellular |
| CaPGIP8 | y | N | y | Extracellular | BoPGIP7 | n | n | y | Extracellular |
| CaPGIP9 | y | Y | n | Extracellular | BoPGIP8 | y | n | y | Extracellular |
| CaPGIP10 | y | N | y | Extracellular | BoPGIP9 | y | n | y | Extracellular |
| CaPGIP11 | y | N | y | Extracellular | BoPGIP10 | y | y | y | Extracellular |
| CaPGIP12 | y | N | y | Extracellular | BoPGIP11 | n | n | y | Cytoplasm |
| CaPGIP13 | y | N | y | Extracellular | BoPGIP12 | y | y | y | Extracellular |
| CaPGIP14 | y | N | y | Extracellular | BoPGIP13 | y | n | y | Extracellular |
| CaPGIP15 | y | N | y | Extracellular | BoPGIP14 | y | y | y | Extracellular |
| Daucus carota | BoPGIP15 | y | n | y | Extracellular | ||||
| DcPGIP1 | y | N | y | Extracellular | BoPGIP16 | y | n | y | Extracellular |
| DcPGIP2 | y | N | y | Extracellular | BoPGIP17 | y | n | n | Extracellular |
| DcPGIP3 | y | N | y | Extracellular | BoPGIP18 | y | y | y | Extracellular |
| DcPGIP4 | n | N | y | Extracellular | BoPGIP19 | y | n | y | Extracellular |
| DcPGIP5 | y | N | y | Extracellular | Brassica rapa | ||||
| DcPGIP6 | y | Y | y | Extracellular | BrPGIP1 | y | n | y | Extracellular |
| DcPGIP7 | y | N | n | Extracellular | BrPGIP2 | y | n | y | Extracellular |
| DcPGIP8 | n | N | y | Not find | BrPGIP3 | y | n | y | Extracellular |
| DcPGIP9 | y | Y | y | Extracellular | BrPGIP4 | y | n | y | Extracellular |
| Helianthus annuus | BrPGIP5 | y | n | y | Extracellular | ||||
| HaPGIP1 | y | Y | y | Extracellular | BrPGIP6 | y | y | y | Extracellular |
| HaPGIP2 | y | Y | y | Extracellular | BrPGIP7 | y | n | y | Extracellular |
| HaPGIP3 | y | N | y | Extracellular | BrPGIP8 | y | y | y | Extracellular |
| HaPGIP4 | y | Y | y | Extracellular | BrPGIP9 | y | n | y | Extracellular |
| HaPGIP5 | y | N | y | Extracellular | BrPGIP10 | y | y | y | Extracellular |
| HaPGIP6 | y | N | y | Extracellular | BrPGIP11 | y | y | y | Extracellular |
| HaPGIP7 | y | N | y | Extracellular | BrPGIP12 | y | n | y | Extracellular |
| HaPGIP8 | y | N | y | Extracellular | BrPGIP13 | y | y | y | Extracellular |
| Lactuca sativa | BrPGIP14 | y | y | y | Extracellular | ||||
| LsPGIP1 | y | Y | y | Extracellular | BrPGIP15 | y | n | y | Extracellular |
| LsPGIP2 | y | Y | y | Extracellular | BrPGIP16 | y | n | n | Extracellular |
| LsPGIP3 | y | Y | y | Extracellular | BrPGIP17 | y | n | y | Extracellular |
| LsPGIP4 | y | N | y | Extracellular | BrPGIP18 | y | y | y | Extracellular |
| LsPGIP5 | y | Y | y | Extracellular | BrPGIP19 | y | y | y | Extracellular |
| LsPGIP6 | y | N | y | Extracellular | Sinapis alba | ||||
| LsPGIP7 | n | N | y | Cytoplasm | SaPGIP1 | y | n | y | Extracellular |
| LsPGIP8 | y | N | y | Extracellular | SaPGIP2 | y | n | y | Extracellular |
| LsPGIP9 | y | N | y | Extracellular | SaPGIP3 | y | n | y | Extracellular |
| LsPGIP10 | y | N | y | Extracellular | SaPGIP4 | y | n | y | Extracellular |
| LsPGIP11 | y | N | y | Extracellular | SaPGIP5 | y | n | y | Extracellular |
| LsPGIP12 | y | N | y | Extracellular | SaPGIP6 | y | y | y | Extracellular |
| LsPGIP13 | y | N | y | Extracellular | SaPGIP7 | n | y | y | Nucleus |
| Mimulus guttatus | SaPGIP8 | y | y | y | Extracellular | ||||
| MgPGIP1 | y | N | y | Extracellular | SaPGIP9 | y | y | y | Extracellular |
| MgPGIP2 | y | Y | y | Extracellular | SaPGIP10 | y | n | y | Extracellular |
| MgPGIP3 | y | N | y | Extracellular | SaPGIP11 | y | y | y | Extracellular |
| MgPGIP4 | y | Y | y | Extracellular | SaPGIP12 | y | n | y | Extracellular |
| MgPGIP5 | y | Y | y | Extracellular | SaPGIP13 | y | y | y | Extracellular |
| MgPGIP6 | y | Y | y | Extracellular | SaPGIP14 | y | n | y | Extracellular |
| MgPGIP7 | n | Y | y | Cytoplasm | SaPGIP15 | y | n | y | Extracellular |
| MgPGIP8 | n | Y | y | Cytoplasm | SaPGIP16 | y | n | y | Extracellular |
| MgPGIP9 | y | Y | y | Extracellular | SaPGIP17 | y | n | y | Extracellular |
| MgPGIP10 | y | Y | y | Cell membrane | SaPGIP18 | y | n | y | Extracellular |
| Olea europaea | N | n | SaPGIP19 | y | n | y | Extracellular | ||
| OePGIP1 | y | N | y | Extracellular | SaPGIP20 | y | y | n | Extracellular |
| OePGIP2 | y | N | y | Extracellular | SaPGIP21 | n | n | y | Extracellular |
| OePGIP3 | y | N | y | Extracellular | SaPGIP22 | n | n | y | Lysosome |
| OePGIP4 | n | N | y | Nucleus | SaPGIP23 | y | n | y | Extracellular |
| OePGIP5 | n | Y | y | Cell membrane | SaPGIP24 | y | n | y | Extracellular |
| Solanum lycopersicum | SaPGIP25 | y | n | y | Extracellular | ||||
| SlPGIP1 | y | N | y | Extracellular | SaPGIP26 | y | y | n | Extracellular |
| SlPGIP2 | y | N | y | Extracellular | Gossypium hirsutum | ||||
| SlPGIP3 | y | N | y | Extracellular | GhPGIP1 | y | n | y | Extracellular |
| SlPGIP4 | y | Y | y | Extracellular | GhPGIP2 | y | y | y | Extracellular |
| SlPGIP5 | y | N | y | Extracellular | GhPGIP3 | y | n | y | Extracellular |
| Solanum tuberosum | GhPGIP4 | y | y | y | Extracellular | ||||
| StPGIP1 | y | N | y | Extracellular | GhPGIP5 | y | n | y | Extracellular |
| StPGIP2 | y | Y | y | Extracellular | GhPGIP6 | y | n | y | Extracellular |
| StPGIP3 | y | Y | y | Extracellular | Citrus sinensis | ||||
| Eucalyptus grandis | CsPGIP1 | y | n | y | Extracellular | ||||
| EgPGIP1 | n | N | y | Nucleus | CsPGIP2 | y | n | y | Extracellular |
| EgPGIP2 | n | Y | y | Cytoplasm | CsPGIP3 | y | y | y | Extracellular |
| EgPGIP3 | y | N | y | Extracellular | CsPGIP4 | y | n | y | Extracellular |
| EgPGIP4 | n | N | y | Extracellular | Ananas comosus | ||||
| EgPGIP5 | n | N | y | Extracellular | AcPGIP1 | y | y | y | Extracellular |
| EgPGIP6 | y | N | y | Extracellular | AcPGIP2 | y | y | y | Extracellular |
| EgPGIP7 | y | N | y | Extracellular | AcPGIP3 | y | n | y | Extracellular |
| EgPGIP8 | y | Y | y | Extracellular | AcPGIP4 | y | n | y | Extracellular |
| EgPGIP9 | y | Y | y | Extracellular | AcPGIP5 | y | n | y | Extracellular |
| Vitis vinifera | Dioscorea alata | ||||||||
| VvPGIP1 | y | N | y | Extracellular | DaPGIP1 | y | y | y | Extracellular |
| VvPGIP2 | y | N | y | Extracellular | DaPGIP2 | y | n | y | Extracellular |
| VvPGIP3 | y | N | y | Extracellular | DaPGIP3 | y | y | y | Extracellular |
| VvPGIP4 | y | N | y | Extracellular | DaPGIP4 | y | n | y | Extracellular |
| Arachis hypogaea | DaPGIP5 | n | y | y | Extracellular | ||||
| AhPGIP1 | y | N | y | Extracellular | DaPGIP6 | y | n | y | Extracellular |
| AhPGIP2 | y | N | y | Extracellular | DaPGIP7 | y | y | y | Extracellular |
| AhPGIP3 | y | N | y | Extracellular | DaPGIP8 | y | y | y | Extracellular |
| AhPGIP4 | y | N | y | Extracellular | DaPGIP9 | y | y | y | Extracellular |
| AhPGIP5 | y | Y | y | Extracellular | Musa acuminata | ||||
| AhPGIP6 | y | Y | y | Extracellular | MaPGIP1 | y | y | y | Extracellular |
| AhPGIP7 | y | N | y | Extracellular | MaPGIP2 | y | n | y | Extracellular |
| AhPGIP8 | y | N | y | Extracellular | MaPGIP3 | n | y | y | Cytoplasm |
| AhPGIP9 | y | Y | y | Extracellular | MaPGIP4 | y | y | y | Extracellular |
| AhPGIP10 | y | Y | y | Extracellular | MaPGIP5 | n | y | y | Extracellular |
| AhPGIP11 | y | Y | y | Extracellular | Hordeum vulgare | ||||
| AhPGIP12 | y | Y | y | Extracellular | HvPGIP1 | y | y | y | Extracellular |
| AhPGIP13 | y | Y | n | Extracellular | HvPGIP2 | y | y | y | Extracellular |
| AhPGIP14 | y | Y | n | Extracellular | HvPGIP3 | n | y | y | Extracellular |
| AhPGIP15 | n | Y | y | Cytoplasm | HvPGIP4 | y | y | y | Extracellular |
| AhPGIP16 | n | Y | y | Cytoplasm | HvPGIP5 | y | y | y | Extracellular |
| AhPGIP17 | y | Y | y | Extracellular | HvPGIP6 | y | y | y | Extracellular |
| Castanea dentate | HvPGIP7 | y | y | y | Extracellular | ||||
| CdPGIP1 | y | Y | y | Extracellular | HvPGIP8 | n | y | y | Extracellular |
| CdPGIP2 | y | N | y | Extracellular | HvPGIP9 | n | y | y | Cytoplasm |
| CdPGIP3 | y | N | y | Extracellular | HvPGIP10 | y | y | y | Extracellular |
| CdPGIP4 | y | Y | y | Extracellular | HvPGIP11 | y | y | y | Extracellular |
| CdPGIP5 | n | N | y | Lysosome | HvPGIP12 | y | y | y | Extracellular |
| Cicer arietinum | HvPGIP13 | y | y | y | Extracellular | ||||
| CiaPGIP1 | y | Y | y | Extracellular | Oryza sativa | ||||
| CiaPGIP2 | y | N | y | Extracellular | OsPGIP1 | y | y | y | Extracellular |
| CiaPGIP3 | y | N | y | Extracellular | OsPGIP2 | y | n | y | Extracellular |
| CiaPGIP4 | n | Y | y | Extracellular | OsPGIP3 | n | n | y | Cell membrane |
| CiaPGIP5 | y | Y | y | Extracellular | OsPGIP4 | y | y | y | Extracellular |
| CiaPGIP6 | y | Y | y | Extracellular | OsPGIP5 | y | y | y | Extracellular |
| CiaPGIP7 | y | N | y | Extracellular | OsPGIP6 | y | y | y | Extracellular |
| CiaPGIP8 | y | N | y | Extracellular | OsPGIP7 | n | y | y | Nucleus |
| Cucumis sativus | OsPGIP8 | y | n | y | Extracellular | ||||
| CusPGIP1 | y | N | y | Extracellular | OsPGIP9 | y | y | y | Extracellular |
| CusPGIP2 | y | N | y | Extracellular | Triticum aestivum | ||||
| CusPGIP3 | y | N | y | Extracellular | TaPGIP1 | y | y | y | Extracellular |
| Fragaria vesca | TaPGIP2 | y | y | y | Extracellular | ||||
| FvPGIP1 | y | Y | y | Extracellular | TaPGIP3 | y | y | y | Extracellular |
| FvPGIP2 | y | N | y | Extracellular | TaPGIP4 | y | y | y | Extracellular |
| FvPGIP3 | y | Y | y | Extracellular | TaPGIP5 | y | y | y | Extracellular |
| FvPGIP4 | y | N | y | Extracellular | TaPGIP6 | y | y | y | Extracellular |
| Malus domestica | TaPGIP7 | y | n | y | Extracellular | ||||
| MdPGIP1 | y | N | y | Extracellular | TaPGIP8 | y | n | y | Extracellular |
| MdPGIP2 | y | Y | y | Extracellular | TaPGIP9 | y | y | y | Extracellular |
| MdPGIP3 | n | N | y | Lysosome | TaPGIP10 | y | y | y | Extracellular |
| MdPGIP4 | y | N | y | Extracellular | TaPGIP11 | y | y | y | Extracellular |
| MdPGIP5 | y | N | y | Extracellular | TaPGIP12 | y | y | y | Extracellular |
| MdPGIP6 | y | N | y | Extracellular | TaPGIP13 | y | y | y | Extracellular |
| MdPGIP7 | y | Y | y | Extracellular | TaPGIP14 | y | y | y | Extracellular |
| MdPGIP8 | y | Y | y | Extracellular | TaPGIP15 | y | y | y | Extracellular |
| Medicago truncatula | TaPGIP16 | y | y | y | Extracellular | ||||
| MtPGIP1 | y | N | y | Extracellular | TaPGIP17 | y | y | y | Extracellular |
| MtPGIP2 | y | Y | y | Extracellular | TaPGIP18 | n | n | y | Cytoplasm |
| MtPGIP3 | y | Y | y | Extracellular | TaPGIP19 | n | n | y | Cytoplasm |
| MtPGIP4 | y | N | y | Extracellular | Brachypodium distachyon | ||||
| MtPGIP5 | y | Y | y | Extracellular | BdPGIP1 | y | y | y | Extracellular |
| MtPGIP6 | y | Y | y | Extracellular | BdPGIP2 | y | y | y | Extracellular |
| MtPGIP7 | y | N | y | Extracellular | BdPGIP3 | y | y | y | Extracellular |
| MtPGIP8 | y | Y | y | Extracellular | BdPGIP4 | y | y | y | Extracellular |
| MtPGIP9 | n | N | y | Not find | BdPGIP5 | y | y | y | Extracellular |
| MtPGIP10 | y | Y | y | Extracellular | BdPGIP6 | y | y | y | Extracellular |
| MtPGIP11 | y | Y | y | Extracellular | BdPGIP7 | y | y | y | Extracellular |
| MtPGIP12 | y | Y | y | Extracellular | Miscanthus sinensis | ||||
| MtPGIP13 | y | Y | y | Extracellular | MsPGIP1 | y | n | y | Extracellular |
| MtPGIP14 | y | N | y | Extracellular | MsPGIP2 | y | y | y | Extracellular |
| MtPGIP15 | y | Y | y | Extracellular | MsPGIP3 | y | y | y | Extracellular |
| MtPGIP16 | y | Y | y | Extracellular | MsPGIP4 | y | n | y | Extracellular |
| MtPGIP17 | y | Y | y | Extracellular | MsPGIP5 | n | y | y | Extracellular |
| MtPGIP18 | n | Y | y | Chloroplast | MsPGIP6 | y | y | y | Extracellular |
| MtPGIP19 | y | Y | y | Extracellular | MsPGIP7 | y | y | y | Extracellular |
| MtPGIP20 | y | N | y | Extracellular | MsPGIP8 | y | n | y | Extracellular |
| MtPGIP21 | y | Y | y | Extracellular | MsPGIP9 | y | n | y | Extracellular |
| MtPGIP22 | y | Y | y | Cell membrane | MsPGIP10 | n | y | y | Cytoplasm |
| MtPGIP23 | y | Y | y | Extracellular | MsPGIP11 | y | n | y | Extracellular |
| Phaseolus vulgaris | MsPGIP12 | n | y | y | Extracellular | ||||
| PvPGIP1 | y | Y | y | Extracellular | MsPGIP13 | y | y | y | Extracellular |
| PvPGIP3 | y | Y | y | Extracellular | Sorghum bicolor | ||||
| PvPGIP4 | y | Y | y | Extracellular | SbPGIP1 | y | y | y | Extracellular |
| PvPGIP5 | y | Y | y | Extracellular | SbPGIP2 | y | y | y | Extracellular |
| PvPGIP6 | y | N | y | Extracellular | SbPGIP3 | y | y | y | Extracellular |
| PvPGIP7 | y | N | y | Extracellular | SbPGIP4 | y | y | y | Extracellular |
| PvPGIP8 | n | Y | y | Extracellular | SbPGIP5 | y | y | n | Extracellular |
| PvPGIP9 | y | N | y | Extracellular | SbPGIP6 | y | y | y | Extracellular |
| Prunus persica | SbPGIP7 | y | y | y | Extracellular | ||||
| PpPGIP1 | y | N | y | Extracellular | SbPGIP8 | y | y | y | Extracellular |
| PpPGIP2 | y | N | y | Extracellular | Zea mays | ||||
| PpPGIP3 | y | N | y | Extracellular | ZmPGIP1 | y | y | y | Extracellular |
| PpPGIP4 | y | Y | y | Extracellular | ZmPGIP2 | y | y | y | Extracellular |
| PpPGIP5 | y | N | y | Extracellular | ZmPGIP3 | y | n | y | Extracellular |
| Quercus rubra | ZmPGIP4 | y | y | y | Extracellular | ||||
| QrPGIP1 | y | Y | y | Extracellular | ZmPGIP5 | y | y | y | Extracellular |
| QrPGIP2 | y | Y | y | Extracellular | ZmPGIP6 | y | y | y | Extracellular |
| QrPGIP3 | y | Y | y | Extracellular | ZmPGIP7 | y | y | y | Extracellular |
| QrPGIP4 | y | Y | y | Nucleus | ZmPGIP8 | y | y | y | Extracellular |
| QrPGIP5 | y | N | y | Extracellular | ZmPGIP9 | y | y | y | Extracellular |
| QrPGIP6 | y | N | y | Extracellular | Panicum hallii | ||||
| QrPGIP7 | y | Y | y | Cytoplasm | PhPGIP1 | y | y | y | Extracellular |
| Trifolium pretense | PhPGIP2 | y | y | y | Extracellular | ||||
| TpPGIP1 | y | Y | y | Extracellular | PhPGIP3 | y | y | y | Extracellular |
| TpPGIP2 | y | Y | y | Extracellular | PhPGIP4 | y | y | y | Extracellular |
| TpPGIP3 | y | Y | y | Extracellular | PhPGIP5 | y | y | y | Extracellular |
| TpPGIP4 | y | Y | y | Extracellular | PhPGIP6 | y | y | y | Extracellular |
| TpPGIP5 | y | Y | y | Extracellular | PhPGIP7 | y | y | y | Cell membrane |
| TpPGIP6 | y | N | y | Extracellular | Vaccinium darrowii | ||||
| TpPGIP7 | y | Y | y | Extracellular | VdPGIP1 | y | y | y | Extracellular |
| TpPGIP8 | y | N | y | Extracellular | VdPGIP2 | y | y | y | Extracellular |
| TpPGIP9 | y | N | y | Extracellular | VdPGIP3 | y | y | y | Extracellular |
| TpPGIP10 | y | Y | y | Extracellular | VdPGIP4 | y | y | y | Extracellular |
| TpPGIP11 | n | Y | y | Cell membrane | VdPGIP5 | y | n | y | Extracellular |
| TpPGIP12 | y | Y | y | Extracellular | VdPGIP6 | y | y | y | Extracellular |
| TpPGIP13 | y | N | y | Extracellular | VdPGIP7 | y | y | y | Extracellular |
| TpPGIP14 | y | Y | y | Cell membrane | VdPGIP8 | y | n | y | Extracellular |
| TpPGIP15 | y | Y | y | Extracellular | VdPGIP9 | n | n | y | Cytoplasm |
| TpPGIP16 | n | Y | y | Extracellular | VdPGIP10 | y | n | y | Extracellular |
| Carya illinoinensis | VdPGIP11 | y | y | y | Extracellular | ||||
| CiPGIP1 | y | Y | n | Extracellular | VdPGIP12 | y | y | y | Extracellular |
| CiPGIP2 | y | Y | n | Extracellular | VdPGIP13 | n | y | y | Extracellular |
| CiPGIP3 | y | Y | n | Extracellular | Glycine max | ||||
| GmPGIP1 | y | y | y | Extracellular | |||||
| GmPGIP2 | n | y | y | Cytoplasm | |||||
| GmPGIP3 | y | y | y | Extracellular | |||||
| GmPGIP4 | y | y | y | Extracellular | |||||
| GmPGIP5 | n | y | y | Nucleus | |||||
| GmPGIP6 | y | y | y | Extracellular | |||||
| GmPGIP7 | y | y | y | Extracellular | |||||
| GmPGIP8 | y | y | y | Extracellular | |||||
| GmPGIP9 | n | y | y | Cell membrane | |||||
| GmPGIP10 | y | y | y | Extracellular | |||||
| GmPGIP11 | y | n | y | Extracellular | |||||
In contrast to the above-presented findings both GmPGIP1 and GmPGIP11 are predicted to be N-glycosylated. However, some of their predicted N-glycosylation sites are not at homologous aa positions (Fig. 1; Supplemental Data File 5). For example, the NPTT site found in GmPGIP1 and starting at aa position 41 is not identified in GmPGIP11 (Fig.). In contrast, a NLSG site found in GmPGIP11 and starting at aa position 101 is not found in GmPGIP1. Similarly, an NLSG predicted N-glycosylation site found in GmPGIP11 and starting at aa position 174 is not found in GmPGIP1 (Fig. 1). Furthermore, an NKTT predicted N-glycosylation site found in GmPGIP11 and starting at aa position 258 is not found in GmPGIP1 (Fig. 1). However, N-glycosylation sites that are in homologous positions between GmPGIP1 and GmPGIP11 do exist (Fig. 1). GmPGIP1 has a NVSG predicted N-glycosylation site starting at aa position 132 while GmPGIP11 has a NVSG predicted N-glycosylation site starting at aa position 150 (Fig. 1). Consequently, while experimentation has not proven that these sites are important to the functional differences occurring between GmPGIP1 and GmPGIP11, they are different and provide a basis for future experimentation.
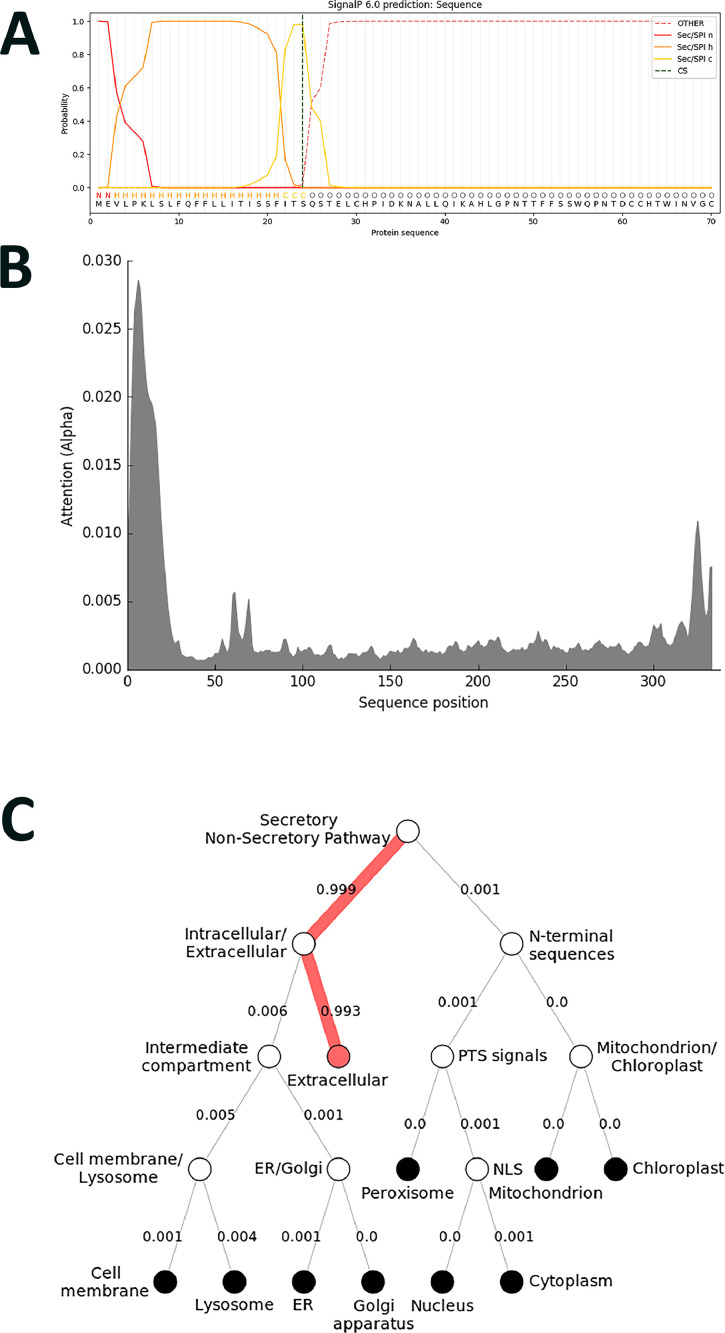
Fig. 1.
Beta vulgaris BvPGIP4 protein analysis. A. Signal peptide prediction. B. Amino acid relative importance. C. Hierarchical tree, showing the localization likelihood in numerical value: Extracellular, 0.9701; Lysosome/Vacuole, 0.0263; Endoplasmic reticulum, 0.0024; Cell membrane, 0.0007; Cytoplasm, 0.0003; Mitochondrion, 0.0001; Golgi apparatus, 0; Plastid, 0; Nucleus, 0; Peroxisome, 0; Soluble, 0.9994; Membrane, 0.0006. Prediction: Extracellular, Soluble.
2.3. Artificial intelligence
The 469 identified PGIP proteins spanning the 51 genomes are assessed by artificial intelligence analyses to produce a sequence position file (Supplemental Data File 6). A second file generates a map to the cellular destination where the predicted protein is predicted to function (Table 3; Supplemental Data File 7). An example for Beta vulgaris BvPGIP4, shown to function in defence to various pathogens in N. tabacum, is presented (Fig. 2) [5].
Fig. 2.
Predicted O- and N-glycosylation sites of G. max PGIP proteins. Cyan, predicted O-glycosylation site. Magenta, N-glycosylation site. Yellow, an aa that overlaps between two predicted N-glycosylation sites. Blue, an aa that overlaps between an O- and N-glycosylation site. Gray, possible mis-annotated N-terminal sequence.
The secretion of plant proteins is an important cellular property used for a variety of processes including development and disease resistance [10]. The data presented here is computational support showing PGIPs identified as belonging to taxa positioned at the base of angiosperm evolution are predicted to have signal peptides, have O- and/or N-glycosylation, and undergo secretion into the apoplast. Further assessment identifies PGIPs from both monocot and dicot lineages with predicted signal peptides and the subcellular or supracellular compartment to which they are targeted [5,11].
2.4. Analysed proteomes
The study analyses the proteomes of 51 plants not including G. max, many important to agriculture. The 51 proteomes span the base of angiosperm evolution (A. trichopoda), a monotypic genus of Amborellaceae and the only member of the Amborellales that has 2 predicted PGIP proteins [12]. Each PGIP is predicted to have signal peptides, experience O- and N-glycosylation, and undergo secretion into the apoplast. The monocots presented here are represented by A. comosus, D. alata, M. acuminata, H. vulgare, O. sativa, T. aestivum, B. distachyon, M. sinensis, S. bicolor, Z. mays and P. hallii with the remaining plants belonging to the Eudicots. All of the studied species have at least one putative PGIP that is predicted to have a signal peptide, have O- and/or N-glycosylation, and are secreted into the apoplast.
Local duplication of plant genes, including PGIPs, results in the generation of genes whose protein products perform an important function in defence [6]. The PGIP proteins identified here also appear to be products of localized gene duplications. Consequently, the identified genes may relate to the birth and death model for PGIPs that is proposed [6]. Possible localized gene duplication is identified from the analysis of the 51 proteomes. Based off the annotations, the analysis, identifying direct tandem duplications for at least one PGIP gene duplication in 29 of the 51 proteomes including A. hypochondriacus, B. vulgaris, C. quinoa, C. arabica, D. carota, M. guttatus, O. europaea, E. grandis, C. arietinum, M. domestica, M. truncatula, P. vulgaris, P. persica, Q. rubra, V. unguiculata, L. usitatissimum, M. esculenta, T. cacao, A. thaliana, S. parvula, B. oleracea capitata, B. rapa, S. alba, D. alata, V. darrowii, O. sativa, M. sinensis, S. bicolor and P. hallii.
2.5. Glycosylation
Computational studies are performed to identify O- and/or N-glycosylation of the PGIP proteins. Glycosylation is an important feature of proteins that imparts new function and in plants is important in both development and defence [13]. Glycosylation is not a random event and occurs on greater than 50% of eukaryote proteins [14]. Pyrus communis (pear) PGIP exhibits heterogeneous glycosylation that relates to pathogen defence [15]. The results presented here provide computational support that plant PGIPs experience glycosylation, broadly.
2.6. Sequence alignments identify glycosylation variation that may explain functional differences
Transgenic studies show GmPGIP11 functions in defence to H. glycines while GmPGIP1 does not. A computational study analysing the O- and N-glycosylation sites show that while GmPGIP1 is predicted to be O-glycosylated that GmPGIP11 is not. Furthermore, GmPGIP11 has predicted N-glycosylation sites that GmPGIP1 does not while GmPGIP1 has predicted N-glycosylation sites that are lacking in GmPGIP11. Glycosylation performs important defence roles [16].
2.7. Alternate spicing of PGIP mRNAs
What has not been presented is the possible importance of alternate RNA splicing in PGIP biology. The analysis presented here identifies 4 proteomes (L. sativa, P. persica, H. vulgare, and T. aestivum) that are annotated to contain products of alternate splicing. Alternatively spliced transcripts of genes encode transcripts that perform important defence functions, including parasitic nematodes [8].
3. Supplemental Data
Supplemental Data Set 1: The PGIP accessions obtained by the blast queries according to the described protocol. Supplemental data file 1 - https://data.mendeley.com/datasets/66r9pkckjz/1.
Supplemental Data Set 2: The protein sequences obtained from Phytozome. Supplemental data file 2- https://data.mendeley.com/datasets/66r9pkckjz/1.
Supplemental Data Set 3: The signal peptide prediction made by SignalP 6.0. Supplemental data file 3- https://data.mendeley.com/datasets/66r9pkckjz/1.
Supplemental Data Set 4: The O-glycosylation prediction made by NetOGlyc - 4.0. Supplemental data file 4- https://data.mendeley.com/datasets/66r9pkckjz/1.
Supplemental Data Set 5: The N-glycosylation prediction made by NetNGlyc - 1.0. Supplemental data file 5 - https://data.mendeley.com/datasets/66r9pkckjz/1.
Supplemental Data Set 6: The amino acid relative importance predicted by DeepLoc-1.0. Supplemental data file 6- https://data.mendeley.com/datasets/66r9pkckjz/1.
Supplemental Data Set 7: The hierarchical trees predicted by DeepLoc-1.0. Supplemental data file 7- https://data.mendeley.com/datasets/66r9pkckjz/1.
4. Experimental Design, Materials and Methods
4.1. Data access
4.2. Analysed proteomes
The 11 G. max PGIP protein sequences are used in Basic Local Alignment Search Tool program (BLAST) searches of the proteomes (BLASTP) using the default parameters at Phytozome (http://www.phytozome.net/) [7]. There are 52 total proteomes analysed [7]. The default BLAST parameters are used in querying, including Target type: Proteome; Program: BLASTP-protein query to protein database; Expect (E) threshold: -1; Comparison matrix: BLOcks SUbstitution Matrix (BLOSUM) 62 (BLOSUM62); Word (W) length: default = 3; number of alignments to show: 100 allowing for gaps and filter query, in order that they appear on the BLAST program. Through these analyses it is possible to extract the genomic DNA, transcript, cDNA, protein accessions, their sequences, and gene family members. The analyses also permit the extraction of protein homologs and splice variants from the selected agricultural crops of international importance, those with importance in the U.S., and those important biologically according to [8].
The identified PGIP proteins are compiled using a Bitscore of 140 as a cutoff. To identify the PGIP proteins, each of the 11 G. max PGIP protein sequences are queried into the studied proteomes. The individual queries for GmPGIP1 through GmPGIP11 are stored in individual tabs in Excel. Then, the PGIPs that have Bitscores of 140 or higher are compiled for all of the queries for the individual GmPGIPs. The duplicate PGIPs then are removed in Excel. The analysis results in a list of PGIP proteins that include the products of alternate splicing so the numbers in some cases are higher than the numbers of genes in some genomes.
4.3. Signal peptide prediction
Signal peptide prediction is done using SignalP 6.0 [9]. SignalP 6.0 is based on protein language models (LMs). The models use information from millions of unannotated protein sequence which are been analysed across all life domains. LMs create logical protein representations capturing their biological structure and properties. SignalP 6.0, thus, predicts additional SP types not possible in earlier iterations of SignalP (e.g., SignalP 5.0) and better extrapolates them to distantly related proteins and ones used to create the model and metagenomic data of unknown origin. SignalP 6.0 also identifies SP subregions. The default parameters are used.
4.4. O-glycosylation determination
O-glycosylation is determined using NetOGlyc - 4.0 [17]. The parameters are set on default. The output format is imported into Excel.
4.5. N-glycosylation determination
N-glycosylation is determined using NetNGlyc - 1.0 set [18]. The parameters are on set default. The output format is imported into Excel.
4.6. Protein alignment
Protein alignment is performed using CLUSTAL Omega, CLUSTAL O(1.2.4) multiple sequence alignment [19]. The analysis is performed using default parameters. The output file is imported into MS Word.
4.7. Artificial intelligence
Prediction of eukaryotic protein subcellular localization using deep learning is done using DeepLoc-1.0 [20]. The DeepLoc-1.0 analysis determines the importance of a particular amino acid along a protein chain that is relevant for prediction (attention) of its subcellular location and is done in default settings. DeepLoc-1.0 then predicts the subcellular localization of eukaryotic proteins, differentiating between 10 different localizations including the nucleus, cytoplasm, extracellular, mitochondrion, cell membrane, endoplasmic reticulum, (ER) chloroplast, Golgi apparatus, lysosome/vacuole, and peroxisome and is done in default settings. The output of the analysis is presented as a graphic that shows the relative importance of each AA along the polypeptide chain as well as a hierarchical tree that shows where the protein is expected to be located withing a cell [20].
Limitations
Not applicable.
Ethics Statement
The authors have read and follow the ethical requirements for publication in Data in Brief and confirm that the current work does not involve human subjects, animal experiments, or any data collected from social media platforms.
CRediT authorship contribution statement
Sudha Acharya: Validation, Formal analysis, Investigation. Hallie A. Troell: Validation, Formal analysis, Investigation. Rebecca L. Billingsley: Validation, Formal analysis, Investigation. Katherine S. Lawrence: Validation, Formal analysis, Investigation, Resources, Writing – original draft, Supervision, Project administration, Funding acquisition. Daniel S. McKirgan: Validation, Formal analysis, Investigation. Nadim W. Alkharouf: Methodology, Validation, Investigation, Resources, Writing – original draft, Supervision, Project administration. Vincent P. Klink: Conceptualization, Methodology, Validation, Investigation, Resources, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Acknowledgements
VK is thankful to the Department of Biological Sciences and the Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology (BMBEPP) at Mississippi State University. Furthermore, VK is thankful to Gary Lawrence (retired) (BMBEPP) for all his support over the years. Robert Nichols and Kater Hake of Cotton Incorporated are thanked for their support during this project. Yixiu (Jan) Pinnix BMBEPP is thanked for her technical support. Jeff Dean (BMBEPP) is thanked for his generosity of providing greenhouse, headhouse, storage, and field space for the experiments and maintenance of plant stocks. The authors thank Scott Willard, Wes Burger, George Hopper, and Reuben Moore, Mississippi Agricultural and Forestry Experiment Station (MAFES), and Mississippi State University for support. The College of Arts and Sciences and MAFES at Mississippi State University have each provided Special Research Initiative (SRI) funding for this work.
USDA-ARS NP301- 8042-21220-233; Cotton Incorporated, grants 17-603, 19-603; MAFES-Special Research Initiative (SRI); SRI-01; Alabama Hatch Grant ALA015-2-14003.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
References
- 1.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Waldron K.W., Faulds C.B. Comprehensive Glycoscience, Elsevier; 2007. Cell Wall Polysaccharides: Composition and Structure, Editor(s): Hans Kamerling; pp. 181–201. [DOI] [Google Scholar]
- 3.Phaff H.J. The production of exocellular pectic enzymes by Penicillium chrysogenum; on the formation and adaptive nature of polygalacturonase and pectinesterase. Arch. Biochem. 1947;13:67–81. [PubMed] [Google Scholar]
- 4.Cervone F., Castoria R., Leckie F., De Lorenzo G. Perception of fungal elicitors and signal transduction. Aducci P., editor. Perception of fungal elicitors and signal transductionSignal Transduction in Plants. 1997 doi: 10.1007/978-3-0348-9183-7_8. [DOI] [Google Scholar]
- 5.Li H., Smigocki A.C. Sugar beet polygalacturonase-inhibiting proteins with 11 LRRs confer Rhizoctonia, Fusarium and Botrytis resistance in Nicotiana plants. Physiol. Mol. Plant Pathol. 2018;102:200–208. doi: 10.1016/J.PMPP.2018.03.001. [DOI] [Google Scholar]
- 6.Kalunke R.M., Cenci A., Volpi C., O'Sullivan D.M., Sella L., Favaron F., Cervone F., De Lorenzo G., D'Ovidio R. The pgip family in soybean and three other legume species: evidence for a birth-and-death model of evolution. BMC Plant Biol. 2014;14:189. doi: 10.1186/s12870-014-0189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodstein D., Shu S., Howson R. Phytosome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klink V.P., Darwish O., Alkharouf N.W., Lawaju B.R., Khatri R., Khatri R., Lawrence K.S. Conserved oligomeric Golgi (COG) complex genes functioning in defense are expressed in root cells undergoing a defense response to a pathogenic infection. PLoS One. 2021 doi: 10.1371/journal.pone.0256472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teufel F., Almagro Armenteros J.J., Johansen A.R., Gíslason M.H., Pihl S.I., Tsirigos K.D., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022 doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 11.Ferrari S., Sella L., Janni M., De Lorenzo G., Favaron F., D'Ovidio R. Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. (Stuttg.) 2012;14(Suppl 1):31–38. doi: 10.1111/j.1438-8677.2011.00449.x. [DOI] [PubMed] [Google Scholar]
- 12.Group Angiosperm Phylogeny. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linnean Soc. 2009;161:105–121. doi: 10.1111/j.1095-8339.2009.00996.x. [DOI] [Google Scholar]
- 13.Tan L., Qiu F., Lamport D.T., Kieliszewski M.J. Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 2004;279:13156–13165. doi: 10.1074/jbc.M311864200. [DOI] [PubMed] [Google Scholar]
- 14.Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica Biophysica Acta. 1999;1473:4–8. doi: 10.1016/0031-9422(81)83083-X. [DOI] [PubMed] [Google Scholar]
- 15.Lim J.M., Aoki K., Angel P., Garrison D., King D., Tiemeyer M., Bergmann C., Wells L. Mapping glycans onto specific N-linked glycosylation sites of Pyrus communis PGIP redefines the interface for EPG-PGIP interactions. J. Proteome Res. 2009;8:673–680. doi: 10.1021/pr800855f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Held M.A., Kaur D., Showalter A.M. CRISPR-Cas9 multiplex genome editing of the hydroxyproline-O-galactosyltransferase gene family alters arabinogalactan-protein glycosylation and function in Arabidopsis. BMC Plant Biol. 2021;21:16. doi: 10.1186/s12870-020-02791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steentoft C., Vakhrushev S.Y., Joshi H.J., Kong Y., Vester-Christensen M.B., Schjoldager K.T., Lavrsen K., Dabelsteen S., Pedersen N.B., Marcos-Silva L., Gupta R., Bennett E.P., Mandel U., Brunak S., Wandall H.H., Levery S.B., Clausen H. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta R., Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002;2002:310–322. [PubMed] [Google Scholar]
- 19.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33:3387–3395. doi: 10.1093/bioinformatics/btx431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.