Highlights
-
•
SLC39A4 expression has significant value in diagnosis and prognosis of cervical cancer.
-
•
SLC39A4 expression promoted the malignant biological behavior of cervical cancer in vitro and in vivo studies.
-
•
SLC39A4 expression was related to the immune microenvironment and immunotherapy response of cervical cancer.
Keywords: Solute carrier family 39 members 4 (SLC39A4), Cervical carcinoma, Prognosis, Immune microenvironment
Abstract
Background
Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) are becoming more common in younger women. Solute carrier family 39 member 4 (SLC39A4) produces a zinc ion transporter involved in metastasis and invasion of tumors.
Methods
The Cancer Genome Atlas RNA-seq data was used to investigate the expression of SLC39A4 and its prognostic potential. The assessment of the effect of SLC39A4 on cell growth and migration in CESC was conducted using MTT, colony formation, and Transwell assays. SLC39A4 was studied in vivo using a xenograft mouse model, and its functional involvement in oncogenesis was investigated by identifying the associated differentially expressed genes (DEGs). We evaluated the relationships among SLC39A4 levels, chemosensitivity, radiosensitivity and immune infiltration.
Results
SLC39A4 was upregulated in CESC samples, and individuals with greater SLC39A4 mRNA expression had shorter overall survival. SLC39A4 has been identified to be a regulator of tumor cell metastasis and proliferation in vivo and in vitro, with an area under the curve of 0.874 for diagnosing CESC. In total, 948 DEGs were discovered to be enriched in key CESC progression-related signaling pathways. Additionally, intratumoral immune checkpoint and infiltration activity were associated with SLC39A4 expression. High SLC39A4 expression exhibited poor chemosensitivity and radiosensitivity profiles.
Conclusion
In conclusion, SLC39A4 is a key regulator of CESC development, prognosis, and the composition of the tumor immune microenvironment. SLC39A4 could be used as a prognostic or diagnostic screening tool and as a potential target for CESC treatment.
Introduction
Cervical cancer (CC) remains the 2nd most common type of gynecological cancer and the 4th most prominent cause of cancer-related mortality in women worldwide [1]. Numerous epidemiological studies conducted over the past few decades have robustly confirmed the causal relationship between persistent infection with high-risk human papillomavirus and the development of CC [2]. Although the human papillomavirus vaccine has contributed to many developed countries reducing the prevalence of CC, its incidence rates are still high in less developed regions, especially among younger women, who are typically associated with an unfavorable prognosis [3,4]. It is necessary to establish improved biomarkers related to CC patient prognosis to promote the diagnosis and treatment of this malignancy.
A specific class of metal ion transporter is the ZRT, IRT-like protein (ZIP) family. Together with the zinc transporter family, its primary activity helps maintain the equilibrium of Zn2+ in cells [5,6]. ZIP's primary role is to transport Zn2+ into cells. The ZIP4 protein, encoded by SLC39A4 and located in the cell membrane, is essential in maintaining internal zinc concentration by absorbing dietary zinc into cells and releasing zinc from cell solute vesicles. Several pathological anomalies, including intestinal inflammation, eyeball ischemia, and growth impairment, are caused by the mutation of SLC39A4 [7,8].
Not only does SLC39A4 participate in a variety of normal physiological functions within the human body, but it also has a close association with the occurrence and development of multiple cancers [[9], [10], [11]]. Inhibiting SLC39A4 expression inhibits the development and invasion of malignant cells, and SLC39A4 overexpression promotes the occurrence and proliferation of various epithelial malignant cancers. Recent studies indicate that SLC39A4 is primarily responsible for tumor invasion and metastasis by promoting processes such as epithelial-mesenchymal transition (EMT), the production of inflammatory mediators, and angiogenesis [12]. Additionally, the degree of malignancy, clinical classification, and cancer treatment resistance correlate with SLC39A4 expression levels. SLC39A4 is overexpressed in gallbladder carcinoma, nasopharyngeal carcinoma, pancreatic cancer, and esophageal squamous cell carcinoma [[13], [14], [15], [16]], although its role in CC is undetermined.
Data from The Cancer Genome Atlas (TCGA) were analyzed and validated by in vivo and in vitro examinations of the involvement of SLC39A4 in cancer to help understand the prognosis and diagnosis implications of SLC39A4 expression and its functional relevance in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC). Correlations between SLC39A4 expression and various other factors were examined, including chemosensitivity, radiosensitivity, immune checkpoint expression, and immune cell invasion. The results of this study will provide a strong framework for further studies investigating the role of SLC39A4 in CESC and its application as a diagnostic or therapeutic candidate.
Materials and methods
The Cancer Genome Atlas (TCGA) analyses
Data on patients with CESC were obtained from the TCGA database. This data included SLC39A4 mRNA levels and clinicopathological characteristics such as age, TNM staging, clinical staging, and other relevant variables.
Patient's sample acquisition
Samples of postoperative tumors and adjacent healthy tissues from CESC patients were collected from our hospital's Department of Obstetrics and Gynecology in 2022. Participants were initially diagnosed with CESC, and all included patients showed a distinct pathological diagnosis before surgery despite not receiving any treatment. The ethical committee of The Second Hospital of Jilin University provided their approval for all human experimentation, and donors submitted their signed and written informed consent for the experiments to be performed.
Immunohistochemical (IHC) analysis
Slices of tissue were rehydrated with an ethanol gradient after being deparaffinized with xylene. After being sliced into 4 μm slices, these tissues were treated with 3 % H2O2 before antigen retrieval at room temperature (RT). Before hematoxylin counterstaining, ethanol gradient dehydration, neutral resin fixation, and light microscopy imaging, sections were blocked for 0.5 h at RT with 5 % BSA, incubated overnight at 4 °C with anti-SLC39A4 (Abcam, UK), probed for 0.5 h with a secondary antibody, and color was developed using DAB.
Western immunoblotting
The tissues were cut into small pieces and homogenized to extract total protein. The protein samples (20 μg/sample) were then extracted using RIPA lysate buffer (Solarbio, China) and separated using 10 % SDS-PAGE gel (Beyotime, China), accompanied by transfer to PVDF membrane (Millipore, USA). Following rapid blocking with 5 % skim milk for 1 hour indoors, the membrane was incubated overnight with the primary antibody SLC39A4 (Abcam) at 4 °C or the internal protein standard β-actin (Abcam). The secondary antibody (Abcam) was added to the membrane for 2 h at RT. Finally, the immunoblots were visualized using BeyoECL Plus Kit (Beyotime). β-actin was used as an internal control.
Cell culture and gene knockdown in vitro
The Procell Life Science and Technology Company (China) provided the human HeLa cells with short tandem repeat qualification. HeLa cells were cultured in MEM medium (Procell) with 10 % fetal bovine serum (FBS) and 5 % CO2 in a humid 37 °C environment. To establish stably transfected cell lines, HeLa cells were individually infected with lentiviral vectors containing either SLC39A4 short hairpin RNA (sh-SLC39A4) or negative control (sh-NC), which were generated by Genepharma (China). The transfection was performed using the Polybrene (Hanbio, China). Cells were collected after 48 h and used in subsequent experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR) assay
Total RNA was extracted using the Triquick Reagent from Solarbio. SLC39A4 was reversely transcribed from RNA employing the SweScript RT I First Strand cDNA Synthesis Kit (Servicebio, China). The obtained cDNA was combined with 2SYBR Green qPCR Master Mix (Low ROX) (Servicebio) with specific primers for qRT-PCR analysis and then amplified on a PCR apparatus. Through the 2−ΔΔCt approach, the relative expression of SLC39A4 was determined, and the GAPDH was used as a standardized internal control. SLC39A4 primer sequences included: 5'-CGAGGTCCCTATGACGCTG-3' forward and 5'-CACTCAGGCATACCGTGTCC-3' reverse primers.
3-(4, 5-Dimethylthiazol-2-yl)−2, 5-diphenyltetrazolium bromide (MTT) assay
The MTT assay was used to identify cell proliferation. Each group's transfected HeLa cells were seeded into 96-well plates at 1 × 105 cells/mL density. MTT (5 mg/mL; 20 μL; Procell) was applied to each well after 48 h, and the cells were maintained for 4 h at 37 °C. After removing the supernatant, 150 μL DMSO (Solarbio) was applied to dissolve the precipitated product. The OD value at 570 nm was then determined using a microplate analyzer (Bio-Rad, model 550, USA).
Colony formation assays
Five hundred logarithmic-growing cells were seeded into 6-well plates and cultured. After 10 days, the cell clones were washed with PBS. Visible colonies were quantified using an optical microscope after the cells were fixed with 4 % paraformaldehyde (Beijing Chemical Plant, China) and stained with 2 % crystal violet (Beyotime).
Scratch assay
Transfected HeLa cells were seeded in 6-well plates and expanded almost to saturation. After 48 h, the sterilized 20 μL pipette tips were used to make longitudinal scratches on cell monolayers. PBS was later used to remove unattached cells. At 0 and 24 h after culture, the width of the scratches was photographed using an optical microscope, and the migratory distance was determined using ImageJ software (NIH).
Transwell assays
In order to determine the invasiveness of HeLa cells, the transwell chambers (8 μm pore size; Corning, USA) were pre-coated with Matrigel (BD Biosciences, USA) under the directions provided by the manufacturer. The top compartment of the chamber was seeded with the transfected HeLa cells in 200 μL serum-free medium in each group, and the bottom compartment was filled with 600 μL MEM complete medium containing 10 % FBS. The invaded cells transferred to the lower chamber were cultured for 24 h before being fixed with 4 % paraformaldehyde (Beijing Chemical Plant) and stained with Giemsa stain reagent (Solarbio). Finally, an inverted microscope was used to count and take pictures of the invasive cells in five randomly selected regions.
In vivo xenograft model
The Animal Care and Use Committee at the College of Basic Medicine of Jilin University approved all animal experiments that were conducted in accordance with the NIH guidelines. For this study, HeLa cells (1 × 107 cells/mouse) were subcutaneously injected into 7-week-old BALB/c nude mice (weight = 18–20 g). Three mice were injected with transfected sh-SLC39A4 and sh-NC cells. These mice were euthanized by cervical dislocation after 21 days, and tumors were removed, analyzed, and measured. Tumor volumes were calculated as (length × width2)/2. Before hematoxylin and eosin (H&E) staining, tumors were stained with a 4 % tissue fixative solution (Beyotime).
Identification and characterization of differentially expressed genes (DEGs)
Data for CESC patients was collected using information from the TCGA and LinkedOmics databases, and DEGs associated with SLC39A4 were identified. Results were utilized to construct volcano plots, and the genes showing a fold-change of > 1.5 and an FDR-corrected P-value of < 0.05 were identified as DEGs. The 50 DEGs with the maximum up- and down-regulation were also combined to construct a heatmap. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment and Gene Ontology (GO) analyses were also performed on these DEGs.
Immune cell infiltration analysis
A single-sample GSEA examination of immune cell infiltration was carried out through the R GSVA package to determine the expression of immune checkpoint molecules such as CD274, CTLA4, and HAVCR2 as well as the infiltration of 24 different immune cell types. Participants with low and high levels of SLC39A4 expression were compared based on these molecule expressions.
Chemosensitivity analysis
In order to evaluate the chemosensitivity, several concentrations of cisplatin (Shandong Qilu, China), paclitaxel (Bristol-Myers Squibb, USA), and doxorubicin (Pharmacia, Italy) were administered to the culture plate for cellular incubation. After 24 h of incubation, the viability of the cells was assessed using the MTT approach, as described earlier. Concentration-effect curves were constructed by plotting drug concentration on the x-axis and cell viability on the y-axis. The SPSS statistical software package employed the Probability Unit Method to determine the IC50, representing the concentration at which the inhibitory effect is half-maximal.
Radiosensitivity analysis
Cells transfected from each experimental group were cultured for 24 h on a 96-well plate while ensuring that the previously described conditions were maintained. Following the replacement of the medium with newly prepared complete MEM, the cells were promptly subjected to X-ray irradiation. The irradiation was carried out at a dosage rate of 1.020 Gy per minute, utilizing an X-ray biological irradiator (X-Rad320, Precision X-Ray, USA). The source-to-skin distance during the irradiation process was maintained at 70 cm. The cells were exposed to X-ray radiation at 0, 6, 10, and 15 Gy dosages. After radiation exposure, the cells were subsequently incubated for 24 h. Following this incubation period, the viability of the cells was evaluated using the MTT assay.
Statistical analysis
For the statistical evaluations, R version 4.0.3 was used. Wilcoxon tests were utilized for group comparisons, and the pROC tool was employed to construct receiver operating characteristic (ROC) curves to establish the significance of SLC39A4 expression levels. The associations between SLC39A4 levels and clinical characteristics in CESC patients were evaluated using Chi-square tests. Kaplan-Meier curves and log-rank tests were used to evaluate overall survival (OS) between groups.The significance threshold was determined at P < 0.05.
Results
Evaluation of the expression pattern of SLC39A4 in cancer
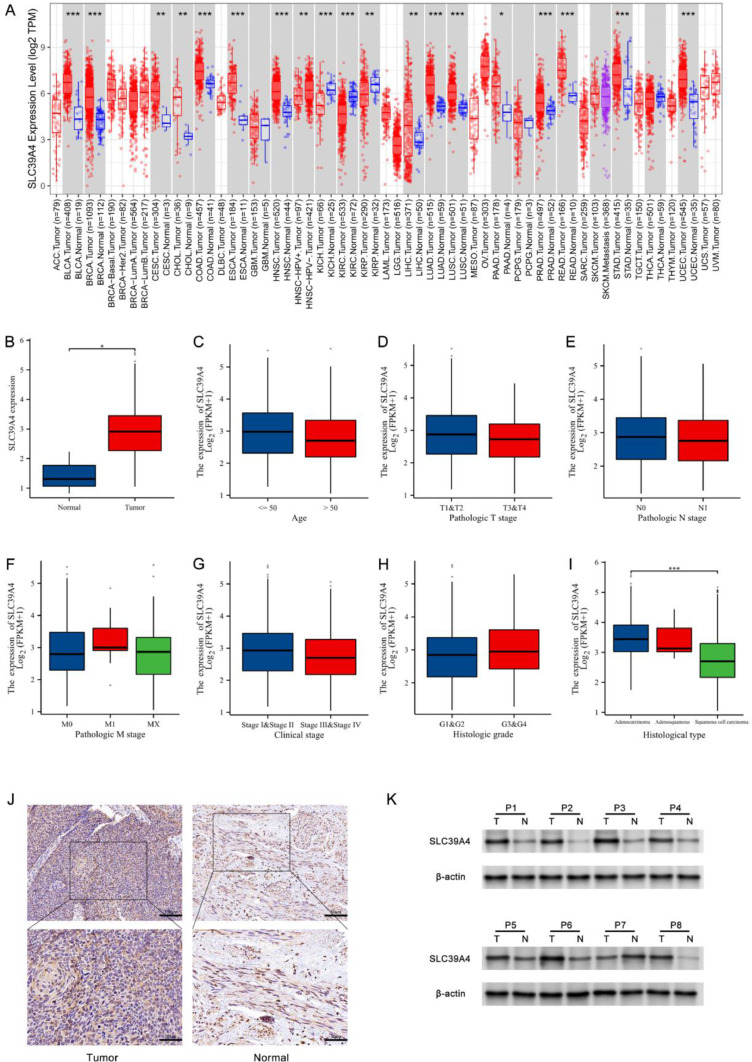
Initially, the expression of SLC39A4 in all tumors in the TIMER 2.0 database was examined; this revealed that SLC39A4 was overexpressed in UCEC, STAD, READ, PRAD, PAAD, LUSC, LUAD, LIHC, HNSC, ESCA, COAD, CHOL, CESC, BRCA, and BLCA. At the same time, SLC39A4 downregulation was found in KICH, KIRC, and KIRP (Fig. 1a). SLC39A4 expression was found to be higher in CESC patient tumors compared to healthy tissues (P < 0.05) (Fig. 1b), and associations between this gene's expression and clinical factors, such as histopathological tumor type, were observed (P < 0.001) (Fig. 1i). SLC39A4 levels, on the other hand, were not correlated with patient age, tumor T, N, M, or G stages, or clinical phases (P > 0.05) (Fig. 1c-h). IHC and Western immunoblotting assays also validated the upregulation of SLC39A4 in tumors compared to corresponding control samples (Fig. 1j-k), which aligns with the TCGA database outcomes.
Fig. 1.
The expression levels of SLC39A4, as determined from analysis of the TCGA database and patients' samples. (a) The examination of SLC39A4 expression was performed in the pan-cancer analyses using the TIMER 2.0 database. This encompassed both samples of tumors and samples without tumors. (b) The expression level of SLC39A4 mRNA in both healthy control tissues and CESC. (c-i) The relationships between the clinicopathological features associated with CESC patients, such as age, histologic grade, TNM staging, clinical staging, histologic type, and the expression of SLC39A4 mRNA. The expression of SLC39A4 was evaluated using (j) IHC and (k) Western immunoblotting in pairs of CESC tumor tissues and neighboring paracancerous tissues. *P < 0.05, **P < 0.01, ***P < 0.001; ns = not significant.
Analyzing the relationship between SLC39A4 expression and clinical data from CESC patients
CESC patients in the TCGA database were then divided into two groups based on whether their samples had low or high levels of SLC39A4 expression, and clinical characteristics such as age, TNM classification, G classification, and clinical classification were compared between these subgroups (Table 1, Supplementary materials). This analysis showed a substantial correlation between SLC39A4 mRNA levels, histological type, and primary therapy outcome (P < 0.0001 and P = 0.0026, respectively).
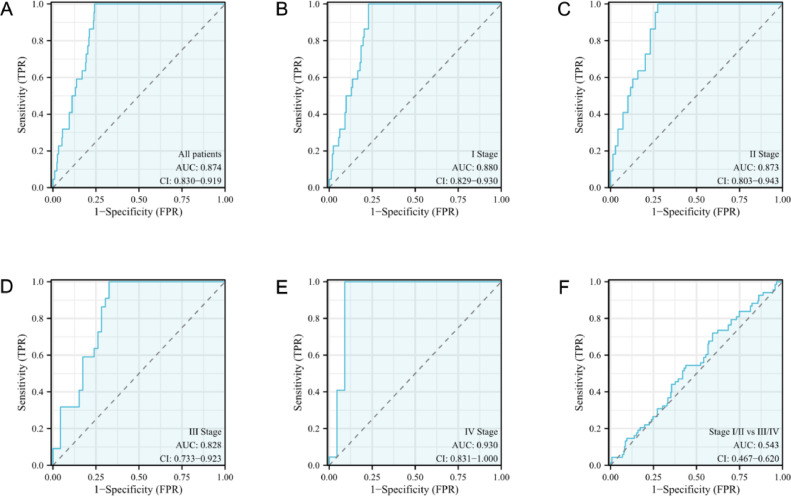
The diagnostic effectiveness of SLC39A4 in CESC
Following the development of ROC curves based on SLC39A4 expression in CESC, a calculated area under the curve (AUC) value of 0.874 was discovered, indicating a better diagnostic performance (Fig. 2a). The AUC values for various CESC tumor stages were also determined, with the corresponding values for stage I, stage II, stage III, and stage IV disease being 0.880, 0.873, 0.828, and 0.930, respectively (Fig. 2b–e). The SLC39A4 level could also differentiate between early and late CESC stages (AUC: 0.543 for stage I/II vs stage III/IV) (Fig. 2f).
Fig. 2.
ROC curve analysis for SLC39A4 in patients with CESC. Comparison of normal samples with (a) tumors, (b) stage I tumors, (c) stage II tumors, (d) stage III tumors, and (e) stage IV tumors. (f) Stage I/II against stage III/IV tumors.
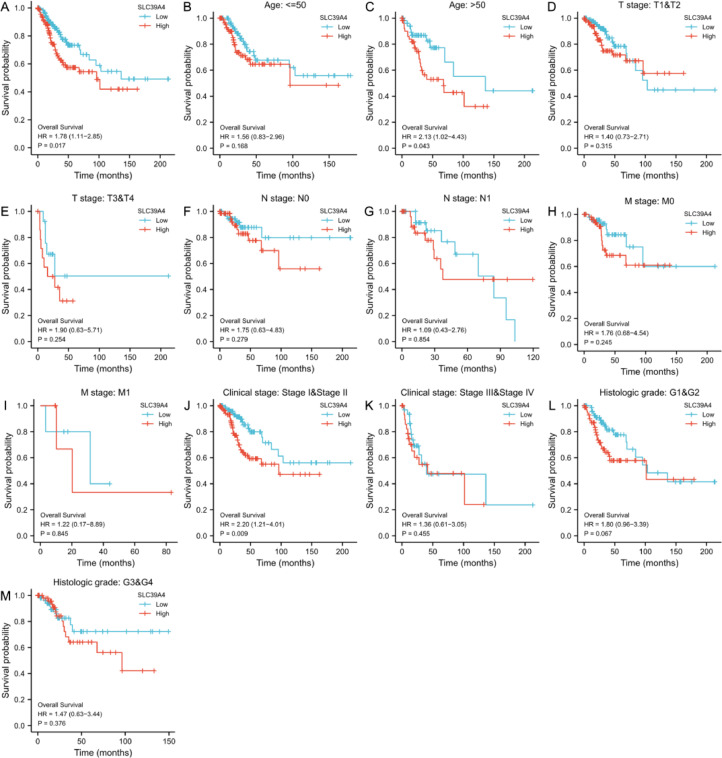
The role of SLC39A4 in CESC prognosis
The prognostic significance of SLC39A4 mRNA levels in CESC was investigated further using Kaplan-Meier curves, revealing a relationship between SLC39A4 overexpression and poorer patient OS (P = 0.017, Fig. 3a). Overexpression of SLC39A4 was also found to be associated with a lower patient OS in the older people (P = 0.043) and stage I/II (P = 0.009) groups in subgroup analysis. In comparison, SLC39A4 levels were unrelated to survival rates in other subgroups (P > 0.05) (Fig. 3b–m).
Fig. 3.
Analysis of the relationship between SLC39A4 expression and CESC patient survival. (a) Kaplan-Meier analyses were performed in the TCGA database on CESC patients to evaluate OS outcomes. (b-m) Survival subgroup analyses were performed based on CESC patient age, histologic grade, TNM, and clinical staging.
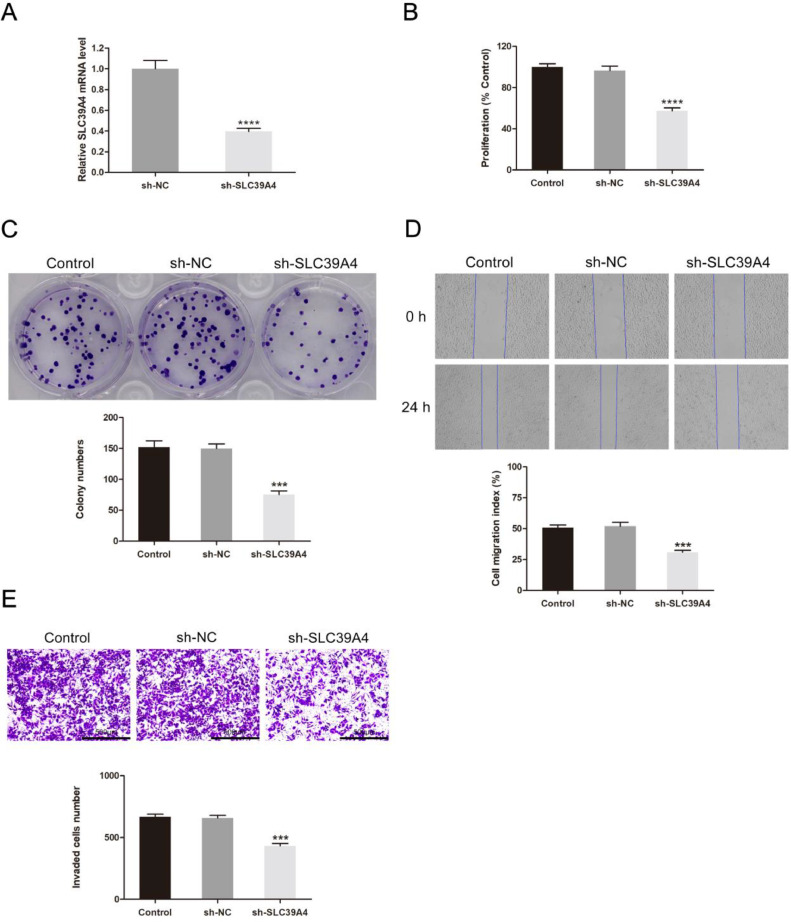
SLC39A4 functional involvement in CESC validated using in vivo and in vitro assays
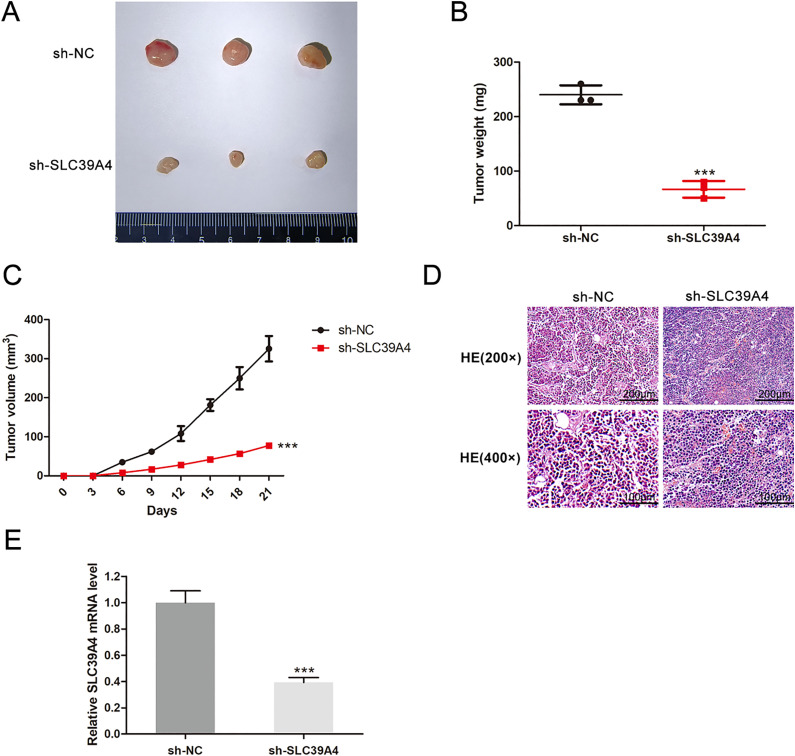
Following that, SLC39A4 silencing was conducted in HeLa cells, and the efficiency of silencing was detected by the qRT-PCR method (Fig. 4a). MTT and colony formation assays revealed that this knockdown resulted in reduced cellular proliferation when compared to control construct transfection (P < 0.0001 and P < 0.001, respectively) (Fig. 4b, c). The migration (P < 0.001) and invasion (P < 0.001) abilities of these cells were significantly reduced when SLC39A4 expression was knocked down (Fig. 4d, e). In vivo xenograft experiments indicated that on day 21 following tumor weights in the sh-NC and sh-SLC39A4 groups were 240.00 ± 17.32 mg and 66.67 ± 15.28 mg (P < 0.001) (Fig. 5a-b), respectively. In contrast, respective tumor volumes were 325.44 ± 32.60 mm3 and 77.44 ± 1.59 mm3 (P < 0.001) (Fig. 5c). In comparison to the tumor in the sh-NC group, the knocked-down SLC39A4 tumors displayed a diminished nuclear portion, a lower intensity of nuclear staining, and a lower number of mitotic images (Fig. 5d). Additionally, knocked-down SLC39A4 tumors demonstrated a significant reduction in SLC39A4 expression levels, as indicated by qRT-PCR results (P < 0.001) (Fig. 5e).
Fig. 4.
SLC39A4 knockdown inhibited CESC cell growth and metastatic potential. (a) qRT-PCR detected the transfection efficiency of sh-SLC39A4. (b) MTT, (c) colony formation, (d) wound healing, and (e) Transwell invasion assays were performed on HeLa cells transfected with sh-NC or sh-SLC39A4 constructs. ***P < 0.001, ****P < 0.0001.
Fig. 5.
SLC39A4 knockdown inhibited tumor development in CESC models. (a) Tumor size, (b) weight, (c) volume, (d) histological grading and (e) SLC39A4 mRNA level. ***P < 0.001.
Exploring the roles of DEGs associated with SLC39A4
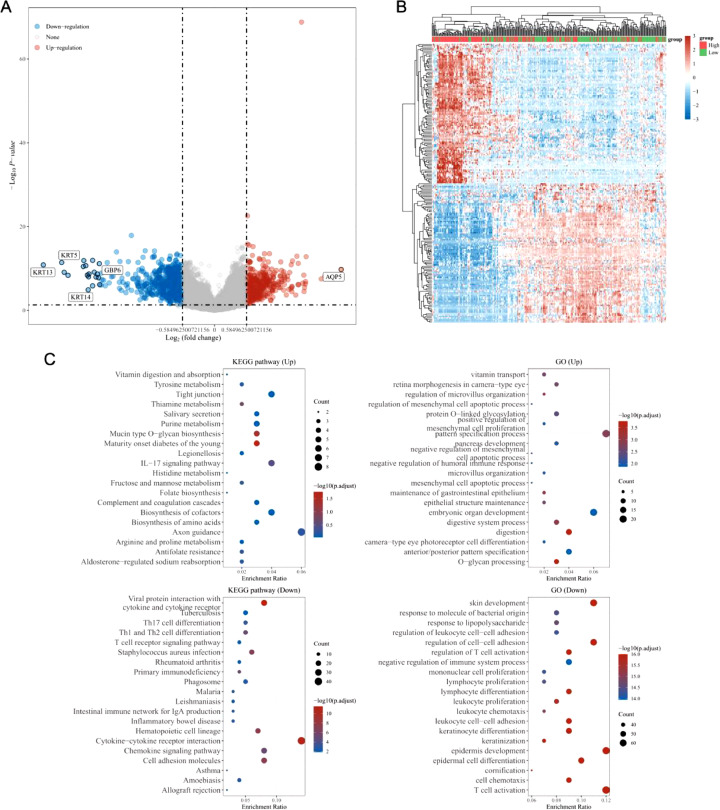
Genes specifically expressed in a way related to SLC39A4 expression were identified using the LinkedOmics database, which includes 367 and 581 DEGs that were either positively or negatively correlated with SLC39A4 expression, respectively (Fig. 6a). The heatmap was constructed using the 50 most favorably and negatively correlated genes (Fig. 6b). According to the KEGG enrichment study, DEGs were primarily involved in cell adhesion molecules, the chemokine signaling pathway, cytokine-cytokine receptor interaction, the IL-17 signaling pathway, and axon guidance. These genes were found to play a significant role in the growth and specialization of skin, epidermis, and keratinocytes, in the control of immune system function and immune response, and in the proliferation, activation, differentiation, and adhesion of leukocytes such as lymphocytes, monocytes, and T cells (Fig. 6c).
Fig. 6.
Enrichment analyses of SLC39A4-associated DEGs. (a) Analysis of identified DEGs using volcano plots, with upregulated and downregulated genes represented in red and blue, respectively. (b) DEGs were plotted in a heatmap, with up-and downregulated genes highlighted in red and blue. (c) DEG KEGG and GO enrichment analyses.
An association between SLC39A4 levels and tumor immune cell invasion
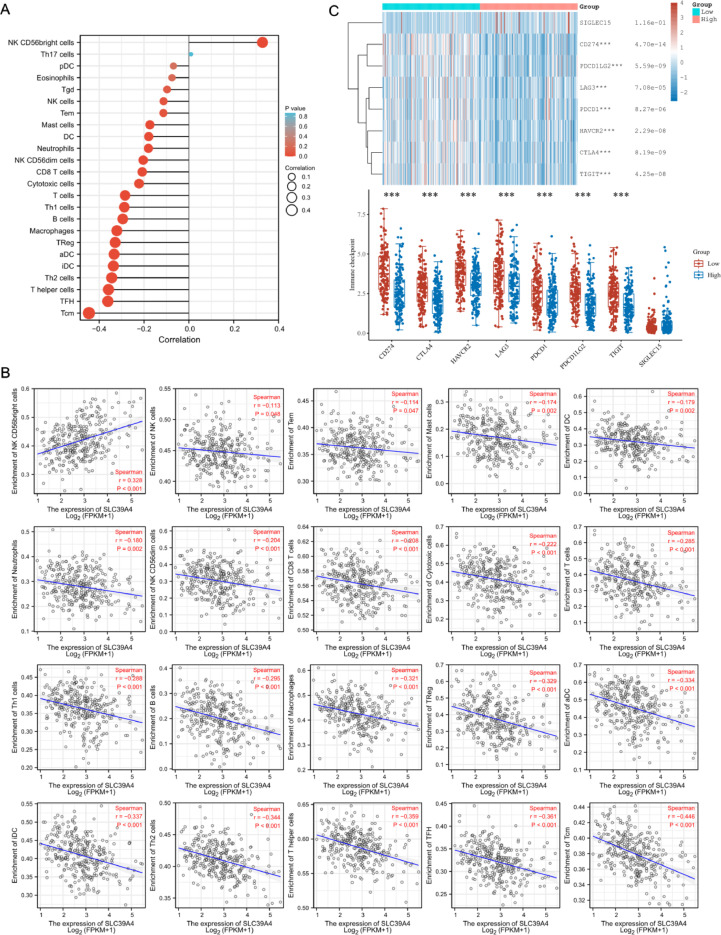
The next step was to examine the connection between intratumoral immune cell invasion and SLC39A4 transcription in CESC patients. SLC39A4 levels and NK CD56bright cell infiltration of these tumors were significantly positively correlated (P < 0.001). In contrast, SLC39A4 levels were negatively associated with infiltration by NK cells (P = 0.048), Tem (P = 0.047), Mast (P = 0.002), DC (P = 0.002), Neutrophils (P = 0.002), NK CD56dim, CD8 T, Cytotoxic, T cells, Th1, B cells, Macrophages, TReg, aDC, iDC, Th2, T helper cells, TFH and Tcm (all P values are < 0.001) (Fig. 7a-b). SLC39A4 expression and immune checkpoints were correlated to examine the predictive value of SLC39A4 regarding immunotherapy responses. These findings showed that CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, and TIGIT were 7 immune checkpoints whose expression was inversely correlated with SLC39A4 expression (Fig. 7c).
Fig. 7.
TME immune cell infiltration analyses. (a) A forest plot. (b) Spearman correlation analyses. (c) The expression of immune checkpoint markers investigated in patient populations expressing low and high levels of SLC39A4. ***P < 0.001.
The relationship between chemosensitivity and SLC39A4 expression
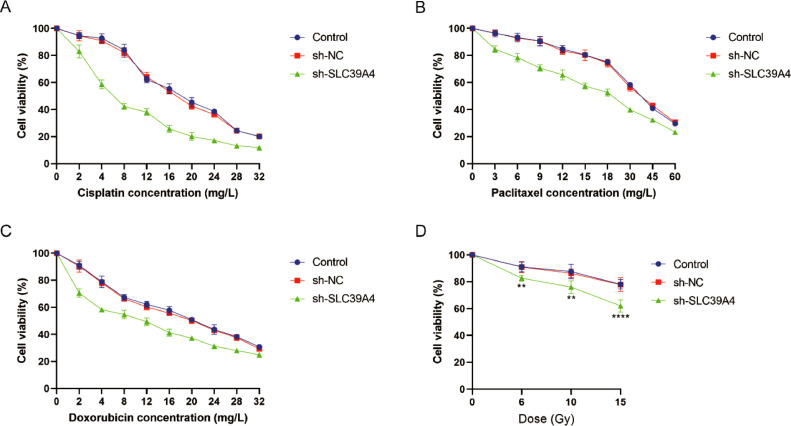
Advanced cases of CESC are commonly managed through chemotherapy, with cisplatin, paclitaxel, and doxorubicin being the first-line chemotherapy drugs utilized in clinical practice. In order to ascertain the sensitivity of transfected CESC cells to chemotherapy, the cells were incubated with various concentrations of these drugs while developing concentration-effect curves (Fig. 8a–c). The IC50 values were determined using the Probabilistic Unit Method. The findings of this study indicate that the IC50 values for cisplatin (P < 0.0001), paclitaxel (P < 0.001), and doxorubicin (P < 0.0001) were substantially decreased in the sh-SLC39A4 group as compared to both the control group and the sh-NC group. There was no statistically significant difference observed between the control group and the sh-NC group (P > 0.05) (Table 2, Supplementary materials).
Fig. 8.
Analyses of chemosensitivity and radiosensitivity. The effect of different concentrations of chemotherapy drugs, including (a) cisplatin, (b) paclitaxel, and (c) doxorubicin, on the cell viability of CESC cells. (d) Cell viability of CESC cells under different radiation doses.
The relationship between radiosensitivity and SLC39A4 expression
At 0, 6, 10, and 15 Gy dosages, X-ray irradiation was applied to the transfected CESC cells. Cell viability was gradually decreased with increasing radiation dose. At dosages of 6, 10, and 15 Gy, the sh-SLC39A4 group had a substantial decrease in cell viability (P < 0.01, P < 0.01 and P < 0.0001, respectively), in comparison to the control group and the sh-NC group. At the same radiation dose, there was no noticeable difference in cell survival between the control and sh-NC groups (P > 0.05) (Fig. 8d).
Discussion
Although CC poses a serious risk to human health worldwide, less developed countries are particularly affected by its impacts. The incidence of CC is unevenly distributed between developed and developing countries [17]. Over 85 % of cases occur in economically challenged regions, with nearly 90 % of CC-related deaths happening in resource-limited areas worldwide [18]. In contrast, in developed countries and regions, both the incidence and mortality rates of CC exhibit a gradual decline [19]. Furthermore, the age of developing CC is gradually shifting to young women, with 60.3 % and 57.38 % of people diagnosed with CC being under the age of 50 and 40 years, respectively [20]. As this type of cancer affects increasing numbers of young people, the importance of surgical intervention in the early stages of the disease to enhance patient outcomes and maintain fertility becomes increasingly essential [21]. As a result, finding new biomarkers to effectively diagnose, treat, and prognostic individuals with CC is imperative.
As a catalytic cofactor of numerous enzymes and a requirement for numerous transcription regulators and enzymes implicated in the growth and metastasis of cancer, zinc is an important trace element and nutrient for human health [22]. Cells have sophisticated mechanisms for regulating zinc absorption, intracellular storage, and excretion. Since zinc can modify tight junction proteins and specifically strengthen the epithelial barrier, it is critical for tumor invasion and metastasis [23]. ZIP4 is the most significant zinc transporter into intestinal epithelial cells as a member of the zinc transporter family [24]. SLC39A4 overexpression in cancer may regulate the expression of other genes or transcription factors by providing enough zinc ions for rapidly proliferating tumor cells. The role of SLC39A4 in the onset and progression of human cancers has long been recognized.
EMT is the first link and a critical stage in tumor cell invasion and distant metastasis. EMT causes epithelial cells to change shape into spindle cells with greater mobility, as well as cytoskeleton rearrangement and extracellular matrix remodeling, all of which cause cancer cells to lose their polarity and adhesion, making them more prone to invasion and distant metastasis [25,26]. According to Liu et al., the ZIP4-ZEB1-integrin axis causes tumor metastasis and chemotherapy tolerance by activating downstream genes ZEB1 and YAP1. This encourages EMT and metastasis of cancer cells [27,28]. It is possible to use ZIP4 as an upstream regulator of ZBE1 expression, which initiates EMT by suppressing the expression of interstitial proteins and promoting that of epithelial marker proteins [29]. The following mechanisms can help ZIP4 support tumor progression in addition to the EMT process. In hepatocellular carcinoma, ZIP4 encourages actin skeleton reorganization, upregulates MMP-2 and MMP-9 expression, and hastens extracellular matrix degradation [30]. In pancreatic cancer, ZIP4 can break down the intercellular linkage structure and influence the secretion of inflammatory factors like IL-6, which activates the carcinogenic pathway [31,32]. In order to promote tumor angiogenesis, ZIP4 can also increase the expression of neurofelin-1 and vascular endothelial growth factor [33].
The results above highlight the necessity to evaluate SLC39A4 as a regulator of tumor development, prognostic outcomes, and the constitution of the tumor microenvironment (TME). This study demonstrated that SLC39A4 overexpression was present in CESC tumors compared to healthy tissues, which another set of clinical samples validated. SLC39A4 levels required further research as a potential biomarker since they demonstrated promise in differentiating between various CESC phases. Furthermore, a correlation between decreased patient survival and increased SLC39A4 levels was observed. In vitro and xenograft studies confirmed SLC39A4's function as a pro-oncogenic factor, emphasizing its significance in this cancerous context.
The KEGG pathways (including the IL-17 signaling pathway, cytokine-cytokine receptor interaction, and chemokine signaling pathway) were all enriched in these DEGs. Among them, IL-17 is connected to the tumorigenic effect brought on by persistent inflammation and to the suppression of the immune system's anti-tumor response [34]. Several chemokines or cytokines also play essential roles in the TME's inflammatory response, immune regulation, and promotion of cancer growth and progression, chemoresistance, and evasion of the body's immune surveillance [35]. The biological processes that regulate immune system function and immune reaction, as well as leukocyte proliferation, activation, differentiation, and adhesion, were identified in the GO analysis that was carried out. As a result, we assume these processes may be closely linked to the crucial function of immune cells in the TME and tumor immune escape. In conclusion, these pathways may play a role in how CESC progression is affected by elevated SLC39A4 expression levels.
Tumor cells are found in the TME, a dynamic, complicated network, and their interactions with the TME are complementary. A range of immune cells and extracellular components like cytokines, chemokines, and extracellular matrix surround tumor cells in the TME. TME is increasingly being shown to be essential to the occurrence and development of tumors, as well as having a significant impact on the effectiveness of therapy and patient outcomes [36]. In this research, 19 out of 24 immune cells negatively correlate with SLC39A4 level. As a result of the current findings, we could hypothesize that elevated SLC39A4 expression in CESC is associated with decreased immune cell infiltration, resulting in tumor immune evasion and poor patient prognosis.
Immunotherapy targeting the proteins cytotoxic T-lymphocyte-associated protein-4, programmed cell death protein-1, or programmed cell death ligand 1 has recently revolutionized how various cancers are treated and offered patients with advanced CESC new hope. Screening patients who might benefit from immunotherapy and combination therapy is extremely important because the major downside of immune checkpoint inhibitor therapy is that it can only help a small number of patients. In this research, we examined the connection between the expression of SLC39A4 and different immune checkpoints. Our findings demonstrated that SLC39A4 expression was negatively associated with almost all immune checkpoints, which is consistent with the discussion above. SLC39A4 may be involved in accelerating the development of cancer by impairing immune responses, and high SLC39A4 expression may also render CESC less amenable to immunotherapy.
Moreover, chemotherapy and radiotherapy are integral components of cancer therapy, and they play pivotal roles in the control and treatment of malignancies in advanced stages. In the present in vitro study, we examined the impact of SLC39A4 expression on the susceptibility to chemotherapy and radiation therapy. The results of our study indicate that in vitro knockdown of SLC39A4 leads to increased chemosensitivity and radiosensitivity. This observation underscores the potential efficacy of SLC39A4 as a tool for assessing the responsiveness of patients with CESC to chemotherapy and radiotherapy, presenting a substantial prospect for clinical implementation and therapeutic advice.
In conclusion, the upregulation of SLC39A4 was associated with advanced stages and a less favorable prognosis in CESC. The upregulation of this gene resulted in the progression of malignancy in CESC, as observed in both in vivo and in vitro experiments. Additionally, it is suggested that this gene's overexpression may have impacted immune system reactions and the sensitivity of CESC cells to radiotherapy and chemotherapy treatments. As mentioned above, the findings have established a crucial basis for identifying pathways responsible for regulating the malignancy and progression of CESC. Consequently, this has opened up avenues for developing innovative targeted therapeutic approaches.
Funding statement
This study was supported by Jilin Provincial Science and Technology Department (YDZJ202301ZYTS069), Jilin Provincial Education Department (JJKH20221084KJ), Jilin Provincial Health Department (The study of Pianzaihuang on alleviating acetaminophen-induced liver damage by inhibiting the TLR4/PI3K/Akt and NLRP3 pathways) and Wu Jie-Ping Medical Foundation (The role of the CNGB1 and its immune relevance in the progression of cervical cancer).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. SLC39A4 transcription profiles of samples from CESC patients with their clinical and other relevant medical information were obtained from the TCGA platform (https://portal.gdc.cancer.gov/).
Ethics approval statement
The study was conducted following the Declaration of Helsinki, and the study protocol was approved through the Medical Ethics Committee (Second Hospital of Jilin University). We state that animal studies were conducted with the approval of the Animal Ethics Committee (College of Basic Medicine of Jilin University).
CRediT authorship contribution statement
Yue-Chen Zhao: Conceptualization, Visualization, Writing – original draft. Tie-Jun Wang: Investigation, Validation, Writing – review & editing. Jie Cui: Methodology, Validation, Visualization, Writing – review & editing. Li-Zhen She: Methodology, Writing – review & editing. Rui-Feng Zhang: Methodology, Writing – review & editing. Chao-He Zhang: Project administration, Conceptualization, Data curation, Funding acquisition, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors have no relevant financial or non-financial interests to disclose.
Acknowledgments
The authors would like to thank MJEditor (www.mjeditor.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101839.
Appendix. Supplementary materials
References
- 1.Arbyn M., Weiderpass E., Bruni L., de Sanjose S., Saraiya M., Ferlay J., Bray F. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob. Health. 2020;8(2):e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okunade K.S. Human papillomavirus and cervical cancer. J. Obstet. Gynaecol. 2020;40(5):602–608. doi: 10.1080/01443615.2019.1634030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Torres-Roman J.S., Ronceros-Cardenas L., Valcarcel B., Bazalar-Palacios J., Ybaseta-Medina J., Carioli G., La Vecchia C., Alvarez C.S. Cervical cancer mortality among young women in Latin America and the Caribbean: trend analysis from 1997 to 2030. BMC Public Health. 2022;22(1) doi: 10.1186/s12889-021-12413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bin B.H., Seo J., Kim S.T. Function, structure, and transport aspects of ZIP and ZnT zinc transporters in immune cells. J. Immunol. Res. 2018;2018 doi: 10.1155/2018/9365747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers K., Srai S.K.S. The trafficking of metal ion transporters of the Zrt- and Irt-like protein family. Traffic. 2018;19(11):813–822. doi: 10.1111/tra.12602. [DOI] [PubMed] [Google Scholar]
- 7.Kambe T., Fukue K., Ishida R., Miyazaki S. Overview of inherited zinc deficiency in infants and children. J. Nutr. Sci. Vitaminol. 2015;61:S44–S46. doi: 10.3177/jnsv.61.S44. (Tokyo)Suppl. [DOI] [PubMed] [Google Scholar]
- 8.Dufner-Beattie J., Weaver B.P., Geiser J., Bilgen M., Larson M., Xu W., Andrews G.K. The mouse acrodermatitis enteropathica gene Slc39a4 (Zip4) is essential for early development and heterozygosity causes hypersensitivity to zinc deficiency. Hum. Mol. Genet. 2007;16(12):1391–1399. doi: 10.1093/hmg/ddm088. [DOI] [PubMed] [Google Scholar]
- 9.Bagchi S., Yuan R., Engleman E.G. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu. Rev. Pathol. 2021;16:223–249. doi: 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 10.Fan Q., Zhang W., Emerson R.E., Xu Y. ZIP4 is a novel cancer stem cell marker in high-grade serous ovarian cancer. Cancers. 2020;12(12) doi: 10.3390/cancers12123692. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong W., Liang L., Gu Y., Qi Z., Qiu H., Yang X., Zeng W., Ma L., Xie J. Immune-related lncRNA to construct novel signature and predict the immune landscape of human hepatocellular carcinoma. Mol. Ther. Nucleic Acids. 2020;22:937–947. doi: 10.1016/j.omtn.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M., Zhang Y., Yang J., Zhan H., Zhou Z., Jiang Y., Shi X., Fan X., Zhang J., Luo W., Fung K.A., Xu C., Bronze M.S., Houchen C.W., Li M. Zinc-dependent regulation of ZEB1 and YAP1 coactivation promotes epithelial-mesenchymal transition plasticity and metastasis in pancreatic cancer. Gastroenterology. 2021;160(5):1771–1783. doi: 10.1053/j.gastro.2020.12.077. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M., Fan K., Zheng B.H., Zekria D., Suo T., Liu H., Shen S., Liu H.B., Ni X.L. Knockdown of SLC39A4 expression inhibits the proliferation and motility of gallbladder cancer cells and tumor formation in nude mice. Cancer Manag. Res. 2021;13:2235–2246. doi: 10.2147/CMAR.S282269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M.Y., Zhang Y.Q., Yang J.X., Cui X.B., Zhou Z.J., Zhan H.X., Ding K., Tian X., Yang Z.B., Fung K.M.A., Edil B.H., Postier R.G., Bronze M.S., Fernandez-Zapico M.E., Stemmler M.P., Brabletz T., Li Y.P., Houchen C.W., Li M. ZIP4 Increases expression of transcription factor ZEB1 to promote integrin α3β1 signaling and inhibit expression of the gemcitabine transporter ENT1 in pancreatic cancer cells. Gastroenterology. 2020;158(3):679. doi: 10.1053/j.gastro.2019.10.038. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia C.M., Chen X., Li J., Chen P. SLC39A4 as a novel prognosis marker promotes tumor progression in esophageal squamous cell carcinoma. Oncotargets Ther. 2020;13:3999–4008. doi: 10.2147/OTT.S245094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Q., Liu Y.M., Liu J., Han J., Guo J.X., Lu S., Huang X.M., Yi P., Lang J.Y., Zhang P., Wang C.T. Inhibition of ZIP4 reverses epithelial-to-mesenchymal transition and enhances the radiosensitivity in human nasopharyngeal carcinoma cells. Cell Death Dis. 2019;10 doi: 10.1038/s41419-019-1807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allemani C., Weir H.K., Carreira H., Harewood R., Spika D., Wang X.S., Bannon F., Ahn J.V., Johnson C.J., Bonaventure A., Marcos-Gragera R., Stiller C., Silva G.A.E., Chen W.Q., Ogunbiyi O.J., Rachet B., Soeberg M.J., You H., Matsuda T., Bielska-Lasota M., Storm H., Tucker T.C., Coleman M.P., Grp C.W. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385(9972):977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bermudez A., Bhatla N., Leung E. Cancer of the cervix uteri. Int. J. Gynecol. Obstet. 2015;131:S88–S95. doi: 10.1016/j.ijgo.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2016. Ca Cancer J. Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 20.Sasieni P., Castanon A., Cuzick J. Screening and adenocarcinoma of the cervix. Int. J. Cancer. 2009;125(3):525–529. doi: 10.1002/ijc.24410. [DOI] [PubMed] [Google Scholar]
- 21.Fleming N.D., Ramirez P.T., Soliman P.T., Schmeler K.M., Chisholm G.B., Nick A.M., Westin S.N., Frumovitz M. Quality of life after radical trachelectomy for early-stage cervical cancer: a 5-year prospective evaluation. Gynecol. Oncol. 2016;143(3):596–603. doi: 10.1016/j.ygyno.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donahue T., Hines O.J. The ZIP4 pathway in pancreatic cancer. Cancer Biol. Ther. 2010;9(3):243–245. doi: 10.4161/cbt.9.3.11064. [DOI] [PubMed] [Google Scholar]
- 23.Unal O., Baltaci A.K., Mogulkoc R., Avunduk M.C. Effect of pinealectomy and melatonin supplementation on metallothionein, ZnT2, ZIP2, ZIP4 and zinc levels in rat small intestine. Biotech. Histochem. 2021;96(8):623–635. doi: 10.1080/10520295.2021.1885738. [DOI] [PubMed] [Google Scholar]
- 24.Xu C., Wallace M.B., Yang J., Jiang L., Zhai Q., Zhang Y., Hong C., Chen Y., Frank T.S., Stauffer J.A., Asbun H.J., Raimondo M., Woodward T.A., Li Z., Guha S., Zheng L., Li M. ZIP4 is a novel diagnostic and prognostic marker in human pancreatic cancer: a systemic comparison between EUS-FNA and surgical specimens. Curr. Mol. Med. 2014;14(3):309–315. doi: 10.2174/1566524013666131217112921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Brabletz S., Schuhwerk H., Brabletz T., Stemmler M.P. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 2021;40(18) doi: 10.15252/embj.2021108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu M., Zhang Y., Yang J., Cui X., Zhou Z., Zhan H., Ding K., Tian X., Yang Z., Fung K.A., Edil B.H., Postier R.G., Bronze M.S., Fernandez-Zapico M.E., Stemmler M.P., Brabletz T., Li Y.P., Houchen C.W., Li M. ZIP4 Increases Expression of Transcription Factor ZEB1 to Promote Integrin alpha3beta1 Signaling and Inhibit Expression of the Gemcitabine Transporter ENT1 in Pancreatic Cancer Cells. Gastroenterology. 2020;158(3):679–692. doi: 10.1053/j.gastro.2019.10.038. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M., Yang J., Zhang Y., Zhou Z., Cui X., Zhang L., Fung K.M., Zheng W., Allard F.D., Yee E.U., Ding K., Wu H., Liang Z., Zheng L., Fernandez-Zapico M.E., Li Y.P., Bronze M.S., Morris K.T., Postier R.G., Houchen C.W., Yang J., Li M. ZIP4 promotes pancreatic cancer progression by repressing ZO-1 and claudin-1 through a ZEB1-dependent transcriptional mechanism. Clin. Cancer Res. 2018;24(13):3186–3196. doi: 10.1158/1078-0432.CCR-18-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H., Wang H., Liu X., Yu T. miR-1271 inhibits migration, invasion and epithelial-mesenchymal transition by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2016;472(2):346–352. doi: 10.1016/j.bbrc.2016.02.096. [DOI] [PubMed] [Google Scholar]
- 30.Xu X., Guo H.J., Xie H.Y., Li J., Zhuang R.Z., Ling Q., Zhou L., Wei X.Y., Liu Z.K., Ding S.M., Chen K.J., Xu Z.Y., Zheng S.S. ZIP4, a novel determinant of tumor invasion in hepatocellular carcinoma, contributes to tumor recurrence after liver transplantation. Int. J. Biol. Sci. 2014;10(3):245–256. doi: 10.7150/ijbs.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmer R., Goumas F.A., Waetzig G.H., Rose-John S., Kalthoff H. Interleukin-6: a villain in the drama of pancreatic cancer development and progression. Hepatobiliary Pancreat. Dis. Int. 2014;13(4):371–380. doi: 10.1016/s1499-3872(14)60259-9. [DOI] [PubMed] [Google Scholar]
- 32.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., Liu J., Freeman G.J., Brown M.A., Wucherpfennig K.W., Liu X.S. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Chen C., Yao Q., Li M. ZIP4 upregulates the expression of neuropilin-1, vascular endothelial growth factor, and matrix metalloproteases in pancreatic cancer cell lines and xenografts. Cancer Biol. Ther. 2010;9(3):236–242. doi: 10.4161/cbt.9.3.10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brevi A., Cogrossi L.L., Grazia G., Masciovecchio D., Impellizzieri D., Lacanfora L., Grioni M., Bellone M. Much more than IL-17A: cytokines of the IL-17 family between microbiota and cancer. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.565470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bule P., Aguiar S.I., Aires-Da-Silva F., Dias J.N.R. Chemokine-directed tumor microenvironment modulation in cancer immunotherapy. Int. J. Mol. Sci. 2021;22(18) doi: 10.3390/ijms22189804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Z., Zhou J., Li L., Liao S., He J., Zhou S., Zhou Y. Pericytes in the tumor microenvironment. Cancer Lett. 2023;556 doi: 10.1016/j.canlet.2023.216074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article. SLC39A4 transcription profiles of samples from CESC patients with their clinical and other relevant medical information were obtained from the TCGA platform (https://portal.gdc.cancer.gov/).