Abstract
The cytoplasmic, NAD-reducing hydrogenase (SH) of Alcaligenes eutrophus H16 is a heterotetrameric enzyme which contains several cofactors and undergoes a complex maturation during biogenesis. HoxH is the Ni-carrying subunit, and together with HoxY it forms the hydrogenase dimer. HoxF and HoxU represent the flavin-containing diaphorase moiety, which is closely related to NADH:ubiquinone oxidoreductase and mediates NADH oxidation. A variety of mutations were introduced into the four SH structural genes to obtain mutant enzymes composed of monomeric and dimeric forms. A deletion removing most of hoxF, hoxU, and hoxY led to the expression of a HoxH monomer derivative which was proteolytically processed at the C terminus like the wild-type polypeptide. While the hydrogenase dimer, produced by a strain deleted of hoxF and hoxU, displayed H2-dependent dye-reducing activity, the monomeric form did not mediate the activation of H2, although nickel was incorporated into HoxH. Deletion of hoxH and hoxY led to the production of HoxFU dimers which displayed NADH:oxidoreductase activity. Mixing the hydrogenase and the diaphorase moieties in vitro reconstituted the structure and catalytic function of the SH holoenzyme.
Hydrogenases are widespread among microorganisms. These redox catalysts mediate oxidation of molecular hydrogen and reduction of protons according to the following equation: H2 ↔ 2 H+ + 2 e−. Depending on the metal content in the hydrogen active site, two major classes of hydrogenases are distinguished. The [Fe]-only hydrogenases contain Fe-S clusters, including a specific catalytic H cluster (1) in one polypeptide. The [NiFe] hydrogenases are composed of two subunits, a large one of about 60 kDa with an Ni center and a small subunit of about 30 kDa, with Fe-S clusters, instrumental in electron transfer. Most members of both classes of hydrogenase are attached to an additional redox protein(s) which provides individual electron acceptor specificity (4).
Alcaligenes eutrophus, a facultative chemolithoautotrophic bacterium, is able to grow on hydrogen as the sole energy source. It contains two [NiFe] hydrogenases. The dimeric, periplasmically exposed, membrane-bound enzyme (MBH) couples H2 oxidation to electron transport-dependent phosphorylation via a cytochrome b-type membrane anchor (6, 11, 36). The cytoplasmic enzyme transfers electrons directly to NAD as the physiological acceptor (16, 40). The genes coding for the cytoplasmic, NAD-reducing hydrogenase (SH) and the MBH are arranged in separate operons located on the endogenous megaplasmid pHG1 (23, 45). The SH, which is the subject of this report, is composed of two dimeric moieties (39, 45). HoxH, the Ni-harboring subunit (20), and HoxY form the hydrogenase dimer. HoxF and HoxU represent the Fe-S-containing flavoprotein, called the diaphorase, which mediates NADH oxidoreductase activity.
Spectroscopic data on various [NiFe] hydrogenases indicate that Ni is directly involved in H2 activation (4). The first crystallographic analysis of a periplasmic [NiFe] hydrogenase from Desulfovibrio gigas uncovered the metal center deep inside the protein and showed that Ni is coordinated by two pairs of N- and C-terminal cysteine-borne thiol groups in the large subunit (46). These four residues are completely conserved in the corresponding 52-kDa subunit (HoxH) of the A. eutrophus soluble hydrogenase (45). Fe was identified as a second metal in the hydrogenase catalytic site of D. gigas, sharing with Ni one cysteine of each pair as a bridging ligand. Moreover, three diatomic nonprotein ligands, most likely one CO and two CN−, are bound to the Fe atom (19, 47). X-ray data on the D. gigas enzyme showed that the small subunit harbors one 3Fe-4S and two 4Fe-4S clusters in a linear arrangement (46). It has not yet been determined whether all of these Fe-S clusters participate in electron transport. An equivalent electron transferring function is assigned to HoxY, the 23-kDa subunit of the SH dimer. This polypeptide, however, is substantially smaller than its standard [NiFe] hydrogenase counterpart; its structure predicts the presence of only one 4Fe-4S cluster proximal to the Ni-Fe center (45). The diaphorase moiety of the SH, consisting of the 67-kDa HoxF and the 26-kDa HoxU polypeptides, harbors three to four Fe-S clusters and one molecule of flavin mononucleotide (FMN) (39, 45). The electron pathway from the hydrogen active site to NAD is barely understood.
Rhodococcus opacus (formerly Nocardia opaca) contains an SH homolog which dissociates in vitro into two dimeric catalytically active derivatives, the hydrogenase and the diaphorase moieties. A precise assignment of Fe-S clusters to the individual subunits, however, was not possible (39, 49). Previous biochemical analysis of a genetically less-defined SH mutant of A. eutrophus led to the isolation of a single-subunit (HoxH) SH derivative which contained Ni and Fe and exhibited low H2-oxidizing activity (20). Nonassembled HoxH variants, devoid of H2-oxidizing activity, were also identified in mutants impaired in maturation of the SH and led to the identification of auxilliary proteins which are essential for the formation of the hydrogen active site. Hyp proteins are essential for insertion of Ni into HoxH and HoxG of the SH and MBH, respectively (10). HoxW is supposed to be a protease which specifically cleaves off the C-terminal extension of HoxH after metal insertion (43). This allows folding of HoxH into its native conformation and oligomerization to the tetrameric holoenzyme (28).
In the present communication, we report on the construction of a set of SH mutants carrying defined deletions in the structural genes hoxF, hoxU, hoxY, and hoxH. The resulting mutant proteins, HoxHY, HoxH, HoxFU, and HoxF, are characterized with respect to catalytic properties, Ni content, and maturation. Moreover, catalytically active SH holoenzyme is reconstituted in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. A. eutrophus H16 is the wild-type strain, harboring the endogenous megaplasmid pHG1. Strains carrying the initials HF are derivatives of A. eutrophus H16. Escherichia coli S17-1 (42) was used as the host in standard cloning procedures and was the donor for conjugative plasmid transfer.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source(s) or reference |
|---|---|---|
| Strains | ||
| A. eutrophus | ||
| H16 | SH+ MBH+ | DSM 428, ATCC 17699 |
| HF14 | SH− MBH+ | 37 |
| HF376 | SH− MBH+ ΔhoxW | 43 |
| HF387 | SH− MBH+ ΔhoxFUYHW | 28 |
| HF402 | SH− MBH+ ΔhoxFUY | This study |
| HF424 | SH− MBH− ΔhoxFUYHW ΔhoxG | This study |
| HF444 | SH− MBH− ΔhoxFUY ΔhoxG | This study |
| E. coli S17-1 | Tra+recA pro thi hsdR chr::RP4-2 | 42 |
| Plasmids | ||
| pBluescript KS+ | AprlacZ′ T7; φ10 promoter, f1 ori | Stratagene, Cloning Systems |
| pTZ18R | AprlacZ′; f1 ori | 31 |
| pVK101 | Kmr Tcr; RP4 ori | 22 |
| pLO1 | KmrsacB; RP4 oriT, ColE1 ori | 24 |
| pCH241 | 4.6-kb HindIII-KpnI fragment of pGE15 in pTZ18R | C. Böcker and B. Friedrich |
| pCH291 | 2.9-kb HindIII-BamHI fragment of pGE15 in pTZ18R | A. Tran-Betcke and B. Friedrich |
| pCH424 | 2.2-kb SalI-SmaI fragment of pCH423 in pLO1; ΔhoxG | 7 |
| pCH455 | 15.0-kb HindIII fragment of pGE15 in pKS+ | This study |
| pCH494 | Derivative of pCH291 deleted for a 2,390-bp XhoI fragment (deletion of hoxFU) | This study |
| pCH549 | Derivative of pCH241 deleted for a 2,230-bp SacII fragment (deletion of hoxUYH) | This study |
| pCH550 | Derivative of pCH241 deleted for a 1,219-bp EcoRI fragment (deletion of hoxYH) | This study |
| pCH551 | Derivative of pCH455 containing a 2.2-kb AatII-KpnI fragment of pCH549 | This study |
| pCH552 | Derivative of pCH455 containing a 1.2-kb AatII-KpnI fragment of pCH550 | This study |
| pCH568 | Derivative of pCH241 deleted for a 2,650-bp NruI fragment (deletion of hoxFUY) | This study |
| pCH569 | 800-bp HindIII-BstEII fragment of pCH568, filled in with Klenow polymerase, ligated in PmeI site of pLO1 | This study |
| pGE15 | 15.0-kb HindIII fragment of pHG1 in pVK101 | 45 |
| pGE346 | Derivative of pGE15 containing a 0.8-kb HindIII-BamHI fragment of pCH494 (ΔhoxFU) | This study |
| pGE347 | Derivative of pGE15 deleted for a 921-bp PstI fragment (ΔhoxH) | 43 |
| pGE348 | 0.7-kb fragment (hoxW) in pGE151 | 43 |
| pGE370 | 12.2-kb HindIII fragment of pCH551 in pVK101 (ΔhoxUYH) | This study |
| pGE371 | 13.2-kb HindIII fragment of pCH552 in pVK101 (ΔhoxYH) | This study |
SH+, SH positive; SH−, SH negative; MBH+, MBH positive; MBH−, MBH negative.
Strains of A. eutrophus were cultivated in mineral salts medium containing 0.4% (wt/vol) fructose or a mixture of 0.2% (wt/vol) fructose and 0.2% (vol/vol) glycerol (FGN medium) (14). The medium was supplemented with 1 μM NiCl2 under standard conditions. Lithoautotrophically cultured cells were grown in mineral salts medium under an atmosphere of hydrogen, carbon dioxide, and oxygen (8:1:1, vol/vol/vol). Strains of E. coli were grown in Luria broth (29). Concentrations of antibiotics for A. eutrophus used were as follows: kanamycin, 350 μg/ml; and tetracycline, 15 μg/ml. Concentrations of antibiotics for E. coli used were as follows: kanamycin, 25 μg/ml; tetracycline, 15 μg/ml; and ampicillin, 100 μg/ml.
Recombinant DNA techniques and plasmid constructions.
Standard DNA techniques were used (34). Defined deletion alleles were constructed in subcloned wild-type DNA fragments. The mutations were verified by double-stranded DNA sequencing of the deletion fusion points, using Sequenase (U.S. Biochemical Corp.) and [35S]dATP, according to the method of Sanger et al. (35). For construction of HF444, plasmid pCH241 was digested with NruI, and religation yielded plasmid pCH568. An 800-bp HindIII-BstEII fragment of pCH568 containing the NruI fusion point was filled in by treatment with the Klenow fragment of DNA polymerase and ligated into the PmeI site of pLO1 (24). The resulting plasmid, pCH569, was transferred to A. eutrophus H16 by conjugation, and allelic exchange was done as described previously (24), yielding strain HF402. Plasmid pCH424 (7), which carries a deletion allele of hoxG, was used for gene replacement in HF402 to generate HF444. Plasmid pCH424 was also used to introduce an in-frame hoxG deletion into the SH mutant strain HF387 (28). The resulting double mutant (ΔhoxFUYHW ΔhoxG) was designated HF424. The identities of resulting strains were verified on the basis of altered electrophoretic mobility of the amplified products. Strain HF424 was used as a recipient for pGE15 (45) and its deletion derivatives.
For construction of pGE346, a 2.3-kb deletion fusing hoxF and hoxU was introduced by digesting pCH291 with XhoI. Religation yielded pCH494. An 0.8-kb HindIII-BamHI fragment was used to replace the corresponding wild-type allele. pCH241 was the starting plasmid for generation of the deletion alleles ΔhoxUYH and ΔhoxYH, yielding plasmids pCH549 and pCH550, respectively. Subsequently, each of the deletion-bearing alleles was transferred to pCH455, resulting in plasmids pCH551 and pCH552, respectively. The HindIII fragments of these two plasmids were introduced into pVK101 (22), yielding pGE370 (ΔhoxUYH) and pGE371 (ΔhoxYH), respectively.
Preparation of soluble and membrane extracts.
A. eutrophus cells were grown in FGN medium to an optical density of 10 to 11 at 436 nm. Cells were harvested by centrifugation, washed once, and resuspended in 50 mM potassium phosphate buffer, pH 7, containing 0.1 mM phenylmethylsulfonyl fluoride. Cells were disrupted, and soluble and membrane fractions were prepared as described previously (14). Protein concentrations were determined by the method of Lowry et al. (25), using bovine serum albumin as the reference.
Autoradiography of 63Ni-labeled proteins.
Proteins were labeled in vivo by growing the cells to an optical density of 10 to 11 at 436 nm in FGN medium containing 150 nM 63NiCl2 (867 mCi/mmol; Amersham Buchler). Soluble extracts were analyzed as described previously (10).
Immunoblot analysis.
Proteins were separated by electrophoresis in polyacrylamide gels and transferred to BA83 nitrocellulose membranes (Schleicher and Schuell) in accordance with a standard protocol (44). Subunits of the SH were detected with rabbit polyclonal antisera and an alkaline phosphatase-labeled goat anti-rabbit immunoglobulin (Jackson ImmunoResearch Laboratories). The antisera were prepared with isolated SH subunit proteins and used either separately or as a mixture as described previously (10).
In vitro reconstitution.
Cells of HF424(pGE346) and HF424(pGE371) were grown in FGN under standard conditions. Soluble extracts from cells of both strains were mixed 1:1 (vol/vol) and incubated for 15 min at 37°C. In a second approach, resuspended cells of both strains were mixed 1:1 (vol/vol) prior to disruption. The crude extract was incubated for 15 min at 37°C, and subsequently a soluble extract was prepared.
Enzyme assays.
Activities of the SH (hydrogen:NAD+ oxidoreductase; EC 1.12.1.2) and the MBH (ferredoxin:H+ oxidoreductase; EC 1.18.99.1) were determined with cells grown in FGN medium to an optical density of 10 to 11 at 436 nm. Hydrogenase activity of the SH in soluble extracts was determined by spectrophotometric measurement of H2-dependent reduction of NAD and benzyl viologen (BV) (40). Diaphorase activity was assayed with the soluble fraction, using BV as the acceptor and NADH as the electron donor (40). MBH activity in the membrane extract was assayed by recording H2-dependent methylene blue reduction (36). Deuterium-water (D2-H2O) exchange experiments were performed with a membrane leak chamber fitted to a mass spectrometer (Mastorr 200 DX quadrupole; VG Quadrupoles Ltd.). There was no gas phase in the chamber, and the contents were mixed with a magnetic stirrer. Details of the chamber construction and of the calibration have been described by Cammack et al. (9). The assays were done as previously reported (28).
RESULTS
Generation of subforms of the NAD-reducing hydrogenase.
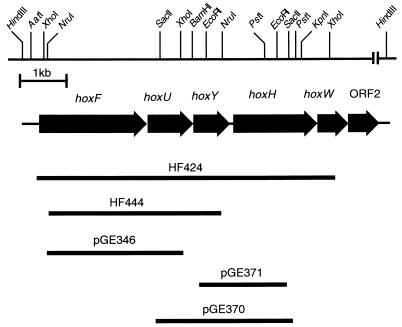
Previous studies showed that the four SH genes (hoxF, hoxU, hoxY, and hoxH) form a transcriptional unit starting from a promoter upstream of hoxF (45) and that the HoxH-specific protease gene hoxW is tightly linked (43). To investigate the formation as well as the structural and catalytic properties of hydrogenase subforms, we constructed various combinations of deletions in the four SH structural genes. The positions and extents of the deleted regions in the SH genes are illustrated in Fig. 1. Synthesis of a monomeric HoxH derivative was achieved by disrupting the promoter-proximal genes hoxF, hoxU, and hoxY. The deleted allele was introduced into the megaplasmid of A. eutrophus by double recombination (24). To avoid interference by the activity of the second hydrogenase, the MBH, the mutation was constructed in a ΔhoxG background. The resulting double mutant, HF444 (Fig. 1), showed the expected phenotype of an SH- and MBH-negative strain: autotrophic growth on H2 was abolished, while heterotrophic growth was unaffected (data not shown).
FIG. 1.
The SH operon of A. eutrophus. The diagram gives a simplified restriction map of a 15-kb segment. The region contains the four SH structural genes (hoxF, -U, -Y, and -H) and two accessory genes (hoxW and ORF2), indicated by solid arrows. The solid bars represents the positions and extents of the deletions. The designation of the respective mutant strain or plasmid is given above each bar.
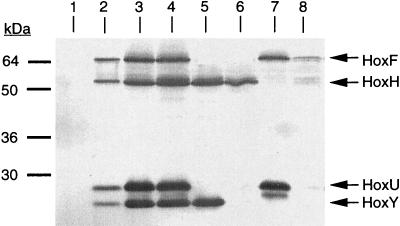
Previous studies with maturation-deficient mutants blocked in SH oligomerization pointed to a rather unstable constitution for three of the four nonassembled subunits (HoxF, HoxU, and HoxY) (10, 43). To compensate for the proteolytic loss of SH protein, we increased the level of SH gene expression by introducing the mutant alleles on broad-host-range plasmids derived from pGE15 (45). The plasmids were transferred into the SH- and MBH-negative recipient HF424, which carries ΔhoxFUYHW and ΔhoxG deletions. SH activity in HF424 was completely restored by plasmid pGE15 (Table 2), which harbors a 15-kb HindIII fragment encompassing all of the structural and SH-specific accessory genes (43). In contrast to pGE15-harboring cells, transconjugants bearing the mutant alleles (Fig. 1) failed to grow autotrophically on H2, indicating that the SH was physiologically inactive. Hence, the mutants were cultivated heterotrophically under hydrogenase-derepressing conditions (14) and examined for the presence of SH protein. Immunoblot experiments were conducted with a mixture of antibodies directed against the four individual SH subunits. Figure 2 demonstrates the lack of SH-specific antigen in soluble extracts of the recipient HF424 (lane 1) and the occurrence of four specific signals obtained with purified SH as the control (lane 2). As expected, the pGE15-harboring transconjugant cells of HF424 (Fig. 2, lane 4) exhibited an immunopattern similar to that of the wild-type strain H16 (Fig. 2, lane 3). Mutants with deletions in hoxF and hoxU produced the large and small subunits of the hydrogenase dimer (Fig. 2, lane 5); conversely, mutants devoid of intact hoxY and hoxH contained the two polypeptides of the diaphorase moiety (Fig. 2, lane 7). The latter was rather unstable, and its intracellular concentration decreased rapidly in the advanced stationary phase of growth (data not shown). A second fast-migrating band (Fig. 2, lane 7) originated from degradation of HoxU, as verified with HoxU-specific antiserum (data not shown). Elimination of HoxU by deletion of the corresponding gene further increased the instability of the HoxF polypeptide (Fig. 2, lane 8). Again, the occurrence of multiple bands pointed to rapid proteolysis. Comparing these data with the behavior of the HoxH monomer, formed by mutant HF444 (Fig. 2, lane 6), shows that the H2-activating subunit is stably maintained.
TABLE 2.
H2-dependent and H2-independent activities in soluble extracts of wild-type and deletion mutant A. eutrophus strains
| Strain | SH subunit(s) | Activity (%) ofa:
|
|||
|---|---|---|---|---|---|
| Hydrogenase
|
Diaphorase (NADH-BV) | ||||
| H2-NAD | H2-BV | D2-H+ exchange | |||
| H16 | HoxFUYH | 87 | 79 | 91 | 68 |
| HF424(pGE15) | HoxFUYH | 100 | 100 | 100 | 100 |
| HF424 | None | 0 | <0.5 | <0.5 | 4 |
| HF424(pGE346) | HoxHY | 0 | 9 | 20 | 4 |
| HF444 | HoxH | 0 | <0.5 | <0.5 | 5 |
| HF424(pGE371) | HoxFU | 0 | <0.5 | ND | 56 |
| HF424(pGE370) | HoxF | 0 | <0.5 | ND | 6 |
Cells were grown on fructose-glycerol medium. Activities are given in percentages; values of strain HF424(pGE15) are taken as 100%. Maximal H2-dependent NAD reduction (H2-NAD) is 3.8 μmol of H2 · min−1 · mg−1, and maximal BV reduction (H2-BV) is 1.5 μmol of H2 · min−1 · mg of protein−1. The maximal D2-H+ exchange rate is 0.91 μmol of HD · min−1 · mg of protein−1. Maximal NADH-dependent BV reduction (NADH-BV) is 5.5 μmol of NADH · min−1 · mg of protein−1. Values give the average of data from at least two independent experiments. The relative error was in the range of 10 to 20%.
FIG. 2.
Detection of SH subunits by immunoblotting. Soluble extracts (20 μg of protein) were applied to lanes 1 and 3 to 8 and separated on a sodium dodecyl sulfate–12.5% polyacrylamide gel. Lanes: 1, HF424; 2, 0.5 μg of purified SH; 3, H16; 4, HF424(pGE15); 5, HF424(pGE346 [ΔhoxFU]); 6, HF444 (ΔhoxFUY); 7, HF424(pGE371 [ΔhoxYH]); 8, HF424(pGE370 [ΔhoxUYH]). The blot was processed with a mixture of antibodies raised against the four SH subunits. The positions of the molecular mass standards are given on the left.
Catalytic properties of the mutant enzymes.
To investigate the catalytic properties of the SH subforms, we used various enzymatic assays which are based on the following reactions potentially catalyzed by SH: (i) H2-dependent reduction of NAD and BV, (ii) electron acceptor-independent D2-H+ exchange, and (iii) BV-coupled oxidation of NADH. Not surprisingly, all of the mutants included in this study were impaired in H2-dependent NAD-reducing activity (Table 2). Using BV as the electron acceptor yielded substantial activity only in extracts of the HoxHY-containing cells. This activity disappeared during freezing and thawing or after storage of the extract for 24 h at 4°C, pointing to a catalytically rather unstable conformer. The HoxH monomer-containing mutant, HF444, showed only the BV-reducing background activity, like other mutant strains, including the hydrogenase-negative control HF424. The HoxHY-containing extract of strain HF424(pGE346) exhibited 20% of the wild-type specific D2-H+ exchange rate, whereas extracts prepared from cells containing the monomeric HoxH derivative (HF444) were inactive (Table 2). Addition of dithionite to the reaction mixture of HoxHY-containing mutant extracts proved to be essential for D2-H+ exchange activity. It is worth noting that the assay conditions were optimized for the wild-type hydrogenase and that they might not have been optimal for the subforms.
Coexpression of HoxF and HoxU led to high diaphorase activity in soluble extracts of mutant HF424(pGE371). A total of 80% of this activity was recovered after storing the extract for 6 days on ice. The residual NADH oxidoreductase activity measured in extracts of the other mutant strains was nonspecific and was even observed in the SH-free strain, HF424 (Table 2).
Proteolytic processing and Ni content of HoxH.
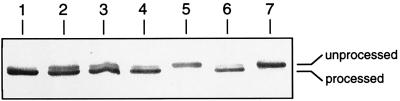
Metal center assembly of [NiFe] hydrogenases requires a series of maturation steps (26). The mutants generated in this study were analyzed for two features: (i) the ability of the HoxH precursor to undergo conversion to the proteolytically processed form and (ii) the capacity to incorporate Ni specifically. C-terminal proteolysis was investigated by immunoblot analysis with an antibody preparation raised against HoxH. The precursor of HoxH was identified in soluble extracts on the basis of electrophoretic mobility. The preform is readily identifiable in the protease mutant HF376 (Fig. 3, lane 7). An increase in the copy number of the wild-type SH genes [HF424(pGE15)] led to the occurrence of both the precursor and the proteolytically processed form of HoxH (Fig. 3, lane 2), indicating that in this strain the maturation process may be rate limiting. Elimination of the HoxH-accompanying subunits HoxF, HoxU, and HoxY scarcely affected the immunoblot pattern (Fig. 3, lanes 3 and 4); most of the HoxH present was in the mature form. It is interesting that the monomeric HoxH which was previously purified from the genetically less-defined SH-negative mutant HF14 and then extensively characterized (20) proved to be defective in C-terminal proteolysis (Fig. 3, lane 5). Complementation by the protease gene hoxW restored maturation (Fig. 3, lane 6) and 73% of H2-dependent NAD-reducing activity (data not shown).
FIG. 3.
Analysis of HoxH processing: Western blot analysis of soluble extracts with antibodies against HoxH. A total of 20 μg of protein was applied to each lane of a sodium dodecyl sulfate–12.5% polyacrylamide gel. Lanes: 1, H16; 2, HF424(pGE15); 3, HF424(pGE346 [ΔhoxFU]); 4, HF444 (ΔhoxFUY); 5, HF14 (hoxW14) (see text); 6, HF14(pGE348 [hoxW]); 7, HF376 (ΔhoxW).
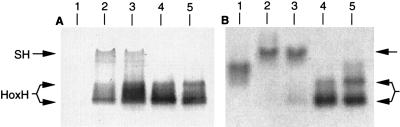
Finally, Ni incorporation into HoxH was examined by growing the cells heterotrophically under hydrogenase-derepressing conditions (14) in the presence of 120 nM 63NiCl2. Soluble extracts were prepared and subjected to native polyacrylamide gel electrophoresis, which allows identification of assembled and nonassembled HoxH conformers (10, 28). Figure 4A shows a Western blot on which are marked the positions of the SH holoenzyme and the HoxH monomer. Figure 4B presents the corresponding 63Ni-labeled proteins resolved by autoradiography. As expected, the SH- and MBH-negative control was devoid of HoxH antigen (Fig. 4A, lane 1); nevertheless, autoradiography resolved a strong Ni signal (Fig. 4B, lane 1), which may result from an Ni-processing protein. The upper bands visible in extracts of H16 and HF424(pGE15) represent the SH holoenzyme (Fig. 4A, lanes 2 and 3), which correlated well with the two slowly migrating Ni-labeled proteins (Fig. 4B, lanes 2 and 3). The weak immunoblot reaction of the SH holoenzyme results from a less sensitive cross-reaction with the HoxH-specific antibody. The nonassembled HoxH occurred exclusively in mutants carrying deletions in the associated subunit genes (Fig. 4A, lanes 4 and 5). Nevertheless, these monomeric forms were capable of binding Ni, as shown in the corresponding autoradiogram (Fig. 4B, lanes 4 and 5). The occurrence of multiple bands may indicate the formation of aggregates. Nonassembled HoxH of H16 and HF424(pGE15) showed only very weak 63Ni labeling (Fig. 4A and B, lanes 2 and 3), indicating that most of this species is present as an Ni-free precursor.
FIG. 4.
Effect of deletions in the SH genes on 63Ni incorporation into HoxH: analyses of soluble extracts separated on native polyacrylamide gels (4 to 15%). Cells were grown in the presence of 150 nM 63NiCl2. SH-specific bands are marked by arrows. (A) Western blotting with antibodies against HoxH. A total of 50 μg of protein each was applied to each lane. Lanes: 1, HF424; 2, H16; 3, HF424(pGE15); 4, HF424(pGE346 [ΔhoxFU]); 5, HF444 (ΔhoxFUY). (B) Corresponding autoradiogram. A total of 100 μg of protein was applied to each lane.
In vitro reconstitution of the SH.
Each of the dimeric forms of the SH (HoxFU and HoxYH) showed the enzymatic characteristics of the diaphorase and the hydrogenase moiety (Table 2). To determine if in vitro reconstitution to the holoenzyme occurs, the two mutant extracts were mixed and incubated for 15 min at 37°C. Subsequently, H2-dependent NAD-reducing activity was monitored. In fact, 72% activity of the HF424(pGE15) extract was recovered. The level of activity upon mixing the cells of both strains prior to disruption amounted to 91%.
Transfer of hoxF, hoxU, and hoxY on plasmid pGE347 into the HoxH monomer-containing mutant HF444 restored 32% of the NAD-reducing activity of H16 (data not shown). In vitro reconstitution of SH by mixing and disrupting cells of HF444 and of HF424(pGE347) was unsuccessful. In this case, no H2-dependent NAD-reducing activity was recovered, although Western blot analysis confirmed the presence of the four subunits in the mixture (data not shown).
DISCUSSION
The enzyme investigated in this study belongs to a subclass of [NiFe] hydrogenases which differs from the prototypic dimeric hydrogenases by two major criteria: (i) the presence of an additional (Fe-S) flavoprotein part which catalyzes a two-electron reduction of the coenzyme acceptor and (ii) localization in the cytoplasm or at the inner face of the membrane. Well-known examples of these multimeric [NiFe] hydrogenases are the F420-reducing hydrogenase present in methanogens and the NAD-reducing hydrogenase initially found in the Knallgas bacterium A. eutrophus (4). For a long period of time, the possession of SH-like hydrogenases appeared to be restricted to strains of Alcaligenes species and the gram-positive actinomycete R. opacus (formerly N. opaca) (15). More recently, an increasing number of SH-containing cyanobacteria have been described in the literature (5, 8, 38), indicating that this multimeric hydrogenase is more abundant than originally envisaged.
The first primary structure of an SH-type hydrogenase (45) received special attention when it became apparent that its dimeric flavoprotein, called the diaphorase, is closely related to three peripheral subunits of NADH:ubiquinone oxidoreductase from mitochondria (32) and bacterial species (48). The FMN-containing subunit HoxF of the SH appears to be a fusion product of the 24- and 51-kDa subunits of bovine complex I, and HoxU is homologous to the N-terminal part of the 75-kDa subunit. Meanwhile, diaphorase-related subunits were also discovered as constituents of the [Fe]-only hydrogenase of Desulfovibrio fructosovorans (27) and in a formate dehydrogenase of A. eutrophus (30). Moreover, sequence similarities of complex I also extend to the hydrogen:acceptor oxidoreductase moiety (3). Thus, the SH is composed of two distinct functional modules that are conserved in a number of redox proteins which may have a common origin. These features make the enzyme an attractive model with which to study complex intramolecular electron transport processes.
To facilitate future structural and functional investigations, an attempt was made to dissect SH subforms genetically. Various combinations of deletions in the SH subunit genes led to the expression of two monomeric protein derivatives (HoxF and HoxH) and two dimeric conformers (HoxFU and HoxHY). As determined by immunological analysis, the HoxH monomer was remarkably stable in intact as well as broken cells, which is consistent with previous results obtained with maturation-deficient mutants (10, 28, 43). Conversely, solely expressed HoxF polypeptide showed typical traces of proteolytic degradation; its stability was significantly enhanced by coexpression of the small diaphorase HoxU subunit. The HoxFU-containing mutant showed 56% of wild-type specific diaphorase activity, which is compatible with the notion that the dimeric form is maintained in a fairly active configuration. This encouraging result invites future investigations on the FU dimer, such as electron spin resonance spectroscopy and site-directed mutagenesis to allocate the number and nature of Fe-S clusters, which have been controversially discussed in the past (12, 15, 39). Moreover, the HoxFU dimer offers an attractive system for studying redox interactions between Fe-S, FMN, and NAD cofactors.
Despite a high degree of protein stability, a stable content of nickel, and an apparently correct conversion to the C-terminally processed mature form, the HoxH monomer proved to be catalytically inactive in various H2:acceptor oxidoreductase assays applied in this study. Loss of H2-dependent NAD and BV reduction was not surprising, but the lack of D2-H+ exchange activity in an extract of HF444 was unexpected. The D2-H+ exchange rate is based on the reversible binding and cleavage of D2 at the hydrogen active site accompanied by the exchange of solvent protons (9). The fact that cells expressing the HoxHY dimer exhibited 20% of the characteristic wild-type exchange rate and 9% of the corresponding BV-reducing activity shows that the dimer has the capacity to oxidize H2, although at a significantly lower rate than the tetrameric enzyme. The decrease in activity may be due to reduced protein stability, to a less-active conformation, and/or to inappropriate assay conditions. The fact that in vitro reconstitution of HoxHY and HoxFU subforms yielded highly active NAD-reducing holoenzyme indicates that the dimer does not undergo irreversible damage. Nevertheless, the significant decrease or even absence of an exchange activity, as observed for the HoxH monomer, indicates that binding and heterolytic cleavage of H2 respond rather sensitively to the removal of connecting subunits. Thus, para-ortho H2 conversion experiments (9), which only rely on heterolytic cleavage of H2, in combination with spectroscopy (4), will certainly be worth undertaking to explore the metal center of the HoxH monomer in more detail. On the basis of the available data, we conclude that the smallest enzymatically active [NiFe] hydrogenase in A. eutrophus consists of a large subunit with a binuclear metal center and a small subunit with a minimum of one 4Fe-4S cluster, as predicted for HoxHY (13). The essentiality of the small subunit for activity was also shown in studies of Azotobacter vinelandii hydrogenase (35a). The results presented here raise doubts about previous reports on the existence of active monomeric [NiFe] hydrogenases (reviewed in reference 33). It seems more likely that in these cases the small subunit was not recognized due to its high instability. Crystal structure analysis of the D. gigas [NiFe] hydrogenase has shown that the small and large subunits have a broad contact interface (46), supporting the interpretation that the small subunit does not exclusively serve as an electron transferring unit but greatly conditions the protein environment of the active Ni center.
Studies of periplasmically exposed enzymes from various organisms showed that maturation of this type of hydrogenase is based on a tight coupling between the large and small subunits. C-terminal proteolysis of the large subunit does not occur in the absence of the small subunit (26). In the case of the cytoplasmic SH protein, however, the situation appears to be different. Removal of the small subunit, HoxY, in mutant HF444 did not prevent HoxH from being processed.
Furthermore, we have demonstrated that separately expressed HoxHY and HoxFU conformers are able to reconstitute in vitro to a highly active NAD-reducing holoenzyme. This result clearly shows that both dimeric subforms contain all necessary cofactors in a well-assembled configuration. Again, this observation supports the view of two independent functional modules. It was previously shown that unlike the A. eutrophus SH, the in vitro hydrogenase activity of R. opacus can be significantly stimulated by the addition of NiCl2 (2). Moreover, upon treatment under nickel deprivation or at low ionic strength in an alkaline pH, the latter enzyme dissociates into two dimeric proteins with different enzymatic activities (18, 41). Attempts to reassociate in vitro-produced subfragments of the A. eutrophus SH to an active protein were unsuccessful (21). The reason for the different behaviors of the two related hydrogenases is not yet understood and is not immediately obvious from an alignment of the primary structures of the four polypeptides, which proved to be almost identical (17). A clearer picture of the dissociation-association process may emerge when genetic studies are extended to well-defined hybrid SH derivatives. This future approach looks promising since preliminary experiments indicate that the R. opacus hydrogenase genes may be functionally expressed in A. eutrophus (17).
ACKNOWLEDGMENTS
We thank V. M. Fernandez and E. Santamaria for encouraging help during this work. We are indebted to A. Strack for expert technical assistance and to E. Schwartz for stimulating discussions on the manuscript.
This work was supported by a grant from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie as well as by Fonds der Chemischen Industrie. A short-term scientific mission of C. Massanz to the lab of V. M. Fernandez was funded by the European Commission Cooperation in Science and Technology action 818.
REFERENCES
- 1.Adams M W W. The structure and mechanism of iron hydrogenases. Biochim Biophys Acta. 1990;1020:115–145. doi: 10.1016/0005-2728(90)90044-5. [DOI] [PubMed] [Google Scholar]
- 2.Aggag M, Schlegel H G. Studies on a gram-positive hydrogen bacterium, Nocardia opaca strain 1b. III. Purification, stability and some properties of the soluble hydrogen dehydrogenase. Arch Mikrobiol. 1974;100:25–39. doi: 10.1007/BF00446303. [DOI] [PubMed] [Google Scholar]
- 3.Albracht S P J. Intimate relationships of the large and the small subunits of all nickel hydrogenases with two nuclear-encoded subunits of mitochondrial NADH:ubiquinone oxidoreductase. Biochim Biophys Acta. 1993;1144:221–224. doi: 10.1016/0005-2728(93)90176-g. [DOI] [PubMed] [Google Scholar]
- 4.Albracht S P J. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1188:167–204. doi: 10.1016/0005-2728(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 5.Appel J, Schulz R. Sequence analysis of an operon of a NAD(P)-reducing nickel hydrogenase from the cyanobacterium Synechocystis sp. PCC6803 gives additional evidence for direct coupling of the enzyme to NAD(P)H dehydrogenase (complex I) Biochim Biophys Acta. 1996;1298:141–147. doi: 10.1016/s0167-4838(96)00176-8. [DOI] [PubMed] [Google Scholar]
- 6.Bernhard M, Benelli B, Hochkoeppler A, Zanoni D, Friedrich B. The membrane-bound hydrogenase (MBH) of Alcaligenes eutrophus H16: functional and structural role of the cytochrome b subunit. Eur J Biochem. 1997;248:179–186. doi: 10.1111/j.1432-1033.1997.00179.x. [DOI] [PubMed] [Google Scholar]
- 7.Bernhard M, Schwartz E, Rietdorf J, Friedrich B. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J Bacteriol. 1996;178:4522–4529. doi: 10.1128/jb.178.15.4522-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boison G, Schmitz O, Mikheeva L, Shestakov S, Bothe H. Cloning, molecular analysis and insertional mutagenesis of the bidirectional hydrogenase genes from the cyanobacterium Anacystis nidulans. FEBS Lett. 1996;394:153–158. doi: 10.1016/0014-5793(96)00936-2. [DOI] [PubMed] [Google Scholar]
- 9.Cammack R, Fernandez V M, Hatchikian E C. Nickel-iron hydrogenase. Methods Enzymol. 1994;234:43–68. [Google Scholar]
- 10.Dernedde J, Eitinger T, Patenge N, Friedrich B. hyp gene products in Alcaligenes eutrophus are part of a hydrogenase-maturation system. Eur J Biochem. 1996;235:351–358. doi: 10.1111/j.1432-1033.1996.00351.x. [DOI] [PubMed] [Google Scholar]
- 11.Eismann K, Mlejnek K, Zipprich D, Hoppert M, Gerberding H, Mayer F. Antigenic determinants of the membrane-bound hydrogenase in Alcaligenes eutrophus are exposed toward the periplasm. J Bacteriol. 1995;177:6309–6312. doi: 10.1128/jb.177.21.6309-6312.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erkens A, Schneider K, Müller A. The NAD-linked soluble hydrogenase from Alcaligenes eutrophus H16: detection and characterisation of EPR signals deriving from nickel and flavin. J Biol Inorg Chem. 1996;1:99–110. [Google Scholar]
- 13.Friedrich B, Bernhard M, Dernedde J, Eitinger T, Lenz O, Massanz C, Schwartz E. Hydrogen oxidation by Alcaligenes. In: Lindstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 110–117. [Google Scholar]
- 14.Friedrich B, Heine E, Finck A, Friedrich C G. Nickel requirement for active hydrogenase formation in Alcaligenes eutrophus. J Bacteriol. 1981;145:1144–1149. doi: 10.1128/jb.145.3.1144-1149.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich B, Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich C G, Schneider K, Friedrich B. Nickel in the catalytically active hydrogenase of Alcaligenes eutrophus. J Bacteriol. 1982;152:42–48. doi: 10.1128/jb.152.1.42-48.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grzesnik C, Lübbers M, Reh M, Schlegel H G. Genes encoding the NAD-reducing hydrogenase of Rhodococcus opacus MR11. Microbiology. 1997;24:1271–1286. doi: 10.1099/00221287-143-4-1271. [DOI] [PubMed] [Google Scholar]
- 18.Grzesnik C, Roß K, Schneider K, Reh M, Schlegel H G. Location, catalytic activity, and subunit composition of NAD-reducing hydrogenases of some Alcaligenes strains and Rhodococcus opacus MR22. Arch Microbiol. 1997;167:172–176. [PubMed] [Google Scholar]
- 19.Happe R P, Roseboom W, Baglay K A, Pierik A J, Albracht S P J. Biological activation of hydrogen. Nature. 1997;385:126. doi: 10.1038/385126a0. [DOI] [PubMed] [Google Scholar]
- 20.Hornhardt S, Schneider K, Schlegel H G. Characterisation of a native subunit of the NAD-linked hydrogenase isolated from a mutant of Alcaligenes eutrophus H16. Biochimie. 1986;68:15–24. doi: 10.1016/s0300-9084(86)81063-x. [DOI] [PubMed] [Google Scholar]
- 21.Johannssen W, Gerberding H, Rohde M, Zaborosch C, Mayer F. Structural aspects of the soluble NAD-dependent hydrogenase isolated from Alcaligenes eutrophus H16 and from Nocardia opaca 1b. Arch Microbiol. 1991;155:303–308. [Google Scholar]
- 22.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 23.Kortlüke C, Horstmann K, Schwartz E, Rohde M, Binsack R, Friedrich B. A gene complex coding for the membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992;174:6277–6289. doi: 10.1128/jb.174.19.6277-6289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.Maier T, Böck A. Nickel incorporation into hydrogenases. In: Hausinger R P, Eichhorn G L, Marzilli L G, editors. Advances in inorganic biochemistry. 11. Mechanisms of metallocenter assembly. New York, N.Y: VCH Publishers Inc.; 1996. pp. 173–192. [Google Scholar]
- 27.Malki S, Saimmaime I, De Luca G, Rousset M, Dermoun Z, Belaich J-P. Characterization of an operon encoding an NADP-reducing hydrogenase in Desulfovibrio fructosovorans. J Bacteriol. 1995;177:2628–2636. doi: 10.1128/jb.177.10.2628-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massanz C, Fernandez V M, Friedrich B. C-terminal extension of the H2-activating subunit, HoxH, directs maturation of the NAD-reducing hydrogenase in Alcaligenes eutrophus. Eur J Biochem. 1997;245:441–448. doi: 10.1111/j.1432-1033.1997.t01-3-00441.x. [DOI] [PubMed] [Google Scholar]
- 29.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 30.Oh J-I, Bowien B. Characterisation of the fds operon encoding the soluble formate dehydrogenase in Ralstonia eutropha. Biospectrum. 1997;1997:127. [Google Scholar]
- 31.Perbal B. A practical guide to molecular cloning. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 278–296. [Google Scholar]
- 32.Pilkington S J, Skehel J M, Gennis R B, Walker J E. Relationship between mitochondrial NADH-ubiquinone reductase and a bacterial NAD-reducing hydrogenase. Biochemistry. 1991;30:2166–2175. doi: 10.1021/bi00222a021. [DOI] [PubMed] [Google Scholar]
- 33.Przybyla A E, Robbins J, Menon N, Peck H D J., Jr Structure/function relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev. 1992;88:109–136. doi: 10.1111/j.1574-6968.1992.tb04960.x. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sanger F, Micklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Sayavedra-Soto L A, Arp D J. In Azotobacter vinelandii hydrogenase, substitution of serine for the cysteine residues at positions 62, 65, 289, and 292 in the small (HoxK) subunit affects H2 oxidation. J Bacteriol. 1993;175:3414–3421. doi: 10.1128/jb.175.11.3414-3421.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schink B, Schlegel H G. The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification and biochemical properties. Biochem Biophys Acta. 1979;567:315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- 37.Schlesier M, Friedrich B. Effect of molecular hydrogen on histidine utilisation by Alcaligenes eutrophus. Arch Mikrobiol. 1982;132:260–265. [Google Scholar]
- 38.Schmitz O, Boison G, Hilscher R, Hundeshagen B, Zimmer W, Lottspeich F, Bothe H. Molecular biological analysis of a bidirectional hydrogenase from cyanobacteria. Eur J Biochem. 1995;233:266–276. doi: 10.1111/j.1432-1033.1995.266_1.x. [DOI] [PubMed] [Google Scholar]
- 39.Schneider K, Cammack R, Schlegel H G. Content and localization of FMN, Fe-S clusters and nickel in the NAD-linked hydrogenase of Nocardia opaca 1b. Eur J Biochem. 1984;142:75–84. doi: 10.1111/j.1432-1033.1984.tb08252.x. [DOI] [PubMed] [Google Scholar]
- 40.Schneider K, Schlegel H G. Purification and properties of the soluble hydrogenase from Alcaligenes eutrophus H16. Biochim Biophys Acta. 1976;452:66–80. doi: 10.1016/0005-2744(76)90058-9. [DOI] [PubMed] [Google Scholar]
- 41.Schneider K, Schlegel H G, Jochim K. Effect of nickel on activity and subunit composition of purified hydrogenase from Nocardia opaca 1b. Eur J Biochem. 1984;138:533–541. doi: 10.1111/j.1432-1033.1984.tb07948.x. [DOI] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:717–743. [Google Scholar]
- 43.Thiemermann S, Dernedde J, Bernhard M, Schroeder W, Massanz C, Friedrich B. Carboxyl-terminal processing of the cytoplasmic NAD-reducing hydrogenase of Alcaligenes eutrophus requires the hoxW gene product. J Bacteriol. 1996;178:2368–2374. doi: 10.1128/jb.178.8.2368-2374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4357. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran-Betcke A, Warnecke U, Böcker C, Zaborosch C, Friedrich B. Cloning and nucleotide sequence of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990;172:2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volbeda A, Charon M-H, Piras C, Hatchikian E C, Frey M, Fontecilla-Camps J C. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 47.Volbeda A, Garcin E, Piras C, de Lacey A L, Fernandez V M, Hatchikian E C, Frey M, Fontecilla-Camps J C. Structure of the [NiFe] hydrogenase active site: evidence for biologically uncommon Fe ligands. J Am Chem Soc. 1996;118:12989–12996. [Google Scholar]
- 48.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 49.Zaborosch C, Köster M, Bill E, Schneider K, Schlegel H G, Trautwein A X. EPR and Mössbauer spectroscopic studies on the tetrameric, NAD-linked hydrogenase of Nocardia opaca 1b and its two dimers. 1. The βδ dimer—a prototype of a simple hydrogenase. BioMetals. 1995;8:149–162. [Google Scholar]