Abstract
The multicopy suppressors of the snf1 defect, Msn2p and Msn4p transcription factors (Msn2/4p), activate genes through the stress-responsive cis element (CCCCT) in response to various stresses. This cis element is also the target for repression by the cyclic AMP (cAMP)-signaling pathway. We analyzed the two-dimensional gel electrophoresis pattern of protein synthesis of the msn2 msn4 double mutant and compared it with that of the wild-type strain during exponential growth phase and at the diauxic transition. Thirty-nine gene products (including those of ALD3, GDH3, GLK1, GPP2, HSP104, HXK1, PGM2, SOD2, SSA3, SSA4, TKL2, TPS1, and YBR149W) are dependent upon Msn2/4p for their induction at the diauxic transition. The expression of all these genes is repressed by cAMP. Thirty other genes identified during this study are still inducible in the mutant. A subset of these genes were found to be superinduced at the diauxic transition, and others were subject to cAMP repression (including ACH1, ADH2, ALD6, ATP2, GPD1, ICL1, and KGD2). We conclude from this analysis that Msn2/4p control a large number of genes induced at the diauxic transition but that other, as-yet-uncharacterized regulators, also contribute to this response. In addition, we show here that cAMP repression applies to both Msn2/4p-dependent and -independent control of gene expression at the diauxic shift. Furthermore, the fact that all the Msn2/4p gene targets are subject to cAMP repression suggests that these regulators could be targets for the cAMP-signaling pathway.
When the yeast Saccharomyces cerevisiae is grown on glucose-based medium in which the glucose becomes exhausted, a transient growth arrest occurs. This arrest, called the diauxic transition, corresponds to the adaptation of the cell to growth on ethanol accumulated in the culture medium as a new carbon source. Major changes in the pattern of gene expression occurring during this transition have been characterized by analysis of the protein pattern obtained by two-dimensional (2-D) gel electrophoresis. Genes encoding components of the glycolytic pathway and cell growth activity are repressed, whereas genes not expressed during growth on glucose are induced (1, 3, 16). At least two overlapping classes of proteins are induced at the diauxic transition: those synthesized during growth on ethanol or glycerol but not on glucose (called ccr) and those induced by heat shock from 26 to 36°C for 25 min (called hs) (5). Down regulation of the cyclic AMP (cAMP)-signaling pathway seems to be an important controlling factor of this transition. A decrease in the level of intracellular cAMP during the consumption of glucose has been reported (15, 34) and is required for subsequent growth on ethanol after the diauxic transition (34). We previously observed that the diauxic shift response is largely prevented when intracellular cAMP is maintained at an artificially high level (7). When cAMP is exogenously added, genes expressed during growth on glucose are still expressed when glucose is exhausted, whereas a large proportion of the genes expressed at the diauxic transition are not induced. These results are consistent with direct control by the cAMP-signaling pathway of one or more transcription factors.
A repressing effect of the cAMP-signaling pathway has been reported for the stress-induced CTT1 (25), UBI4 (38), SSA3 (2, 44), and SSA1, HSP12, HSP26, HSC82, and HSP104 (12). In the case of CTT1 and HSP12, a stress-responsive cis element (STRE), whose sequence is CCCCT, has been shown to mediate both stress induction and repression by the cAMP-signaling pathway (25, 42). STRE is also important for the induction of DDR2 (21), TPS2 (18), GSY2 (30), and SOD2 (14) and has been found upstream of a large number of stress-inducible genes (24). The transcription factor Msn2p and its homolog Msn4p (called Msn2/4p in this study) bind to STRE and appear to mediate gene activation in response to nutritional starvation, heat shock, oxidative stress, DNA damage, and osmotic shock (26, 36). These two transcription factors appear to be functionally redundant (13).
We decided to characterize the gene targets which are controlled by Msn2/4p for their induction at the diauxic transition by 2-D gel electrophoresis. We show here that Msn2/4p control a large number of genes induced at the diauxic transition. We further characterize the functional link between the cAMP-signaling pathway and the Msn2/4 regulators by comparing the genes induced at the diauxic transition: those dependent upon Msn2/4p with those repressed by exogenous cAMP. We observed that the cAMP repressive effect applies to all the Msn2/4p gene targets and also to Msn2/4p-independent gene targets. These results suggest that Msn2/4 regulators could be targets for the cAMP-signaling pathway.
MATERIALS AND METHODS
Yeast strains.
W303-1A (a ade2 can1 his3 leu2 trp1 ura3) and Wmsn2-msn4 (a ade2 can1 his3 leu2 trp1 ura3 msn2-D3::HIS3 msn4-1::TRP1) are isogenic strains (13). Strain OL556-STRE is derived from the diploid strain OL556 (a/α cdc25-5/cdc25-5 his3/his3 leu2/leu2 trp1/TRP1 rca1/rca1 ura3/ura3) (7) by genomic integration into the URA3 locus of the PMM2 plasmid (26). This plasmid, linearized at the unique ApaI site, contains four copies of the oligonucleotide with the HSP12 sequence from −221 to −241, including the STRE motif, in tandem at the EcoRI site upstream of the LEU2-lacZ gene fusion of the PLS9 plasmid (35). The single chromosomal integration at the URA3 locus has been controlled by PCR analysis (data not shown).
Culture conditions.
YNBS medium is a 2% glucose-based minimal medium (7) supplemented with the required bases and amino acids. The cultures were performed at 28°C.
Glucose measurement.
Glucose measurement was performed with Sigma diagnostic glucose reagent kit no. 510-A.
Protein synthesis analysis.
Radioactive labelling of proteins, preparation of cell extracts, and 2-D gel electrophoresis were performed as described previously (4). Quantitative analysis of the synthesis of the polypeptides separated on the 2-D gel was performed as follows. After drying, gels were exposed to phosphor screens which were scanned in a Molecular Dynamics PhosphorImager. Image files were then exported into BioImage software for image analysis and spot quantification. The spot intensities on the different images were standardized with regard to the actin spot. For proteins which are present as several distinct polypeptides with different pI values, the spot intensities were added. Spot intensities are expressed in arbitrary units.
β-Galactosidase measurement.
Yeast protein extracts and assay of β-galactosidase activity were performed as described previously (33). Units of β-galactosidase activity are nanomoles of O-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolyzed per minute at 37°C. Protein concentration in the extracts was measured by the dye-binding method (8).
RESULTS
Gene products not expressed in the absence of Msn2/4p.
The contribution of the transcription factors encoded by MSN2 and MSN4 in the change of gene expression induced during diauxic transition was monitored by comparing the pattern of proteins synthesized in a strain with a double deletion of both msn2 msn4 and in the isogenic wild-type strain. We used a strain with a double deletion rather than a strain with a single deletion in order to avoid partial phenotypic complementation. Cells grown on a 2% glucose medium were pulse labelled with [35S]methionine in early exponential growth (samples W1 and M1) and when glucose became limited (Fig. 1, W3, W4, and W5 and M3, M4, and M5). Proteins were separated by 2-D gel electrophoresis. Quantification of protein synthesis rates is based upon the radioactivity measurement of each spot with a PhosphorImager. No significant difference was found between wild-type and mutant cells during exponential growth on glucose (data not shown). This result is consistent with the lack of phenotype of the msn2 msn4 mutant on glucose-based medium.
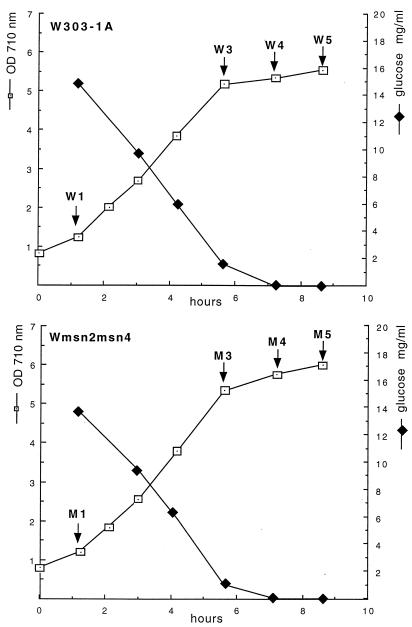
FIG. 1.
Growth of strains Wmsn2-msn4 and W303-1A. Growth in YNBS medium was monitored by turbidimetry at 710 nm (open squares). Glucose concentration was monitored in the medium during growth (filled squares). Arrows indicate the withdrawal of samples for labelling proteins with [35S]methionine.
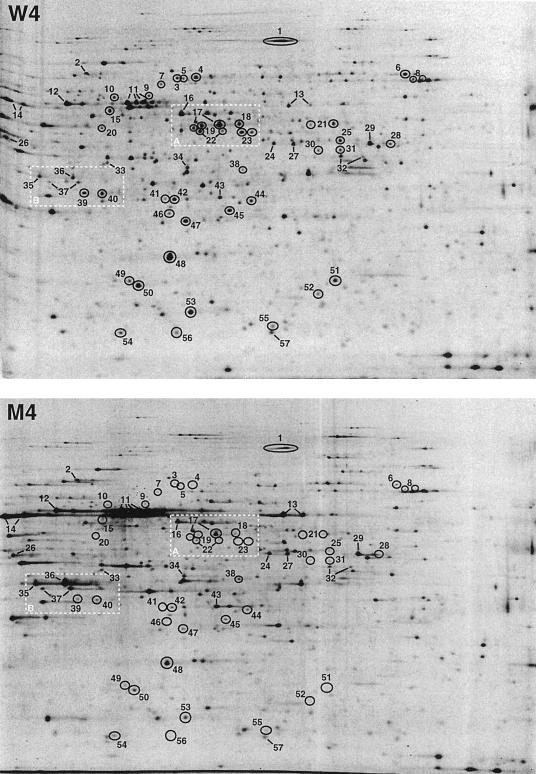
In contrast, large differences in rates of protein synthesis were observed when glucose became limited. Spots which consistently display at least a twofold difference in mutant and wild-type cells in the three sets of samples analyzed (W3 and M3, W4 and M4, and W5 and M5) were considered to be Msn2/4p-dependent proteins. Among the 61 proteins induced at the diauxic transition, 39 exhibited a significantly reduced synthesis rate in the mutant (Fig. 2 and 3 and Table 1). As shown in Table 1, 30 of these 39 proteins were induced by heat shock, transfer from glucose- to ethanol-based medium, or both these treatments (5). Twelve of these 39 proteins are the products of known genes. Identification of the two spot 23 polypeptides as Ald3 and/or 5p was performed on the basis of their amino acid compositions and did not allow distinction between the products of the 92% identical ALD3 and ALD5/ALD2/YMR170C (later called ALD5). The intensity of the gene induction defect in the msn2 msn4 mutant is different for different proteins (Table 1). Eleven proteins were not detectable. Nineteen other proteins exhibited a 3- to 10-fold decrease in synthesis rate, and 9 other proteins exhibited a decrease in synthesis rate that was less than threefold.
FIG. 2.
Comparison between proteins of W303-1A and Wmsn2-msn4 synthesized at the diauxic transition by 2-D gel protein pattern analysis. Sample cultures (0.5 ml) from W303-1A (W3, W4, and W5) and Wmsn2-msn4 (M3, M4, and M5) (see Fig. 1) were labelled for 60 min with 3.7 × 106 mBq of [35S]methionine (>3.7 × 1013 mBqm · mol−1; Amersham). After the labelling period, the cells were washed and extracted in 200 μl of extraction buffer. An aliquot of 15 μl was loaded onto the 1-D gel. Radioactivity on the gels was revealed with a PhosphorImager (Molecular Dynamics Inc.). Image intensities were adjusted so that the actin spot on the different images would have approximately the same intensity. Images from W4 (upper panel) and M4 (lower panel) are presented. Polypeptides are numbered from 1 to 57. Polypeptides which correspond to different isoforms of the same protein were assigned the same number. Their characteristics are reported in Tables 1 and 2. Polypeptides which are dependent upon Msn2/4p for the induction of their synthesis at the diauxic transition are indicated by circles, and those whose induction is independent of Msn2/4p and repressed by cAMP are shown by lines. The two framed regions are enlarged in Fig. 3.
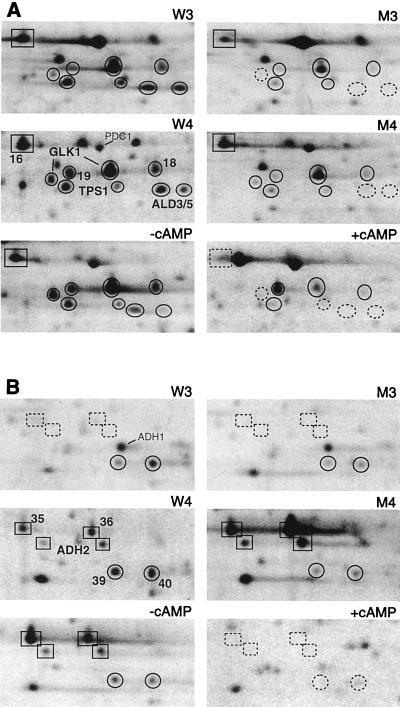
FIG. 3.
Enlargement of the two regions framed in Fig. 2, from maps obtained with samples W3, M3, W4, and M4 and from maps S1 and S1+cAMP, previously published (7), obtained with samples from strain OL556 at the diauxic transition without (−) or with (+) cAMP. Glk1p, Tps1p, and Ald3/5p (A) and Adh2p (B) correspond to spots 17, 22, 23, 37, respectively, on the map for sample W4 (Fig. 2). The superinduction of Adh2p (B) and the polypeptides 16 (A) and 35 and 36 (B) in the msn2 msn4 mutant are illustrated.
TABLE 1.
Msn2/4p-dependent proteins
| Spot no.a | Gene
|
Protein | Spot intensity for indicated sampled
|
Ratio (%)
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Nameb | Regulation treatmentc | W4 | M4 | −cAMP | +cAMP | M4/W4 | −cAMP/+cAMP | ||
| 41 | 1,530 | 0 | 2,564 | 0 | 0 | 0 | |||
| 52 | 1,875 | 0 | 3,643 | 0 | 0 | 0 | |||
| 25 | 3,928 | 0 | 6,149 | 0 | 0 | 0 | |||
| 55 | 3,059 | 0 | 1,292 | 0 | 0 | 0 | |||
| 46 | HS | 1,730 | 0 | 2,139 | 0 | 0 | 0 | ||
| 28 | HS | 1,815 | 0 | 3,070 | 0 | 0 | 0 | ||
| 51 | HS | 6,102 | 0 | 6,434 | 0 | 0 | 0 | ||
| 23* | ALD3/5 | HS | Aldehyde dehydrogenase-like | 6,904 | 0 | 25,920 | 0 | 0 | 0 |
| 56 | HS, CCR | 2,720 | 0 | 5,300 | 0 | 0 | 0 | ||
| 15 | HS, CCR | 3,956 | 0 | 2,596 | 0 | 0 | 0 | ||
| 4 | TKL2 | HS, CCR | Transketolase 2 | 6,083 | 0 | 6,088 | 0 | 0 | 0 |
| 30 | 1,982 | 191 | 4,634 | 0 | 10 | 0 | |||
| 20 | CCR | 1,707 | 196 | 1,435 | 0 | 11 | 0 | ||
| 3 | CCR | 2,475 | 319 | 2,993 | 0 | 13 | 0 | ||
| 5 | 1,708 | 253 | 1,716 | 0 | 15 | 0 | |||
| 19* | HS | 4,904 | 854 | 8,526 | 3,042 | 17 | 36 | ||
| 21 | HXK1 | HS, CCR | Hexokinase 1 | 5,848 | 1,051 | 5,334 | 0 | 18 | 0 |
| 6 | SSA3 | HS | Heat shock protein (Hsp70) | 6,397 | 1,220 | 5,896 | 0 | 19 | 0 |
| 22* | TPS1 | HS | Trehalose-6-phosphate synthase | 6,892 | 1,608 | 15,656 | 0 | 23 | 0 |
| 18* | 5,061 | 1,233 | 7,002 | 1,657 | 24 | 24 | |||
| 45 | CCR | 5,575 | 1,403 | 6,255 | 0 | 25 | 0 | ||
| 7 | CCR | 1,446 | 369 | ND | ND | 26 | ND | ||
| 31 | GDH3 | CCR | Glutamate dehydrogenase (NADP+) | 2,679 | 715 | 4,294 | 0 | 27 | 0 |
| 42 | HS, CCR | 5,207 | 1,421 | 6,311 | 0 | 27 | 0 | ||
| 47 | HS | 6,069 | 1,666 | 7,886 | 842 | 27 | 11 | ||
| 10 | HS | 2,531 | 737 | 1,611 | 0 | 29 | 0 | ||
| 40* | HS | 4,674 | 1,405 | 5,016 | 0 | 30 | 0 | ||
| 39* | 4,289 | 1,346 | 3,757 | 0 | 31 | 0 | |||
| 8 | SSA4 | HS | Heat shock protein (Hsp70) | 2,762 | 894 | 6,805 | 4,807 | 32 | 71 |
| 48 | GPP2 | HS | Glycerol phosphate phosphatase | 30,787 | 10,116 | 27,593 | 1,698 | 33 | 6 |
| 44 | YBR149W | CCR | Aldehyde dehydrogenase-like | 3,028 | 1,133 | 3,030 | 0 | 37 | 0 |
| 1 | HSP104 | HS | Heat shock protein (Hsp104) | 29,851 | 11,597 | 37,130 | 18,258 | 39 | 49 |
| 50 | HS | 8,854 | 3,558 | 14,030 | 0 | 40 | 0 | ||
| 17* | GLK1 | HS | Glucokinase | 16,123 | 6,506 | 32,635 | 8,957 | 40 | 27 |
| 53 | HS, CCR | 9,895 | 4,131 | 18,096 | 7,250 | 42 | 40 | ||
| 38 | 2,538 | 1,067 | 1,301 | 0 | 42 | 0 | |||
| 49 | HS, CCR | 3,528 | 1,525 | 5,900 | 3,645 | 43 | 62 | ||
| 9 | PGM2 | HS, CCR | Phosphoglucomutase | 1,782 | 826 | 1,968 | 0 | 46 | 0 |
| 54 | SOD2 | HS, CCR | Manganese super oxide dismutase | 3,735 | 1,748 | 8,986 | 0 | 47 | 0 |
Spot number of each polypeptide in Fig. 2. The asterisks indicate polypeptides which are present in Fig. 3. Additional data on these polypeptides (isoelectric point, molecular weight, values of spot intensity) on maps from samples W3, M3, W5, and M5 can be accessed on the yeast protein map server (www.ibgc.u-bordeaux2.fr/YPM).
References for the identification of the gene products on the map can be accessed on the yeast protein map server.
HS, heat shock; CCR, transfer from glucose- to ethanol-based medium.
Correlation of the effects of cAMP and Msn2/4p.
We have previously shown by 2-D gel analysis of strain OL556 that induction of a large number of proteins at the diauxic transition can be repressed by exogenous cAMP (7). The same set of proteins was induced at the diauxic shift in the two strains OL556 and W303-1A with a similar level of induction, except for spot 7, which is not detected in OL556 (Table 1). This experiment allowed us to compare the effect of Msn2/4p observed in W303-1A with that of cAMP in OL556. Proteins whose induction at the diauxic transition is decreased in the msn2 msn4 mutant are also repressed by cAMP in OL556 (Table 1).
Proteins repressed by cAMP but still induced in the msn2 msn4 mutant.
Thirty of the 69 proteins induced at the diauxic transition were still normally induced in the msn2 msn4 mutant. Eighteen of these 30 proteins were subject to cAMP repression (Table 2). Thus, all Msn2/4p gene targets are subject to cAMP repression, but cAMP also represses other Msn2/4p-independent targets. The cAMP repressing effect was heterogeneous (Table 2). Interestingly, most of these 18 proteins identified here are induced only after glucose exhaustion (sample W4), in contrast to the Msn2/4p-dependent proteins, which are induced earlier when glucose is still present (sample W3) (data not shown). All these cAMP-repressed proteins are already known as proteins expressed in ethanol but not in glucose medium. These data clearly suggest that this set of proteins is controlled by pathway(s) other than Msn2/4p and may respond to a different signal.
TABLE 2.
Msn2/4p-independent proteins repressed by cAMP
| Spot no.a | Gene
|
Protein | Spot intensity for indicated sampled
|
||||
|---|---|---|---|---|---|---|---|
| Nameb | Regulation treatmentc | W4 | M4 | −cAMP | +cAMP | ||
| 33 | CCR | 2,154 | 1,865 | 663 | 0 | ||
| 57 | HS, CCR | 5,038 | 3,100 | 3,624 | 0 | ||
| 2 | CCR | 3,214 | 3,538 | 4,005 | 0 | ||
| 37* | ADH2 | CCR | Alcohol dehydrogenase II | 10,461 | 30,806 | 8,955 | 0 |
| 32 | GPD1 | HS, CCR | Glycerol-3-phosphate dehydrogenase | 5,468 | 4,279 | 9,315 | 0 |
| 43 | CCR | 4,150 | 7,067 | 9,349 | 0 | ||
| 36* | CCR | 3,517 | 14,343 | 9,834 | 0 | ||
| 16* | HS, CCR | 10,775 | 6,357 | 10,953 | 0 | ||
| 35* | CCR | 3,459 | 7,843 | 14,582 | 0 | ||
| 14 | CCR | 23,408 | 87,100 | 121,323 | 0 | ||
| 11 | ICL1 | CCR | Isocitrate lyase | 31,906 | 87,028 | 131,020 | 0 |
| 24 | CCR | 3,790 | 3,924 | 4,298 | 804 | ||
| 26 | CCR | 6,140 | 4,658 | 6,629 | 927 | ||
| 13 | ALD6 | CCR | Acetaldehyde dehydrogenase | 6,246 | 22,663 | 7,017 | 1,573 |
| 34 | CCR | 3,275 | 3,170 | 4,728 | 1,707 | ||
| 29 | ATP2 | CCR | F1-beta ATP synthase | 9,338 | 9,703 | 8,513 | 2,245 |
| 12 | ACH1 | CCR | Acetyl-coenzyme A hydrolase | 13,553 | 13,219 | 13,987 | 2,855 |
| 27 | KGD2 | CCR | 2-Oxoglutarate dehydrogenase complex E2 component | 4,969 | 6,292 | 8,583 | 2,977 |
Finally, the syntheses of 12 proteins were found to be independent of Msn2/4p for their induction and insensitive to cAMP (among these proteins, Hsp60, Hsp78, Hsp82, Cit2p, Cor1p, and Mdh1p are known gene products) (data not shown).
Several gene products are superinduced in the absence of Msn2/4p.
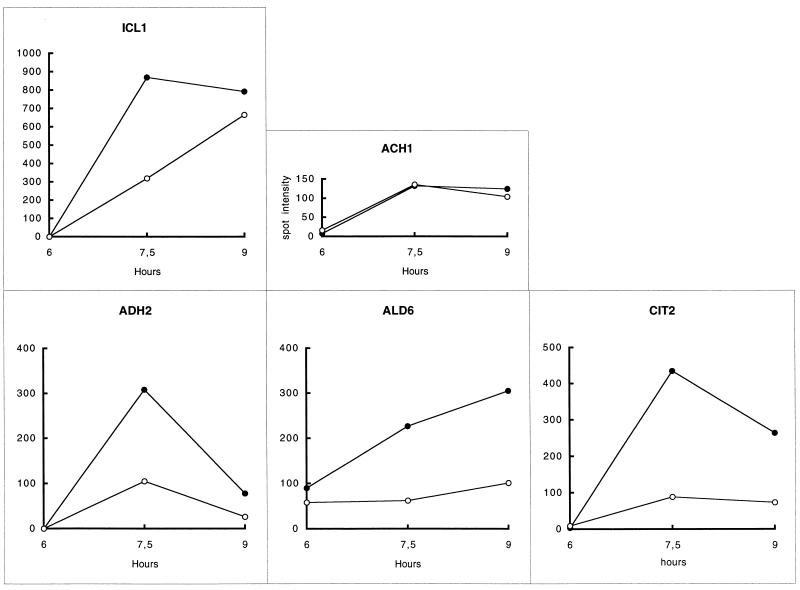
Seven proteins were found to be superinduced in the msn2 msn4 mutant. These include the products of ADH2, ALD6, CIT2, and ICL1 and spots 14, 35, and 36. The kinetics of induction of ADH2, ALD6, CIT2, and ICL1 showed that except for ALD6, this superinduction was transient (Fig. 4).
FIG. 4.
Gene products superinduced in the msn2 msn4 mutant. The kinetics of induction at the end of growth (Fig. 1) was monitored for each gene product by measuring the intensities of the spots on maps for samples W3, W4, and W5 and M3, M4, and M5. Open circles, W303-1A; closed circles, Wmsn2-msn4.
STRE is activated at the diauxic transition.
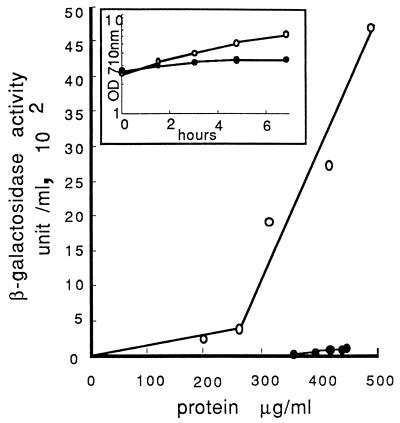
These data demonstrated that Msn2/4p control a large number of genes at the diauxic transition, presumably by activating these genes through their STREs. We thus monitored the activation of STRE at the diauxic transition with an STRE-lacZ reporter gene. Strain OL556-STRE, carrying the integrated STRE-lacZ reporter gene, was grown in glucose medium with or without cAMP until diauxic transition, and the rate of accumulation of β-galactosidase was determined (Fig. 5). The β-galactosidase synthesis rate was low during exponential growth and increased dramatically (12-fold) at the end of this phase. When cAMP was present in the culture, the level of β-galactosidase activity in the cells remained very low and no significant induction could be observed when glucose was exhausted. Thus, an induction of the STRE-driven lacZ reporter gene is observed at the diauxic transition. In excess, cAMP has a negative effect on this STRE-dependent induction, as previously described in the case of N starvation and heat shock response (25).
FIG. 5.
Induction mediated by STRE and the cAMP-negative effect. Strain OL556-STRE was grown in YNBS medium without uracil and without or with cAMP (3 mM) at 28°C. Growth was monitored by turbidimetry at 710 nm, and samples were withdrawn from the culture without cAMP (open circles) and with cAMP (filled circles). β-Galactosidase activity and protein concentration measurements were performed in the cellular extracts. For each sample, units of β-galactosidase activity accumulated per milliliter of culture, without (open circles) or with (filled circles) cAMP, were plotted against the protein concentration accumulated in the cultures (milligrams/milliliter). Protein concentration was calculated from the turbidity values for each sample, from the cell concentration per unit of turbidity, and from the cell-soluble protein content (7). For each sample, β-galactosidase activity per milliliter of culture was calculated by multiplying the β-galactosidase specific activity of each extract (nanomoles of ONPG hydrolyzed per minute and per milligram of protein at 37°C) by the protein concentration in the culture. OD, optical density.
Promoter analysis of the Msn2/4p-dependent genes.
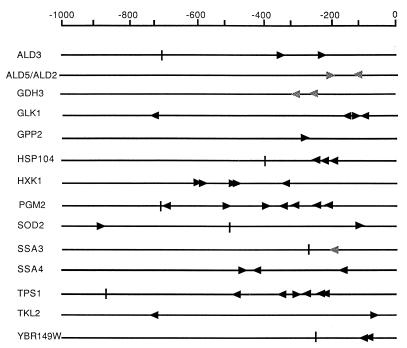
STREs were searched in the promoter region of 12 Msn2/4p-dependent genes (Fig. 6). Except for GDH3 and SSA3, all these genes contain one or several STREs. The importance of the STRE for gene induction at the stationary phase was demonstrated only for SOD2 (14). For SSA3, there is a STRE-related sequence, CCCT, which is part of a 35-bp postdiauxic shift upstream activating sequence. This sequence, called UASPDS, was shown to be important for gene activation under starvation conditions and to be subject to cAMP repression (2). In addition, this sequence and STRE are probably bound by the same factor as that shown in gel retardation experiments (25). Together, these results suggest that PDS is also a target for Msn2/4p. Two PDS elements are present in the GDH3 promoter region and probably could serve as Msn2/4p binding sites. A PDS element is also present in ALD5 and may be important for induction of this gene by stress conditions (29).
FIG. 6.
STRE sites on the promoter regions of Msn2/4p-dependent genes. The STRE sites (CCCCT, black arrowheads) found up to 1,000 bp upstream of the ATG of each gene in the Saccharomyces genome database from the genomic DNA of strain S288C are indicated. Each vertical bar indicates the end of the intergenic region when it is less than 1,000 bp. PDS sites (CCCT) present upstream of the GDH3 and ALD5 open reading frames and the functional PDS site of the SSA3 gene are indicated by grey arrowheads.
DISCUSSION
Msn2/4p provide a major contribution to the diauxic transition.
From the analysis presented here, it can be seen that a large proportion of the proteins induced at the diauxic transition are dependent upon Msn2/4p. Regulation very likely occurs at the transcriptional level. However, the possibility of posttranscriptional control cannot be excluded for some of these gene products. Indeed, from the recent results obtained by multiple RNA analysis, products of 10 of the 13 known genes dependent upon Msn2/4p (ALD5, PGM2, HXK1, HSP104, GLK1, YBR149W, TPS1, TKL2, SSA3, and SSA4) are transcriptionally induced at the diauxic transition (10). These transcription factors were first discovered as multicopy suppressors of the snf1 mutation (13), suggesting their involvement in the regulation of glucose-repressed genes. When it was later found that they bind to STRE, they were considered mostly to be transcription factors involved in the general stress response (26). In this work, we have not attempted to analyze the respective contributions of Msn2p and Msn4p, which are known to be at least partially redundant. We confirm the role of these transactivators in controlling the expression of genes known to be induced by stresses such as heat shock and of genes expressed in ethanol- or glycerol-containing medium but not in glucose-containing medium. Among the proteins involved in stress response which we found to be induced less in the msn2 msn4 double mutant are several that are known. The gene products Hsp104p, Ssa3p, and Ssa4p are chaperon proteins induced by heat shock. Sod2p has a protective function in response to oxidative stress and was previously described as essential to osmotic stress response (23, 31). Two other genes dependent upon Msn2/4p, TPS1 and PGM2, are involved in carbohydrate storage, a cellular response to various stresses (19, 32). In the case of TPS1, the expression of a lacZ fusion with the TPS1 promoter region is dependent upon Msn2/4p, confirming the data from the 2-D gel analysis and indicating transcriptional control (37a). In the case of SSA3, it should be emphasized that its induction in response to glucose starvation has been previously reported to be independent of Msn2/4p (26). The differences in the physiological conditions used to test the effect of Msn2/4p or an indirect effect of Msn2/4p at the translational level of SSA3 expression could explain these conflicting results.
Interestingly, all genes encoding metabolic enzymes that we have found to be Msn2/4p dependent (GDH3, GLK1, HXK1, TKL2, YBR149W, and ALD3 or ALD5) belong to families. HXK2 and GDH1 are preferentially expressed when glucose is present and repressed when glucose is exhausted (5, 37), and the Tkl1p/Tkl2p ratio varies with physiological state (22). Moreover, the abundance of the transcripts from the related isoenzymes in exponential growth on glucose medium estimated from the yeast transcriptome data (43) indicates that GDH1, HXK2, and TKL1 are expressed at significantly higher levels than their related genes, GDH3, HXK1, and TKL2. The polypeptides identified as products of ALD3 or ALD5 have an aldehyde dehydrogenase function. These two genes have already been reported to be induced by osmotic stress, and the induction of ALD5 is impaired in a bcy1 mutant (29). Products of ALD3 and ALD5 and of YPR149W, which also encodes a putative aldehyde dehydrogenase, have the same function as those of ALD1 and ALD6, and ALD6 is transcribed more during exponential growth on glucose medium than ALD3 and ALD5 (43). The Msn2/4p-dependent induction of different isoenzymes can reflect a role of these transcription factors in adjusting the types and levels of isoenzymes involved in carbon and nitrogen metabolism in response to glucose limitation.
Two-thirds of the gene products whose induction is reduced in the absence of Msn2/4p are still induced but at a lower level in the msn2 msn4 mutant. This Msn2/4p-independent induction indicates the complexity of the regulation of these genes at the diauxic transition.
The Msn2/4p defect stimulates the induction of a class of gene products.
The observation of a large collection of gene products by the 2-D gel analysis allowed us to discover an unexpected effect of the msn2 msn4 deletion. Some genes which do not require Msn2/4p for their induction at the diauxic transition are even more induced in the mutant than in the wild type, an effect that is transient for the majority of the genes. This phenomenon may be an indirect effect of the lack of Msn2/4p and may be related to the inability of the mutant to respond normally to nutrient limitation. A more direct effect of Msn2/4p can also be hypothesized. The possibility of a direct repressing effect of Msn2/4p on some of these genes cannot be excluded, although it does not seem likely for genes such as ICL1, which does not have a STRE site in its promoter region. The Msn2/4p-dependent activation of a repressor is a more likely explanation. It could also be that the induction of these genes by their transcription factors depends on a limiting factor which could be associated with Msn2/4p.
Msn2/4p could be targets for the cAMP-signaling pathway.
In a previous study, we have shown that a high level of cAMP, artificially maintained, prevents the induction of a large number of genes at the diauxic transition. Since Msn2p acts on STRE (26), which has been shown to be a target for the cAMP-signaling pathway (25, 42; also this study), it was important to compare the patterns of the proteins regulated by Msn2/4p with those of the proteins regulated by cAMP at the diauxic transition. The good reproducibility of the protein patterns at the diauxic transition for strains W3031-A and OL556 allowed us to perform this comparison. The fact that all the genes which are dependent upon Msn2/4p for their induction are repressed by excessive cAMP argues in favor of Msn2/4p mediating the cAMP regulation of these genes. The effect of cAMP on SSA4 is weak; nevertheless, the absolute value is in the same range of order as the Msn2/4p effect. Moreover, a repressing effect of cAMP on SSA4 transcription has already been described (12).
Transcriptional regulators other than Msn2/4p are also controlled by the cAMP-signaling pathway.
Some of the gene products whose induction at the diauxic transition is not completely dependent upon Msn2/4p are still completely repressed by cAMP. This difference between the effects of Msn2/4p and cAMP may indicate sensitivity to the cAMP pathway of regulators other than Msn2/4p involved in the induction of the synthesis of these gene products, although the possibility of experimental variations cannot be excluded.
Moreover, a class of proteins which is not dependent on Msn2/4p for induction at the diauxic transition is repressed by cAMP, arguing strongly for other cAMP-sensitive transcription factors. Notably, all these have been previously classified as genes expressed in ethanol but not in glucose medium, suggesting that they could have similar regulatory properties. As previously shown (40), an effect of the cAMP pathway on the Snf1 kinase, a major regulatory element involved in glucose repression, is unlikely. In the case of ADH2, the regulatory factor Adr1p has been shown to be partially sensitive to the cAMP pathway (9, 11). For the others, the regulatory proteins sensitive to cAMP remain to be identified. Sequence analysis of the upstream region of the known genes does not allow the formulation of any predictions. However, the fact that they are induced at the diauxic transition and repressed by glucose might lead to some known transcription factors such as Adr1p, Hap2p, Cat8p, and Mig1p.
The biological significance of Msn2/4p gene control.
MSN2 and MSN4 are dispensable for exponential growth on glucose; their deletion does not give a detectable growth defect. Indeed, we show here that the pattern of gene expression of the msn2 msn4 double mutant is similar to the pattern of the wild type under these conditions. In contrast, a large number of proteins normally induced at the diauxic transition fail to be induced in the mutant. This result indicates that Msn2p and/or Msn4p are required at this transition to activate the transcription of a large number of genes whose products could be important for adaptation to new growth conditions. A reduced ability to adaptation could explain the various phenotypes described for the msn2 msn4 double mutant: the hypersensitivity to glucose starvation (26) of exponentially growing cells, the impaired growth on galactose in anaerobic conditions, and the deleterious effect on growth of the overexpression of the DNA binding domain of Msn2p (13).
Since Msn2p has been shown to be produced constitutively during growth on glucose-based medium (16a), its function can be inferred to be activated when glucose for cell growth becomes limited. The activation of Msn2/4p is parallel with the drop in intracellular cAMP which occurs at the diauxic transition (34), and all Msn2/4p-dependent genes are repressed by cAMP, as already noted. The activation of Msn2/4p could be directly controlled by the cAMP-signaling pathway, since it is known that this pathway can relay a glucose signal (28). Msn2/4p could be direct targets for the protein kinase A, since the two transcription factors present putative cAMP-dependent phosphorylation sites. If Msn2/4p were directly controlled by the cAMP-signaling pathway, then they would have a larger role in cell physiology than stress-induced transactivation, considering the critical role of this pathway in environmental adaptation and differentiation (6, 17, 20, 27, 39, 41). We therefore propose, on the basis of the large spectrum of genes regulated by these transcription factors, that their activity is increased by a drop in the cAMP-dependent protein kinase activity in the cell, thereby stimulating the transcription of the genes that are required for adaptation to new environmental conditions.
ACKNOWLEDGMENTS
This work was supported by grants from the Association pour la Recherche sur le Cancer, la Ligue Nationale Française contre le Cancer, and the MENESR within ACCSV1 9501040.
We thank Francisco Estruch for providing strains W303-1A and Wmsn2msn4 and plasmid PMM2 and for critical reading of the manuscript. We are grateful to Christelle Monribot and Karine Paquier for their excellent technical assistance. We thank Monique Bolotin-Fukuhara, Michel Toledano, and Jean Labarre for critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Bataillé N, Régnacq M, Boucherie H. Induction of a heat-shock-type response in Saccharomyces cerevisiae following glucose limitation. Yeast. 1991;7:367–378. doi: 10.1002/yea.320070407. [DOI] [PubMed] [Google Scholar]
- 2.Boorstein W R, Craig E A. Regulation of a yeast HSP70 by a cAMP responsive transcriptional control element. EMBO J. 1990;9:2543–2553. doi: 10.1002/j.1460-2075.1990.tb07435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucherie H. Protein synthesis during transition and stationary phases under glucose limitation. J Bacteriol. 1985;161:385–392. doi: 10.1128/jb.161.1.385-392.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucherie H, Dujardin G, Kermogant M, Monribot C, Slonimsky P, Perrot M. Two dimensional protein map of Saccharomyces cerevisiae: construction of a gene-protein index. Yeast. 1995;11:601–613. doi: 10.1002/yea.320110702. [DOI] [PubMed] [Google Scholar]
- 5.Boucherie H, Sagliocco F, Joubert R, Maillet I, Labarre J, Perrot M. Two-dimensional gel protein data of Saccharomyces cerevisiae. Electrophoresis. 1996;17:1683–1699. doi: 10.1002/elps.1150171106. [DOI] [PubMed] [Google Scholar]
- 6.Boy-Marcotte E, Garreau H, Jacquet M. Cyclic AMP controls the switch between division cycle and resting state programs in response to ammonium availability in Saccharomyces cerevisiae. Yeast. 1987;3:85–93. doi: 10.1002/yea.320030205. [DOI] [PubMed] [Google Scholar]
- 7.Boy-Marcotte E, Tadi D, Perrot M, Boucherie H, Jacquet M. High cAMP levels antagonize the reprogramming of gene expression that occurs at the diauxic shift in Saccharomyces cerevisiae. Microbiology. 1996;141:459–467. doi: 10.1099/13500872-142-3-459. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Cherry J R, Johnson T R, Dollard C, Shuster J R, Denis C L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1989;56:409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- 10.DeRisi J L, Vishwanath R I, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1998;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 11.Dombek K M, Young E T. Cyclic AMP-dependent protein kinase inhibits ADH2 expression in part by decreasing expression of the transcription factor gene ADR1. Mol Cell Biol. 1997;17:1450–1458. doi: 10.1128/mcb.17.3.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelberg D, Zandi E, Parker C S, Karin M. The yeast and mammalian Ras pathways control transcription of heat shock genes independently of heat shock transcription factor. Mol Cell Biol. 1994;14:4929–4937. doi: 10.1128/mcb.14.7.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flattery-O’Brien J A, Grant C M, Dawes I A. Stationary phase regulation of the Saccharomyces cerevisiae SOD2 gene is dependent on additive effects of HAP2/3/4/5- and STRE-binding elements. Mol Microbiol. 1997;23:303–312. doi: 10.1046/j.1365-2958.1997.2121581.x. [DOI] [PubMed] [Google Scholar]
- 15.François J M, Eraso P, Gancedo C. Changes in the concentration of cAMP, fructose 2,6-biphosphate and related metabolites and enzymes in Saccharomyces cerevisiae during growth on glucose. Eur J Biochem. 1987;164:369–373. doi: 10.1111/j.1432-1033.1987.tb11067.x. [DOI] [PubMed] [Google Scholar]
- 16.Fuge E K, Braun E L, Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J Bacteriol. 1994;176:5802–5813. doi: 10.1128/jb.176.18.5802-5813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Garreau, H. Unpublished results.
- 17.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 18.Gounalaki N, Thireos G. Yap1, a yeast transcriptional activator that mediates multidrug resistance, regulates the metabolic stress response. EMBO J. 1994;13:4036–4041. doi: 10.1002/j.1460-2075.1994.tb06720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hottiger T, Boller T, Wiemken A. Rapid changes of heat and dessication tolerance correlated with changes of trehalose content in Saccharomyces cerevisiae. FEBS Lett. 1987;220:113–115. doi: 10.1016/0014-5793(87)80886-4. [DOI] [PubMed] [Google Scholar]
- 20.Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi N, McEntee K. Evidence for a heat shock transcription factor-independent mechanism for heat shock induction of transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:6550–6554. doi: 10.1073/pnas.87.17.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuimov A, Filippov M, Kochetov G. Multiple forms of transketolase. Biochem Int. 1990;21:1081–1087. [PubMed] [Google Scholar]
- 23.Larsson T, Norbeck J, Karlsson H, Karlsson K A, Blomberg A. Identification of two-dimensional gel electrophoresis resolved yeast proteins by matrix-assisted laser desorption ionization mass spectrometry. Electrophoresis. 1997;18:418–423. doi: 10.1002/elps.1150180316. [DOI] [PubMed] [Google Scholar]
- 24.Mager W H, Kruijff A J J D. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto K, Uno I, Ishikawa T. Genetic analysis of the role of the cAMP in yeast. Yeast. 1985;1:15. doi: 10.1002/yea.320010103. [DOI] [PubMed] [Google Scholar]
- 28.Mbonyi K, Beullens M, Detremerie K, Geerts L, Thevelein J M. Requirement of one functional RAS gene and inability of an oncogenic ras variant to mediate the glucose-induced cyclic AMP signal in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3051–3057. doi: 10.1128/mcb.8.8.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirrales V J, Serrano R. A genomic locus in Saccharomyces cerevisiae with four genes up-regulated by osmotic stress. Mol Microbiol. 1995;17:653–662. doi: 10.1111/j.1365-2958.1995.mmi_17040653.x. [DOI] [PubMed] [Google Scholar]
- 30.Ni H T, LaPorte D C. Response of a yeast glycogen synthase gene to stress. Mol Microbiol. 1995;16:1197–1205. doi: 10.1111/j.1365-2958.1995.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 31.Norbeck J, Blomberg A. Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 M NaCl. Evidence for osmotic induction of glycerol dissimilation via the dihydroxyacetone pathway. J Biol Chem. 1997;272:5544–5554. doi: 10.1074/jbc.272.9.5544. [DOI] [PubMed] [Google Scholar]
- 32.Parrou J L. Effect of various stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- 33.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 34.Russel M, Bradshaw-Rouse J, Markwardt D, Heideman W. Changes in gene expression in the Ras/adenylate cyclase system of S. cerevisiae: correlation with cAMP levels and growth. Mol Biol Cell. 1993;4:757–765. doi: 10.1091/mbc.4.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarokin L, Carlson M. Upstream region of the SUC2 gene confers regulated expression to a heterologous gene in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2521–2526. doi: 10.1128/mcb.5.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sierkstra L N, Verkabel J M A, Venips C T. Analysis of transcription and translation of glycolytic enzymes in glucose-limited continuous cultures of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:2559–2566. doi: 10.1099/00221287-138-12-2559. [DOI] [PubMed] [Google Scholar]
- 37a.Tadi, D. Unpublished results.
- 38.Tanaka K, Matsumoto K, Toh-e A. Dual regulation of the expression of the polyubiquitin gene by cAMP and heat shock in yeast. EMBO J. 1988;7:495–502. doi: 10.1002/j.1460-2075.1988.tb02837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatchell K D, Robinson L C, Breitenbach M. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc Natl Acad Sci USA. 1985;82:3785–3789. doi: 10.1073/pnas.82.11.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson-Jaeger S, François J, Gaughran J P, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, Ras proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 42.Varela J C S, Praekelt U M, Meacock P A, Planta R J, Mager W H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velculescu V E, Zhang L, Zhou W, Vogellstein J, Basrai M A, Bassett D E, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 44.Werner-Washburne M, Becker J, Kosic-Smithers J, Craig E A. Yeast HSP70 RNA levels vary in response to the physiological status of the cell. J Bacteriol. 1989;171:2680–2688. doi: 10.1128/jb.171.5.2680-2688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]