Abstract
We have analyzed the core RNA polymerase (RNAP) binding activity of the purified products of nine defective alleles of the rpoH gene, which encodes ς32 in Escherichia coli. All mutations studied here lie outside of the putative core RNAP binding regions 2.1 and 2.2. Based on the estimated Kss for the mutant sigma and core RNAP interaction determined by in vitro transcription and by glycerol gradient sedimentation, we have divided the mutants into three classes. The class III mutants showed greatly decreased affinity for core RNAP, whereas the class II mutants’ effect on core RNAP interaction was only clearly seen in the presence of ς70 competitor. The class I mutant behaved nearly identically to the wild type in core RNAP binding. Two point mutations in class III altered residues that were distant from one another. One was found in conserved region 4.2, and the other was in a region conserved only among heat shock sigma factors. These data suggest that there is more than one core RNAP binding region in ς32 and that differences in contact sites probably exist among sigma factors.
Transcription in bacteria requires the interaction of sigma factors (ς) and core RNA polymerase (RNAP), which is composed of β, β′, and α2 subunits. This interaction creates a holoenzyme which initiates transcription via direct contacts between the RNA polymerase and specific promoter DNA. At least one primary sigma factor (ς70) and six alternative sigma factors (ς32, ςE, ςF, ςS, ς54, and FecI) are present in Escherichia coli, all promoting the expression of different sets of genes (1, 15). The primary sigma factor controls the expression of the housekeeping genes and exists as the most abundant sigma factor during the exponential phase of growth. The activities and the level of the other six alternative sigma factors become more crucial to the viability of the cell in response to certain stress conditions. Depending on the stimuli, an alternative sigma factor may preferentially bind to core RNAP in lieu of the primary sigma factor to initiate transcription of genes that are under its control. This interchange of sigma factors on core RNAP causes an efficient switching of gene expression in response to the internal and external environment.
A few studies of sigma-core RNAP interaction have provided information about the location of core RNAP binding regions on sigma factors. Amino acid sequence alignment of sigma factors in the ς70 family has revealed that region 2.2 is the most highly conserved region (12, 22). Some have speculated that the most conserved region is probably the core RNAP binding region, based on the idea that sigma factors contact the same surface on core RNAP (9, 27, 32). Recently, we reported that a single amino acid change in region 2.2 of ς32, the heat shock sigma factor in E. coli encoded by rpoH, reduced its affinity for core RNAP (17). This result suggests that at least one residue in this most highly conserved region is directly involved in sigma factor-core RNAP interaction. Another highly conserved region, 2.1, has also been implicated in core RNAP binding, based on the deletion analysis of ς70 (20) and, subsequently, a single amino acid substitution of ςE in Bacillus subtilis (30). The crystal structure of the proteolytically stable fragment of E. coli ς70, which includes regions 2.1 and 2.2, further implicates these two regions as important for protein-protein interaction (23). Using ς32 with 24 amino acids deleted, Zhou et al. (35) have proposed that region 3 may also be involved in core RNAP binding. This region, however, is weakly conserved, especially among alternative sigma factors.
In this communication, we report our analysis of the products of nine rpoH alleles, each carrying a mutation downstream of region 2.2. These alleles suppress the temperature sensitivity of rpoD285 by preventing the proteolytic degradation of ς70 that contains a small internal deletion (10). In order to study the activities of these mutant sigma factors in vitro, we have purified the products of nine rpoH alleles. Subsequent biochemical examination of these purified products revealed that most of the mutants may exhibit reduced affinity for core RNAP. Interestingly, not all mutations were located in conserved regions, nor did they affect conserved residues. These results suggest that there are residues outside of regions 2.1 and 2.2 that may also participate in core RNAP interaction and that the mutated nonconserved residues may represent unique core RNAP binding sites for ς32.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. rpoH alleles have been selected by plating the E. coli strain carrying the partial deletion mutant allele rpoD285 at high temperature (11, 14). The strains that carry rpoD285 and rpoH mutant alleles are called PM111–113, PM161–163, PM173, PM174, PM176, PM181, and PM182. The rpoH allele number corresponds to the strain number.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Characteristics | Source or reference |
|---|---|---|---|
| Strains | |||
| 285c | rpoD285 | 10 | |
| CG410 | dnaK756 | 34 | |
| RL721 | rpoC3531(His6) zja::kan | Gift of R. Landick | |
| NUT20 | dnaK756/pUHE212-1 pDMI,1 | This laboratory | |
| NUT21 | dnaK756/pUHE211-1 pDMI,1 | This laboratory | |
| NUT26 | dnaK756/phis176 pDMI,1 | pUHE211-1 derivative (this laboratory) | |
| NUT29 | dnaK756/phis163 pDMI,1 | pUHE212-1 derivative (this laboratory) | |
| NUT33 | dnaK756/phis112 pDMI,1 | pUHE212-1 derivative (this laboratory) | |
| NUT34 | dnaK756/phis174 pDMI,1 | pUHE212-1 derivative (this laboratory) | |
| NUT35 | dnaK756/phis182 pDMI,1 | pUHE211-1 derivative (this laboratory) | |
| NUT48 | dnaK756/phis111 pDMI,1 | pUHE211-1 derivative (this laboratory) | |
| NUT49 | dnaK756/phis161 pDMI,1 | pUHE212-1 derivative (this laboratory) | |
| NUT50 | dnaK756/phis162 pDMI,1 | pUHE212-1 derivative (this laboratory) | |
| NUT53 | dnaK756/phis113 pDMI,1 | pUHE211-1 derivative (this laboratory) | |
| Plasmids | |||
| pUHE211-1 | ς32 C-his, Apr | 5 | |
| pUHE212-1 | ς32 N-his, Apr | 5 | |
| pDMI,1 | lacIq Kmr | 5 | |
| pJET40 | dnaK-P1 promoter | Gift of J. Erickson |
Cloning of rpoH alleles.
New rpoH alleles were cloned by a previously described procedure (3). The DNA was cut with HindIII and HpaI, and 1.5-kbp fragments were isolated. They were cloned into the large HindIII-PvuII fragment of plasmid pBR322. Plasmids with these cloned fragments were used to transform K165, which carries an rpoH amber mutation and a temperature-sensitive ochre suppressor. The selection in transformation was for resistance to 50 μg of ampicillin per ml and the ability to grow at 37°C. The location of each rpoH mutation was identified by DNA sequence analysis. These plasmids were called propH, with the allele number appended (e.g., prpoH174).
The rpoH alleles in the propH plasmids were subcloned into an rpoH His-tagged expression vector by fragment exchange. Subcloning of rpoH111, rpoH113, rpoH176, and rpoH182 was performed by the same procedure described for rpoH173 (17). The remaining alleles were subcloned into pUHE212-1 in the following manner. pUHE212-1 was cut with HindIII and blunt ended with the Klenow fragment. The linearized vector was digested with PstI, which cleaves within the rpoH gene. The resulting 4,278 bp was purified by gel electrophoresis, followed by elution. Each prpoH plasmid was treated with EcoRI, PstI, and XmnI. The fragments were electrophoresed. The 664-bp PstI-XmnI fragments, which contained the mutations in the rpoH gene, were eluted and ligated with the 4,278-bp fragment from pUHE212-1. The ligated plasmids were transformed into dnaK756 to reduce DnaK contamination during ς32 purification (21). Plasmids were isolated and used for DNA sequence analysis to confirm the site of each mutation.
Protein purification.
All proteins described in this paper were purified according to the previously described method (17) with modifications only for the His-tagged ς32 purification protocol. The His-tagged ς32 proteins eluted from the nickel-nitrilotriacetic acid-agarose column were dialyzed against a liter of buffer Z (50 mM KH2PO4 [pH 7.9] at 4°C, 150 mM KCl, 5% glycerol). The dialyzed solution was then loaded onto a 1-ml HiTrap Q column (Pharmacia), preequilibrated with buffer Z, at a rate of 0.4 ml/min. The column was then subjected to a 50-ml step of a 150 to 400 mM linear gradient of KCl and a 10-ml step gradient at 1 M KCl.
Purified ς32 proteins were dialyzed against two changes of 1 liter of a mixture containing 50 mM KH2PO4 (pH 7.9) at 4°C, 300 mM KCl, and 50% glycerol. Purified core RNAP and ς32 proteins were dialyzed against two changes of a mixture containing 10 mM Tris-HCl (pH 7.9) at 20°C, 0.1 mM EDTA, 0.1 mM dithiothreitol, 100 mM NaCl, and 50% glycerol. The protein concentrations were determined with the following molar coefficient extinctions: 43,100 M−1 cm−1 for ς32, 41,745 M−1 cm−1 for ς70, and 198,500 M−1 cm−1 for core RNAP (7, 28).
In vitro transcription assay.
The experiments were performed as previously described (17). RNA transcripts were visualized with a STORM PhosphorImager and quantitated with IMAGEQUANT software (Molecular Dynamics).
Glycerol gradient sedimentation and immunoblot analysis.
Sedimentation analyses were performed as described previously (17). Aliquots from sedimentation assays were subjected to electrophoresis in a sodium dodecyl sulfate–10% polyacrylamide gel (19). The proteins in the gel were transferred to a nitrocellulose membrane with a Trans-blot SD apparatus according to the manufacturer’s instructions (Bio-Rad). The membrane was blocked overnight in 5% nonfat milk solution and treated for 1 h with a 1:5,000 dilution of polyclonal ς32 antiserum and then with a 1:15,000 dilution of horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Bio-Rad) for 30 min. The sedimentation pattern of ς32 was detected with chemiluminescent reagents (Pierce). Polyclonal antiserum was obtained after injecting a rabbit with a mixture of gel-purified ς32 proteins and RIBI adjuvant, followed by periodic booster injections according to the manufacturer’s directions (RIBI ImmunoChem Research). Chemiluminescent blots were exposed to autoradiographic film for 30 s to 4 min, which was determined to be within the linear range of the film. Bands representing sigma factors were quantitated by densitometry with a scanner (ScanMan II; Logitech) and image analysis software (Sigmagel; Jandel).
RESULTS
Most of the amino acid changes in ς32 mutants affect conserved residues.
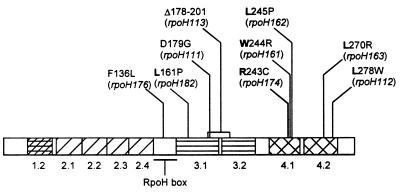
The locations of the changes in the following mutants have been reported: D179G (rpoH111), L278W (rpoH112), and Δ178–201 (rpoH113) (3, 10). Using methods described in references 3 and 10, we have isolated, cloned, and identified the mutations of other ς32 alleles (Fig. 1). Each mutation, except that of F136L, occurs in a conserved region determined by analyzing the amino acid sequence alignment of primary and alternative sigma factors in various bacteria (9, 12, 22). More significantly, most of the changes in mutants with point mutations affect conserved residues.
FIG. 1.
Schematic diagram of the conserved regions of ς32 and the location of the mutations studied in this report. The allele number of each mutation is listed in parentheses below the amino acid changes of the mutants. The regions are separated according to amino acid sequence similarity (9, 12, 22). The RpoH box is a conserved region only found among heat shock homologs (26). The conserved residues, according to the alignment analysis of Lonetto et al. (22), are depicted in boldface.
The mutant F136L is centered in the RpoH box, where the region and the affected residue are highly conserved only among heat shock sigma factors (26). This unique region from heat shock homologs overlaps with region C, a segment of the polypeptide implicated in the regulation of ς32 as a DnaK binding domain (24, 25).
Three mutations are found in region 3, a weakly conserved region, especially among alternative sigma factors. L161P and D179G are in region 3.1. Of these two residues, only L161 is well conserved and lies within the first helix of the putative helix-turn-helix (HTH) motif in region 3.1. The residue at position 179 is not conserved among alternative sigma factors, but the analogous residue in primary sigma factors, proline, is invariant. Recently, the P504L mutation of ς70, which is the residue corresponding to D179 of ς32, has been suggested to affect the rate of promoter escape (12a). The process of promoter escape has yet to be studied on ς32 or in other alternative sigma factors. It is plausible that region 3.1 may be involved in promoter escape and that the analysis of D179G may reveal interesting aspects of this step of transcription. The deletion mutant Δ178–201 is missing 24 amino acids spanning parts of regions 3.1 and 3.2. Bacteria carrying this mutation show an increase in the level of ς32 that is not bound to core RNAP (35).
We found a cluster of ς32 mutants affecting three contiguous amino acids, R243C, W244R, and L245P, in region 4.1. The role of this conserved region is not currently known, but the alterations affect conserved residues. The mutations in region 4.2, L270R and L278W, modify two of the most highly conserved residues among sigma factors. L270 is a part of the putative recognition domain for the −35 region of the promoter (6, 18, 31). This residue is placed in the DNA binding helix (the second helix) of the HTH unit. Although this residue was not implicated in contact with a specific DNA base (6, 31), it may be a key element in maintaining the structural integrity of the alpha helix. L278 is surrounded by highly conserved basic residues, which were suggested to stabilize the contact between the upstream HTH motif and DNA by neutralizing the negative charges of the nucleotides’ phosphate backbone (12). L278W may prevent the proper placement of the basic amino acids for ionic interactions.
In vitro transcriptional activities of purified ς32 proteins.
To investigate biochemically the possible defects of the mutant sigma factors, we purified the histidine-tagged products of each rpoH allele by nickel affinity chromatography. Six histidine residues were placed at the carboxyl terminus for F136L, L161P, D179G, and Δ178–201. The remaining mutants possessed the His tag at the amino terminus. The different positions of the metal affinity tag were chosen to simplify the cloning procedure. As positive controls, the wild-type sigma factors with histidine residues at either of the protein termini (designated ς32 C-his and ς32 N-his) were purified and characterized.
The activities of each purified protein were determined with an in vitro transcription assay. Holoenzyme reconstitutions were performed with variable concentrations of sigma factors and fixed levels of core RNAP and the dnaK-P1 promoter template. Transcription was restricted to one round by the addition of rifampin immediately after the beginning of elongation. The supercoiled DNA template contained a terminator, causing the reconstituted RNAP to produce a transcript with a size of 290 nucleotides. We were able to estimate the activity of the reconstituted RNAP by measuring the maximum yield of transcripts and the equilibrium constant (Ks), which includes the sigma-core RNAP (Eς) complex formation (17).
Our analysis of the two differently tagged wild-type ς32 proteins, ς32 C-his and ς32 N-his, revealed similar Kss and maximum yields of transcripts (Table 2). Because 200 fmol of core RNAP was present in each reaction, and because rifampin prevents multiple rounds of transcription, the maximum level of transcripts would be 200 fmol. Both types of wild-type proteins produced approximately 160 fmol of transcripts when reconstituted with core RNAP. At an equimolar ratio of sigma factor and core RNAP, approximately 100 fmol was produced. These yields from the in vitro transcription assays were in close agreement with our previously published analysis of purified core RNAP and ς32 C-his (17). Furthermore, these results showed that there was no significant difference between the activities of C-terminally or N-terminally His-tagged ς32. A similar observation was also reported regarding the activities of ς32 C-his and ς32 N-his in vivo (5).
TABLE 2.
Summary of in vitro transcription and glycerol gradient sedimentation resultsa
| ς32 protein (allele)b | Region affected | Ks (nM) | Maximum yield of transcripts (fmol) | % of core RNAP bindingc:
|
|
|---|---|---|---|---|---|
| Without ς70 | With ς70 | ||||
| Wild type | |||||
| ς32 C-his | None | ∼1.5 | ∼160 | >99 | ∼57 |
| ς32 N-his | None | ∼1.7 | ∼162 | >99 | ∼53 |
| Mutants | |||||
| Class I | |||||
| L161P (rpoH182) | 3.1 | ∼3 | ∼120 | >99 | ∼47 |
| Class II | |||||
| D179G (rpoH111) | 3.1 | ∼8 | ∼70 | ∼94 | ∼6 |
| R243C (rpoH174) | 4.1 | ∼6 | ∼108 | ∼99 | ∼30 |
| W244R (rpoH161) | 4.1 | ∼7 | ∼160 | ∼99 | ∼28 |
| L245P (rpoH162) | 4.1 | ∼8 | ∼157 | >99 | ∼4 |
| L270R (rpoH163) | 4.2 | ∼6 | ≥141 | >99 | ∼22 |
| Class III | |||||
| F136L (rpoH176) | RpoH box | ≥21 | ≥81 | ∼33 | ∼3 |
| L278W (rpoH112) | 4.2 | ≥40 | ≥157 | ∼41 | ∼1 |
| Δ178–201 (rpoH113) | 3 | NDd | ND | ∼28 | ∼1 |
In addition to the errors obtained from the experimental procedure in in vitro transcription and in immunoblot assays, there may be additional errors (±20%) from determination of protein concentrations by molar coefficient of extinction.
The mutants are classed according to their Kss and glycerol gradient sedimentation data (see Results).
The estimated percentage of core RNAP-bound ς32 was calculated by densitometry.
ND, not determined.
The transcriptional activity of each mutant differed from that of the wild type (Fig. 2). Although most of the mutants showed significant levels of activity, all of the mutants apparently exhibited a higher Ks and/or a lower maximum yield of transcripts (Table 2). Based on the estimated Kss (and the subsequent results of glycerol gradient sedimentation assays), we divided the mutants into three classes: class I, a mutant (L161P) with a minor effect (a 2-fold increase in Ks); class II, mutants (D179G, R243C, W244R, L245P, and L270R) with moderate effects (a range of 3- to 5-fold increases in Ks); and class III, mutants (F136L, Δ178–201, and L278W) with substantial effects (at least 14-fold higher in Ks).
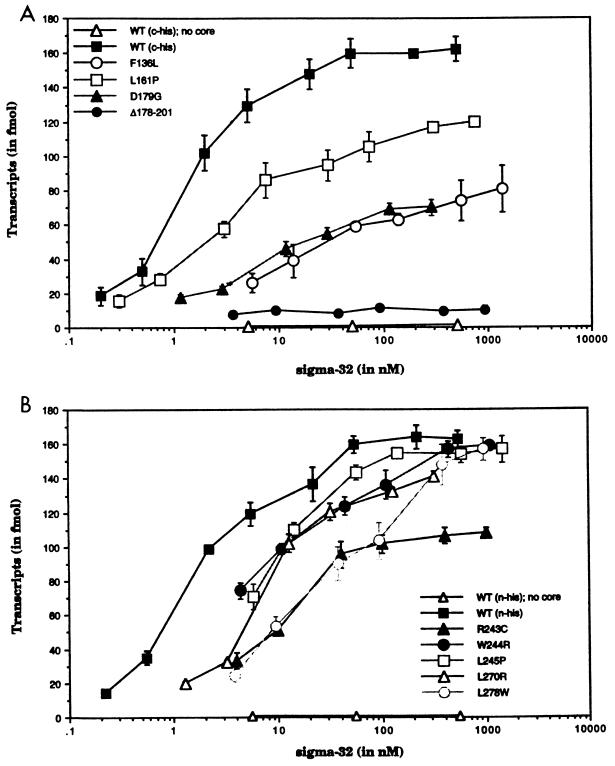
FIG. 2.
Graphic representation of transcripts produced from mutant and wild-type ς32 factors containing RNAP. Core RNAP (200 fmol) was incubated with increasing concentrations of sigma factors at 30°C. Supercoiled dnaK-P1 promoter templates (1.6 nmol) with a terminator from an E. coli rRNA transcription unit were used to generate transcripts of 290 nucleotides. The reaction occurred at 30°C for a single round of transcription. Transcripts were visualized and quantified with a PhosphorImager. Data from at least three experiments are expressed as means ± standard deviations. In vitro transcription results are displayed in panel A for the C-terminally His-tagged ς32 proteins and in panel B for the N-terminally His-tagged ς32 proteins. WT, wild type.
L161P was the only mutant that showed a relatively small increase in its Ks, which was estimated to be 3 nM. The maximum yield of transcripts was decreased to approximately 120 fmol, which represented a 25% decrease from the wild-type level. Although the differences in the in vitro transcriptional activities of L161P and the wild type were small, these results were quite reproducible.
The class II mutants revealed a more moderate increase in Ks. The range of their Kss was narrow, from a low of 6 nM to a high of 8 nM. R243C, W244R, and L245P, the mutants that affected the contiguous amino acids in region 4.1, were members of this group. Although their Kss were similar, R243C, the mutant that altered the positively charged residue, had the lowest maximum yield of transcripts. The other two mutants’ maximum yields were as high as those of the wild types, and they both affected a bulky hydrophobic amino acid. The exact Ks for L270R could not be determined, because it was not possible to define the maximum yield of transcripts. However, we estimated that the Ks would be equal to or greater than the half-highest point of transcripts produced in the reaction, which was 141 fmol. By this approach, the Ks was calculated to be greater than or equal to 6 nM. D179G revealed the second lowest maximum yield of transcripts at 80 fmol. The causes of the difference in the transcript level are yet to be determined, but we explore the potential effects of the mutations in Discussion.
The class III mutants exhibited a dramatic increase in their Kss. The most defective mutant was the mutant with the in-frame deletion (Δ178–201) in region 3. Although the concentration of this mutant sigma factor was increased to 1 μM in the reaction mixture, only a small increase in the level of transcripts was observed. The Ks of Δ178–201 and core RNAP complex could not be determined, but it was clear from the curve that the Ks would be much higher than that of the wild type. However, we believe that this result is in part due to the aggregation of the mutant sigma factors, which can be alleviated with a higher concentration of glycerol. Two additional mutants displayed significantly higher Kss. Using the same approach to estimate the equilibrium constant of L270R, the Kss were calculated to be greater than or equal to 21 and 40 nM for F136L and L278W, respectively. In terms of transcript production, L278W was more efficient than F136L. At a sigma factor concentration of greater than 1 μM, L278W generated almost as many transcripts as the wild type. F136L produced about half as many transcripts at a similar concentration of sigma factor.
Class III mutants are defective for core RNAP interaction.
To confirm the core RNAP binding defects for the mutants, we performed glycerol gradient sedimentation analysis. This technique separates free ς32 from that which is bound to core RNAP because of the significant difference in the molecular weight of one sigma subunit and all subunits in RNAP. In our experiments, equimolar concentrations of ς32 proteins and core RNAP were incubated to allow holoenzyme formation. The mixture was loaded onto a 15 to 35% glycerol gradient, followed by centrifugation to separate proteins. If ς32 was not bound to core RNAP, or if core RNAP was absent (Fig. 3B), the sigma factor was found closer to the top of the gradient. However, if ς32 associated with core RNAP, it sedimented to the bottom of the gradient (Fig. 3C).
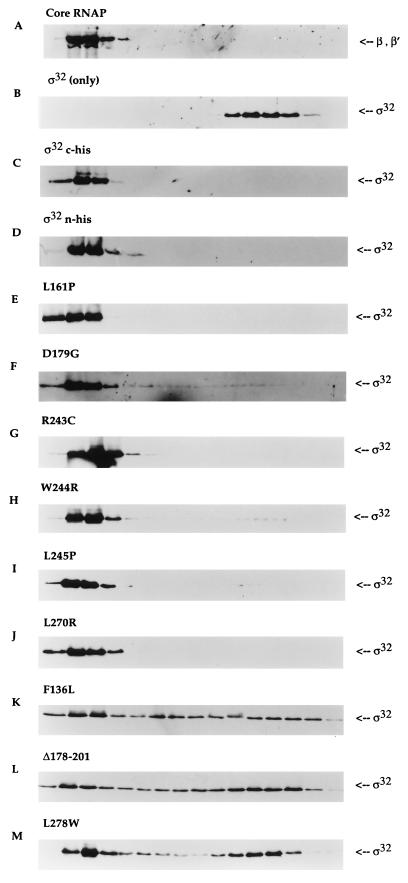
FIG. 3.
Core RNAP binding analysis of ς32 proteins by glycerol gradient sedimentation. Equimolar concentrations of core RNAP and different ς32 proteins (final concentration of 100 nM) were allowed to reconstitute at 30°C for 15 min in a buffer containing 50 mM HEPES (pH 7.9) at 4°C, 0.1 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, and 10 mM MgCl2. The mixture was loaded onto the top of a 5-ml 15 to 35% glycerol gradient and centrifuged at 48,000 rpm for 24 h at 4°C in a Beckman SW50.1 rotor. Sixteen fractions were collected from the bottom of the tube and subjected to immunoblot analysis. The sedimentation patterns of each ς32 protein were detected with anti-ς32 serum. The positions of β and β′ subunits were determined with polyclonal antibodies to core RNAP. The leftmost region of each panel represents the bottom of the tube (35% glycerol). The positions of core RNAP were indistinguishable with (A) or without (data not shown) ς32. The sedimentation patterns are shown for ς32 only (B), for two different wild-type proteins with core RNAP (C and D), and (as indicated above each panel) for the mutant proteins with core RNAP (E through M).
Based on our in vitro transcription analysis, we expected class I and II mutants to be almost completely bound to core RNAP. Because of their relatively small increases in Ks, these mutants would not exhibit reduced core RNAP affinity at the 100 nM level used in the experiment, even after taking into consideration the dilution effect during sedimentation. As predicted, our results showed a near complete interaction with core RNAP for class I and II mutants, except for D179G, which displayed approximately 94% binding (Fig. 3D to I and Table 2). A much higher Ks was seen in the class III mutants compared to those in the other classes of mutants (Table 2). Their core RNAP affinity might be reduced enough to be detected under the conditions used in our sedimentation assays. When glycerol gradient sedimentation was performed with these proteins with mutations in three different regions, the sedimentation patterns were distinctly different (Fig. 3J to L and Table 2). Most of ς32 sedimented closer to the top of the gradient, indicating that ς32 was dissociated from core RNAP. The percentages of unbound sigma factors were 67% in F136L, 72% in Δ178–201, and 59% in L278W. In addition, they all displayed broad sedimentation behavior, showing the instability of the interaction between the sigma factor and core RNAP. The above results for all classes of mutants were thus in close agreement with the data from in vitro transcription. Interestingly, these mutants also showed a large increase in free ς32 according to glycerol gradient analysis of crude extract (35) (data not shown). These observations further support the in vitro data that multiple residues in different regions of ς32 may be important for core RNAP interaction.
Core RNAP binding deficiency of class II mutants becomes evident in the presence of ς70.
Our previous report showed that an equimolar concentration of ς70 caused a nearly complete displacement of ς32 mutants Q80R and Q80N and a partial displacement of ς32 mutant E81G that was larger than what was observed for the wild-type ς32 (17). To better determine the reduction of core RNAP affinity in the mutants, we added an equimolar amount of the core RNAP competitor ς70 to the reaction mixture. We first tested the effects of the competitor on the two different wild-type ς32 proteins, ς32 C-his and ς32 N-his. The results revealed that both proteins were nearly equal to one another in their interaction with core RNAP (Fig. 4A and B). Approximately equal numbers of sigma factors were found bound and unbound to core RNAP (Table 2). Because the Kd of the Eς70 complex has been determined to be 2 nM (7), we expected ς32 to compete for core RNAP with at least equal efficiency.
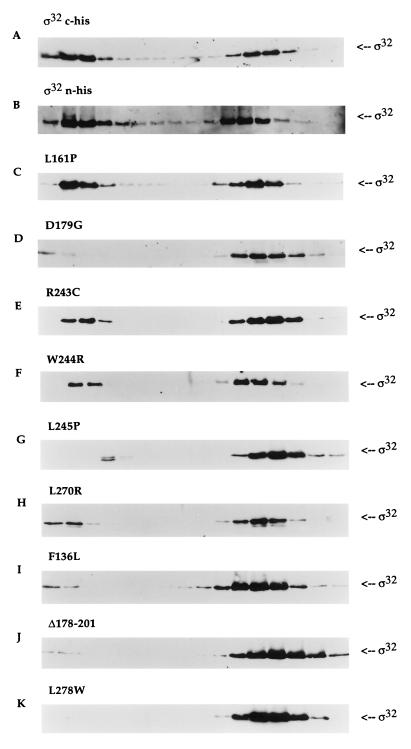
FIG. 4.
Competition between ς70 and ς32 for core RNAP. The same conditions were used as described in the legend to Fig. 3, except that core RNAP was added to a mixture that contained equimolar concentrations of ς32 and ς70 (all proteins at a final concentration of 100 nM). The sedimentation patterns of two wild-type proteins (A and B) and of the mutant proteins (C through K) are presented.
The class I mutant, L161P, was able to bind to core RNAP with high affinity. Almost half of the mutant sigma factor was able to interact with core RNAP even with the addition of ς70 (Fig. 4C and Table 2). This is quite comparable to the wild type’s sedimentation pattern. This result suggested that the mutant was not defective in core RNAP interaction. However, because of the noticeable decrease in the maximum yield of transcripts, L161P was probably defective at some other stage of transcription. The class II mutants provided various results in the presence of ς70 (Fig. 4D to H and Table 2). A significant population of R243C, W244R, and L270R sedimented with core RNAP to levels of 30, 25, and 22%, respectively. These yields from the mutant ς32-containing holoenzymes were definitely reduced, indicating that these mutants are defective for core RNAP binding. The remaining class II mutants, D179G and L245P, were almost completely displaced from core RNAP. This sedimentation behavior was somewhat surprising, because the estimated Ks for these mutants was 8 nM, which was not very much different from that of the previous three class II mutants. We predicted that a higher percentage of D179G and L245P would interact with core RNAP. Nevertheless, the competition assay did show that all class II mutants exhibited reduced affinity for core RNAP. Finally, as predicted, the class III mutants could not compete for core RNAP with ς70. All three mutants in this group were almost completely displaced (Fig. 4I to K and Table 2).
DISCUSSION
Our investigation of nine ς32 mutants suggests regions other than 2.1 and 2.2 may be involved in core RNAP interaction. While only certain mutants demonstrated a reduced maximum yield of transcripts, most mutants exhibited some degree of defect in their interaction with core RNAP. The significant level of in vitro transcriptional activity suggests that the mutations, except for the deletion mutation, do not impart deleterious effects to the structure of ς32. In addition, we were able to purify all ς32 mutants in this study, whereas G82S, which was used in our previous study, was too unstable for purification (17). The analogous residue of G82 in ς70, G408, has been shown to be located in a space-restricted hydrophobic environment imposed by neighboring alpha helices (23). Any alteration of this critical residue may destabilize the structure of the polypeptide, which may be a reason for our inability to purify G82S. However, because we were able to purify the other mutants containing point mutations, the affected residue in the mutant polypeptides probably does not contribute significantly to the stability of its structure. Furthermore, we have examined the melting curve of the class III mutants by circular dichroism spectroscopy (data not shown). Both F136L and L278W produced the cooperative melting behavior and melting temperature of 57°C that are observed for wild-type ς32. On the other hand, Δ178–201 exhibited noncooperative melting behavior, with a melting temperature of 50°C. These findings further suggest that the structural integrity of these two mutants has not been compromised by the single amino acid change. Therefore, a number of the mutants may perturb residues that are in physical contact with core RNAP.
Based on the Kss of the mutants, as determined by in vitro transcription, as well as by the results of glycerol gradient sedimentation, we have categorized the mutants into three classes: class I, with a minor increase in Ks; class II, with a moderate increase; and class III, with a dramatic increase.
The class I mutant.
Among the nine ς32 mutants examined in this report, L161P showed only a slight increase in its Ks. Glycerol gradient sedimentation confirmed the in vitro transcription result. Although the sedimentation pattern of this mutant was very similar to that of the wild type, the activity curve from in vitro transcription was distinguishable. The reduction in the maximum yield of transcripts was small but reproducible. Because the affected residue is located within the segment of the polypeptide showing a weak resemblance to an HTH motif (9), we considered the possibility that L161 may be involved in DNA binding. A preliminary investigation into the mutant’s ability to bind promoter DNA at 30°C with the addition of core RNAP indicates that the mutant is not defective up to the steps leading to promoter melting (data not shown). Hernandez et al. have shown that two mutants with point mutations of ς70 in region 3.1 produced different patterns of abortive transcripts (13). This result suggests a role for L161P in abortive transcription and provides a framework for investigation of the events that occur between transcription initiation and elongation.
Class II mutants.
The mutants R243C, W244R, and L245P, which contain mutations that alter contiguous conserved residues in region 4.1, and L270R, which contains a mutation that affects a residue in the HTH motif of region 4.2, are in class II. Their moderate core RNAP binding defects can be observed under glycerol gradient sedimentation only in the presence of a core RNAP competitor. Additionally, R243C may possess other defects because of its lowered maximum yield of transcripts. The proximity of R243 to the putative HTH motif in region 4.2 supports the idea that the residues in region 4.1 may also be involved, probably indirectly, in the interaction of ς32 with the −35 region of the promoter.
Class III mutants.
The rpoH113 deletion mutant has already been shown to exhibit an increase in free ς32 by glycerol gradient sedimentation of a crude extract of the rpoH mutant strain (35). Using purified proteins, we have demonstrated that the product of the deletion allele, Δ178–201, binds poorly to core RNAP. Our in vitro transcription results suggest that this mutant polypeptide may be improperly folded. Δ178–201 was more prone to aggregation when it was purified. In vitro transcription assays, which typically contained less than 2.5% glycerol in the reaction mixture, revealed a profound reduction of activity for the deletion mutant, which may not be an inherent characteristic of the protein but may be due to its aggregation. In contrast, the glycerol concentration used to reconstitute holoenzyme prior to sedimentation analysis for the deletion mutant was 10%, and the concentration of glycerol in the gradient ranges from 15 to 35%. Thus, there may be less aggregation of the mutant protein in our gradient experiments. We believe that Δ178–201 is defective for core RNAP binding mostly because the protein is misfolded, and not necessarily because of the deficiency of direct contacts with core RNAP.
F136L and L278W exhibited dramatic reductions in core RNAP affinity and, unlike Δ178–201, displayed significant transcriptional activities. Residues F136 and L278 are likely to contact core RNAP. Taken with Q80, another residue that is needed for high affinity for binding of core RNAP (17), the critical residues for efficient core RNAP interaction are quite distant from one another in the polypeptide chain. These results suggest that at least three domains of ς32, represented by region 2.2, region 4.2, and the RpoH box, may comprise the major core RNAP binding sites. Furthermore, because L278 is so highly conserved, the corresponding residue in other sigma factors may also modulate core RNAP interaction.
F136 is not conserved among all sigma factors but is situated in an extremely well-conserved segment of the heat shock sigma factors, designated the RpoH box (26). The lack of conservation among all sigma factors suggests that this binding site is characteristic of ς32 and possibly of all heat shock sigma factors. Furthermore, this additional core RNAP binding domain may be a recognition site for the regulators of ς32. The RpoH box is contained within a regulatory domain called region C. Using a ς32–β-galactosidase fusion protein, Nagai et al. (25) observed an increase in the stability of the fusion protein when region C was truncated or altered. They have proposed that this region controls the degradation of ς32, because ς32 is normally unstable (33). Subsequent biochemical study has revealed that region C may regulate the activity of ς32 by serving as a target for binding of DnaK (24). DnaK is a chaperone protein involved in the degradation of ς32 (34) and has been reported to interact preferentially with peptides containing hydrophobic amino acids (8) and with individual hydrophobic residues (29). F136L is a change from an aromatic hydrophobic residue to an aliphatic hydrophobic residue, and both of these amino acids are good recognition residues for DnaK. Such a change may have altered the binding site of core RNAP without modifying the DnaK-interacting surface on ς32. In support of this analysis, coelution of DnaK with F136L has been observed during the purification of this mutant protein in a dnaK+ strain (16). Based on the published reports on DnaK and on our study, we propose that the RpoH box may serve as a binding site for both DnaK and core RNAP. DnaK-bound ς32 would then inhibit core RNAP from interacting with the sigma subunit to initiate transcription on heat shock promoters, and, conversely, core RNAP-bound ς32 would deter DnaK from tagging the sigma factor for rapid degradation.
Another sigma factor with an antagonist that may share its binding site with core RNAP may be ςF of B. subtilis (4). One of the contact sites of the anti-sigma factor SpoIIAB on ςF is region 2.1, a putative core RNAP binding region. The interaction of SpoIIAB with ςF may block core RNAP from associating with the sigma subunit. In addition to SpoIIAB and DnaK, other antagonists that directly interact with sigma factors have been found (2). Although their exact mechanistic action is unknown, they may behave similarly to DnaK and SpoIIAB. Therefore, the process of blocking the activity of sigma factors by occupying the core RNAP binding site may be a common theme in prokaryotic transcription.
ACKNOWLEDGMENTS
We thank V. James Hernandez for showing us unpublished data. In particular we are indebted to Dick Burgess for critical reading and suggestions.
This work was supported by research grants MV 484 from the American Cancer Society and AI-08722 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. FecI belongs to a new subfamily of ς70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;18:163–174. doi: 10.1111/j.1365-2958.1995.mmi_18010163.x. [DOI] [PubMed] [Google Scholar]
- 2.Brown K L, Hughes K T. The role of anti-sigma factors in gene regulation. Mol Microbiol. 1995;16:397–404. doi: 10.1111/j.1365-2958.1995.tb02405.x. [DOI] [PubMed] [Google Scholar]
- 3.Calendar R, Erickson J W, Halling C, Nolte A. Deletion and insertion mutations in the rpoH gene of Escherichia coli that produce functional ς32. J Bacteriol. 1988;170:3479–3484. doi: 10.1128/jb.170.8.3479-3484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decatur A L, Losick R. Three sites of contact between the Bacillus subtilis transcription factor ςF and its antisigma factor SpoIIAB. Genes Dev. 1996;10:2348–2358. doi: 10.1101/gad.10.18.2348. [DOI] [PubMed] [Google Scholar]
- 5.Gamer J, Bujard H, Bukau B. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor ς32. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-m. [DOI] [PubMed] [Google Scholar]
- 6.Gardella T, Moyle H, Susskind M. A mutant Escherichia coli ς70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- 7.Gill S C, Weitzel S, von Hippel P H. Escherichia coli ς70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol. 1991;220:307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 8.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman M E. Specificity of the DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 9.Gribskov M, Burgess R R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986;14:6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman A D, Zhou Y-N, Gross C, Heilig J, Christie G E, Calendar R. Mutations in the rpoH (htpR) gene of Escherichia coli K-12 phenotypically suppress a temperature-sensitive mutant defective in the ς70 subunit of RNA polymerase. J Bacteriol. 1985;161:939–943. doi: 10.1128/jb.161.3.939-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris J D, Heilig J S, Martinez I I, Calendar R, Isaksson L A. Temperature-sensitive Escherichia coli mutant producing a temperature-sensitive ς subunit of DNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1978;75:6177–6181. doi: 10.1073/pnas.75.12.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 12a.Hernandez, V. J. Personal communication.
- 13.Hernandez V J, Hsu L M, Cashel M. Conserved region 3 of Escherichia coli ς70 is implicated in the process of abortive transcription. J Biol Chem. 1996;271:18775–18779. doi: 10.1074/jbc.271.31.18775. [DOI] [PubMed] [Google Scholar]
- 14.Hu J C, Gross C A. Marker rescue with plasmids bearing deletions in rpoD identifies a dispensable part of Escherichia coli sigma factor. Mol Gen Genet. 1985;191:492–498. doi: 10.1007/BF00425768. [DOI] [PubMed] [Google Scholar]
- 15.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joo D M. Ph.D. thesis. Berkeley: University of California; 1997. [Google Scholar]
- 17.Joo D M, Ng N, Calendar R. A ς32 mutant with a single amino acid change in the highly conserved region 2.2 exhibits reduced core RNA polymerase affinity. Proc Natl Acad Sci USA. 1997;94:4907–4912. doi: 10.1073/pnas.94.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenney T J, York K, Youngman P, Moran C P., Jr Genetic evidence that RNA polymerase associated with ςA uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci USA. 1989;86:9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lesley S A, Burgess R R. Characterization of the Escherichia coli transcription factor ς70: localization of a region involved in the interaction with core RNA polymerase. Biochemistry. 1989;28:7728–7734. doi: 10.1021/bi00445a031. [DOI] [PubMed] [Google Scholar]
- 21.Liberek K, Galitski T P, Zylicz M, Georgopoulos C. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the ς32 transcription factor. Proc Natl Acad Sci USA. 1992;89:3516–3520. doi: 10.1073/pnas.89.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malhotra A, Severinova E, Darst S A. Crystal structure of a ς70 fragment from Escherichia coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 24.McCarty J S, Rudiger S, Schonfeld H-J, Schneider-Mergener J, Nakahigashi K, Yura T, Bukau B. Regulatory region C of the E. coli heat shock transcription factor, ς32, constitutes a DnaK binding site and is conserved among eubacteria. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 25.Nagai H, Yuzawa H, Kanemori M, Yura T. A distinct segment of the ς32 polypeptide is involved in DnaK-mediated negative control of the heat shock response in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:10280–10284. doi: 10.1073/pnas.91.22.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding ς32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- 27.Neidhardt F C, VanBogelen R A, Lau E T. Molecular cloning and expression of a gene that controls the high-temperature regulon of Escherichia coli. J Bacteriol. 1983;153:597–603. doi: 10.1128/jb.153.2.597-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pace C N, Vajados F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richarme G, Kohiyama M. Specificity of the Escherichia coli chaperone DnaK (70-kDa heat shock protein) for hydrophobic amino acids. J Biol Chem. 1993;268:24074–24077. [PubMed] [Google Scholar]
- 30.Shuler M F, Tatti K M, Wade K H, Moran C P., Jr A single amino acid substitution in ςE affects its ability to bind core RNA polymerase. J Bacteriol. 1995;177:3687–3694. doi: 10.1128/jb.177.13.3687-3694.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegele D A, Hu J C, Walter W A, Gross C A. Altered promoter recognition by mutant forms of the ς70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 32.Stragier P, Parsot C, Bouvier J. Two functional domains conserved in major and alternate bacterial sigma factors. FEBS Lett. 1985;187:11–15. doi: 10.1016/0014-5793(85)81203-5. [DOI] [PubMed] [Google Scholar]
- 33.Straus D B, Walter W A, Gross C A. The heat shock response of E. coli is regulated by changes in the concentration of ς32. Nature. 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 34.Tilly K, McKittrick N, Zylicz M, Georgopoulos C. The DnaK protein modulates the heat-shock response of Escherichia coli. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y N, Walter W A, Gross C A. A mutant ς32 with a small deletion in conserved region 3 of ς has reduced affinity for core RNA polymerase. J Bacteriol. 1992;174:5005–5012. doi: 10.1128/jb.174.15.5005-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]