Abstract
Transmembrane proteins have exhibited a significant correlation with glioblastoma multiforme (GBM). The current study elucidates the roles of transmembrane protein 150A (TMEM150A) in GBM. Data on patients with GBM were collected from The Cancer Genome Atlas and Xena databases. The objective was to identify the expression levels of TMEM150A in patients with GBM, and evaluate its diagnostic and prognostic values, accomplished using the receiver operating characteristic and survival analyses. On a cellular level, Cell Counting Kit-8, Wound healing, and Transwell experiments were performed to gauge the impact of TMEM150A on cell growth and migration. The study further investigated the correlation between TMEM150A expression and immune status, along with ribonucleic acid (RNA) modifications in GBM. The findings demonstrated TMEM150A overexpression in the cancerous tissues of patients with GBM, with an area under the curve value of 0.95. TMEM150A overexpression was significantly correlated with poor prognostic indicators. TMEM150A overexpression and isocitrate dehydrogenase (IDH) mutation status were predictive of poor survival time among patients with GBM. In vitro experiments indicated that suppressing TMEM150A expression could inhibit GBM cell proliferation, migration, and invasion. Moreover, TMEM150A overexpression was associated with stromal, immune, and estimate scores, immune cells (such as the T helper (Th) 17 cells, Th2 cells, and regulatory T cells), cell markers, and RNA modifications. Therefore, TMEM150A overexpression might serve as a promising biomarker for predicting poor prognosis in patients with GBM. Inhibiting TMEM150A expression holds the potential for improving the survival time of patients with GBM.
Introduction and preliminaries
Glioblastoma multiforme (GBM) is a highly malignant astrocytoma with limited treatment options. Currently, the preferred approaches for managing GBM involve surgical resection of tumour tissues followed by radiotherapy and chemotherapy. However, the prognosis of patients with GBM patients is significantly poor, with less than 6% of patients surviving beyond 5 years [1]. Improving the survival rate for patients with GBM remains a pressing concern demanding resolution. In recent years, several biomarkers have been identified for their pivotal roles in GBM progression [2–4]. For instance, Shen et al. reported cardiotrophin-like cytokine factor 1 (CLCF1) overexpression in GBM tissues, which was associated with an unfavourable prognosis. Inhibiting CLCF1 expression significantly reduced cell proliferation and migration, induced apoptosis, and prompted cell cycle arrest in GBM [4]. These findings demonstrate the need for identifying new biomarkers to predict the survival time of patients with GBM and improve the overall prognosis of patients with cancer.
Transmembrane proteins serve as channels that control the entry and exit of various substances across cellular membranes. Several investigations have linked transmembrane proteins with cancer [5–8]. For instance, in non-small cell lung cancer (NSCLC), transmembrane protein (TMEM) 229A (TMEM229A) is significantly underexpressed, and this underexpression is associated with poor prognosis in patients with cancer. Conversely, TMEM229A overexpression can inhibit NSCLC cell proliferation, migration, and invasion while promoting E-cadherin overexpression and N-cadherin and matrix metalloproteinase 2 underexpression [8]. Similarly, TMEM168 is overexpressed in GBM and is associated with shorter survival in patients with GBM. Inhibiting TMEM168 expression can impede the viability of GBM cancer cells, induce cell cycle arrest, and enhance apoptosis, which are mechanisms associated with the obstruction of the WNT/β-catenin pathway [7]. Another member of the transmembrane protein family, TMEM150A, plays a role in cell homeostasis by regulating cytokine secretion and transcription. Decreased TMEM150A expression could result in increased lipopolysaccharide (LPS)-induced cytokine secretion [9]. Furthermore, the relationship between ribonucleic acid (RNA) modifications and GBM progression was inseparable [10–12]. For instance, long-chain non-coding RNA LINC00839 was overexpressed in glioma stem cells (GSCs). LINC00839 overexpression was strongly associated with GBM progression and radiation resistance. Methyltransferase-like protein (METTL) 3 could mediate the m6A-modified LINC00839 activation of WNT/β-catenin signalling, thereby enhancing tumour progression and radiation resistance [10]. In addition, a significant correlation exists between the immune microenvironment and GBM progression [13]. Therefore, this study aims to analyse the role of TMEM150A in GBM progression, alongside assessing the relationship between TMEM150A and the immune microenvironment and RNA modifications to provide a new candidate molecule for treating patients with GBM.
Methods
The cancer genome atlas (TCGA) and Xena databases
TCGA stands as an open-access cancer database comprising transcriptional data from patients with cancer. In contrast, the Xena database comprises transcription data from normal tissues of patients. In this study, transcription data was obtained from five normal tissues and 166 cancer tissues of patients with GBM, within the confines of TCGA. In parallel, the Xena database yielded access to transcription data from 1152 normal tissues, sourced from the genome-tissue expression (GTEx) database. Furthermore, data pertaining to TMEM150A expression, clinicopathological features, and prognostic data of the patients with GBM were obtained from TCGA.
The relationship between TMEM150A expression and the clinical features of patients with GBM
TMEM150A expression levels were analysed in normal and cancer tissues. Subsequently, the TMEM150A expression data was integrated with clinical data from patients with GBM, encompassing variables such as age, sex, tumour grade, and survival status. The clinical characteristics of the patient groups were assessed and patients were categorised based on median TMEM150A expression levels, comparing the differences between high- and low-TMEM150A expression groups.
Identification of TMEM150A expression association with diagnosis and patient prognosis
Receiver operating characteristic (ROC) analysis was used to determine the diagnostic significance of TMEM150A in cancerous and normal tissues obtained from patients with GBM. The area under the ROC curve was used as the evaluation index. Additionally, data on TMEM150A expression and prognosis of patients with GBM were merged to examine the association between high- and low-TMEM150A expression levels and overall survival (OS) time, disease-specific survival (DSS), and progression-free interval (PFI) among patients with GBM. The patient groups were categorised based on median TMEM150A expression levels.
Association between TMEM150A expression and survival time in various subgroups of patients with GBM
TMEM150A expression data was combined with clinical characteristics and prognostic information from patients with cancer. This investigation was undertaken to explore the relationship between TMEM150A expression levels and OS, DSS, and PFI of patients with GBM across subgroups based on age, sex, race, isocitrate dehydrogenase (IDH) status, and Karnofsky performance score (KPS). Patient groups were categorised based on median TMEM150A expression levels.
TMEM150A associated nomograms
Univariate Cox regression analysis was performed to assess the correlation between sex, age (≤60 or >60 years), KPS (<80 or ≥80), IDH status, TMEM150A expression level, and poor prognostic indicators in patients with GBM. Variables with a significance level of P<0.05 were further subjected to multivariate Cox regression analysis. Additionally, nomograms were used as a tool to evaluate the prognosis of patients with cancer [14–16]. Thus, nomograms were developed based on the results of the multivariate Cox regression analysis, enabling the prediction of prognosis among patients with GBM.
Tumour immune estimation resource (TIMER) database
The TIMER database, an online cancer immune database, facilitates the visualisation of the correlation between gene expression and cancer immune cells, along with cellular markers. In this study, the TIMER database was used to explore TMEM150A expression across various cancer tissues. This aimed to elucidate the relationship between TMEM150A expression and tumour purity, immune cells, and immune cell markers.
Identification of TMEM150A expression levels in relation to the tumour immune status
The single-sample Gene Set Enrichment Analysis (GSEA) and ESTIMATE methods were used to analyse the tumour microenvironment within the cancerous tissues obtained from patients with GBM sourced from TCGA. TMEM150A expression data and immune cell markers were extracted from these cancer tissues using the Perl programming language, and correlation analysis was performed to investigate the association between TMEM150A expression and the infiltration of various immune cells and specific immune markers. Patients with GBM were classified into two groups based on the median TMEM150A expression to further examine the relationship between TMEM150A expression and immune infiltration.
Gene expression profiling interactive analysis (GEPIA) database
The GEPIA database comprised tissue samples from patients with cancer and healthy individuals obtained from TCGA and GTEx databases. This study used the GEPIA database to establish the correlation between TMEM150A expression and poor prognosis in patients with GBM.
GSEA
Using the CAMOIP database, the cancerous tissues obtained from patients with GBM in TCGA were subjected to GSEA [17]. Additionally, GSEA could elucidate the functions and signalling mechanisms of genes [18, 19]. Therefore, GSEA of the CAMOIP database was performed to investigate the signalling mechanisms of high and low expressions of TMEM150A, with a significance threshold set at P < 0.05.
Identification of the relationship between TMEM150A expression and RNA modifications
The RM2TARGET database systematically collected relevant literature and publicly available RNA modification gene datasets, encompassing informative target gene associations and annotations such as m6A, m1A, m5C, and m7G modifications for human genes. From this repository, the human target gene TMEM150A was specifically identified and the RM2TARGET database was used to identify its associated RNA modifications. Subsequently, using the TIMER database, the correlation between TMEM150A expression levels and RNA modifications was explored. This was corroborated through an examination of RNA sequencing data sourced from tumour tissues of patients with GBM in TCGA.
Cell culture and construction of the TMEM150A inhibition model
GBM U118 cells were cultured in Dulbecco’s Modified Eagle Medium with 10% foetal bovine serum. Following this, the cells were transfected within a six-well plate, and cell proteins and RNA were collected after 24 h. Subsequently, the TMEM150A levels in the control group and the group with inhibited TMEM150A expression were examined using standard reverse transcription-polymerase chain reaction (RT-PCR) and Western blotting [20].
Cell counting kit (CCK)-8
Ninety-six-well plates were arranged with cells in an optimal growth state. Following cell adhesion, a 10μL solution of CCK-8 was added, and the cells were incubated for 1 h to measure cell activity, denoting this time point as 0 h. The cell activity of the control group and the group with inhibited TMEM150A expression was measured and statistically analysed at 24, 48, and 72 h.
Wound healing
The U118 cell suspension was introduced into a six-well plate, and upon cell adhesion, a linear scratch was created across the cell monolayer using the tip of a 200 μL pipette. The subsequent step involved the removal of any cellular debris. Following this, the culture medium was supplemented, and micrographs were taken to document the state. Finally, the process of cellular migration in response to the scratch was closely monitored under a microscope.
Transwell assay
The lower compartment of the Transwell plate was filled with the culture medium, while the Transwell membranes were positioned in the upper compartment. A serum-free suspension of U118 cells was then prepared, and the Transwell apparatus was incubated at a controlled temperature to culture the cells under optimal conditions. Subsequently, the cell invasion capabilities were assessed and compared at distinct time intervals. Following this, the cells were fixed with an appropriate volume of formaldehyde. After thorough washing, staining and visualisation were performed.
Statistical analysis
The Wilcoxon signed-rank test was used to identify differences in TMEM150A expression levels between normal and cancerous tissues. The t-test was used to determine the statistical significance of the growth and migration capabilities of two distinct U118 cell groups. The chi-square test was used to examine the relationship between TMEM150A levels and the clinical characteristics of cancer patients. Additionally, the diagnostic and prognostic capability of TMEM150A was explored using ROC and survival analyses. Spearman correlation analysis was performed to understand the relationship between TMEM150A, immune microenvironment, and RNA modifications. Statistical significance was set at P<0.05.
Results
TMEM150A overexpression was associated with age, diagnosis, and poor prognosis among patients with GBM
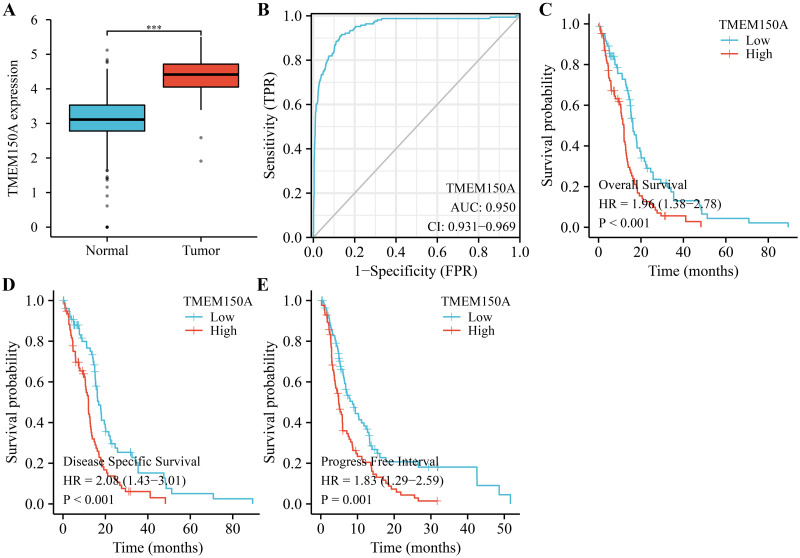
TMEM150A expression significantly increased in GBM tissues compared with normal tissues (Fig 1A). Using ROC analysis, the area under the curve for TMEM150A was 0.95, indicating that TMEM150A overexpression serves as an effective diagnostic biomarker for GBM (Fig 1B). Analysis of data from patients with GBM patient data from TCGA revealed that TMEM150A overexpression was correlated with poor survival rates and disease progression in patients with GBM (Fig 1C–1E). Additionally, data derived from the GEPIA database revealed that TMEM150A overexpression was associated with poor OS and disease-free survival (DFS) in patients with GBM (S1 Fig). Moreover, TMEM150A overexpression levels were associated with age, OS, DSS, and PFI in patients with GBM (Fig 2 and Table 1).
Fig 1. Increased TMEM150A expression level was associated with poor prognosis and diagnosis in patients with GBM.
(A) TMEM150A expression in GBM from the Xena database; (B) the diagnostic value of TMEM150A using ROC analysis; (C-E) the prognostic values of TMEM150A in GBM using survival analysis. Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; ROC, operating characteristic.
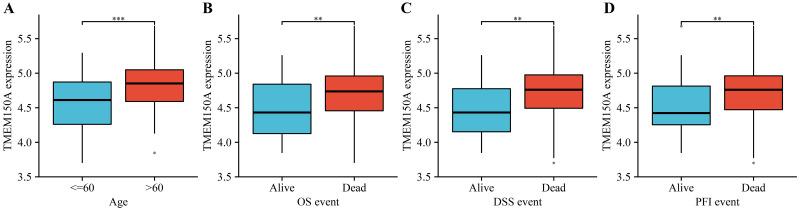
Fig 2. Increased TMEM150A expression level was associated with poor prognosis and age in patients with GBM.
(A) Age; (B) OS event; (C) DSS event; (D) PFI event. Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval.
Table 1. The relationship between TMEM150A levels and clinical indices in patients with GBM.
| Characteristic | TMEM150A | P | |
|---|---|---|---|
| Low expression | High expression | ||
| N | 84 | 84 | |
| Gender | 0.196 | ||
| Female | 34 (20.2%) | 25 (14.9%) | |
| Male | 50 (29.8%) | 59 (35.1%) | |
| Race | 0.925 | ||
| Asian | 3 (1.8%) | 2 (1.2%) | |
| Black or African American | 5 (3%) | 6 (3.6%) | |
| White | 74 (44.6%) | 76 (45.8%) | |
| Age | 0.013 | ||
| < = 60 | 52 (31%) | 35 (20.8%) | |
| >60 | 32 (19%) | 49 (29.2%) | |
| KPS | 0.712 | ||
| <80 | 20 (15.6%) | 16 (12.5%) | |
| > = 80 | 46 (35.9%) | 46 (35.9%) | |
| IDH status | 0.311 | ||
| WT | 70 (43.5%) | 79 (49.1%) | |
| Mut | 8 (5%) | 4 (2.5%) | |
| OS event | 0.077 | ||
| Alive | 21 (12.5%) | 11 (6.5%) | |
| Dead | 63 (37.5%) | 73 (43.5%) | |
| DSS event | 0.013 | ||
| Alive | 24 (15.5%) | 10 (6.5%) | |
| Dead | 54 (34.8%) | 67 (43.2%) | |
| PFI event | 0.011 | ||
| Alive | 23 (13.7%) | 9 (5.4%) | |
| Dead | 61 (36.3%) | 75 (44.6%) | |
Note: TMEM, transmembrane protein; KPS, Karnofsky performance score; GBM, glioblastoma multiforme; OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval.
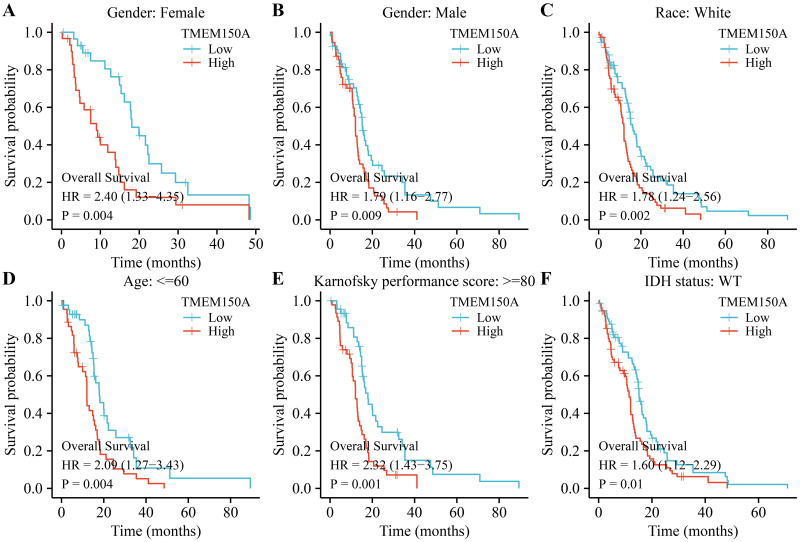
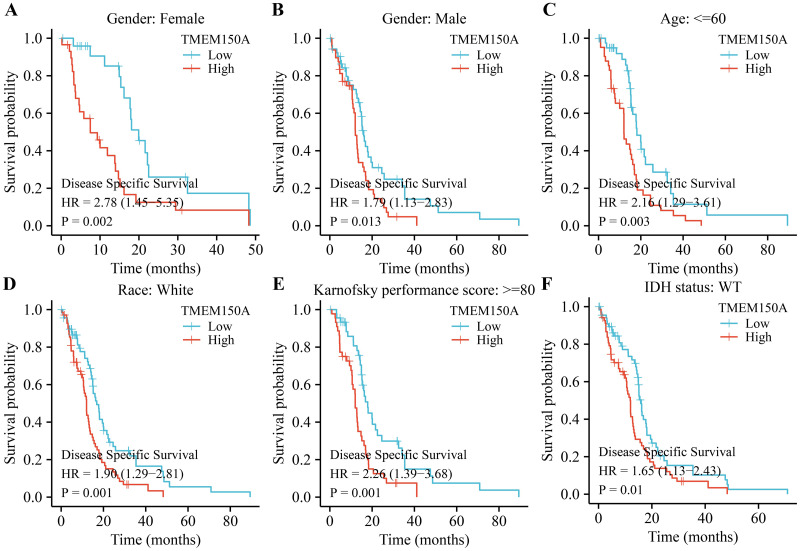
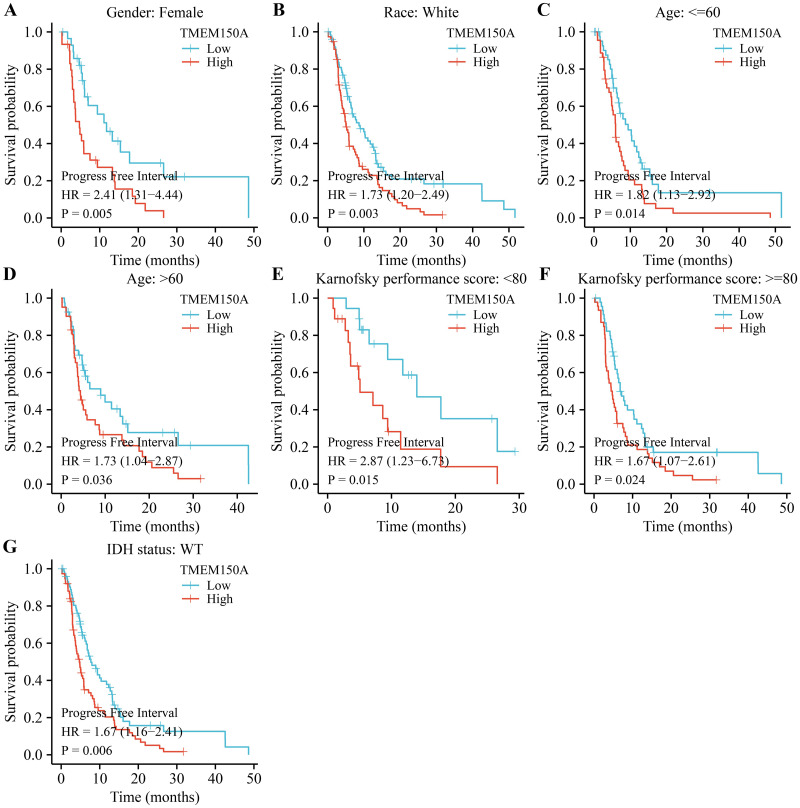
TMEM150A overexpression was associated with poor prognosis specifically in certain subgroups of patients with GBM
Survival analysis revealed that TMEM150A overexpression was associated with poor OS and DSS in male and female patients, regardless of their race, age ≤60, KPS≥80, or IDH wild-type (WT) status in patients with GBM (Figs 3 and 4). TMEM150A overexpression was associated with poor PFI in female and white patients, patients aged <60 years, patients aged >60 years, those with a KPS score of <80, those with a KPS score greater ≥80, and those with an IDH WT status, as presented in Fig 5.
Fig 3. Elevated TMEM150A expression was associated with short OS in a subgroup of patients with GBM.
(A) Female; (B) Male; (C) White; (D) Age ≤60; (E) KPS ≥80; (F) IDH status.Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; OS, overall survival; KPS, Karnofsky performance score; IDH, isocitrate dehydrogenase.
Fig 4. Elevated TMEM150A expression was associated with short DSS in a subgroup of patients with GBM.
(A) Female; (B) Male; (C) Age ≤60; (D) White; (E) KPS ≥80; (F) IDH status. Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; DSS, disease-specific survival; KPS, Karnofsky performance score; IDH, isocitrate dehydrogenase.
Fig 5. Elevated TMEM150A expression was associated with short PFI in a subgroup of patients with GBM.
(A) Female; (B) White; (C) Age ≤60; (D) Age >60; (E) KPS < 80; (F) KPS ≥80; (G) IDH status. Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; PFI, progression-free interval; KPS, Karnofsky performance score; IDH, isocitrate dehydrogenase.
TMEM150A overexpression serves as an indicator of poor prognosis in patients with GBM
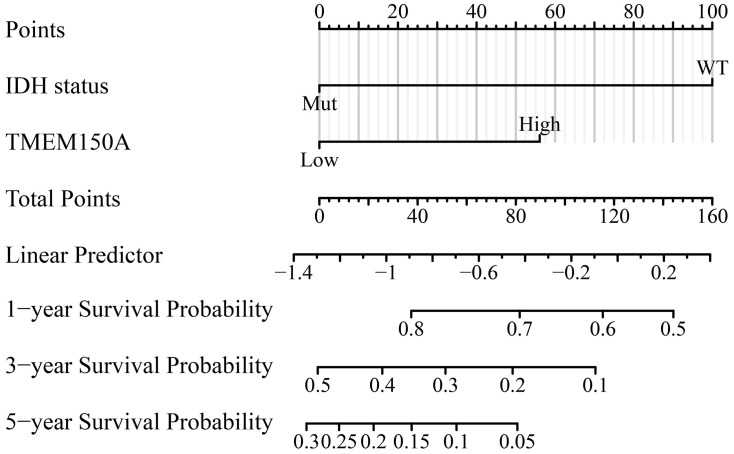
Univariate Cox regression analysis revealed that TMEM150A and IDH WT status were significant predictors of poor OS, DSS, and PFI among patients with GBM, as detailed in Tables 2–4. The outcomes of multivariate Cox regression analysis further confirmed that IDH WT status and TMEM150A overexpression were significant predictors of poor prognosis in patients with GBM (Tables 2–4). The nomograms effectively helped visualise the prognosis and survival outcomes of patients with cancer [14–16]. Therefore, nomograms were constructed for OS, DSS, and PFI in patients with GBM based on the IDH status and TMEM150A expression levels (Fig 6 and S2 Fig).
Table 2. Cox analysis showing the factors influencing OS in patients with GBM.
| Characteristics | N | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | 168 | ||||
| Female | 59 | Reference | |||
| Male | 109 | 1.026 (0.719–1.466) | 0.887 | ||
| Age | 168 | ||||
| < = 60 | 87 | Reference | |||
| >60 | 81 | 1.365 (0.973–1.915) | 0.072 | ||
| KPS | 128 | ||||
| <80 | 36 | Reference | |||
| > = 80 | 92 | 0.838 (0.538–1.305) | 0.434 | ||
| IDH status | 161 | ||||
| WT | 149 | Reference | |||
| Mut | 12 | 0.301 (0.138–0.654) | 0.002 | 0.345 (0.158–0.756) | 0.008 |
| TMEM150A | 168 | ||||
| Low | 84 | Reference | |||
| High | 84 | 1.962 (1.384–2.782) | <0.001 | 1.813 (1.269–2.592) | 0.001 |
Note: KPS, Karnofsky performance score; GBM, glioblastoma multiforme; OS, overall survival.
Table 4. Cox analysis showing the factors influencing the PFI of patients with GBM.
| Characteristics | N | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | 168 | ||||
| Female | 59 | Reference | |||
| Male | 109 | 1.223 (0.855–1.750) | 0.269 | ||
| Age | 168 | ||||
| < = 60 | 87 | Reference | |||
| >60 | 81 | 1.000 (0.710–1.409) | 1.000 | ||
| KPS | 128 | ||||
| <80 | 36 | Reference | |||
| > = 80 | 92 | 1.544 (0.970–2.457) | 0.067 | ||
| IDH status | 161 | ||||
| WT | 149 | Reference | |||
| Mut | 12 | 0.395 (0.183–0.852) | 0.018 | 0.447 (0.206–0.968) | 0.041 |
| TMEM150A | 168 | ||||
| Low | 84 | Reference | |||
| High | 84 | 1.831 (1.294–2.590) | <0.001 | 1.771 (1.237–2.535) | 0.002 |
Note: KPS, Karnofsky performance score; GBM, glioblastoma multiforme; PFI, progression-free interval.
Fig 6. TMEM150A correlating the nomogram in terms of the OS in patients with GBM.
Note: TMEM, transmembrane protein; OS, overall survival; GBM, glioblastoma multiforme.
Table 3. Cox analysis showing the factors influencing DSS of patients with GBM.
| Characteristics | N | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | 155 | ||||
| Female | 54 | Reference | |||
| Male | 101 | 0.975 (0.667–1.425) | 0.894 | ||
| Age | 155 | ||||
| < = 60 | 83 | Reference | |||
| >60 | 72 | 1.340 (0.935–1.920) | 0.111 | ||
| KPS | 119 | ||||
| <80 | 29 | Reference | |||
| > = 80 | 90 | 0.919 (0.552–1.529) | 0.745 | ||
| IDH status | 148 | ||||
| WT | 136 | Reference | |||
| Mut | 12 | 0.315 (0.144–0.686) | 0.004 | 0.370 (0.168–0.814) | 0.013 |
| TMEM150A | 155 | ||||
| Low | 78 | Reference | |||
| High | 77 | 2.076 (1.429–3.014) | <0.001 | 1.922 (1.309–2.822) | <0.001 |
Note: KPS, Karnofsky performance score; GBM, glioblastoma multiforme; DSS, disease-specific survival.
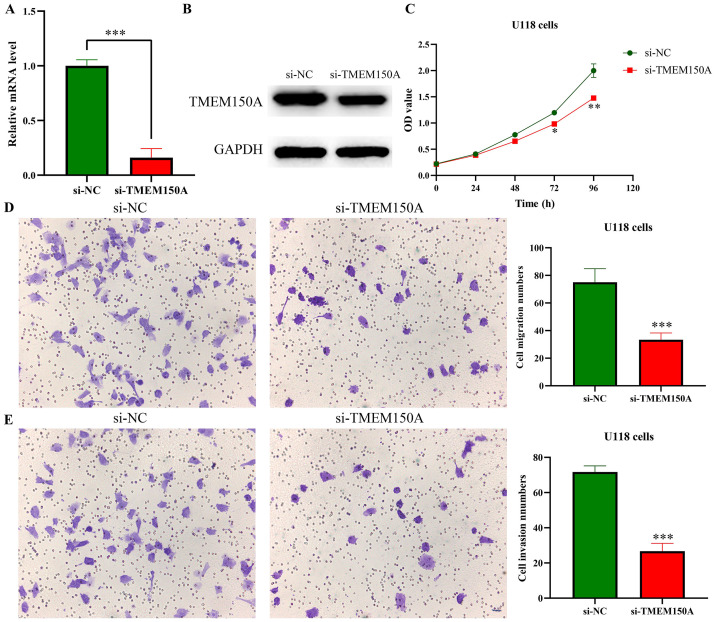
Decreased TMEM150A expression inhibited GBM cell growth and metastasis
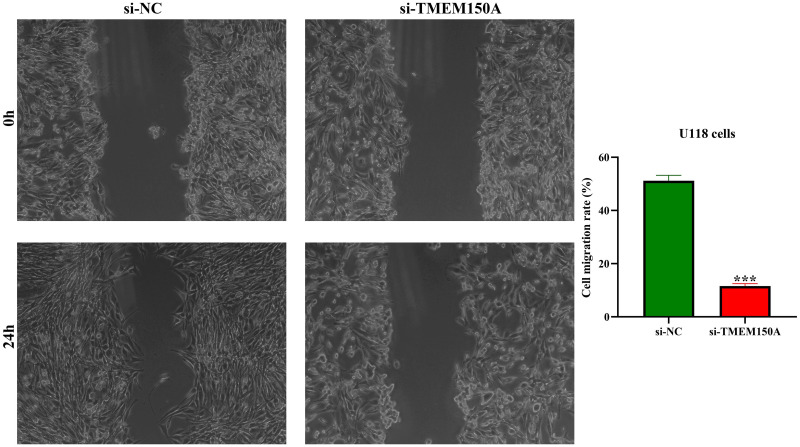
The efficacy of the cell model for inhibiting TMEM150A expression was confirmed through RT-PCR and Western blotting (Fig 7A and 7B). CCK-8 revealed that inhibiting TMEM150A expression could delay GBM cell proliferation (Fig 7C). Furthermore, assessments conducted via wound healing and Transwell experiments demonstrated that inhibiting TMEM150A expression could inhibit GBM cell migration and invasion (Figs 7D–7F and 8).
Fig 7. Decreased TMEM150A expression inhibits cell proliferation, migration, and invasion in GBM using CCK-8 and Transwell assays.
(A-B) Successfully constructed inhibited TMEM150A expression model using RT-PCR and Western blotting; (C) cell proliferation by CCK-8; (D) cell migration by Transwell assay; (E) cell invasion by Transwell assay. Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; CCK-8, Cell Counting Kit-8.
Fig 8. Decreased expression of TMEM150A inhibits cell migration in GBM using wound healing.
Note: TMEM, transmembrane protein.
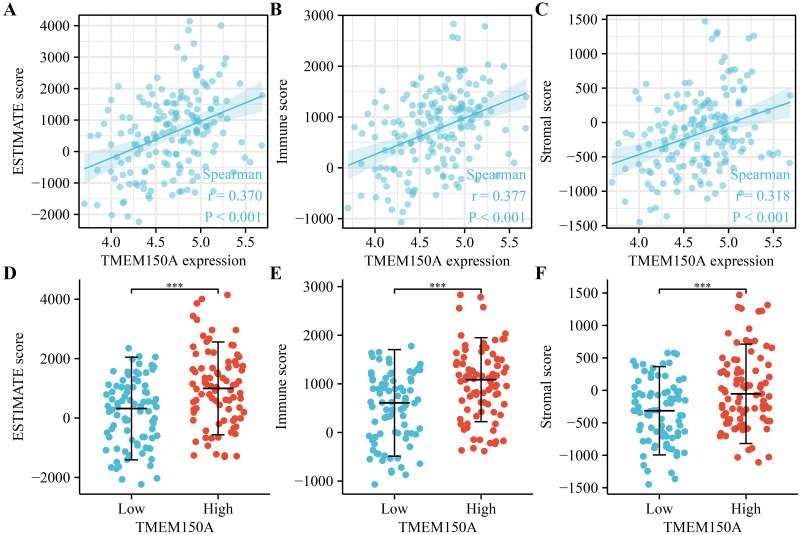
TMEM150A overexpression was associated with the tumour immune microenvironment
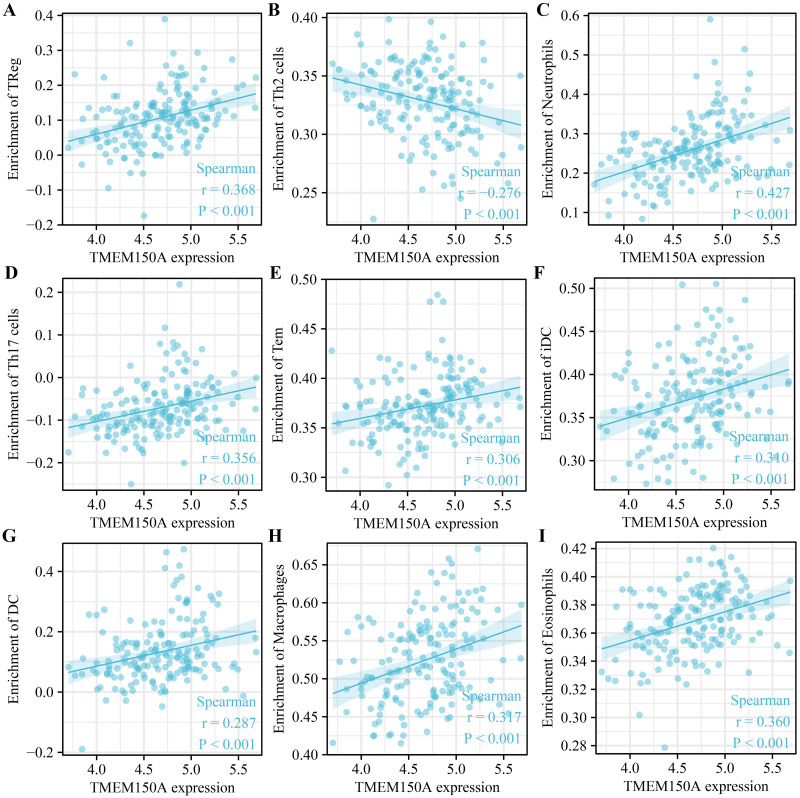
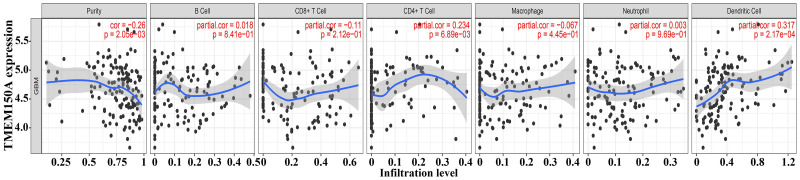
Increased TMEM150A expression within GBM tissues sourced from TCGA was positively correlated with immune, estimate, and stromal scores (Fig 9A–9C). Significant differences were observed between groups with TMEM150A overexpression and low expression (Fig 9D–9F). TMEM150A overexpression was found to be associated with various immune cell types such as T helper (Th) 1 cells, natural killer (NK) cells, NK cells expressing cluster of differentiation (CD) 56 surface antigen in low density (NK CD56dim cells), cytotoxic cells, T cells, activated dendritic cells (aDCs), dendritic cells (DCs), eosinophils, interstitial DCs (iDCs), macrophages, neutrophils, memory T cells (Tems), Th17 cells, Th2 cells, and regulatory T cells (Tregs) (Fig 10). Moreover, the levels of 24 immune cell types between TMEM150A overexpression and low-TMEM150A expression groups in the patients with GBM were showed (S3 Fig). Additionally, the TIMER database revealed a significant correlation between TMEM150A overexpression and the levels of tumour purity, CD4+ T cells, and DCs (Fig 11).
Fig 9. TMEM150A expression was associated with the immune status in GBM.
Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme.
Fig 10. TMEM150A expression was associated with immune cells in GBM using TCGA.
Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; TCGA, The Cancer Genome Atlas.
Fig 11. TMEM150A expression was associated with tumour purity and GBM immune cells in TIMER.
Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; TIMER, Tumour IMmune Estimation Resource.
TMEM150A overexpression was significantly correlated with immune cell markers
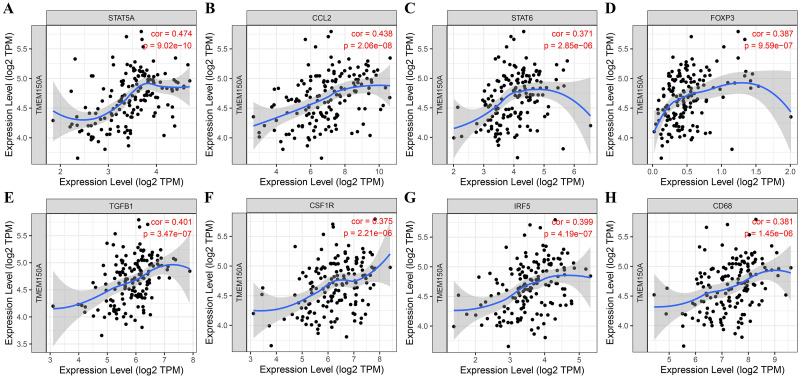
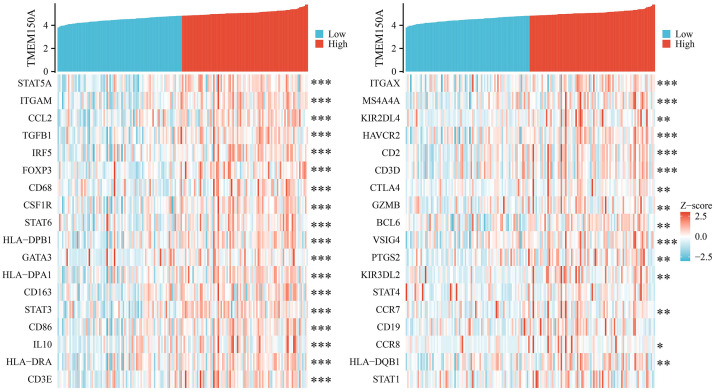
Results from the TIMER database demonstrated that TMEM150A overexpression was significantly correlated with immune cell markers, including signal transducer and activator of transcription (STAT) 5A, integrin alpha M chain (ITGAM), chemokine ligand 2 (CCL2), transforming growth factor beta 1 (TGFB1), interferon regulatory factor 5 (IRF5), forkhead box protein 3 gene (FOXP3), CD68, colony stimulating factor 1 receptor (CSF1R), STAT6, human leukocyte antigen (HLA)-class II histocompatibility antigen, DP(W2) beta chain (DPB1), GATA binding protein 3 (GATA3), HLA-class II DP alpha 1 (DPA1), CD163, STAT3, CD86, interleukin (IL)-10, HLA-class II histocompatibility antigen, DR alpha chain (DRA), CD3-epsilon CD-3E, integrin subunit alpha X (ITGAX), membrane spanning 4-domains A4A (MS4A4A), killer cell immunoglobulin like receptor, two immunoglobulin domains and long cytoplasmic tail 4 (KIR2DL4), hepatitis A virus cellular receptor 2 (HAVCR2), CD2, CD3-delta (CD3D), and cytotoxic T-lymphocyte-associated protein 4 (CTLA4) (Fig 12 and Table 5). Moreover, TCGA verified the relationship between most immune cell markers and TMEM150A levels, as indicated in Fig 13.
Fig 12. TMEM150A expression was associated with GBM immune cell markers in TIMER.
Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; TIMER, Tumour Immune Estimation Resource.
Table 5. TMEM150A expression was associated with GBM cell markers in the TIMER database.
| Markers | Cor | P |
|---|---|---|
| STAT5A | 0.474210258 | 9.02E-10 |
| ITGAM | 0.447512498 | 9.72E-09 |
| CCL2 | 0.438365298 | 2.06E-08 |
| TGFB1 | 0.401350971 | 3.47E-07 |
| IRF5 | 0.398720732 | 4.19E-07 |
| FOXP3 | 0.386906437 | 9.59E-07 |
| CD68 | 0.38083846 | 1.45E-06 |
| CSF1R | 0.374509134 | 2.21E-06 |
| STAT6 | 0.370632464 | 2.85E-06 |
| HLA-DPB1 | 0.366243148 | 3.79E-06 |
| GATA3 | 0.364936711 | 3.52E-06 |
| HLA-DPA1 | 0.335085709 | 2.56E-05 |
| CD163 | 0.323770657 | 4.89E-05 |
| STAT3 | 0.312334982 | 9.18E-05 |
| CD86 | 0.309322772 | 0.000107915 |
| IL10 | 0.307058009 | 0.000113091 |
| HLA-DRA | 0.306390977 | 0.000126114 |
| CD3E | 0.301066838 | 0.000166701 |
| ITGAX | 0.291051827 | 0.000277902 |
| MS4A4A | 0.28283275 | 0.000417141 |
| KIR2DL4 | 0.276280632 | 0.0005464 |
| HAVCR2 | 0.27002332 | 0.000767399 |
| CD2 | 0.266391245 | 0.000907538 |
| CD3D | 0.263283483 | 0.001008514 |
| CTLA4 | 0.259312374 | 0.001208938 |
| GZMB | 0.254419927 | 0.001505702 |
| BCL6 | 0.251082251 | 0.001795947 |
| VSIG4 | 0.243838205 | 0.002447221 |
| PTGS2 | 0.242836369 | 0.002552488 |
| KIR3DL2 | 0.236533159 | 0.003243681 |
| STAT4 | 0.210027073 | 0.009275964 |
| CCR7 | 0.193736346 | 0.016538728 |
| CD19 | 0.188578766 | 0.019572342 |
| CCR8 | 0.186847867 | 0.020742863 |
| HLA-DQB1 | 0.171327383 | 0.034328901 |
| STAT1 | 0.159399166 | 0.049143464 |
Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; TIMER, Tumour IMmune Estimation Resource.
Fig 13. TMEM150A expression was associated with GBM immune cell markers in TCGA.
Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme; TCGA, The Cancer Genome Atlas.
TMEM150A overexpression was associated with RNA modifications
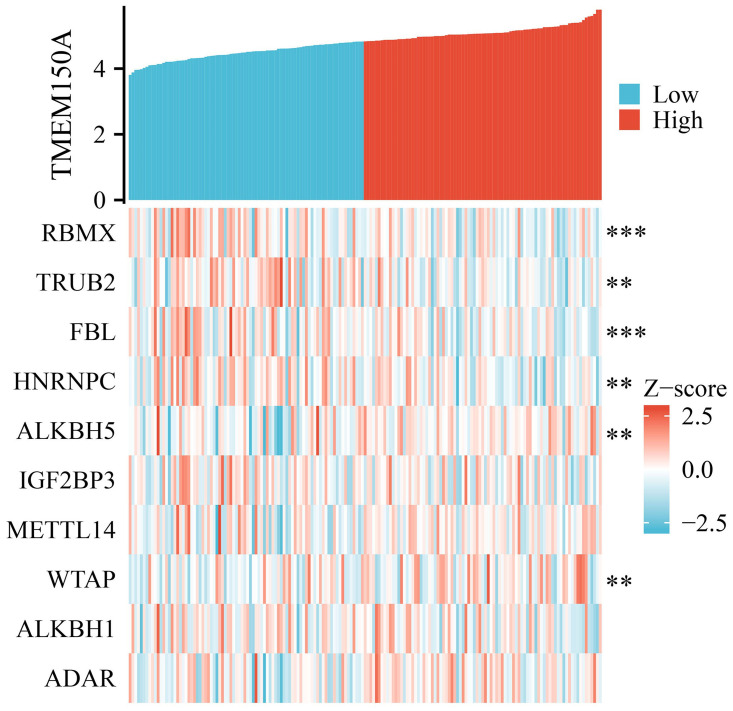
The RM2Target database revealed several RNA modification factors that were associated with TMEM150A, including alkB homolog (ALKBH) 5, METTL16, METTL14, Wilms tumour 1 associated protein (WTAP), adenosine deaminase, adenosine deaminase RNA specific B1, zinc finger CCHC-type containing 4, fat mass and obesity-associated protein, vir-like m6A methyltransferase associate, nucleolar protein 58, heterogeneous nuclear ribonucleoproteins A2/B1, insulin-like growth factor 2 mRNA-binding protein (IGF2BP) 1, YTH N6-methyladenosine RNA binding protein C1, METTL3, zinc finger CCCH-type containing 13, WD repeat domain 4, METTL5, METTL1, YTH N6-methyladenosine RNA binding protein 2, ALKBH1, IGF2BP3, THO complex subunit 4, heterogeneous nuclear ribonucleoproteins C1/C2 (HNRNPC), fibrillarin (FBL), TruB pseudouridine synthase family member 2 (TRUB2), and RNA binding motif protein X-linked (RBMX). Fig 14 and Table 6 reveal that TMEM150A expression levels were correlated with RBMX, TRUB2, FBL, HNRNPC, ALKBH5, and WTAP in TIMER and TCGA databases.
Fig 14. TMEM150A expression was associated with the RNA modification genes of GBM in TCGA.
Note: TMEM, transmembrane protein; RNA, ribonucleic acid; GBM, glioblastoma multiforme; TCGA, The Cancer Genome Atlas.
Table 6. TMEM150A expression was associated with RNA modifications of GBM in the TIMER database.
| Gene | Cor | P |
|---|---|---|
| ALKBH5 | 0.327171538 | 4.04E-05 |
| METT10D | 0.245248817 | 0.002305679 |
| METTL14 | 0.193176792 | 0.016858428 |
| WTAP | 0.175736949 | 0.029790943 |
| ADAR | 0.170787932 | 0.034905916 |
| ADARB1 | 0.154011365 | 0.057403529 |
| ZCCHC4 | 0.104050233 | 0.200330021 |
| FTO | 0.053006849 | 0.514797858 |
| KIAA1429 | 0.019249327 | 0.813101356 |
| NOP58 | -0.017121681 | 0.833442001 |
| HNRNPA2B1 | -0.02793414 | 0.731498583 |
| IGF2BP1 | -0.028988371 | 0.722064809 |
| YTHDC1 | -0.036300645 | 0.655629531 |
| METTL3 | -0.043474328 | 0.593240358 |
| ZC3H13 | -0.04978355 | 0.540712813 |
| WDR4 | -0.058257274 | 0.473999156 |
| METTL5 | -0.065380698 | 0.421586685 |
| METTL1 | -0.13600512 | 0.09366591 |
| YTHDF2 | -0.138678917 | 0.087345835 |
| ALKBH1 | -0.174771823 | 0.030832093 |
| IGF2BP3 | -0.196098535 | 0.015246626 |
| THOC4 | -0.235277365 | 0.003488882 |
| HNRNPC | -0.334053997 | 2.44E-05 |
| FBL | -0.339220377 | 2.01E-05 |
| TRUB2 | -0.342497286 | 1.66E-05 |
| RBMX | -0.393091686 | 6.24E-07 |
Note: TMEM, transmembrane; RNA, ribonucleic acid; GBM, glioblastoma multiforme; TIMER, Tumour IMmune Estimation Resource.
TMEM150A was involved in the signalling mechanisms of GBM progression
GSEA revealed that TMEM150A was associated with several key signalling pathways such as IL-17, tumour necrosis factor (TNF), nuclear factor-kappa B (NF-κB), nucleotide oligomerisation domain-like receptor, chemokine, Toll-like receptor, Th17 cell differentiation, cell cycle, B cell receptor, Janus kinase/STAT (JAK/STAT), cyclic adenosine monophosphate (cAMP), Th1 and Th2 cell differentiation, ferroptosis, deoxyribonucleotide replication, calcium, and T cell receptor pathways (Table 7).
Table 7. Signalling mechanisms involved in TMEM150A expression in GBM.
| Description | NES | P |
|---|---|---|
| Viral protein interaction with cytokine and cytokine receptor | 2.918131509 | 2.65E-09 |
| IL-17 signaling pathway | 2.691572812 | 2.65E-09 |
| Hematopoietic cell lineage | 2.700735102 | 2.65E-09 |
| Rheumatoid arthritis | 2.631918951 | 2.65E-09 |
| Leishmaniasis | 2.507757236 | 2.65E-09 |
| TNF signaling pathway | 2.608121523 | 2.65E-09 |
| NF-kappa B signaling pathway | 2.472338151 | 2.65E-09 |
| Osteoclast differentiation | 2.516315394 | 2.65E-09 |
| Cytokine-cytokine receptor interaction | 2.690593331 | 2.65E-09 |
| NOD-like receptor signaling pathway | 2.41667302 | 2.65E-09 |
| Tuberculosis | 2.397307684 | 2.65E-09 |
| Chemokine signaling pathway | 2.365771712 | 2.65E-09 |
| Phagosome | 2.277445328 | 2.65E-09 |
| Pertussis | 2.453274141 | 4.28E-09 |
| Coronavirus disease—COVID-19 | 2.109315771 | 6.66E-09 |
| Lipid and atherosclerosis | 2.092986955 | 1.54E-08 |
| Staphylococcus aureus infection | 2.360198294 | 2.84E-08 |
| Toll-like receptor signaling pathway | 2.308287324 | 4.07E-08 |
| Th17 cell differentiation | 2.274581192 | 1.01E-07 |
| Inflammatory bowel disease | 2.408088779 | 1.35E-07 |
| Influenza A | 2.16671301 | 1.53E-07 |
| Legionellosis | 2.357755156 | 3.94E-07 |
| Asthma | 2.242844292 | 1.34E-06 |
| Neuroactive ligand-receptor interaction | -1.897368767 | 1.74E-06 |
| Allograft rejection | 2.249788516 | 1.85E-06 |
| Cell cycle | -2.044202048 | 1.85E-06 |
| Neutrophil extracellular trap formation | 2.116953929 | 2.37E-06 |
| Synaptic vesicle cycle | -2.139161712 | 4.58E-06 |
| Toxoplasmosis | 2.033618751 | 1.04E-05 |
| Intestinal immune network for IgA production | 2.227591328 | 1.04E-05 |
| Complement and coagulation cascades | 2.116809353 | 2.34E-05 |
| Systemic lupus erythematosus | 2.139565241 | 5.32E-05 |
| C-type lectin receptor signaling pathway | 2.025220109 | 5.32E-05 |
| B cell receptor signaling pathway | 1.99992968 | 6.93E-05 |
| Malaria | 2.096515564 | 8.85E-05 |
| Cholesterol metabolism | 2.076531232 | 0.000119109 |
| Necroptosis | 1.877213863 | 0.00020778 |
| Type I diabetes mellitus | 2.075710698 | 0.000240009 |
| JAK-STAT signaling pathway | 1.838299559 | 0.000240009 |
| Graft-versus-host disease | 2.053999416 | 0.000320573 |
| Kaposi sarcoma-associated herpesvirus infection | 1.756135432 | 0.000374 |
| Amoebiasis | 1.831397893 | 0.00071861 |
| Arachidonic acid metabolism | 2.035296477 | 0.00071861 |
| Salmonella infection | 1.64750994 | 0.000916268 |
| Nicotine addiction | -1.943873936 | 0.000953128 |
| Glutamatergic synapse | -1.792271003 | 0.00095578 |
| GABAergic synapse | -1.810799653 | 0.001002667 |
| Epstein-Barr virus infection | 1.633550379 | 0.001002673 |
| Retrograde endocannabinoid signaling | -1.703743542 | 0.001051722 |
| cAMP signaling pathway | -1.63403988 | 0.001402037 |
| Yersinia infection | 1.739903151 | 0.00155358 |
| Morphine addiction | -1.757092293 | 0.00179734 |
| Chagas disease | 1.807735773 | 0.002006058 |
| Primary immunodeficiency | 1.902723023 | 0.002982821 |
| Th1 and Th2 cell differentiation | 1.801444316 | 0.003156869 |
| Shigellosis | 1.570944177 | 0.003671997 |
| Pathogenic Escherichia coli infection | 1.552440012 | 0.003698618 |
| Fluid shear stress and atherosclerosis | 1.664620804 | 0.004758112 |
| Lysosome | 1.651441043 | 0.004867696 |
| Epithelial cell signaling in Helicobacter pylori infection | 1.76135419 | 0.005133051 |
| MicroRNAs in cancer | -1.606615384 | 0.005488737 |
| Insulin secretion | -1.700088671 | 0.005488737 |
| Cytosolic DNA-sensing pathway | 1.840016283 | 0.006573315 |
| Fc epsilon RI signaling pathway | 1.728539303 | 0.006989543 |
| Chemical carcinogenesis—DNA adducts | 1.807477135 | 0.007266373 |
| Human T-cell leukemia virus 1 infection | 1.482135158 | 0.009919705 |
| Ferroptosis | 1.75844298 | 0.01045654 |
| Cholinergic synapse | -1.615373511 | 0.01143614 |
| Leukocyte transendothelial migration | 1.637821436 | 0.012136711 |
| alpha-Linolenic acid metabolism | 1.75619479 | 0.012660616 |
| Fanconi anemia pathway | -1.669390129 | 0.013806105 |
| Regulation of lipolysis in adipocytes | -1.711142378 | 0.013948814 |
| Cell adhesion molecules | 1.536562471 | 0.016724539 |
| Antigen processing and presentation | 1.682618797 | 0.016724539 |
| Dopaminergic synapse | -1.519581617 | 0.01732848 |
| Axon guidance | -1.449983659 | 0.01732848 |
| DNA replication | -1.759069927 | 0.01732848 |
| Taste transduction | -1.599031028 | 0.018329191 |
| Histidine metabolism | 1.74684501 | 0.018329191 |
| Spliceosome | -1.517631277 | 0.018785267 |
| Chemical carcinogenesis—reactive oxygen species | 1.423267802 | 0.021412104 |
| Measles | 1.533851005 | 0.022087845 |
| Viral myocarditis | 1.657168464 | 0.022204176 |
| Antifolate resistance | 1.731641631 | 0.02242012 |
| Calcium signaling pathway | -1.399632268 | 0.024309667 |
| Circadian entrainment | -1.531725405 | 0.030308598 |
| Progesterone-mediated oocyte maturation | -1.514185109 | 0.030866525 |
| RIG-I-like receptor signaling pathway | 1.658796131 | 0.035614091 |
| Autoimmune thyroid disease | 1.674049819 | 0.03658195 |
| Arrhythmogenic right ventricular cardiomyopathy | -1.484657271 | 0.040558878 |
| Glutathione metabolism | 1.630019787 | 0.042215687 |
| Cushing syndrome | -1.437641372 | 0.044865218 |
| Linoleic acid metabolism | 1.606399927 | 0.046362916 |
Note: TMEM, transmembrane protein; GBM, glioblastoma multiforme.
Discussion
GBM is characterised by its high malignancy and frequent recurrence, even after surgery, resulting in poor patient prognosis. The survival duration of patients with GBM depends on various factors, with less than 20% surviving beyond 2 years with surgery-based comprehensive treatment. Targeted therapies have shown promise in treating these cancer types [21]. Several studies have demonstrated a significant correlation between glioblastoma progression and transmembrane proteins [5–8]. For instance, TMEM230 expression levels are crucial in preserving normal vascular structural integrity and promoting vascular network formation. Conversely, TMEM230 downregulation could result in compromised cell migration, matrix adhesion, and delivery in GBM. Increased TMEM230 levels might promote cell migration, extracellular scaffold remodelling, vascular hyperplasia, and abnormal vascular formation in GBM. Targeting TMEM230 could inhibit GBM cell proliferation, tumour-driven angiogenesis, and antiangiogenic therapies [5]. In contrast, decreased TMEM150A expression could result in LPS-induced cytokine secretion, indicating that TMEM150A could play a crucial role in cell homeostasis [9]. However, its implications within GBM have not been revealed. Our study has uncovered that TMEM150A expression was significantly elevated in GBM and serves as a diagnostic marker. TMEM150A overexpression was associated with poor OS, DFS, DSS, and PFI in patients with GBM, which was consistent in the subgroup analysis. Cox analysis revealed that TMEM150A overexpression was an independent risk factor for poor survival in patients with GBM. Furthermore, prognostic nomograms related to TMEM150A expression were associated with the prognosis of patients with cancer. Our findings suggest that TMEM150A overexpression could potentially serve as a prognostic biomarker for patients with GBM.
It was well established that IL-17, TNF, NF-κB, Toll-like receptor, cell cycle, JAK-STAT, cAMP, ferroptosis, and others are closely related mechanisms of cancer [22–29]. For instance, Jiang et al. reported that circKPNB1 was overexpressed in GBM and was correlated with poor survival time. Their findings revealed that the TNF-α/NF-κB signalling mechanism resulted in circKPNB1 overexpression, consequently promoting the viability, proliferation, invasion, and stemness of GBM stem cells [23]. Similarly, Zhang et al. reported that the inhibition of COPI coat complex subunit zeta 1 (COPZ1) expression inhibited GBM cell proliferation and tumour formation in nude mice. COPZ1 knockdown resulted in increased nuclear receptor coactivator 4, ferritin degradation, and elevated intracellular ferrous levels, resulting in ferroptosis [8]. It was observed that inhibiting TMEM150A expression could delay U118 cell proliferation, migration, and invasion in GBM. Additionally, TMEM150A was strongly associated with IL-17, TNF, NF-κB, Toll-like receptor, cell cycle, JAK-STAT, cAMP, ferroptosis, and other pathways. However, our study did not investigate the relationship between TMEM150A and these mechanisms, which warrants further exploration in the future.
Diverse strategies exist for treating patients with cancer, and among these, immunotherapy has emerged as a crucial aspect of cancer treatment [30, 31]. This study analysed the relationship between TMEM150A overexpression and the immune microenvironment in GBM. Our findings revealed that TMEM150A overexpression in GBM tissues was strongly correlated with immune, estimate, and stromal scores, and significantly correlated with tumour purity, Th1 cells, NK cells, NK CD56dim cells, cytotoxic cells, CD4+ T cells, T cells, aDCs, DCs, eosinophils, iDCs, macrophages, neutrophils, Tmems, Th17 cells, Th2 cells, and Tregs. Moreover, TMEM150A expression levels were correlated with immune cell markers such as STAT5A, ITGAM, CCL2, TGFB1, IRF5, FOXP3, CD68, CSF1R, STAT6, HLA-DPB1, GATA3, HLA-DPA1, CD163, STAT3, CD86, IL10, HLA-DRA, CD3-E, ITGAX, MS4A4A, KIR2DL4, HAVCR2, CD2, CD3-D, CTLA4, and others. Additionally, RNA modification genes, such as RBMX, HNRNPC, ALKBH5, and WTAP, play a crucial role in cancer biology [32–38]. For instance, the inhibition of ALKBH5 expression in GSCs significantly down-regulated stem cell survival and invasion after irradiation and mediated the radioresistant properties of stem cells [33]. Similarly, long non-coding RNA SRY-box 2 (SOX2) overlapping transcript (SOX2OT) expression levels were significantly increased in temozolomide (TMZ)-resistant cells and tissue samples, concomitant with GBM progression. Increased SOX2OT levels were closely associated with recurrence risk and poor prognosis. Down-regulating SOX2OT expression could inhibit cell proliferation, promote cell apoptosis, and increase sensitivity to TMZ in GBM. SOX2OT could recruit ALKBH5 and ALKBH6 to bind to SOX2 and demethylate the SOX2 transcript and subsequent SOX2 up-regulation, thereby regulating tumour cell apoptosis, cell proliferation, and TMZ resistance [34]. Our study revealed that TMEM150A expression levels were significantly correlated with RNA modification genes, namely RBMX, TRUB2, FBL, HNRNPC, ALKBH5, and WTAP, suggesting that TMEM150A plays crucial biological roles in GBM progression.
This study revealed that TMEM150A plays a crucial role in GBM and offers significant advantages concerning high reliability and large sample sizes in GBM data. However, moving forward, the verification of TMEM150A expression in GBM tissues through clinical sampling remains a crucial avenue to explore, thereby substantiating its potential clinical significance. Furthermore, the establishment of cell models designed to inhibit TMEM150A expression to investigate its roles and signalling mechanisms involved in GBM cell growth and migration. In summary, TMEM150A was overexpressed in GBM tissues and was significantly correlated with diagnosis, poor prognosis, immune status, and RNA modifications in GBM. Inhibition of TMEM150A expression could inhibit GBM cell proliferation, migration, and invasion. Thus, TMEM150A overexpression could be a potential biomarker for poor prognosis in patients with GBM.
Conclusions
TMEM150A overexpression was associated with diagnosis, poor prognosis, immune status, and RNA modifications in GBM. Inhibiting TMEM150A expression could inhibit GBM cell proliferation, migration, and invasion. TMEM150A might serve as a biomarker of poor prognosis in patients with GBM.
Supporting information
Note: TMEM, transmembrane protein; OS, overall survival; DFS, disease-free survival; GBM, glioblastoma multiforme.
(TIF)
Note: TMEM, transmembrane protein; DSS, disease-specific survival; PFI, progression-free interval; GBM, glioblastoma multiforme.
(TIF)
Note: TMEM, transmembrane protein.
(TIF)
Acknowledgments
Thank you for the help and support of Xiantao Academic Website.
Data Availability
Publicly available datasets can be accessed in TCGA (https://www.cancer.gov/) and XENA (http://xena.ucsc.edu/) databases.
Funding Statement
The present study was financially supported by the college-level youth talent project (Sinopharm Dongfeng General Hospital) (2022Q11).
References
- 1.Liu D, Wang X, Liu Y, Li C, Zhang Z, Lv P. Actin-Binding LIM 1 (ABLIM1) Inhibits Glioblastoma Progression and Serves as a Novel Prognostic Biomarker. Dis Markers. 2022; 2022: 9516808. doi: 10.1155/2022/9516808 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Wang X, Xu L, Lin Y, Zhang J, et al. Low expression of CDHR1 is an independent unfavorable prognostic factor in glioma. J Cancer. 2021; 12(17): 5193–5205. doi: 10.7150/jca.59948 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng S, Zhou C, Yang DH, Xu LS, Yang HJ, Xu MH, et al. LEF1-AS1 is implicated in the malignant development of glioblastoma via sponging miR-543 to upregulate EN2. Brain Res. 2020; 1736: 146781. doi: 10.1016/j.brainres.2020.146781 . [DOI] [PubMed] [Google Scholar]
- 4.Shen SH, Guo JF, Huang J, Zhang Q, Cui Y. Bromodomain-containing protein 4 activates cardiotrophin-like cytokine factor 1, an unfavorable prognostic biomarker, and promotes glioblastoma in vitro. Ann Transl Med. 2022; 10(8): 475. doi: 10.21037/atm-22-1164 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocola C, Magnaghi V, Abeni E, Pelucchi P, Martino V, Vilardo L, et al. Transmembrane Protein TMEM230, a Target of Glioblastoma Therapy. Front Cell Neurosci. 2021; 15: 703431. doi: 10.3389/fncel.2021.703431 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiraishi T, Ikeda K, Tsukada Y, Nishizawa Y, Sasaki T, Ito M, et al. High expression of TMEM180, a novel tumour marker, is associated with poor survival in stage III colorectal cancer. BMC Cancer. 2021; 21(1): 302. doi: 10.1186/s12885-021-08046-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Su Z, Ding Q, Shen L, Nie X, Pan X, et al. Inhibition of Proliferation by Knockdown of Transmembrane (TMEM) 168 in Glioblastoma Cells via Suppression of Wnt/β-Catenin Pathway. Oncol Res. 2019; 27(7): 819–826. doi: 10.3727/096504018X15478559215014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, He Y, Jiang Y, Bao Y, Chen Q, Xie D, et al. TMEM229A suppresses non‑small cell lung cancer progression via inactivating the ERK pathway. Oncol Rep. 2021; 46(2): 176. doi: 10.3892/or.2021.8127 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romanet JL, Cupo KL, Yoder JA. Knockdown of Transmembrane Protein 150A (TMEM150A) Results in Increased Production of Multiple Cytokines. J Interferon Cytokine Res. 2022; 42(7): 336–342. doi: 10.1089/jir.2022.0063 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin J, Ding F, Cheng Z, Ge X, Li Y, Zeng A, et al. METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling. Cell Death Dis. 2023; 14(7): 417. doi: 10.1038/s41419-023-05933-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Zhang P, Dong X, Yuan J, Li Y, et al. METTL3 knockdown promotes temozolomide sensitivity of glioma stem cells via decreasing MGMT and APNG mRNA stability. Cell Death Discov. 2023; 9(1): 22. doi: 10.1038/s41420-023-01327-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Wang YL, Qiu K, Cao YQ, Zhang FJ, Zhao HB, et al. YTHDF2 promotes temozolomide resistance in glioblastoma by activation of the Akt and NF-κB signalling pathways via inhibiting EPHB3 and TNFAIP3. Clin Transl Immunology. 2022; 11(5): e1393. doi: 10.1002/cti2.1393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litak J, Mazurek M, Grochowski C, Kamieniak P, Roliński J. PD-L1/PD-1 Axis in Glioblastoma Multiforme. Int J Mol Sci. 2019; 20(21): 5347. doi: 10.3390/ijms20215347 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q, Wu CY, Jiang N, Tong S, Wan JH, Xiao XY, et al. Downregulation of T-cell cytotoxic marker IL18R1 promotes cancer proliferation and migration and is associated with dismal prognosis and immunity in lung squamous cell carcinoma. Front Immunol. 2022; 13: 986447. doi: 10.3389/fimmu.2022.986447 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan F, Cong Z, Cai X, Zhu J, Yuan L, Wang Y, et al. BACH1 as a potential target for immunotherapy in glioblastomas. Int Immunopharmacol. 2022; 103: 108451. doi: 10.1016/j.intimp.2021.108451 . [DOI] [PubMed] [Google Scholar]
- 16.Shi K, Liu XL, Guo Q, Zhang YQ, Fan ST, Dai L, et al. TMEM41A overexpression correlates with poor prognosis and immune alterations in patients with endometrial carcinoma. PLoS One. 2023; 18(7): e0285817. doi: 10.1371/journal.pone.0285817 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin A, Qi C, Wei T, Li M, Cheng Q, Liu Z, et al. CAMOIP: a web server for comprehensive analysis on multi-omics of immunotherapy in pan-cancer. Brief Bioinform. 2022; 23(3): bbac129. doi: 10.1093/bib/bbac129 . [DOI] [PubMed] [Google Scholar]
- 18.Jiang N, Guo Q, Luo Q. Inhibition of ITGB1-DT expression delays the growth and migration of stomach adenocarcinoma and improves the prognosis of cancer patients using the bioinformatics and cell model analysis. J Gastrointest Oncol. 2022; 13(2): 615–629. doi: 10.21037/jgo-22-233 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Q, Wang SH, Ji YM, Tong S, Li D, Ding XC, et al. The Roles and Mechanisms of TRAT1 in the Progression of Non-Small Cell Lung Cancer. Curr Med Sci. 2022; 42(6): 1186–1200. doi: 10.1007/s11596-022-2625-1 . [DOI] [PubMed] [Google Scholar]
- 20.Liu XS, Zhou LM, Yuan LL, Gao Y, Kui XY, Liu XY, et al. NPM1 Is a Prognostic Biomarker Involved in Immune Infiltration of Lung Adenocarcinoma and Associated With m6A Modification and Glycolysis. Front Immunol. 2021; 12: 724741. doi: 10.3389/fimmu.2021.724741 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K, Wang X, Yang L, Chen Z. The Anti-PD-1/PD-L1 Immunotherapy for Gastric Esophageal Cancer: A Systematic Review and Meta-Analysis and Literature Review. Cancer Control. 2021; 28: 1073274821997430. doi: 10.1177/1073274821997430 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Q, Diao S, Wang Q, Zhu C, Sun X, Yin B, et al. IL-17A promotes cell migration and invasion of glioblastoma cells via activation of PI3K/AKT signalling pathway. J Cell Mol Med. 2019; 23(1): 357–369. doi: 10.1111/jcmm.13938 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Zhao J, Liu Y, Hu J, Gao L, Wang H, et al. CircKPNB1 mediates a positive feedback loop and promotes the malignant phenotypes of GSCs via TNF-α/NF-κB signaling. Cell Death Dis. 2022; 13(8): 697. doi: 10.1038/s41419-022-05149-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Yan J, Huang L, Araujo C, Peng J, Gao L, et al. INT-777 attenuates NLRP3-ASC inflammasome-mediated neuroinflammation via TGR5/cAMP/PKA signaling pathway after subarachnoid hemorrhage in rats. Brain Behav Immun. 2021; 91: 587–600. doi: 10.1016/j.bbi.2020.09.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moretti IF, Lerario AM, Trombetta-Lima M, Sola PR, da Silva Soares R, Oba-Shinjo SM, et al. Late p65 nuclear translocation in glioblastoma cells indicates non-canonical TLR4 signaling and activation of DNA repair genes. Sci Rep. 2021; 11(1): 1333. doi: 10.1038/s41598-020-79356-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z, Zhong Y, Chen J, Sun S, Liu W, Han Y, et al. Disruption of β-catenin-mediated negative feedback reinforces cAMP-induced neuronal differentiation in glioma stem cells. Cell Death Dis. 2022. May 24;13(5):493. doi: 10.1038/s41419-022-04957-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Zhu M, Zou X, Mao Y, Niu J, Jiang J, et al. CCL2-targeted ginkgolic acid exerts anti-glioblastoma effects by inhibiting the JAK3-STAT1/PI3K-AKT signaling pathway. Life Sci. 2022; 311(Pt B): 121174. doi: 10.1016/j.lfs.2022.121174 . [DOI] [PubMed] [Google Scholar]
- 28.Visa A, Alza L, Cantí C, Herreros J. Tetralol derivative NNC-55-0396 induces glioblastoma cell death by activating IRE1α, JNK1 and calcium signaling. Biomed Pharmacother. 2022; 149: 112881. doi: 10.1016/j.biopha.2022.112881 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Kong Y, Ma Y, Ni S, Wikerholmen T, Xi K, et al. Loss of COPZ1 induces NCOA4 mediated autophagy and ferroptosis in glioblastoma cell lines. Oncogene. 2021; 40(8): 1425–1439. doi: 10.1038/s41388-020-01622-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YQ, Yuan Y, Zhang J, Lin CY, Guo JL, Liu HS, et al. Evaluation of the roles and regulatory mechanisms of PD-1 target molecules in NSCLC progression. Ann Transl Med. 2021; 9(14): 1168. doi: 10.21037/atm-21-2963 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bausart M, Préat V, Malfanti A. Immunotherapy for glioblastoma: the promise of combination strategies. J Exp Clin Cancer Res. 2022; 41(1): 35. doi: 10.1186/s13046-022-02251-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Geng X, Li Q, Xu J, Tan Y, Xiao M, Song J, Liu F, Fang C, Wang H. m6A modification in RNA: biogenesis, functions and roles in gliomas. J Exp Clin Cancer Res. 2020. Sep 17;39(1):192. doi: 10.1186/s13046-020-01706-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowalski-Chauvel A, Lacore MG, Arnauduc F, Delmas C, Toulas C, Cohen-Jonathan-Moyal E, et al. The m6A RNA Demethylase ALKBH5 Promotes Radioresistance and Invasion Capability of Glioma Stem Cells. Cancers (Basel). 2020; 13(1): 40. doi: 10.3390/cancers13010040 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B, Zhou J, Wang C, Chi Y, Wei Q, Fu Z, et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020; 11(5): 384. doi: 10.1038/s41419-020-2540-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Q, Zeng P, Zhou X, Zhao X, Chen R, Qiao J, et al. RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. Oncogene. 2021; 40(15): 2635–2650. doi: 10.1038/s41388-021-01666-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang N, Liu L, Liu X, Chen Y, Lu J, Wang Z. hnRNPC Promotes Malignancy in Pancreatic Cancer through Stabilization of IQGAP3. Biomed Res Int. 2022; 2022: 6319685. doi: 10.1155/2022/6319685 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D, Li K, Zhang W, Yang KW, Mu DA, Jiang GJ, et al. The m6A/m5C/m1A Regulated Gene Signature Predicts the Prognosis and Correlates with the Immune Status of Hepatocellular Carcinoma. Front Immunol. 2022; 13: 918140. doi: 10.3389/fimmu.2022.918140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019; 18(1): 127. doi: 10.1186/s12943-019-1053-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: TMEM, transmembrane protein; OS, overall survival; DFS, disease-free survival; GBM, glioblastoma multiforme.
(TIF)
Note: TMEM, transmembrane protein; DSS, disease-specific survival; PFI, progression-free interval; GBM, glioblastoma multiforme.
(TIF)
Note: TMEM, transmembrane protein.
(TIF)
Data Availability Statement
Publicly available datasets can be accessed in TCGA (https://www.cancer.gov/) and XENA (http://xena.ucsc.edu/) databases.