Abstract
The metabotropic glutamate receptors (mGluRs) are family C, dimeric G protein–coupled receptors (GPCRs), which play critical roles in synaptic transmission. Despite an increasing appreciation of the molecular diversity of this family, how distinct mGluR subtypes are regulated remains poorly understood. We reveal that different group II/III mGluR subtypes show markedly different beta-arrestin (β-arr) coupling and endocytic trafficking. While mGluR2 is resistant to internalization and mGluR3 shows transient β-arr coupling, which enables endocytosis and recycling, mGluR8 and β-arr form stable complexes, which leads to efficient lysosomal targeting and degradation. Using chimeras and mutagenesis, we pinpoint carboxyl-terminal domain regions that control β-arr coupling and trafficking, including the identification of an mGluR8 splice variant with impaired internalization. We then use a battery of high-resolution fluorescence assays to find that heterodimerization further expands the diversity of mGluR regulation. Together, this work provides insight into the relationship between GPCR/β-arr complex formation and trafficking while revealing diversity and intricacy in the regulation of mGluRs.
Homo- and hetero-dimeric mGluR complexes show diverse modes of regulation by beta-arrestins.
INTRODUCTION
Cells respond to a diverse array of extracellular signals to mediate intracellular signaling pathways that, in turn, control physiological processes. Beyond the breadth of the extracellular signals themselves, further diversity exists in the corresponding repertoire of membrane receptors. Often, families of receptors evolve to respond to the same ligand with a range of responses. In the case of the metabotropic glutamate receptors (mGluRs), eight different receptor subtypes respond to the excitatory neurotransmitter glutamate (1). The mGluRs are family C G protein–coupled receptors (GPCRs), which can signal via either the Gɑq (group I: mGluR1 and mGluR5) or Gɑi/o (group II: mGluR2 and mGluR3; group III: mGluR4, mGluR6, mGluR7, and mGluR8) families of heterotrimeric G proteins. Beyond differential signaling to broad classes of G protein pathways, diversity exists in the range of glutamate concentrations sensed by different subtypes, as well as their basal activity and cooperativity (2, 3). Furthermore, the family of eight mGluRs is greatly expanded by heterodimerization, which can occur between Gɑq- or Gɑi/o-coupled subtypes, with 16 potential combinations with variable assembly preferences (4–9).
A key aspect of receptor diversity is not just the types of signals that are initiated but the temporal dynamics of such signals. In the case of GPCRs, these temporal dynamics are often regulated via agonist-evoked endocytosis and intracellular trafficking, which, together, control the density of receptors on the cell surface available for ligand sensing. This form of desensitization is typically mediated by the beta-arrestins (β-arr1 and β-arr2), which recognize receptors phosphorylated by GPCR kinases (GRKs) and initiate internalization by recruiting endocytic machinery. Studies on a range of GPCRs have revealed two broad patterns of trafficking following β-arr–mediated internalization (10). “Class A” GPCRs transiently interact with β-arrs, which drive targeting to clathrin-coated pits but dissociate during the endocytosis process (11). In contrast, “class B” GPCRs remain bound to β-arrs along the endocytic pathway (12–14). Typically, the class A pattern leads to receptor recycling back to the plasma membrane (15), while the class B pattern is associated with lysosomal targeting and receptor degradation (16–19). However, the molecular determinants of these two distinct modes of β-arr coupling are not well understood despite recent progress (20).
We recently found that a subset of mGluRs is regulated by β-arrs, while other subtypes are resistant to this form of regulation (21). We focused our study on a comparison of the group II mGluRs, mGluR2, and mGluR3, which show a lack of β-arr coupling and robust β-arr–mediated internalization, respectively. While mGluR2 remains on the cell surface following glutamate treatment, mGluR3 shows a class A profile of β-arr coupling with dissociation upon clathrin-mediated endocytosis and subsequent recycling via the endosomal recycling compartment. This distinct behavior of two very closely related receptors with ~70% sequence identity is encoded within the intracellular C-terminal domains (CTDs), which contain distinct patterns of phosphorylatable residues within a variable Ser/Thr (ST)–rich domain. Notably, we found that the understudied group III mGluR, mGluR8, also undergoes β-arr–mediated internalization. mGluR8, despite receiving less attention compared to other mGluR subtypes, is typically found presynaptically where it can serve as an inhibitory autoreceptor (1, 22–25) and has been strongly implicated in the pathophysiology and potential treatment of pain, anxiety disorders, neurodegeneration, and cancer (26–33).
The finding that a subset of presynaptic mGluRs can undergo agonist-driven β-arr–dependent internalization while others cannot raises many questions: Following internalization, do different receptors have different trafficking itineraries and fates? Which aspects of the receptor determine differential β-arr coupling properties? Does heterodimerization between subtypes further tune the β-arr coupling and trafficking of mGluRs? Here, we use a battery of live cell and single-molecule optical microscopy–based assays to reveal that, unlike mGluR3, mGluR8 undergoes cointernalization with β-arr1 or β-arr2, which leads to lysosomal targeting and receptor degradation. Chimera analysis reveals combined roles for the transmembrane core and the CTD in determining the unique β-arr complex stability and trafficking profile of each mGluR. We pinpoint residues in the mGluR8 CTD, which mediate β-arr coupling, including the identification of a splice variant with strongly impaired internalization. Using a single-molecule pulldown (SiMPull) assay, we reveal that mGluR8 shows preferential heterodimerization with mGluR2 and mGluR3 motivating an analysis of the endocytosis and trafficking of heterodimers. Notably, we develop a cleavable fluorophore approach that allows us to isolate internalized receptors at the single-molecule level and determine that mGluR heterodimers undergo glutamate-dependent internalization. Last, we find that, depending on the subunit combination, mGluR heterodimers can either undergo recycling or lysosomal degradation. Overall, this work reveals previously unappreciated diversity in the regulation of a key family of neuromodulatory receptors, provides mechanistic insight into family C GPCR/arrestin coupling, and has major implications for both the neurophysiological roles and therapeutic targeting properties of mGluRs.
RESULTS
mGluR subtypes show distinct β-arr–mediated internalization properties
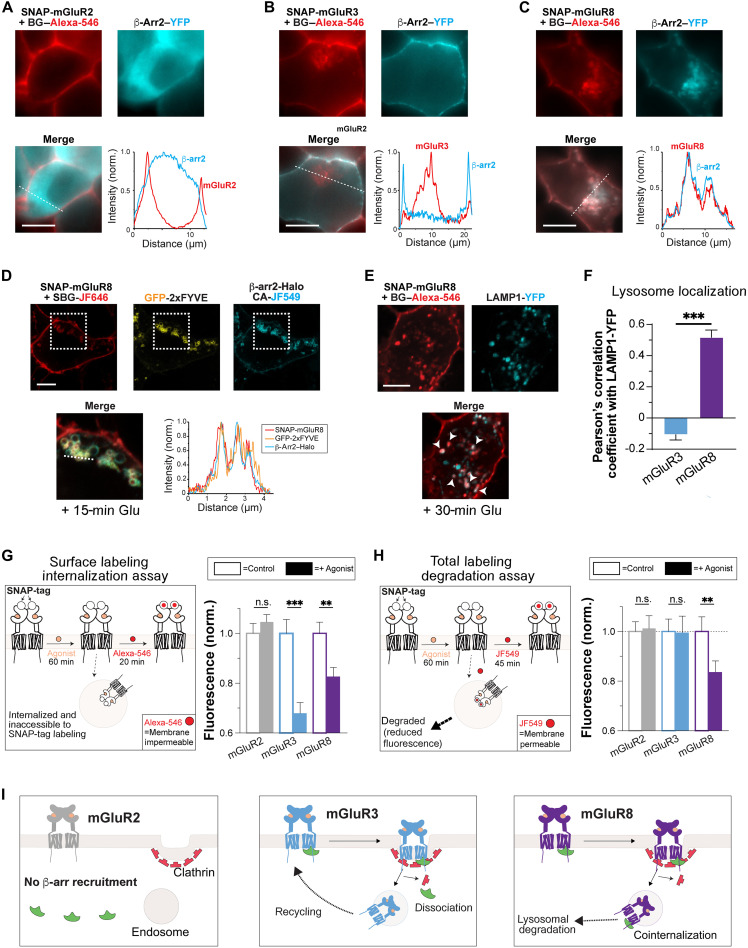
To visualize mGluR/β-arr coupling during internalization, we used N-terminally SNAP-tagged mGluR constructs that can be labeled with membrane-impermeable fluorophores and imaged simultaneously with yellow fluorescent protein (YFP)–tagged β-arr1 or β-arr2 in live cells during glutamate treatment. We performed all experiments in human embryonic kidney (HEK) 293 cells, as these are a well-established platform for studying GPCR trafficking and have shown in many cases to accurately predict the β-arr coupling and trafficking properties of receptors in native systems (15, 34–39). As we previously reported (21), following 1 mM glutamate (Glu) treatment for 30 min, mGluR2 remained on the plasma membrane, while mGluR3 showed robust internalization (Fig. 1, A and B, and fig. S1, A and B). Accordingly, β-arr1–YFP or β-arr2–YFP remained diffusely localized in the cytosol in mGluR2-expressing cells but was relocalized to the plasma membrane in mGluR3-expressing cells (Fig. 1, A and B, and fig. S1, B and E). Notably, while mGluR3 showed both fluorescence on the plasma membrane, from a receptor subpopulation that remained on the surface, and intracellular puncta, from internalized receptor pools, β-arr fluorescence accumulation was restricted to the plasma membrane (Fig. 1B and fig. S1B). This is consistent with mGluR3/β-arr dissociation upon clathrin-mediated endocytosis as reported for many class A GPCRs (10, 11, 13). In stark contrast, mGluR8 displayed strong cointernalization with β-arr1–YFP or β-arr2–YFP (Fig. 1C and fig. S1, C and F). We confirmed with confocal imaging and Pearson’s correlation coefficient (PCC) analysis that internalized mGluR8, but not mGluR2 or mGluR3, shows substantial colocalization with β-arr2 (fig. S1G). Using three-color confocal imaging in live cells, we confirmed that SNAP-mGluR8 and β-arr2–Halo colocalize with an endosomal marker (GFP-2xFYVE; Fig. 1D and fig. S1H). These results suggest that mGluR8 follows a class B arrestin coupling pattern where complexes persist along the endocytic pathway.
Fig. 1. mGluR8, but not mGluR2 or mGluR3, cointernalizes with β-arrs and undergoes lysosomal targeting and degradation.
(A to C) Representative live HEK 293T cell images and line scan profiles (from dotted lines) showing SNAP-mGluR2 (A), SNAP-mGluR3 (B), or SNAP-mGluR8 (C) in red with β-arr2–YFP in green after 30 min with 1 mM glutamate. (D) Scanning confocal images of live cells showing colocalization of SNAP-mGluR8 (red) with endosomal marker, GFP-2xFYVE (green), and β-arr2–Halo (cyan) in live cells. Zoomed in, merged image is from the dotted box area. (E) Scanning confocal images of fixed cells showing colocalization of SNAP-mGluR8 (red) and Lamp1-YFP (green) in cells fixed after 30 min of treatment with 1 mM Glu. (F) PCC analysis comparing the top 10% of pixels between SNAP-mGluR3 and SNAP-mGluR8 with Lamp1-YFP. (G) Schematic of surface labeling internalization assay (left). Surface fluorescence intensity levels from live cells expressing SNAP-tagged mGluRs treated with agonist (1 mM Glu) for 60 min or control conditions (antagonist; see Materials and Methods) before labeling with membrane impermeable fluorophore, BG–Alexa-546. Values are normalized to the fluorescence of a given receptor under control/antagonist conditions. (H) Schematic of total labeling degradation assay (left). Total fluorescence levels from live cells expressing SNAP-tagged mGluRs treated with agonist (1 mM Glu) for 60 min or control (antagonist; see Materials and Methods) before labeling with membrane permeable fluorophore, BG-JF-549. Values are normalized to the fluorescence of a given receptor under control/antagonist conditions. (I) Schematics summarizing the major differences between subtypes. Upon agonist treatment, mGluR2 does not recruit β-arrs and stays on the plasma membrane. mGluR3 recruits β-arrs but dissociates during endocytosis and later recycles back to the membrane. mGluR8 cointernalizes with β-arrs and traffics to lysosomes for degradation. Scale bars, 10 μm. Data are represented as means ± SEM. Unpaired t tests, **P < 0.01 and ***P < 0.001. n.s., not significant.
On the basis of the typical trafficking fate of class B GPCRs (16–19), we hypothesized that mGluR8 traffics to lysosomes where it can be degraded. Internalized mGluR8, but not mGluR3, showed colocalization with the lysosomal marker, Lamp1 (Fig. 1, E and F, and fig. S1H). In contrast, mGluR3 showed a much higher degree of colocalization with Cy3-labeled transferrin (“Tf-Cy3”; fig. S1, I and J), a marker of the endosomal recycling compartment (40). We thus sought to quantify the proportion of each mGluR population that gets internalized and degraded following agonist treatment. We first used our previously established surface labeling internalization assay (21) where SNAP-tagged receptors are treated with ligands before the remaining surface receptors are labeled with a membrane-impermeable fluorophore [BG–Alexa Fluor 546 (Alexa-546)], imaged, and quantified (Fig. 1G). In this assay, the ligand-induced drop in fluorescence intensity reports on the proportion of receptors that are internalized. Consistent with prior imaging results, in this assay, mGluR2 shows no drop in fluorescence, while mGluR3 and mGluR8 show a clear glutamate-induced 20 to 30% reduction (Fig. 1G).
We then modified the surface labeling assay to label both surface and internalized receptors with a membrane-permeable fluorophore (BG-JF549; Fig. 1H) (41). We reasoned that if receptors are degraded during agonist incubation, a drop in fluorescence would be observed. While mGluR2 and mGluR3 showed constant fluorescence intensity levels, mGluR8 showed a ~20% decrease in fluorescence, suggesting that only this subtype is degraded after internalization (Fig. 1H and fig. S1L). Glutamate-induced mGluR8 internalization and degradation were dose dependent, as measured in the surface labeling and total labeling assays, respectively (fig. S1M). We compared our results for mGluRs with two prototypical GPCRs: the μ opioid receptor (MOR), a class A GPCR (10, 42, 43), and the vasopressin receptor 2 (V2R), a class B GPCR (10, 44). Consistent with our expectations, both MOR and V2R showed an agonist-induced fluorescence drop in the surface labeling internalization assay, but only V2R showed a decrease in total fluorescence intensity in the total labeling degradation assay (fig. S1, N and O). Last, we confirmed that mGluR8 degradation is dependent on both internalization and lysosomal targeting by applying either a blocker of GRK2/3, cmpd101 (45), or a blocker of lysosomal proteases, leupeptin (46). Using our SNAP-labeling assays, we found that preincubation with cmpd101 abolished glutamate-induced mGluR8 internalization and degradation, while leupeptin blocked degradation without altering internalization (fig. S1, P to R). Together, these experiments establish the distinct β-arr coupling and trafficking behavior of mGluR2, mGluR3, and mGluR8 (Fig. 1I).
CTD composition determines the endocytic trafficking fate of mGluRs
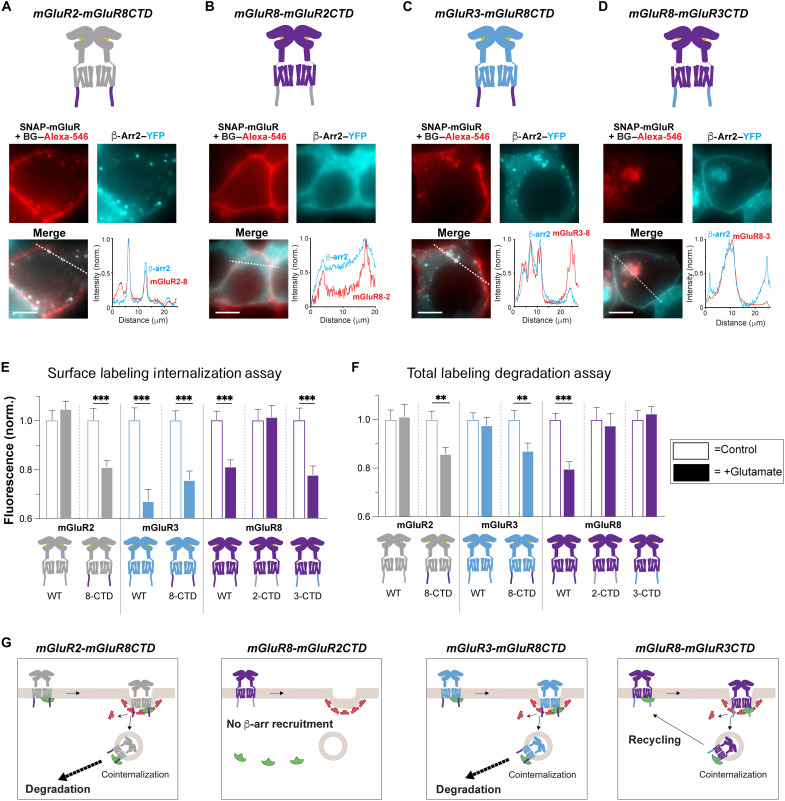
Given the unique trafficking patterns observed upon glutamate-induced internalization of mGluR2 versus mGluR3 versus mGluR8, we investigated the underlying molecular determinants differentiating these receptors. We first generated chimeric mGluR2, mGluR3, and mGluR8 constructs having the SNAP-tagged extracellular and transmembrane domains of one mGluR subtype with the CTD of other mGluR subtypes. All chimeras showed similar surface expression levels to their wild-type counterpart (fig. S2A). Two-color imaging of each chimera with β-arr–YFP revealed a dominant role of the CTD. Unlike wild-type mGluR2, mGluR2-mGluR8CTD showed clear glutamate-induced internalization along with cointernalization of β-arr1–YFP or β-arr2–YFP (Fig. 2A and fig. S2, B and C), revealing that the mGluR8 CTD is sufficient to drive mGluR8-like behavior. In contrast, the reverse construct, mGluR8-mGluR2CTD showed no internalization and β-arr1–YFP or β-arr2–YFP remained cytosolic (Fig. 2B and fig. S2, B and C), indicating that the mGluR8 CTD is required for mGluR8/β-arr cointernalization. In the case of mGluR3-mGluR8CTD, β-arr1–YFP or β-arr2–YFP displayed clear cointernalization unlike the transient association seen with wild-type mGluR3 (Fig. 2C and fig. S2, B and C). mGluR8-mGluR3CTD also showed β-arr1–YFP or β-arr2–YFP cointernalization (Fig. 2D and fig. S2, B and C), suggesting that the transmembrane core of mGluR8 also plays a role in determining the stability of mGluR8/β-arr complexes. We quantified the extent of internalization for all mGluR-CTD chimeras using our surface labeling assay and observed a consistent pattern where constructs containing the mGluR3-CTD or mGluR8-CTD, but not the mGluR2-CTD, showed a clear 20 to 30% internalization (Fig. 2E).
Fig. 2. CTDs determine the unique internalization and trafficking properties of each mGluR subtype.
(A to D) Top: Schematics of mGluR CTD chimeras. Bottom: Fluorescence images and line scan profiles (from dotted lines) of live cells expressing SNAP-mGluR chimeras (red) and β-arr2–YFP (green) following 30 min of Glu treatment. (E) Quantification of surface fluorescence with and without 1 mM Glu treatment across constructs. (F) Quantification of total fluorescence with and without 1 mM Glu across constructs. (G) Schematics summarizing the main findings. mGluR2-mGluR8CTD internalizes with β-arrs and undergoes degradation, while mGluR8-mGluR2CTD does not recruit β-arrs or internalize. mGluR3-mGluR8CTD shows cointernalization with β-arrs and undergoes degradation, while mGluR8-mGluR3CTD shows cointernalization with β-arrs but recycles to the plasma membrane. Scale bars, 10 μm. Data are represented as means ± SEM. Unpaired t tests, **P < 0.01 and ***P < 0.001.
Next, we asked what the trafficking fates are for each chimera. It has previously been hypothesized that class B GPCRs, which cointernalize with β-arrs are prone to degradation (16–19), implying that mGluR3-mGluR8CTD, mGluR2-mGluR8CTD, and mGluR8-mGluR3CTD may all undergo proteolysis. However, only constructs containing the mGluR8-CTD showed clear colocalization to the lysosome (fig. S2, D and E) and a drop in total fluorescence in our degradation assay (Fig. 2F). Notably, despite β-arr cointernalization, mGluR8-mGluR3CTD showed endosomal recycling compartment (ERC) colocalization (fig. S2, D and E) and a lack of degradation (Fig. 2F). Together, these data reveal that cointernalization with β-arr is not itself sufficient to drive degradation and that the receptor CTD composition is the critical determinant of mGluR trafficking.
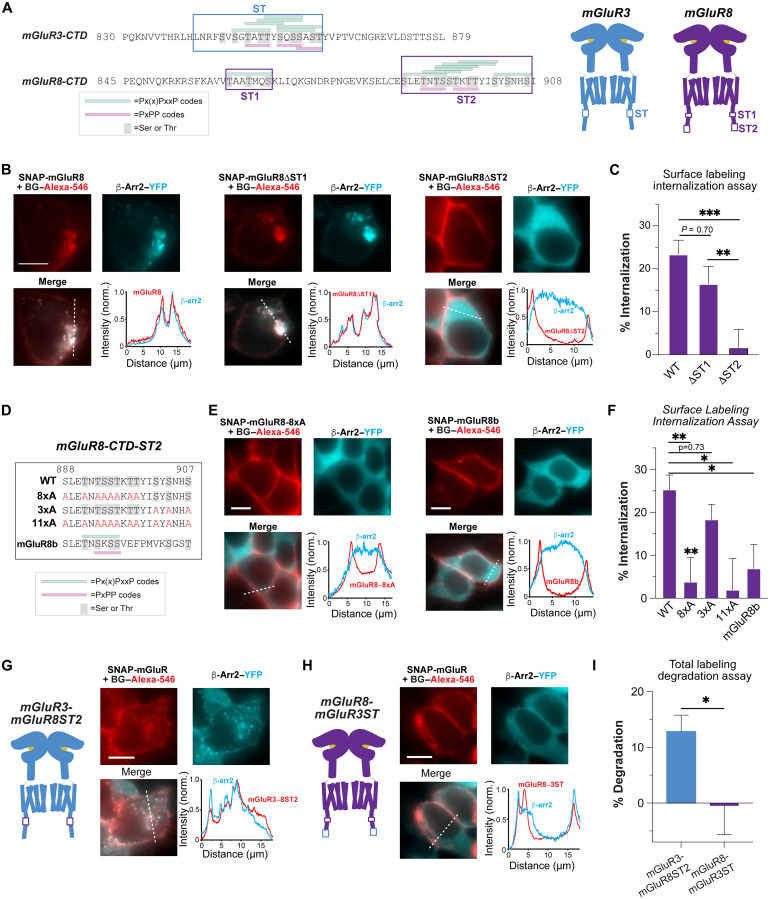
We then aimed to pinpoint which aspects of the CTD determine the unique internalization and trafficking pathway of mGluR8. While mGluR3 has a ST-rich region, which we previously reported to be required for β-arr coupling (21), mGluR8 contains two ST-rich regions, which we termed “ST1” and “ST2” (Fig. 3A). ST1 contains three ST residues that match the proposed Px(x)PxxP/E/D phospho-codes (P, serine or threonine; E, glutamate; and D, aspartate) as identified from an arrestin-bound rhodopsin crystal structure (47), while ST2 region contains 11 ST three residues, which produce seven overlapping Px(x)PxxP/E/D phospho-codes, as well as two of the recently proposed PxPP phospho-code (48). We deleted either the ST1 or ST2 region in mGluR8 to produce “mGluR8ΔST1” and “mGluR8ΔST2” and examined the properties of each construct (fig. S3A). While mGluR8ΔST1 showed similar glutamate-induced internalization and β-arr cointernalization to wild-type mGluR8, mGluR8ΔST2 remained on the plasma membrane without any β-arr recruitment (Fig. 3B and fig. S3B). Consistent with this, mGluR8ΔST1 showed internalization and degradation in SNAP-labeling assays, while internalization and degradation were abolished for mGluR8ΔST2 (Fig. 3C and fig. S3, B and C), indicating that this subregion is the critical driver of β-arr coupling. A small drop in internalization and degradation was observed for mGluR8ΔST1, suggesting that there may be a secondary contribution from this ST1 subregion.
Fig. 3. Pinpointing C-terminal residues on mGluR8, which control internalization and trafficking properties.
(A) Sequence alignment highlighting ST-rich regions of mGluR3 and mGluR8. Two different phospho-code classes are marked in green (Px(x)PxxP) or pink (PxPP). mGluR schematics (right) show the relative positions of ST-rich regions. (B) Fluorescence images and line scan profiles (from dotted lines) of live cells expressing SNAP-mGluR8 wild-type (left), ΔST1 (center), or ΔST2 (right) along with β-arr2–YFP following 30 min of 1 mM Glu treatment. Only ΔST2 prevents interaction with β-arrs and internalization. (C) Quantification of the extent of internalization across constructs determined from the surface labeling assay. (D) Amino acid sequence alignments of the mGluR8-ST2 region showing residues mutated to Ala (shown in red) and splice variant mGluR8b. (E) Representative fluorescence images of live cells expressing SNAP-mGluR8-8xAla (left) or SNAP-mGluR8b (right) showing strongly impaired β-arrs recruitment and internalization. (F) Quantification of the extent of internalization across constructs determined from the surface labeling assay. (G and H) Fluorescence images of live cells expressing SNAP-mGluR3-mGluR8ST2 (G) or SNAP-mGluR8-mGluR3ST (H) with β-arr2–YFP following 15 min of 1 mM Glu treatment. (I) Total labeling fluorescence assay data showing that mGluR3-mGluR8ST2, but not mGluR3-mGluR8-mGluR3ST, undergoes degradation. Scale bars, 10 μm. One-way analysis of variance (ANOVA) (C and F) or unpaired t test (I), *P < 0.05, **P < 0.01, and ***P < 0.001. Data are represented as means ± SEM.
Having identified the ST2 region as crucial for mGluR8 internalization and trafficking, we mutated Ser and Thr residues in this region to test their potential role in phosphorylation-dependent β-arr coupling (Fig. 3D and fig. S3D). Mutating either all 11 (mGluR8-11xA) or the first 8 ST residues (mGluR8-8xA) abolished mGluR8 internalization, β-arr recruitment, and degradation (Fig. 3, E and F, and fig. S3, E to H). However, mutating the last three ST residues in the ST2 region (“mGluR8-3xA”) had no effect (Fig. 3F and fig. S3, E, G, and H). The mGluR8b splice variant, which is observed throughout the brain at comparable but slightly reduced levels compared to mGluR8a (49), differs within the ST2 region on which we have focused. mGluR8b contains only one Px(x)PxxP/E/D and one PxPP phospho-code. Despite comparable surface expression to mGluR8a (fig. S3D), mGluR8b showed substantially reduced internalization (Fig. 3, E and F, and fig. S3, F and G) and degradation (fig. S3H), revealing markedly different trafficking behavior between the two mGluR8 variants.
Because our results show that the ST2 region is critical for the initial β-arr–dependent internalization of mGluR8, we asked whether this CTD subregion is sufficient to drive degradation by producing chimeras that swap the ST-rich region of mGluR3 with the ST2 region of mGluR8 (Fig. 3, G and H, and fig. S3, J and K). mGluR3 with the mGluR8-ST2 (“mGluR3-mGluR8ST2”) showed β-arr cointernalization and degradation, while mGluR8 with the mGluR3-ST (“mGluR8-mGluR3ST”) showed receptor endocytosis without β-arr cointernalization or degradation (Fig. 3, G to I, and fig. S3, J and K). These results reveal that the ST2-rich region includes all residues necessary to drive β-arr–mediated lysosomal targeting and degradation.
mGluR8 efficiently heterodimerizes with group II mGluRs
mGluR2, mGluR3, and mGluR8 homodimers undergo distinct modes of β-arr–mediated regulation (Figs. 1 to 3). However, heterodimerization has the possibility of altering the properties of each subtype to increase molecular and functional diversity among the presynaptic, Gi/o-coupled mGluR subtypes. We and others have shown that group II (mGluR2 and mGluR3) and group III (mGluR4, mGluR6, mGluR7, and mGluR8) mGluRs form heterodimers both under heterologous expression conditions (4–6, 8, 50–53) and natively in rodent tissue (6, 9, 52–55). Recently, we performed analysis of single-cell RNA sequencing data to show that mGluRs are highly coexpressed throughout the diverse cell types of the frontal cortex (6). This included analysis of mGluR8, which we found is highly coexpressed with the other group II/III mGluRs across a range of excitatory and inhibitory neurons.
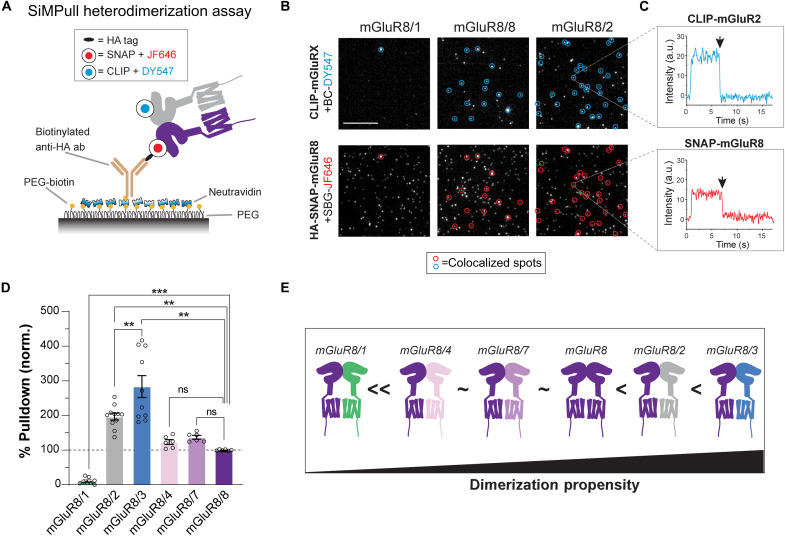
We previously performed quantitative imaging-based analysis of mGluR heterodimerization to assess the relative propensities for different combinations to assemble (6). Our analysis confirmed prior studies that group II and III mGluRs can undergo intra- and intergroup heterodimerization but do not heterodimerize with group I mGluRs. Furthermore, we found that many receptor combinations, including mGluR2 with mGluR3, show preferential heterodimerization compared to homodimerization. We did not include mGluR8 in this analysis, raising the question of its relative dimerization preferences. We assessed this using a SiMPull assay where an N-terminal hemagglutinin (HA)–tag on SNAP-mGluR8 was used to immobilize individual receptors on a passivated glass surface and two-color imaging can be used to assess the relative proportion of mGluR8 complexes containing a coexpressed clasCLIP-tagged mGluR (Fig. 4A). We first established expression conditions where SNAP-mGluR8 is coexpressed with comparable amounts of CLIP-tagged mGluRs (mGluR1, mGluR2, mGluR3, mGluR4, mGluR7, and mGluR8; fig. S4, A and B). We then performed SiMPull for each combination and quantified the ability of HA-SNAP-mGluR8 to pull down CLIP-mGluRs compared to background levels observed in the absence of HA-SNAP-mGluR8 (Fig. 4B and fig. S4, C to E). All CLIP-tagged mGluRs other than mGluR1 showed clear pulldown via HA-SNAP-mGluR8 with many colocalized spots. For all combinations, nearly all colocalized spots showed one-step photobleaching in each channel, consistent with the formation of strict heterodimers and not higher-order complexes (fig. S4F). We quantified the efficiency of pulldown for each combination (see Materials and Methods) and found comparable efficiency for mGluR8/8, mGluR8/7, and mGluR8/4 but higher efficiency for formation of mGluR8/2 and mGluR8/3 heterodimers (Fig. 4, D and E). Together, these data suggest that mGluR8 likely forms a substantial population of heterodimers with mGluR2 and mGluR3, motivating an analysis of the unique properties of these complexes.
Fig. 4. Defining the heterodimerization propensity of mGluR8.
(A) Schematics of two-color SiMPull experiments. Fresh HEK 293T cell lysate from cells expressing HA-SNAP-mGluR8 with CLIP-mGluR1, mGluR2, mGluR3, mGluR4, mGluR7, or mGluR8 is added to a PEG-passivated cover glass containing immobilized anti-HA antibody. SNAP and CLIP tags are labeled with SBG-JF646 and BC-DY547, respectively. (B) Representative single-molecule fluorescence images of HA-SNAP-mGluR8 with CLIP-mGluR1, CLIP-mGluR2, or CLIP-mGluR8. Colocalized spots are circled in green for CLIP-mGluRs and red for SNAP-mGluR8. (C) Representative single-molecule fluorescence traces of mGluR2 (top) and mGluR8 (bottom) from the colocalized spot marked in yellow arrow in (B). Black arrows mark where the photobleaching occurs. (D) Quantification of pulldown efficiency via HA-SNAP-mGluR8 normalized to the homodimer condition of HA-SNAP-mGluR8 with CLIP-mGluR8. (E) Summary of SiMPull results showing the relative dimerization propensities of mGluR8 homo- and heterodimers. One-way ANOVA test, **P < 0.01 and ***P < 0.001. Scale bars, 10 μm. Data are represented as means ± SEM. a.u., arbitrary units.
mGluR heterodimers show distinct internalization and trafficking properties
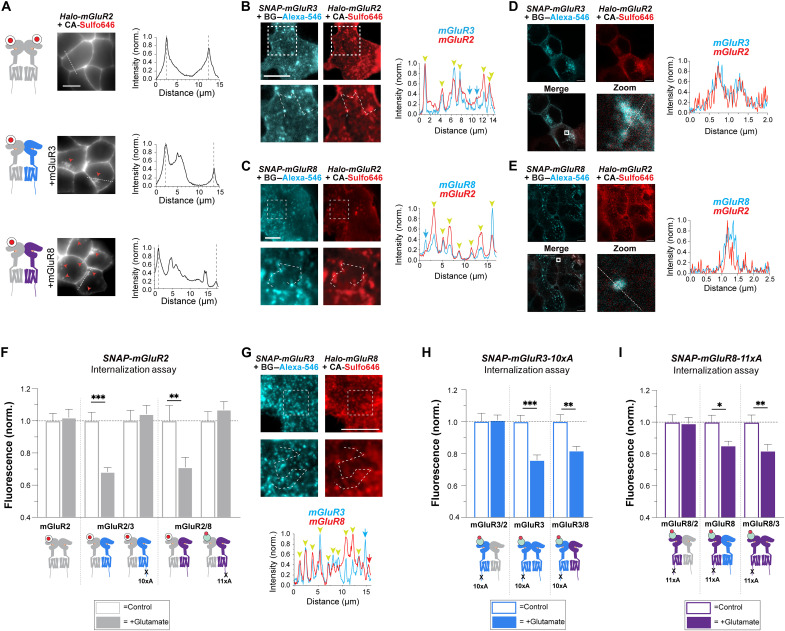
Having established that mGluR8 is frequently coexpressed and readily coassembles with group II mGluRs (Fig. 4 and fig. S4) and on the basis of our prior work showing the efficient formation of mGluR2/3 heterodimers (5, 6, 56), we probed the β-arr–coupling and trafficking properties of heterodimeric combinations of mGluR2, mGluR3, and mGluR8. We first asked whether coassembly with mGluR3 or mGluR8 would enable internalization of mGluR2. Halo-tagged mGluR2 (“Halo-mGluR2”) was coexpressed with untagged mGluR2, mGluR3, or mGluR8, and cells were treated with glutamate. While no internalization was seen upon coexpression of untagged mGluR2, Halo-mGluR2 displayed clear intracellular puncta when coexpressed with untagged mGluR3 or mGluR8 (Fig. 5A). When coexpressed with the MOR and treated with both glutamate and [D-Ala2, N-MePhe4, Gly-ol5]-enkephalin (DAMGO), mGluR2 did not show any internalization, suggesting that mGluR2 internalization is due to specific interactions with other mGluR subtypes (fig. S5, A and B). Two-color confocal imaging with Halo-mGluR2 and SNAP-mGluR3 or SNAP-mGluR8 revealed a high degree of intracellular puncta colocalization (fig. S5, C and D), which was not observed with SNAP-MOR (fig. S5, E and F).
Fig. 5. Functional evidence for internalization of mGluR heterodimers.
(A) Fluorescence live cell images and line scan profiles (from dotted lines) of Halo-mGluR2 alone (top) or coexpressed with either untagged mGluR3 (middle) or mGluR8 (bottom) 30 min after 1 mM Glu application. Red arrows indicate internalized mGluR2 fluorescence. (B and C) TIRF images of cells expressing Halo-mGluR2 with either SNAP-mGluR3 (B) or SNAP-mGluR8 (C) following 10 to 15 min of Glu treatment. Zoomed images are from the dotted box area. Line scan profiles show the colocalization of receptor puncta. (D and E) STED images of cells expressing Halo-mGluR2 with either SNAP-mGluR3 (D) or SNAP-mGluR8 (E) following 30 min of Glu treatment. Zoomed images are from the dotted box area. Line scan profiles show the colocalization of receptor puncta. Scale bars, 5 μm. (F) Quantification of SNAP-mGluR2 internalization using surface labeling assay upon coexpression with other mGluR subunits. When coexpressed with mGluR3 or mGluR8, SNAP-mGluR2 showed decrease in surface fluorescence. When mGluR3-10xAla or mGluR8-11xAla was coexpressed, SNAP-mGluR2 did not show internalization. (G) TIRF images of cells expressing Halo-mGluR3 with SNAP-mGluR8 following 10 to 15 min of Glu treatment. Zoomed images are from the dotted box area. Line scan profiles show the colocalization of receptor puncta. (H and I) Surface labeling assay data showing evidence for mGluR3/8 heterodimers. Internalization deficient mGluR3 (10xA) or mGluR8 (11xA) constructs show a clear drop in surface fluorescence upon coexpression with wild-type mGluR3 or mGluR8 but not mGluR2. One-way ANOVA test, *P < 0.05, **P < 0.01, and ***P < 0.001. Data are represented as means ± SEM.
For higher resolution, we performed images of cell surface receptor clusters using total internal reflection fluorescence (TIRF) microscopy and found colocalized puncta of Halo-mGluR2 with SNAP-mGluR3 or SNAP-mGluR8 (Fig. 5, B and C), consistent with heterodimer targeting to clathrin-coated pits and early endosomes. In contrast, no surface puncta were observed for Halo-mGluR2 upon coexpression with SNAP-MOR and treatment with glutamate and DAMGO (fig. S5, G and H). We confirmed that mGluR2 puncta represent receptors targeted for clathrin-mediated endocytosis, as Halo-mGluR2 showed puncta that were colocalized with clathrin-mCherry only when the receptor was coexpressed with either mGluR3 or mGluR8 and imaged via TIRF microscopy (fig. S5, I and J). To further substantiate our findings with high definition and better resolve intracellular fluorescence patterns, we also performed two-color stimulated emission depletion (STED) superresolution imaging and observed a strong colocalization of Halo-mGluR2 with both SNAP-mGluR3 and SNAP-mGluR8 at the level of individual endosomal puncta following 30-min glutamate treatment (Fig. 5, D and E, and fig. S5F).
We then quantified the internalization of mGluR2-containing heterodimers using our surface labeling assay. SNAP-mGluR2 coexpression with untagged mGluR3 or mGluR8, but not mGluR2, led to a 20 to 30% decrease in surface fluorescence upon glutamate application (Fig. 5F). mGluR3- or mGluR8-dependent internalization of mGluR2 was GRK- and β-arr–dependent based on the ability of the surface fluorescence drop to be blocked by cmpd101 or coexpression of a dominant negative β-arr1 (fig. S5K) (57). Coexpression of MOR and treatment with DAMGO did not drive an effect on mGluR2 surface fluorescence (fig. S5L). When either mGluR3-10xAla (21) or mGluR8-11xA mutants with ablated GRK phospho-sites were coexpressed, mGluR2 did not show any internalization (Fig. 5F and fig. S5M). Coexpression of untagged mGluR2 with SNAP-mGluR3 or SNAP-mGluR8 did not clearly reduce the glutamate-induced drop in surface fluorescence for either receptor (fig. S5, N and O), suggesting that coassembly with mGluR2 does not strongly impair internalization of mGluR3 or mGluR8. Last, we also found evidence for mGluR3/8 internalization, as two-color confocal, TIRF, and STED imaging revealed colocalization of internalized SNAP-mGluR3 and Halo-mGluR8 (Fig. 5G and fig. S5, P to R) and coexpression of mGluR3 or mGluR8 enabled a glutamate-induced drop in surface fluorescence for internalization-deficient mGluR3-10xA and mGluR8-11xA constructs (Fig. 5, H and I).
The experiments described above strongly point to heterodimerization mediating the altered internalization properties of mGluR2 in the presence of coexpressed mGluR3 or mGluR8. However, these experiments do not involve direct and specific detection of mGluR heterodimers, leaving the possibility that indirect forms of cross-talk drive the observed effects. Furthermore, the constitutive dimerization of mGluRs, which has typically been assessed for surface receptors (4–6), raises the question of whether internalized receptors remain dimeric or dissociate, as has been proposed for family A GPCRs (58). To assess these questions, we developed an experimental, SiMPull-based approach to decipher the stoichiometry of internalized receptors. First, we devised a means of specifically detecting internalized receptors by designing and synthesizing a membrane-impermeable Halo-reactive fluorophore that can be rapidly cleaved with a membrane-impermeable reducing agent (Fig. 6A). We incorporated a disulfide group into an Alexa-488 Halo ligand to produce CA-SS–Alexa-488 which remains impermeable because of the polarity of Alexa-488 while being cleavable in response to reducing agents. In vitro, CA-SS–Alexa-488 shows full labeling of recombinantly expressed Halo-tag, and the fluorophore can be removed by adding 100 mM 2-mercaptoethanesulfonic acid sodium salt (“MESNA”), as validated by full-length protein mass spectrometry (fig. S6, A and B). In live HEK 293T cells, CA-SS–Alexa-488 shows efficient surface labeling of Halo-mGluR2 and, upon brief (<5 min) treatment with 100 mM MESNA, shows a near-complete drop in fluorescence (fig. S6C), similar to what has previously been shown with cleavable, SNAP-targeting fluorophores (59, 60). In cells expressing Halo-mGluR3 or Halo-mGluR8 and labeled with CA-SS–Alexa-488, following treatment with glutamate, MESNA abolished surface fluorescence, but fluorescence signal from internalized receptors remained (Fig. 6B and fig. S6, D and E). As a control to test for any effects of MESNA on receptor function, we found that 5 min of 100 mM MESNA treatment did not prevent the ability of mGluR3 or mGluR8 to undergo internalization (fig. S6, F and G) and did not alter glutamate-driven, G protein–dependent potassium currents in a patch-clamp electrophysiology assay (fig. S6H; see Methods).
Fig. 6. Single-molecule, direct detection of internalized mGluR heterodimers.
(A) Schematic of labeling scheme used to visualize only internalized receptors. Surface receptors are labeled with a cleavable fluorophore (CA-SS–Alexa-488) and internalized by agonist incubation. MESNA treatment (100 mM) is then used to cleave the remaining surface fluorophores. (B) Fluorescence cell images of HA-Halo-mGluR3 labeled with CA-SS–Alexa-488 followed by 1 mM glutamate treatment before and after MESNA application. (C) Representative SiMPull images showing HA-Halo-mGluR2 (left) and HA-Halo-mGluR3 (right) after MESNA treatment in control (i.e., antagonist) versus Glu conditions. Single-molecule spots are circled in green. A representative fluorescence intensity trace is plotted with arrows marking photobleaching steps. (D) Quantification of total spots from control or Glu-treated conditions. mGluR3 and mGluR8, but not mGluR2, show glutamate-driven internalized receptors. (E) Photobleaching step distribution for immobilized spots from either untreated or cells treated with glutamate and MESNA. A similar distribution is seen for surface and internalized mGluR3 or mGluR8. (F) Two-color SiMPull images of HA-Halo-mGluR2 coexpressed with either SNAP-mGluR3 or SNAP-mGluR8 labeled with CA-SS–Alexa-488 and SBG-JF646, followed by 1 mM Glu incubation and MESNA treatment. A representative fluorescence intensity trace is plotted in each color for a colocalized spot with arrows marking photobleaching steps. (G) Quantification from two-color SiMPull experiments showing total number of HA-Halo-mGluR2 spots when coexpressed with SNAP-mGluR2, mGluR3, or mGluR8 treated with either antagonist or Glu. (H) Photobleaching step distribution for colocalized spots, indicating strict 1:1 heterodimerization. (I) Two-color SiMPull images of HA-Halo-mGluR8 coexpressed with SNAP-mGluR3 labeled with CA-SS–Alexa-488 and SBG-JF646, followed by 1 mM Glu incubation and MESNA treatment. A fluorescence intensity trace is plotted for a colocalized spot with arrows marking photobleaching steps. (J) Photobleaching step distribution for colocalized HA-Halo-mGluR8/SNAP-mGluR3 spots, indicating strict 1:1 heterodimerization. One-way ANOVA test, ***P < 0.001. Data represented as means ± SEM.
Following CA-SS–Alexa-488 labeling, cells expressing HA-Halo-mGluR2, HA-Halo-mGluR3, or HA-Halo-mGluR8 were treated with either antagonist [LY341495 for mGluR2 and mGluR3; (RS)-α-Cyclopropyl-4-phosphonophenylglycine (CPPG) for mGluR8] or glutamate for 30 min, followed by MESNA treatment, lysis, and SiMPull via an anti-HA antibody. All receptors showed minimal spots following antagonist treatment, but mGluR3 and mGluR8 showed substantial single-molecule immobilization after glutamate treatment (Fig. 6, C and D, and fig. S6I). This matches results from cell imaging assays (Figs. 1 to 3) and validates that this approach can be used for quantitative assessment of receptor internalization. To assess the stoichiometry of internalized receptors, we analyzed fluorophore bleaching steps, as we have previously used to reveal strict dimerization of surface mGluR2 (5) and incomplete dimerization of intracellular mGluR2 (41). We observed that ~40 to 50% of internalized receptors showed two-step bleaching (Fig. 6, C and E) comparable to what was seen for surface receptors immobilized from cells not treated with MESNA (Fig. 6E) and in line with maintained dimerization upon internalization.
Having confirmed that mGluRs remain largely dimeric following endocytosis, we aimed to use the cleavable fluorophore approach to verify that mGluRs can internalize and traffic as heterodimers. We developed a two-color SiMPull approach where HA-Halo-mGluR2 is coexpressed with SNAP-mGluR2, SNAP-mGluR3, or SNAP-mGluR8. Following labeling of Halo with CA-SS–Alexa-488 and SNAP with SBG-JaneliaFluor646 (SBG-JF646) (fig. S6, J and K), cells were treated with antagonist or glutamate, followed by MESNA treatment, lysis, and SiMPull. As expected, on the basis of the cellular imaging data, coexpression with SNAP-mGluR3 or SNAP-mGluR8 lead to the clear immobilization of fluorescent Halo-mGluR2 molecules (Fig. 6, F and G). A large proportion of Halo-mGluR2 spots were colocalized with SNAP-mGluR3 or SNAP-mGluR8 (Fig. 6F). Notably, because SNAP-targeting fluorophores are not cleavable, the spots in the JF646 channel represent either internalized or surface mGluR2-containing heterodimers. There was no clear glutamate-dependent pulldown of fluorescent HA-Halo-mGluR2 upon coexpression of SNAP-mGluR2 (Fig. 6G). Colocalized spots showed almost exclusively one-step bleaching in both the Alexa-488 and JF646 channels (Fig. 6, F and H), confirming that these are indeed internalized, heterodimeric mGluR2/3 and mGluR2/8. We also used this approach to validate internalization of mGluR3/8 heterodimers. Coexpression and labeling of HA-Halo-mGluR8 and SNAP-mGluR3 led to the immobilization of colocalized spots (7.7 ± 0.5% of mGluR8 spots; 26.2 ± 0.7% of mGluR3 spots) following glutamate and MESNA treatment (Fig. 6I). Colocalized spots bleached in one step per channel (Fig. 6J), confirming that they represent internalized mGluR3/8 heterodimers.
We next asked what the trafficking fates are for mGluR2/3, mGluR2/8, and mGluR3/8 subunit combinations after glutamate-driven, β-arr–mediated endocytosis. First, we performed three-color confocal imaging in live cells expressing two receptor subunits along with β-arr2–YFP. Coexpression of mGluR2 or mGluR3 with mGluR8 led to colocalization of all three channels in intracellular puncta, while coexpression of mGluR2 with mGluR3 led to colocalized receptor puncta but β-arr2–YFP fluorescence accumulation exclusively on the cell surface (fig. S7, A to D). On the basis of this, we hypothesized that mGluR8-containing heterodimers may traffic to lysosomes and undergo proteolysis. We first tested this with three-color confocal imaging of receptors with Lamp1-YFP. Coexpression of mGluR2 or mGluR3 with mGluR8, but not mGluR2 with mGluR3, led to clear colocalization of both receptor types with Lamp1-YFP (Fig. 7, A to C, and fig. S7E).
Fig. 7. Heterodimerization reshapes the trafficking fates of mGluRs.
(A and B) Confocal images of fixed HEK 293T cells expressing SNAP-mGluR8 with Halo-mGluR2 (A) or Halo-mGluR3 (B) and treated with glutamate (30 min) showing colocalization with Lamp1-YFP. SNAP and Halo tags are labeled with BG–Alexa-546 and CA-Sulfo646, respectively. Merged images are from area in dotted boxes. Scale bar, 5 μm. (C) Summary of PCC analysis showing colocalization of coexpressed mGluR constructs with Lamp1-YFP. Substantial colocalization is seen for all subtypes only in conditions including mGluR8 expression. (D) Total fluorescence measurement of SNAP-mGluR2 when coexpressed with unlabeled mGluR2, mGluR3, or mGluR8. Only mGluR8 coexpression enables a glutamate-driven reduction in total mGluR2 fluorescence intensity (i.e., degradation). (E) Total fluorescence measurement of SNAP-mGluR3 when coexpressed with unlabeled mGluR3 or mGluR8. Only mGluR8 coexpression enables a glutamate-driven reduction in total mGluR3 fluorescence intensity. (F) Total fluorescence measurement of SNAP-mGluR8 when coexpressed with unlabeled Halo-mGluR8, mGluR3, or mGluR2. All coexpression conditions show a glutamate-driven reduction in total fluorescence intensity. (G) Schematic summarizing the main findings regarding the β-arrs coupling, internalization, and trafficking of mGluR2-, mGluR3-, and mGluR8-containing homo- and heterodimers. One-way ANOVA test, *P < 0.05, **P < 0.01, and ***P < 0.001. Data are represented as means ± SEM.
We then performed our total fluorescence degradation assay to assess degradation when we coexpressed SNAP-mGluR2 with either untagged mGluR2, mGluR3, or mGluR8 and observed a glutamate-induced drop in fluorescence only in the presence of mGluR8 (Fig. 7D). Similarly, SNAP-mGluR3 showed a drop in total fluorescence in the presence of untagged mGluR8, but not mGluR3 (Fig. 7E). As a control, we found that mGluR8 coexpression and coactivation did not drive degradation of SNAP-MOR (fig. S7F) and V2R coexpression and coactivation did not drive degradation of SNAP-mGluR2 (fig. S7G), indicating that a direct physical interaction between subunits is needed for one receptor to drive degradation of another. Last, confirming that mGluR8-containing heterodimers undergo efficient degradation, we observed a drop in SNAP-mGluR8 total fluorescence when coexpressed with mGluR8, mGluR2, or mGluR3 (Fig. 7F). Together, these experiments reveal a wide range of molecular diversity among the mGluR family with regards to internalization and trafficking properties (Fig. 7G).
DISCUSSION
Despite great, ongoing effort to understand β-arr coupling across the members of the GPCR superfamily, unexpectedly, little is known about the biologically critical family C GPCRs. Here, we find unexpected diversity in the modes of β-arr–mediated regulation of mGluRs, the prototypical family C GPCR subfamily. Building from our previous work showing that some subtypes readily undergo β-arr–mediated internalization while others do not (21), we now show that different mGluR subtypes can have markedly different β-arr complex lifetimes. While mGluR3 rapidly dissociates from β-arrs upon endocytosis and recycles back to the plasma membrane, mGluR8 forms stable complexes that persist along the endocytic pathway and, ultimately, enable receptor degradation in lysosomes. mGluR8 represents an example of a family C GPCR with a class B pattern of β-arr coupling and complements the class A pattern seen with mGluR3. Unexpectedly, the mGluR8b splice variant shows very minimal β-arr coupling and internalization.
We also find that the quaternary structure of mGluRs opens a further avenue for molecular diversity via heterodimerization. While prior work on mGluR heterodimerization has focused on deciphering the rules of assembly (4–6), the structural/conformational properties of heterodimers (5, 8, 52, 61), or the G protein–mediated signaling response to different ligands (7, 50, 51, 53, 54, 62, 63), we show here that heterodimerization can also profoundly reshape the endocytosis and trafficking properties of mGluR subtypes. We find that mGluR2, which is highly resistant to β-arr–mediated desensitization when assembled as a homodimer, can undergo β-arr–mediated endocytosis when coassembled into an mGluR2/3 or mGluR2/8 heterodimer with the latter enabling lysosomal degradation. In addition, coassembly with mGluR8 enables mGluR3 to undergo lysosomal targeting and degradation rather than the endosomal recycling seen with homodimers. Together, these comparative analyses between mGluR subtypes have major implications for understanding mechanistic aspects of GPCR/β-arr coupling and trafficking, as well as for defining the physiological properties of mGluRs.
Physiological relevance/implications
The diversity of mGluR/β-arr coupling suggests that, along with variability in glutamate sensitivity, G protein transduction, and basal activity, different desensitization properties enable tuning of specific receptor dimers to their physiological roles. In the presynaptic context where mGluR2, mGluR3, and mGluR8 typically operate, this may manifest in different modes of metaplasticity where extended periods of high activity and extrasynaptic glutamate levels drive changes in the levels of presynaptic autoreceptors. For example, mGluR8 has been shown to be down-regulated following acute stress (64) or hemorrhage (65), which is consistent with our findings of rapid agonist-driven degradation. Similarly, β-arr coupling may be relevant in the context of drug treatment where extended activation may lead to down-regulation of mGluR3 and, especially, mGluR8, potentially driving tolerance and other forms of synaptic compensation that can limit therapeutic efficacy. This is important to consider as mGluR3 and mGluR8 agonists have been proposed for the treatment of anxiety disorders (66, 67), schizophrenia (68), pain (69), and neurodegenerative disorders (31, 70). An important question for future work is if internalized mGluR3 or mGluR8 are capable of initiating intracellular signaling cascades as has been shown for many GPCRs, including mGluR5 (71) and a variety of Gi/o-coupled GPCRs (72, 73). Ultimately, extensive future work is needed to directly probe the β-arr–mediated internalization, trafficking, and signaling properties of mGluRs in the synaptic context where little is known about GPCR regulation. Notably, two recent studies of opioid receptors have shown that GRKs and β-arrs can drive rapid endocytosis and long-term down-regulation of receptor function in presynaptic boutons (74, 75). However, little is known about lysosomal targeting and potential proteolysis of axonal/presynaptic GPCRs, motivating future work on mGluR8.
Our finding that heterodimerization further tunes mGluR/β-arr coupling properties suggests that the complement of mGluR subtypes and the corresponding response to glutamate release can be highly tuned from cell to cell and even from synapse to synapse. For example, in cells coexpressing mGluR3 and mGluR8, there may be a mix of boutons containing primarily mGluR3 homodimers, mGluR8 homodimers, and mGluR3/8 heterodimers, each with a characteristic desensitization and recovery-from-desensitization profile. On the basis of electron microscopy and superresolution imaging studies, group II and III mGluRs are thought to be localized differently within presynaptic boutons with group II mGluRs extrasynaptic and freely diffusing and group III mGluRs immobilized within active zones (25, 76, 77). This raises the questions of both where heterodimers may be localized and how their localization, both within surface microdomains and intracellular sites, is altered by activation and β-arr coupling. Along these lines, it will be interesting to determine whether mGluR8a and mGluR8b heterodimers can form and what trafficking properties they display. Ultimately, techniques to detect and manipulate heterodimers with subcellular precision will be needed to decipher the relative prevalence and roles of each combination. Along these lines, critical future directions will be to understand how subtype-specific ligands alter the β-arr coupling of mGluR heterodimers and to identify how other accessory or scaffold proteins alter the β-arr coupling and trafficking of mGluR homo- and heterodimers.
Molecular mechanisms of mGluR/β-arr coupling
Our data indicate that mGluR3 and mGluR8 are capable of directly coupling with β-arrs, albeit with very different complex lifetimes. Chimera analysis shows that the CTD is the major determinant of arrestin-coupling propensity and stability. We previously showed that swapping the mGluR2 and mGluR3 CTDs is sufficient to swap the ability to strong couple to β-arrs (21). Here, we show that swapping the mGluR8-CTD onto mGluR2 or mGluR3 can confer mGluR8-like β-arr coupling and cointernalization and, ultimately, degradation. The differences in β-arr coupling between the mGluR2-CTD and the mGluR3-CTD are likely explained by the simple fact that mGluR2 does not contain ST residue clusters organized into phospho-codes. However, both the mGluR3-CTD and mGluR8-CTD contain comparable numbers of phospho-codes (six versus nine Px(x)PxxP codes; three versus two PxPP codes) yet drive distinct β-arr coupling. Notably, despite the higher apparent affinity for β-arrs and subsequent degradation, mGluR8 does not internalize more rapidly or completely than mGluR3. A reasonable hypothesis for the distinct apparent complex stability is that the unique sequences in these regions confer sufficiently different β-arr affinities between phosphorylated mGluR3 and mGluR8 CTDs. Alternatively, different CTDs may have a higher or lower propensity for GRK phosphorylation leading to different degrees of phosphorylation. In vitro biophysical and structural work is needed to further dissect the interaction between CTDs and β-arrs. mGluR7 has also been proposed to undergo β-arr–dependent internalization and degradation (78) but shows a far reduced number of phospho-codes (two Px(x)PxxP codes; one PxPP) compared to mGluR3 and mGluR8.
The mGluR transmembrane core/β-arr interactions also appear to mediate subtype differences. Β-arr cointernalization is seen for mGluR8-mGluR3-CTD, implying that mGluR8 core interactions are more long-lived than mGluR3, although this cointernalization does not lead to lysosomal targeting and degradation. This result shows that merely cointernalizing with β-arrs is not sufficient to drive GPCR trafficking to lysosomes. A possible explanation for this is that different CTDs stabilize different β-arr conformations, consistent with the conformational complexity that has been observed in spectroscopic studies of β-arrs (48, 79–82). Β-arr conformation likely controls the ability of receptors to interact with key trafficking factors such as ubiquitin, GPCR associated sorting protein-1 (GASP-1), or ALG-2 interacting protein X (ALIX), as has been proposed for other GPCRs (18, 83–85). Future work is needed to identify the proteins and/or protein complexes that facilitate the distinct trafficking itineraries of mGluR subtypes.
This study also provides insight into β-arr coupling and trafficking in dimeric GPCRs. Our cleavable fluorophore SiMPull experiments show that mGluRs remain dimeric upon internalization, as similar rates of two-step bleaching are seen for internalized and surface receptor. Given that dimerization is required for mGluR activation (86), intracellular dimerization is potentially important both for maintaining functional receptors that can be recycled back to surface or for G protein coupling from the endosome. The unique dimeric arrangement of mGluRs raises many biophysical questions for future studies regarding the stoichiometry of mGluR/β-arr complexes, the orientation and structure of bound β-arrs within the dimeric transmembrane domain (TMD) core, and the possibility of simultaneous coupling to β-arrs and G proteins. Our finding that heterodimeric mGluRs containing one subunit that does not typically couple to β-arrs as a homodimer (mGluR2) can still couple efficiently to β-arrs suggests that a single CTD is sufficient for β-arr coupling and efficient transport to clathrin-coated pits. This raises the question of whether CTD binding to one subunit enables coupling to the TMD of the same subunit or the other subunit in trans. This has potential implications for how β-arr coupling is altered in response to subtype-specific orthosteric and allosteric ligands within heterodimers.
Along these conceptual lines, our data shows that mGluR3/8 heterodimers primarily undergo β-arr cointernalization, lysosomal targeting, and proteolysis. Why does the mGluR8 subunit seem to dominate in this heterodimeric context where both subunits, in principle, can couple to β-arrs? Structural and biophysical work is needed to better understand these complexes and to determine whether one or two β-arrs can simultaneously bind to a receptor dimer. It is possible that there is a competition between receptor subunits and that the apparent higher affinity of mGluR8 allows it to dominate such that asymmetric mGluR3/mGluR8-β-arr complexes exist. Alternatively, two β-arrs could bind to the heterodimer on the plasma membrane, but the longer lifetime of mGluR8/β-arr complexes allows just one to persist into the endosomal pathway and dominate the subsequent trafficking properties. The ability of mGluR8 to override the normal trafficking of mGluR3 is consistent with the notion that recycling is the default pathway for internalized membrane proteins (40, 87), although further studies are needed to understand the post-endocytic trafficking and sorting mechanisms of mGluRs.
Ultimately, our study reveals great diversity between homo- and heterodimeric mGluR subtypes in their modes of β-arr coupling and intracellular trafficking that should pave the way for many future mechanistic and physiological studies with major clinical relevance for harnessing mGluRs as therapeutics. Beyond our focus on mGluR2, mGluR3, and mGluR8, future work should extend this analysis to other widely expressed group III subtypes, mGluR7 and mGluR4, which are both also capable of forming a variety of heterodimers (4, 9, 51, 52, 61–63, 88) and have unique trafficking properties (21, 78, 89–91).
MATERIALS AND METHODS
Molecular biology
All SNAP- and CLIP-tagged mGluR1 (human), mGluR2 (rat), mGluR3 (rat), mGluR4 (human), mGluR7 (rat), and mGluR8 (human) were generated with N-terminal signal peptide from rat mGluR5 followed by HA- and SNAP-tagged mGluRs or FLAG- and CLIP-tagged mGluRs in pRK5 vector as previously described (4, 6). SNAP-tagged mGluR8b was cloned by introducing an insertion and deletion into the mGluR8a plasmid. Halo-tagged mGluR2, mGluR3, and mGluR8 were additionally cloned by replacing the SNAP tag. For cloning mGluR CTD chimeras, mGluR2-CTD (amino acids 821 to 872), mGluR3-CTD (amino acids 830 to 879), and mGluR8-CTD (amino acids 845 to 908) were replaced with the respective CTD sequences of other subtypes. mGluR3 ST region (amino acids 841 to 860), mGluR8 ST1 (aa861–867), and ST2 (aa888–907) regions were used for cloning ST-rich region deletions or chimeras using a polymerase chain reaction (PCR)–based DNA ligation method using primers with 5′ phosphates and the enzyme T4 DNA Ligase (Invitrogen) or a Gibson Assembly Cloning Kit (New England Biolabs), respectively. Point mutations were made using PCR-based site-directed mutagenesis. β-Arr2–Halo was cloned by replacing YFP from β-arr2–YFP (Addgene, #36917) with Halo tag using a Gibson Assembly Cloning Kit.
Cell culture and transfection
HEK 293T (American Type Culture Collection; negative for mycoplasma) cells were seeded on poly-l-lysine–coated cover glass in a 12-well plate using Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and cultured at 37°C and 5% CO2. Cells were transfected using Lipofectamine 2000 (Thermo Scientific) and maintained in antagonist (5 μM LY34 for group II mGluRs, 20 μM CPPG for mGluR8, or 10 μM naloxone for MOR-transfected cells). All experiments were performed 24 to 48 hours after transfection.
Wide-field fluorescence imaging and quantification of receptor surface or total levels
To visualize the internalization and colocalization of SNAP-tagged receptors and β-arrs (Figs. 1 to 4), HEK 293T cells were cotransfected with 0.2 to 0.5 μg of SNAP-tagged mGluRs with 0.2 μg of GRK2 and either 0.2 μg of β-arr1–YFP or β-arr2–YFP. For receptor coexpression (Figs. 5 to 7), 0.3 μg of Halo-tagged mGluR2 was coexpressed with 0.3 to 0.6 μg of SNAP-tagged or untagged mGluR8, mGluR3, or MOR; twenty-four to 36 hours after transfection, cells were labeled with 1 μM BG–Alexa-546 (cell membrane impermeable) for SNAP and CA-Sulfo646 (cell membrane impermeable) (41, 92) for Halo in extracellular buffer (EX) containing 10 mM Hepes, 135 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, and 1 mM MgCl2 (pH 7.4) for 30 min at 37°C. For two-color cell imaging (Figs. 5 to 7), SNAP- or Halo-tagged receptors are coexpressed together and labeled simultaneously with fluorophores for SNAP and Halo tags. Labeled cells were then incubated in agonist (1 mM glutamate, 1 μM DAMGO, or both) for 10 to 30 min at 37°C. Live cells were imaged on an inverted microscopy (Olympus IX83) using 100× oil immersion [numerical aperture (NA) 1.45] objective. YFP from β-arr1/2 was excited with a 488-nm laser, Alexa-546, and JF549 fluorophores were excited with a 561-nm laser, and Sulfo646 or JF646 fluorophore was excited with a 640-nm laser.
For the surface labeling internalization assay (Fig. 1G), we followed our previously described protocol (21, 93). Twenty-four hours after transfection, HEK 293T cells expressing SNAP-tagged receptors (0.2 to 0.5 μg alone or in combination with untagged receptors) were washed with fresh media and incubated in saturating antagonist (negative control; 10 μM LY34 for mGluR2 or mGluR3, 20 μM CPPG for mGluR8, 10 μM naloxone for MOR, and no drug for V2R) or agonist (1 mM Glu, 10 μM DAMGO, and 100 nM vasopressin) in media for 1 hour at 37°C. Saturating antagonist was used as a control condition to maximize reproducibility and dynamic range as HEK 293T cells can release glutamate. All compounds were purchased from Tocris. To label remaining surface receptors after drug exposure, cells were incubated in 1 μM BG–Alexa-546 (New England Biolabs) for 20 min at room temperature in EX. For the total labeling degradation assay (Fig. 1H), after antagonist or agonist treatment, cells were incubated in 1 μM BG-JF549 (cell membrane permeable) (41) for 45 min at 37°C in EX to label both the plasma membrane and the intracellular receptor populations. For both internalization and degradation assays, after fluorophore labeling, cells were washed and imaged with a 60× 1.49 NA objective. A minimum of 10 snapshots were acquired per day per condition. Fluorescence cells were analyzed using ImageJ (Fiji). Mean intensity of fluorescent intensity was calculated and normalized to the antagonist incubated condition. For the GRK activity block experiment, cells were preincubated with 30 μM cmpd101 for >30 min and maintained in the presence of agonist and antagonist treatment. For the dominant negative β-arr1 (mutant S412D), we cotransfected 0.3 μg together with the receptor. For lysosomal degradation inhibition, 60 μM leupeptin was preincubated for 2 hours at 37°C before treatment with agonist or antagonist and maintained during the entire experiment. All conditions were tested on at least two separate experimental days. Fluorescence intensity values were normalized to the antagonist control condition on the same day and pooled into summary bar graphs.
Confocal microscopy
Scanning confocal microscopy was used to visualize internalized receptor colocalization with various markers: GFP-2xFYVE (Addgene, #140047), β-arr2–Halo, Lamp1-YFP (Addgene, #1816), or Tf-Cy3 (Jackson ImmunoResearch Inc.). For three-color imaging of SNAP-tagged mGluR3 or mGluR8 with GFP-2xFYVE β-arr2–Halo, cells were labeled with 1 μM BG–Alexa-546 and CA-Sulfo646 in EX for 30 min at 37°C followed by 1 mM Glu incubation in media for 15 to 30 min at 37°C and imaged as live cells. For colocalization imaging with Lamp1-YFP, SNAP-mGluR8, SNAP-mGluR3, SNAP-mGluR2, or SNAP-MOR was coexpressed with GRK2. Unlabeled β-arr2 was additionally coexpressed for mGluR8 to enhance internalization. Cells were incubated in 1 μM BG–Alexa-546 in EX solution for ~30 min at 37°C followed by 1 mM glutamate incubation for 30 min at 37°C. For colocalization with transferrin-Cy3, cells were incubated in 1 μM BG–Alexa-488 in EX solution for ~30 min at 37°C followed by incubation with Tf-Cy3 (5 μg/ml) and 1 mM glutamate in EX for 30 min at 37°C. For heterodimer experiment using SNAP- and Halo-tagged mGluRs, 1 μM CA-Sulfo646 fluorophore was additionally added to the labeling solution to label Halo-tagged mGluRs. Fixation of labeled cells was done by incubating in 4% paraformaldehyde/4% sucrose for 10 min. Cells were imaged using a 63× objective with 1.5 to 2× zoom on a Zeiss LSM880 scanning confocal microscope and the ZEN Black software. Fluorophores were excited using 488-, 561-, and/or 640-nm lasers. The plane of imaging was chosen on the basis of the organelle marker channel. Images were captured using one-directional scanning and a pinhole of 1 Airy unit. For Pearson’s correlation coefficient analysis, regions of interest (ROIs) were drawn only within the cells to exclude surface receptors using ImageJ software. The pixels with the top 10% highest intensity (FT) in each channel were used to measure the Pearson’s correlation coefficient via the plugin EzColocalization (94) on ImageJ. All data come from two or three separate transfections and include at least 12 cells per condition.
TIRF imaging
HEK 293T cells expressing Halo- and SNAP-tagged receptors were imaged via TIRF microscopy as previously described (21). Briefly, agonists (1 mM Glu and 10 μM DAMGO) were applied in the imaging bath, and TIRF images were captured with a 100× 1.49 NA objective after 5 to 15 min of drug incubation at 37°C. For colocalization analysis, positions of individual puncta from each channel were compared using the multipoint function in ImageJ. All data come from at least two separate transfections and include at least 10 cells per condition.
Single-molecule pulldown
Heterodimerization propensity SiMPull measurements (Fig. 5) where performed as previously described (6). Cells were transfected with HA-SNAP-mGluR8 with CLIP-mGluR1, mGluR2, mGluR3, mGluR4, mGluR7, and mGluR8 at a DNA ratio (1:2 to 1:5) optimized to produce similar expression levels for each receptor pair (fig. S4, A and B). Forty-eight hours after transfection, cells were labeled in EX at 37°C with 1.5 μM SBG-JF646 (41) for 45 min, followed by 1.5 μM BC-DY547 (New England Biolabs) for 45 min 37°C. For SiMPull measurements with a cleavable fluorophore, HA-Halo-mGluR2, HA-Halo-mGluR3, or HA-Halo-mGluR8 was expressed alone or coexpressed with SNAP-mGluR2, SNAP-mGluR3, or SNAP-mGluR8 without an HA tag. Cells were labeled with 1.5 μM CA-SS–Alexa-488 (and SBG-JF646 for heterodimer conditions) in EX for 45 min, 37°C. Cells were then washed with EX twice and incubated with 100 mM MESNA for 10 min at room temperature to cleave the surface labeled CA-SS–Alexa-488 followed by another wash with EX twice. Labeled cells were then gently harvested in phosphate-buffered saline (0 Ca2+ and 0 Mg2+), pelleted at 10,000 rpm at 4°C, and lysed using 0.5% Lauryl Maltose Neopentyl Glycol (LMNG) + 0.05% Cholesteryl hemisuccinate (CHS) (Anatrace) plus protease inhibitor cocktail (Thermo Fisher Scientific) at 4°C for 1 hour. After lysis, samples were centrifuged at 17,000g for 20 min at 4°C, and supernatants were collected. A microflow chamber was prepared using a glass coverslip and quartz slide passivated with mPEG-SVA and biotinylated polyethylene glycol (PEG; molecular weight, 5000; 50:1 molar ratio; Laysan Bio). Before each experiment, each chamber was incubated with NeutrAvidin (0.2 mg/ml) in T50 buffer [50 mM NaCl and 10 mM tris (pH 8.0)] followed by biotinylated anti-HA antibody (0.002 mg/ml; ab26228, Abcam) in T50 buffer. Fresh lysate containing fluorescently labeled receptors was then diluted using buffer containing 0.05% LMNG and 0.005% CHS in EX buffer and added to the flow chamber. When a desired single-molecule spot density (~0.2 spots/μm2) was obtained, unbound receptors were washed with the dilution buffer. Single-molecule movies were recorded using a 100× oil immersion objective (NA 1.49) on an inverted microscope (Olympus IX83) with TIR mode at 20 Hz with 50-ms exposure time using two scientific Complementary Metal-Oxide Semiconductor (sCMOS) cameras (Hamamatsu ORCA-Flash4v3.0). Labeled receptors were excited with 640- and 561-nm to visualize CA-Sulfo646 and BC-DY547, respectively. Movies were recorded until >90% of molecules were bleached in the field of each movie. Data were analyzed using a custom-built LabVIEW program (95). Briefly, each movie from different channels was concatenated and loaded on the analysis program to visualize each channel for identification of colocalized molecules. Bleaching steps were assigned by inspecting the fluorescence traces manually for each molecule and plotting to show the bleaching step distribution. Data were obtained from at least two separate experimental days and averaged across multiple movies using Origin Pro 2017 (OriginLab).
STED microscopy
STED microscopy was performed on fixed HEK 293T cells using a Leica SP8 TCS STED FALCON (Leica Microsystems) equipped with a pulsed white-light excitation laser (80-MHz repetition rate; NKT Photonics), a 100× objective (HC PL APO CS2 100×/1.40 NA oil), a temperature-controlled chamber and operated by LAS X. Alexa-546 was excited using λ = 546 nm, and emission signals were captured at λ = 556 to 650 nm. Sulfo646 was excited using λ = 646 nm, and emission signals were captured at λ = 656 to 750 nm. For STED images, 775-nm depletion of 10 to 20% for Sulfo646 and 100% for Alexa-546 and 16× line averaging were used. Images were collected using a time-gated Hybrid detector (0.5 to 6 ns). Images of 2048 × 2048 pixel had a pixel size of 18.9 nm.
Synthesis and in vitro characterization of cleavable fluorophores
3-((2-((((9H-Fluoren-9-yl)methoxy)carbonyl)amino)ethyl)disulfaneyl)propanoic acid (2)
A round-bottom flask was charged with 3-((2-aminoethyl)disulfaneyl)propanoic acid (1) (50 mg, 276 μmol, and 1.0 equiv) dissolved in 1 ml of Na2CO3 (20%) and stirred for 5 min. The mixture was cooled to 0°C and FmocCl (78.7 mg, 304 μmol, and 1.1 equiv) in 1,4-dioxane (1 ml) was added dropwise over 5 min and then stirred for 5 hours at room temperature. The reaction was quenched with acetic acid (AcOH) and taken up in acetonitrile (MeCN):H2O = 1:1, high-performance liquid chromatography [HPLC; MeCN:H2O + 0.1% trifluoroacetic acid (TFA) = 30:90 to 90:10 over 46 min] provided 35 mg (86.8 μmol, 31%) as a white solid after lyophilization [1H nuclear magnetic resonance (NMR) (600 MHz, CDCl3): δ (parts per million) = 7.76 (d, J = 7.56 Hz, 2H), 7.59 (d, J = 7.44 Hz, 2H), 7.40 (t, J = 7.47 Hz, 2H), 7.31 (t, J = 7.38 Hz, 2H), 5.16 (s, 1H), 4.41 (dd, J = 6.78 Hz, 2H), 4.22 (t, J = 6.67 Hz, 1H), 3.52 (q, J = 5.84 Hz, 2H), 2.93 (t, J = 6.84, 2H), and 2.79 (p, J = 6.52 Hz, 4H); 13C NMR (150 MHz, CDCl3): δ (parts per million) = 175.8, 156.4, 143.9, 141.3, 127.7, 127.1, 125.0 120.0 66.8, 47.2, 39.7, 38.0, 33.7, and 32.9; HRMS (ESI): calc; for C20H21NO4S2+ [M + Na]+: 426.0804, found: 426.0836].
(9H-Fluoren-9-yl)methyl(20-chloro-7-oxo-11,14-dioxa-3,4-dithia-8-azaicosyl)carbamate (3)
A round-bottom flask was charged with 2 (10 mg, 276 μmol, and 1.0 equiv) dissolved in 1 ml of dimethyl sulfoxide (DMSO) and N, N-Diisopropylethylamine (DIPEA; 34.5 μl, 198.4 μmol, and 8.0 equiv), and N,N,N′,N′-Tetramethyl-O-(N-succinimidyl)uronium tetrafluoroborate (TSTU; 11.2 mg, 37.2 μmol, and 1.5 equiv) was added, and the reaction was stirred for 15 min at room temperature. CA-NH2 (11.1 mg, 49.6 μmol, and 2.0 equiv) was added to the mixture and stirred for 15 min. The reaction was quenched in MeCN:H2O = 1:1 (2 ml), and HPLC purification (MeCN:H2O + 0.1% TFA = 30:90 to 90:10 over 46 min) provided 15 mg (276 μmol, quant.) of the desired product as a colorless oil after lyophilization [1H NMR (600 MHz, CDCl3): δ (parts per million) = 7.77 (d, J = 7.55 Hz, 2H), 7.61 (d, J = 7.44 Hz, 2H), 7.40 (t, J = 7.47 Hz, 2H), 7.31 (t, J = 7.52 Hz, 2H), 4.41 (d, J = 6.95 Hz, 2H), 4.22 (t, J = 6.77 Hz, 1H), 3.62 to 3.59 (m, 2H), 3.58 to 3.54 (m, 4H), 3.53 (t, J = 6.66 Hz, 4H), 3.50 to 3.44 (m, 4H), 2.99 (t, J = 6.79 Hz, 2H), 2.82 (t, J = 6.07 Hz, 2H), 2.61 (t, J = 6.81 Hz, 2H), 1.77 (p, J = 7.08, 2H), 1.60 (p, J = 7.15, 2H), 1.48 to 1.42 (m, 2H), and 1.40 to 1.34 (m, 2H); 13C NMR (150 MHz, CDCl3): δ (parts per million) = 143.9, 141.3, 127.7, 127.0, 125.1, 120.0, 71.3, 70.2, 70.0, 69.6, 66.8, 47.2, 45.0, 39.7, 39.4, 38.3, 36.2, 33.7, 32.5, 29.4, 26.7, and 25,4; HRMS (ESI): calc; for C30H41ClN2O5S2+ [M + Na]+: 631.2038, found: 631.2055].
2-(6-Amino-3-iminio-4,5-disulfo-3H-xanthen-9-yl)-4-((20-chloro-7-oxo-11,14-dioxa-3,4-dithia-8-azaicosyl)carbamoyl)benzoate (CA-SS–Alexa-488)
A round-bottom flask was charged with 3 (0.73 mg, 1.20 μmol, and 4.0 equiv) dissolved in 500 μl of MeCN + 1% 1,8-Diazabicyclo[5.4.0]undec-7-ene (DBU) and stirred for 1 hour, before being quenched with AcOH. The mixture was concentrated in vacuo, dissolved in 500 μl of DMSO, and DIPEA (0.8 μl, 4.80 μmol, and 16.0 equiv) was added, before Alexa-488–N-hydroxysuccinimide (189 μg, 300 nmol, and 1.0 equiv; Thermo Fisher Scientific, #A20000), and the reaction stirred for 15 min at room temperature. The reaction was quenched in MeCN:H2O = 1:1 (2 ml), HPLC (MeCN:H2O + 0.1% TFA = 10:90 to 90:10 over 46 min) provided 130 μg (144 nmol and 48%) of the desired product as a yellow powder over two steps after lyophilization [HRMS (ESI): calc; for C36H43ClN4O13S4+ [M + Na]+: 925.1290, found: 925.1312].
High-resolution mass spectrometry was performed using an Agilent Technologies 6230 series accurate mass time-of-flight liquid chromatography–mass spectrometry (LC-MS) linked to an Agilent Technologies 1290 Infinity Series machine with a Thermo Accucore RP-MS column, 2.6-μm pore size, 30 × 2.1 mm, and a 3-min gradient from 5 to 99% aqueous MeCN with 0.1% TFA and MeCN with 0.1% TFA (flow rate: 0.8 ml/min; ultraviolet detection: 220, 254, and 300 nm). Intact proteins were analyzed using a Waters H-class instrument equipped with a quaternary solvent manager, a Waters sample manager-FTN, a Waters PDA detector, and a Waters column manager with an ACQUITY UPLC protein BEH C4 column (300 Å, 1.7 μm, and 2.1 mm by 50 mm). Proteins were eluted with a flow rate of 0.3 ml/min at a column temperature of 80°C. The following gradient was used: A, 0.01% formic acid (FA) in H2O and B, 0.01% FA in MeCN (gradient 5 to 95% B from 0 to 6 min). Mass analysis was conducted with a Waters Xevo G2-XS QToF analyzer. Proteins were ionized in positive ion mode applying a one voltage of 40 kV. Raw data were analyzed with MaxEnt 1. After deconvolution of the crude spectra, no single or nonlabeled Halo-tag was observed, indicating complete reaction.
NMR spectra were recorded at 300 K in deuterated solvents on a Bruker AV-III spectrometers using either a cryogenically cooled 5-mm TCI-triple resonance probe equipped with one-axis self-shielded gradients or room temperature 5-mm broadband probe and calibrated to residual solvent peaks (1H/13C in parts per million): CDCl3-d6 (7.26/77.16). Multiplicities are abbreviated as follows: s, singlet; d, doublet; t, triplet; q, quartet; p, pentet; h, heptet; br, broad; and m, multiplet. Coupling constants J are reported in hertz. Spectra are reported on the basis of appearance, not on theoretical multiplicities derived from structural information. This also concerns the report on 13C NMR.
HPLC was performed on an Agilent 1260 Infinity II LC System equipped with columns as follows: preparative column: Reprospher 100 C18 columns (10 μm: 50 mm by 30 mm at flow rate of 20 ml/min; semipreparative column: 5 μm, 250 mm by 10 mm at flow rate of 4 ml/min). Eluents A (0.1% TFA in H2O) and B (0.1% TFA in MeCN) were applied as a linear gradient. Peak detection was performed at maximal absorbance wavelength.
LC-MS was performed on an Agilent 1260 Infinity II LC System equipped with Agilent SB-C18 column (1.8 μm; 2.1 mm by 50 mm; buffer A: 0.1% FA in H2O; buffer B: 0.1% FA in acetonitrile). The typical gradient was from 10% B for 1.0 min → gradient to 90% B over 6.0 min → 90% B for 1.0 min with flow rate of 0.8 ml/min. Retention times (tR) are given in minutes. Chromatograms were imported into GraphPad Prism 8. See fig. S6 (A and B) and Supplemental Chemical Characterization for further information.
Patch-clamp electrophysiology
To test the effects of MESNA treatment on the ability of mGluRs to couple to G proteins (fig. S6), we used an established whole cell patch-clamp assay to measure agonist-evoked G protein-coupled inwardly rectifying potassium channels (GIRK) currents as previously described (96). Briefly, cells were seeded at low density on poly-l-lysine–coated coverslips and transfected with SNAP-mGluR3 (0.7 μg), GIRK1-F137S (0.7 μg) (97), and tdTomato (0.15 μg). tdTomato positive cells were selected for the recordings. Whole-cell patch-clamp recordings were performed in a bath solution containing 120 mM KCl, 25 mM NaCl, 10 mM Hepes, 2 mM CaCl2, and 1 mM MgCl2. Pipettes of 3 to 5 megohm were filled with intracellular solution containing 140 mM KCl, 10 mM Hepes, 5 mM EGTA, 3 mM MgCl2, 3 mM Na2ATP (Sigma-Aldrich, #A7699), and 0.2 mM Na2GTP. Recordings were made using an Axopatch 200B amplifier coupled to a Digidata 1550B (Molecular Devices) using Clampex acquisition software. Glutamate dose response curves were acquired after treating cells with MESNA for 5 min at room temperature or under control conditions in gap-free mode at −60 mV. Glutamate-evoked potassium currents were measured at the peak, normalized to the saturating dose of glutamate (1 mM), and fitted to a curve using Clampfit (Molecular Devices) and Prism 9 (GraphPad).
Acknowledgments
We thank the Levitz laboratory and M. Lehmann (FMP) for helpful discussion and technical support.
Funding: This work was supported by NIH grant R35GM124731 (J.Lev.), NIH grant R01NS129904 (J.Lev.), NIH grant F32GM148001 (D.C.M.), the Rohr Family Research Scholar Award (J.Lev.), the Monique Weill-Caulier Award (J.Lev.), the Margarita Salas Fellowship from the Ministry of Universities of Spain (A.J.G.-H.), and NSF Graduate Research Fellowship (N.A.).
Author contributions: Conceptualization: J. Lee, A.J.G.-H., M.K., N.A., D.C.M., and J.Lev. Methodology: J. Lee, A.J.G.-H., K.R., J.B., and J.Lev. Investigation: J. Lee, A.J.G.-H., M.K., N.A., A.A., D.C.M., K.R., and J.B. Supervision: J.B. and J.Lev. Writing (original draft): J. Lee and J.Lev. Writing (review and editing): J. Lee, A.J.G.-H., D.C.M., J.B., and J.Lev.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Requests for materials should be submitted to jtl2003@med.cornell.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
NMR spectra
LCMS data
REFERENCES AND NOTES
- 1.C. M. Niswender, P. J. Conn, Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. Reiner, J. Levitz, Glutamatergic signaling in the central nervous system: Ionotropic and metabotropic receptors in concert. Neuron 98, 1080–1098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. Ellaithy, J. Gonzalez-Maeso, D. A. Logothetis, J. Levitz, Structural and biophysical mechanisms of Class C G protein-coupled receptor function. Trends Biochem. Sci. 45, 1049–1064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.E. Doumazane, P. Scholler, J. M. Zwier, E. Trinquet, P. Rondard, J. P. Pin, A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 25, 66–77 (2011). [DOI] [PubMed] [Google Scholar]
- 5.J. Levitz, C. Habrian, S. Bharill, Z. Fu, R. Vafabakhsh, E. Y. Isacoff, Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron 92, 143–159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. Lee, H. Munguba, V. A. Gutzeit, D. R. Singh, M. Kristt, J. S. Dittman, J. Levitz, Defining the homo- and heterodimerization propensities of metabotropic glutamate receptors. Cell Rep. 31, 107605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.T. W. McCullock, P. J. Kammermeier, The evidence for and consequences of metabotropic glutamate receptor heterodimerization. Neuropharmacology 199, 108801 (2021). [DOI] [PubMed] [Google Scholar]
- 8.J. Du, D. Wang, H. Fan, C. Xu, L. Tai, S. Lin, S. Han, Q. Tan, X. Wang, T. Xu, H. Zhang, X. Chu, C. Yi, P. Liu, X. Wang, Y. Zhou, J.-P. Pin, P. Rondard, H. Liu, J. Liu, F. Sun, B. Wu, Q. Zhao, Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature 594, 589–593 (2021). [DOI] [PubMed] [Google Scholar]
- 9.J. Meng, C. Xu, P.-A. Lafon, S. Roux, M. Mathieu, R. Zhou, P. Scholler, E. Blanc, J. A. J. Becker, J. Le Merrer, J. González-Maeso, P. Chames, J. Liu, J.-P. Pin, P. Rondard, Nanobody-based sensors reveal a high proportion of mGlu heterodimers in the brain. Nat. Chem. Biol. 18, 894–903 (2022). [DOI] [PubMed] [Google Scholar]
- 10.R. H. Oakley, S. A. Laporte, J. A. Holt, M. G. Caron, L. S. Barak, Differential affinities of visual arrestin, βarrestin1, and βarrestin2 for G protein-coupled receptors delineate two major classes of receptors. J. Biol. Chem. 275, 17201–17210 (2000). [DOI] [PubMed] [Google Scholar]
- 11.R. H. Oakley, S. A. Laporte, J. A. Holt, L. S. Barak, M. G. Caron, Association of β-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization. J. Biol. Chem. 274, 32248–32257 (1999). [DOI] [PubMed] [Google Scholar]
- 12.S. S. G. Ferguson, J. Zhang, L. S. Barakt, M. G. Caron, Molecular mechanisms of G protein-coupled receptor desensitization and resensitization. Life Sci. 62, 1561–1565 (1998). [DOI] [PubMed] [Google Scholar]
- 13.J. Zhang, L. S. Barak, P. H. Anborgh, S. A. Laporte, M. G. Caron, S. S. G. Ferguson, Cellular trafficking of g protein-coupled receptor/β-arrestin endocytic complexes. J. Biol. Chem. 274, 10999–11006 (1999). [DOI] [PubMed] [Google Scholar]
- 14.R. H. Oakley, S. A. Laporte, J. A. Holt, L. S. Barak, M. G. Caron, Molecular determinants underlying the formation of stable intracellular G protein-coupled receptor-β-arrestin complexes after receptor endocytosis*. J. Biol. Chem. 276, 19452–19460 (2001). [DOI] [PubMed] [Google Scholar]
- 15.M. von Zastrow, B. K. Kobilka, Ligand-regulated internalization and recycling of human beta 2-adrenergic receptors between the plasma membrane and endosomes containing transferrin receptors. J. Biol. Chem. 267, 3530–3538 (1992). [PubMed] [Google Scholar]
- 16.P. H. Anborgh, J. L. Seachrist, L. B. Dale, S. S. G. Ferguson, Receptor/beta-arrestin complex formation and the differential trafficking and resensitization of beta2-adrenergic and angiotensin ii type 1A receptors. Mol. Endocrinol. 14, 2040–2053 (2000). [DOI] [PubMed] [Google Scholar]
- 17.J. Trejo, S. R. Coughlin, The cytoplasmic tails of protease-activated receptor-1 and substance P receptor specify sorting to lysosomes versusrecycling. J. Biol. Chem. 274, 2216–2224 (1999). [DOI] [PubMed] [Google Scholar]
- 18.A. Marchese, M. M. Paing, B. R. S. Temple, J. Trejo, G protein–coupled receptor sorting to endosomes and lysosomes. Annu. Rev. Pharmacol. Toxicol. 48, 601–629 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.P. I. Tsao, M. von Zastrow, Type-specific sorting of G protein-coupled receptors after endocytosis. J. Biol. Chem. 275, 11130–11140 (2000). [DOI] [PubMed] [Google Scholar]
- 20.J. Janetzko, R. Kise, B. Barsi-Rhyne, D. H. Siepe, F. M. Heydenreich, K. Kawakami, M. Masureel, S. Maeda, K. C. Garcia, M. von Zastrow, A. Inoue, B. K. Kobilka, Membrane phosphoinositides regulate GPCR-β-arrestin complex assembly and dynamics. Cell 185, 4560–4573.e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.N. Abreu, A. Acosta-Ruiz, G. Xiang, J. Levitz, Mechanisms of differential desensitization of metabotropic glutamate receptors. Cell Rep. 35, 109050 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. S. Julie, J. M. Kinzie, M. S. Michi, P. S. Thomas, L. W. Gary, Cloning and expression of rat metabotropic glutamate receptor 8 reveals a distinct pharmacological profile. Mol. Pharmacol. 51, 119–125 (1997). [DOI] [PubMed] [Google Scholar]
- 23.P. Koulen, R. Kuhn, H. Wässle, J. H. Brandstätter, Modulation of the intracellular calcium concentration in photoreceptor terminals by a presynaptic metabotropic glutamate receptor. Proc. Natl. Acad. Sci. U.S.A. 96, 9909–9914 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. E. Ayala, C. M. Niswender, Q. Luo, J. L. Banko, P. J. Conn, Group III mGluR regulation of synaptic transmission at the SC-CA1 synapse is developmentally regulated. Neuropharmacology 54, 804–814 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. Shigemoto, A. Kinoshita, E. Wada, S. Nomura, H. Ohishi, M. Takada, P. J. Flor, A. Neki, T. Abe, S. Nakanishi, N. Mizuno, Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J. Neurosci. 17, 7503–7522 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.F. Nicoletti, J. Bockaert, G. L. Collingridge, P. J. Conn, F. Ferraguti, D. D. Schoepp, J. T. Wroblewski, J. P. Pin, Metabotropic glutamate receptors: From the workbench to the bedside. Neuropharmacology 60, 1017–1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. M. Duvoisin, C. Zhang, T. F. Pfankuch, H. O'Connor, J. Gayet-Primo, S. Quraishi, J. Raber, Increased measures of anxiety and weight gain in mice lacking the group III metabotropic glutamate receptor mGluR8. Eur. J. Neurosci. 22, 425–436 (2005). [DOI] [PubMed] [Google Scholar]
- 28.R. M. Duvoisin, L. Villasana, T. Pfankuch, J. Raber, Sex-dependent cognitive phenotype of mice lacking mGluR8. Behav. Brain Res. 209, 21–26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.I. Marabese, V. de Novellis, E. Palazzo, M. A. Scafuro, D. Vita, F. Rossi, S. Maione, Effects of (S)-3,4-DCPG, an mGlu8 receptor agonist, on inflammatory and neuropathic pain in mice. Neuropharmacology 52, 253–262 (2007). [DOI] [PubMed] [Google Scholar]
- 30.E. Palazzo, Y. Fu, G. Ji, S. Maione, V. Neugebauer, Group III mGluR7 and mGluR8 in the amygdala differentially modulate nocifensive and affective pain behaviors. Neuropharmacology 55, 537–545 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. S. Woo, F. Ufer, N. Rothammer, G. Di Liberto, L. Binkle, U. Haferkamp, J. K. Sonner, J. B. Engler, S. Hornig, S. Bauer, I. Wagner, K. Egervari, J. Raber, R. M. Duvoisin, O. Pless, D. Merkler, M. A. Friese, Neuronal metabotropic glutamate receptor 8 protects against neurodegeneration in CNS inflammation. J. Exp. Med. 218, e20201290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T. D. Prickett, X. Wei, I. Cardenas-Navia, J. K. Teer, J. C. Lin, V. Walia, J. Gartner, J. Jiang, P. F. Cherukuri, A. Molinolo, M. A. Davies, J. E. Gershenwald, K. Stemke-Hale, S. A. Rosenberg, E. H. Margulies, Y. Samuels, Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat. Genet. 43, 1119–1126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Koda, J. Hu, X. Ju, G. Sun, S. Shao, R.-X. Tang, K.-Y. Zheng, J. Yan, The role of glutamate receptors in the regulation of the tumor microenvironment. Front. Immunol. 14, 1123841 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Z. Fang, S. Chen, Y. Manchanda, S. Bitsi, P. Pickford, A. David, M. M. Shchepinova, I. R. Corrêa Jr., D. J. Hodson, J. Broichhagen, E. W. Tate, F. Reimann, V. Salem, G. A. Rutter, T. Tan, S. R. Bloom, A. Tomas, B. Jones, Ligand-specific factors influencing GLP-1 receptor post-endocytic trafficking and degradation in pancreatic beta cells. Int. J. Mol. Sci. 21, 8404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D. E. Keith, B. Anton, S. R. Murray, P. A. Zaki, P. C. Chu, D. V. Lissin, G. Monteillet-Agius, P. L. Stewart, C. J. Evans, M. von Zastrow, μ-Opioid receptor internalization: opiate drugs have differential effects on a conserved endocytic Mechanism in vitro and in the mammalian brain. Mol. Pharmacol. 53, 377–384 (1998). [PubMed] [Google Scholar]
- 36.C. Sternini, M. Spann, B. Anton, D. Keith Jr., N. W. Bunnett, M. Von Zastrow, C. Evans, N. C. Brecha, Agonist-selective endocytosis of mu opioid receptor by neurons in vivo. Proc. Natl. Acad. Sci. U.S.A. 93, 9241–9246 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.G. Scherrer, P. Tryoen-Tóth, D. Filliol, A. Matifas, D. Laustriat, Y. Q. Cao, A. I. Basbaum, A. Dierich, J.-L. Vonesh, C. Gavériaux-Ruff, B. L. Kieffer, Knockin mice expressing fluorescent δ-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 9691–9696 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R. G. Vickery, M. von Zastrow, Distinct dynamin-dependent and -independent mechanisms target structurally homologous dopamine receptors to different endocytic membranes. J. Cell Biol. 144, 31–43 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Odley, H. S. Hahn, R. A. Lynch, Y. Marreez, H. Osinska, J. Robbins, G. W. Dorn, Regulation of cardiac contractility by Rab4-modulated β2-adrenergic receptor recycling. Proc. Natl. Acad. Sci. U.S.A. 101, 7082–7087 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.F. R. Maxfield, T. E. McGraw, Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5, 121–132 (2004). [DOI] [PubMed] [Google Scholar]
- 41.P. Poc, V. A. Gutzeit, J. Ast, J. Lee, B. J. Jones, E. D'Este, B. Mathes, M. Lehmann, D. J. Hodson, J. Levitz, J. Broichhagen, Interrogating surface versus intracellular transmembrane receptor populations using cell-impermeable SNAP-tag substrates. Chem. Sci. 11, 7871–7883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. Tanowitz, M. von Zastrow, A novel endocytic recycling signal that distinguishes the membrane trafficking of naturally occurring opioid receptors. J. Biol. Chem. 278, 45978–45986 (2003). [DOI] [PubMed] [Google Scholar]
- 43.J. Cuitavi, L. Hipólito, M. Canals, The life cycle of the mu-opioid receptor. Trends Biochem. Sci. 46, 315–328 (2021). [DOI] [PubMed] [Google Scholar]
- 44.N. P. Martin, R. J. Lefkowitz, S. K. Shenoy, Regulation of V2 vasopressin receptor degradation by agonist-promoted ubiquitination *. J. Biol. Chem. 278, 45954–45959 (2003). [DOI] [PubMed] [Google Scholar]
- 45.D. L. Janet, S. S. Helen, E. C. Alexandra, O. Mehrnoosh, T. Elena, L. W. Sarah, C. Charles, M. H. Stephen, K. Eamonn, H. Graeme, P. B. Chris, Role of G protein–coupled receptor kinases 2 and 3 in μ-opioid receptor desensitization and internalization. Mol. Pharmacol. 88, 347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R. H. Moore, A. Tuffaha, E. E. Millman, W. Dai, H. S. Hall, B. F. Dickey, B. J. Knoll, Agonist-induced sorting of human β2-adrenergic receptors to lysosomes during downregulation. J. Cell Sci. 112, 329–338 (1999). [DOI] [PubMed] [Google Scholar]
- 47.X. E. Zhou, Y. He, P. W. de Waal, X. Gao, Y. Kang, N. Van Eps, Y. Yin, K. Pal, D. Goswami, T. A. White, A. Barty, N. R. Latorraca, H. N. Chapman, W. L. Hubbell, R. O. Dror, R. C. Stevens, V. Cherezov, V. V. Gurevich, P. R. Griffin, O. P. Ernst, K. Melcher, H. E. Xu, Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell 170, 457–469.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.P. Isaikina, I. Petrovic, R. P. Jakob, P. Sarma, A. Ranjan, M. Baruah, V. Panwalkar, T. Maier, A. K. Shukla, S. Grzesiek, A key GPCR phosphorylation motif discovered in arrestin2•CCR5 phosphopeptide complexes. bioRxiv 2022.2010.2010.511578 [Preprint] (2022). [DOI] [PubMed]
- 49.C. Corti, S. Restituito, J. M. Rimland, I. Brabet, M. Corsi, J. P. Pin, F. Ferraguti, Cloning and characterization of alternative mRNA forms for the rat metabotropic glutamate receptors mGluR7 and mGluR8. Eur. J. Neurosci. 10, 3629–3641 (1998). [DOI] [PubMed] [Google Scholar]
- 50.D. Moreno Delgado, T. C. Møller, J. Ster, J. Giraldo, D. Maurel, X. Rovira, P. Scholler, J. M. Zwier, J. Perroy, T. Durroux, E. Trinquet, L. Prezeau, P. Rondard, J. P. Pin, Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells. eLife 6, e25233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.P. J. Kammermeier, Functional and pharmacological characteristics of metabotropic glutamate receptors 2/4 heterodimers. Mol. Pharmacol. 82, 438–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.C. H. Habrian, J. Levitz, V. Vyklicky, Z. Fu, A. Hoagland, I. McCort-Tranchepain, F. Acher, E. Y. Isacoff, Conformational pathway provides unique sensitivity to a synaptic mGluR. Nat. Commun. 10, 5572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R. C. Werthmann, M. Tzouros, J. Lamerz, A. Augustin, T. Fritzius, L. Trovò, M. Stawarski, A. Raveh, C. Diener, C. Fischer, M. Gassmann, L. Lindemann, B. Bettler, Symmetric signal transduction and negative allosteric modulation of heterodimeric mGlu1/5 receptors. Neuropharmacology 190, 108426 (2021). [DOI] [PubMed] [Google Scholar]
- 54.S. Yin, M. J. Noetzel, K. A. Johnson, R. Zamorano, N. Jalan-Sakrikar, K. J. Gregory, P. J. Conn, C. M. Niswender, Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J. Neurosci. 34, 79–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.P. Scholler, D. Nevoltris, D. de Bundel, S. Bossi, D. Moreno-Delgado, X. Rovira, T. C. Møller, D. El Moustaine, M. Mathieu, E. Blanc, H. McLean, E. Dupuis, G. Mathis, E. Trinquet, H. Daniel, E. Valjent, D. Baty, P. Chames, P. Rondard, J.-P. Pin, Allosteric nanobodies uncover a role of hippocampal mGlu2 receptor homodimers in contextual fear consolidation. Nat. Commun. 8, 1967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. K. Thibado, J. Y. Tano, J. Lee, L. Salas-Estrada, D. Provasi, A. Strauss, J. M. L. Ribeiro, G. Xiang, J. Broichhagen, M. Filizola, M. J. Lohse, J. Levitz, Differences in interactions between transmembrane domains tune the activation of metabotropic glutamate receptors. eLife 10, e67027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.F.-T. Lin, K. M. Krueger, H. E. Kendall, Y. Daaka, Z. L. Fredericks, J. A. Pitcher, R. J. Lefkowitz, Clathrin-mediated endocytosis of the β-adrenergic receptor is regulated by phosphorylation/dephosphorylation of β-arrestin1. J. Biol. Chem. 272, 31051–31057 (1997). [DOI] [PubMed] [Google Scholar]
- 58.T.-H. Lan, S. Kuravi, N. A. Lambert, Internalization dissociates β2-adrenergic receptors. PLOS ONE 6, e17361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Z. Fang, S. Chen, P. Pickford, J. Broichhagen, D. J. Hodson, I. R. Corrêa Jr., S. Kumar, F. Görlitz, C. Dunsby, P. M. W. French, G. A. Rutter, T. Tan, S. R. Bloom, A. Tomas, B. Jones, The influence of peptide context on signaling and trafficking of glucagon-like peptide-1 receptor biased agonists. ACS Pharmacol. Transl. Sci. 3, 345–360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.V. Bitsikas, I. R. Corrêa Jr., B. J. Nichols, Clathrin-independent pathways do not contribute significantly to endocytic flux. eLife 3, e03970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.T. Kukaj, C. Sattler, T. Zimmer, R. Schmauder, K. Benndorf, Kinetic fingerprinting of metabotropic glutamate receptors. Commun. Biol. 6, 104 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.X. Lin, N. M. Fisher, S. Dogra, R. K. Senter, C. W. Reed, J. J. Kalbfleisch, C. W. Lindsley, W. B. Asher, Z. Xiang, C. M. Niswender, J. A. Javitch, Differential activity of mGlu7 allosteric modulators provides evidence for mGlu7/8 heterodimers at hippocampal Schaffer collateral-CA1 synapses. J. Biol. Chem. 298, 102458 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.J. Liu, Z. Zhang, D. Moreno-Delgado, J. A. R. Dalton, X. Rovira, A. Trapero, C. Goudet, A. Llebaria, J. Giraldo, Q. Yuan, P. Rondard, S. Huang, J. Liu, J.-P. Pin, Allosteric control of an asymmetric transduction in a G protein-coupled receptor heterodimer. eLife 6, e26985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.H. B. Gosnell, Y. Silberman, B. A. Grueter, R. M. Duvoisin, J. Raber, D. G. Winder, mGluR8 modulates excitatory transmission in the bed nucleus of the stria terminalis in a stress-dependent manner. Neuropsychopharmacology 36, 1599–1607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.J. B. Kuzmiski, Q. J. Pittman, J. S. Bains, Metaplasticity of hypothalamic synapses following in vivo challenge. Neuron 62, 839–849 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R. M. Duvoisin, T. Pfankuch, J. M. Wilson, J. Grabell, V. Chhajlani, D. G. Brown, E. Johnson, J. Raber, Acute pharmacological modulation of mGluR8 reduces measures of anxiety. Behav. Brain Res. 212, 168–173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.F. Ferraguti, Metabotropic glutamate receptors as targets for novel anxiolytics. Curr. Opin. Pharmacol. 38, 37–42 (2018). [DOI] [PubMed] [Google Scholar]
- 68.D. Shalini, P. J. Conn, Metabotropic glutamate receptors as emerging targets for the treatment of schizophrenia. Mol. Pharmacol. 101, 275–285 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.S. Chiechio, F. Nicoletti, Metabotropic glutamate receptors and the control of chronic pain. Curr. Opin. Pharmacol. 12, 28–34 (2012). [DOI] [PubMed] [Google Scholar]
- 70.S. H. Li, K. S. Abd-Elrahman, S. S. G. Ferguson, Targeting mGluR2/3 for treatment of neurodegenerative and neuropsychiatric diseases. Pharmacol. Ther. 239, 108275 (2022). [DOI] [PubMed] [Google Scholar]
- 71.C. A. Purgert, Y. Izumi, Y.-J. I. Jong, V. Kumar, C. F. Zorumski, K. L. O’Malley, Intracellular mGluR5 can mediate synaptic plasticity in the hippocampus. J. Neurosci. 34, 4589–45898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. Stoeber, D. Jullié, B. T. Lobingier, T. Laeremans, J. Steyaert, P. W. Schiller, A. Manglik, M. von Zastrow, A genetically encoded biosensor reveals location bias of opioid drug action. Neuron 98, 963–976.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R. Rozenfeld, L. A. Devi, Regulation of CB1 cannabinoid receptor trafficking by the adaptor protein AP-3. FASEB J. 22, 2311–2322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D. Jullié, M. Stoeber, J.-B. Sibarita, H. L. Zieger, T. M. Bartol, S. Arttamangkul, T. J. Sejnowski, E. Hosy, M. von Zastrow, A discrete presynaptic vesicle cycle for neuromodulator receptors. Neuron 105, 663–677.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.D. Jullié, C. Benitez, T. A. Knight, M. S. Simic, M. von Zastrow, Endocytic trafficking determines cellular tolerance of presynaptic opioid signaling. eLife 11, e81298 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.S. Siddig, S. Aufmkolk, S. Doose, M.-L. Jobin, C. Werner, M. Sauer, D. Calebiro, Super-resolution imaging reveals the nanoscale organization of metabotropic glutamate receptors at presynaptic active zones. Sci. Adv. 6, eaay7193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.A. Bodzęta, F. Berger, H. D. MacGillavry, Subsynaptic mobility of presynaptic mGluR types is differentially regulated by intra- and extracellular interactions. Mol. Biol. Cell 33, ar66 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.S. Lee, S. Park, H. Lee, S. Han, J.-m. Song, D. Han, Y. H. Suh, Nedd4 E3 ligase and beta-arrestins regulate ubiquitination, trafficking, and stability of the mGlu7 receptor. eLife 8, e44502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.W. B. Asher, P. Geggier, M. D. Holsey, G. T. Gilmore, A. K. Pati, J. Meszaros, D. S. Terry, S. Mathiasen, M. J. Kaliszewski, M. D. McCauley, A. Govindaraju, Z. Zhou, K. G. Harikumar, K. Jaqaman, L. J. Miller, A. W. Smith, S. C. Blanchard, J. A. Javitch, Single-molecule FRET imaging of GPCR dimers in living cells. Nat. Methods 18, 397–405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Y. Shiraishi, Y. Kofuku, T. Ueda, S. Pandey, H. Dwivedi-Agnihotri, A. K. Shukla, I. Shimada, Biphasic activation of β-arrestin 1 upon interaction with a GPCR revealed by methyl-TROSY NMR. Nat. Commun. 12, 7158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.N. R. Latorraca, M. Masureel, S. A. Hollingsworth, F. M. Heydenreich, C.-M. Suomivuori, C. Brinton, R. J. L. Townshend, M. Bouvier, B. K. Kobilka, R. O. Dror, How GPCR phosphorylation patterns orchestrate arrestin-mediated signaling. Cell 183, 1813–1825.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.F. Yang, X. Yu, C. Liu, C.-X. Qu, Z. Gong, H.-D. Liu, F.-H. Li, H.-M. Wang, D.-F. He, F. Yi, C. Song, C.-L. Tian, K.-H. Xiao, J.-Y. Wang, J.-P. Sun, Phospho-selective mechanisms of arrestin conformations and functions revealed by unnatural amino acid incorporation and 19F-NMR. Nat. Commun. 6, 8202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.J. L. Whistler, J. Enquist, A. Marley, J. Fong, F. Gladher, P. Tsuruda, S. R. Murray, M. von Zastrow, Modulation of postendocytic sorting of G protein-coupled receptors. Science 297, 615–620 (2002). [DOI] [PubMed] [Google Scholar]
- 84.A. Marchese, J. L. Benovic, Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J. Biol. Chem. 276, 45509–45512 (2001). [DOI] [PubMed] [Google Scholar]
- 85.M. R. Dores, H. Lin, N. J. Grimsey, F. Mendez, J. Trejo, The α-arrestin ARRDC3 mediates ALIX ubiquitination and G protein–coupled receptor lysosomal sorting. Mol. Biol. Cell 26, 4660–4673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D. El Moustaine, S. Granier, E. Doumazane, P. Scholler, R. Rahmeh, P. Bron, B. Mouillac, J.-L. Banères, P. Rondard, J.-P. Pin, Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc. Natl. Acad. Sci. 109, 16342–16347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.M. von Zastrow, A. Sorkin, Mechanisms for regulating and organizing receptor signaling by endocytosis. Annu. Rev. Biochem. 90, 709–737 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.J. Haubrich, J. Font, R. B. Quast, A. Goupil-Lamy, P. Scholler, D. Nevoltris, F. Acher, P. Chames, P. Rondard, L. Prézeau, J.-P. Pin, A nanobody activating metabotropic glutamate receptor 4 discriminates between homo- and heterodimers. Proc. Natl. Acad. Sci. U.S.A. 118, e2105848118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.C. H. Kim, J. Lee, J. Y. Lee, K. W. Roche, Metabotropic glutamate receptors: Phosphorylation and receptor signaling. J. Neurosci. Res. 86, 1–10 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Y. H. Suh, K. Chang, K. W. Roche, Metabotropic glutamate receptor trafficking. Mol. Cell. Neurosci. 91, 10–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.A. Bodzęta, N. Scheefhals, H. D. MacGillavry, Membrane trafficking and positioning of mGluRs at presynaptic and postsynaptic sites of excitatory synapses. Neuropharmacology 200, 108799 (2021). [DOI] [PubMed] [Google Scholar]
- 92.R. Birke, J. Ast, D. A. Roosen, J. Lee, K. Roßmann, C. Huhn, B. Mathes, M. Lisurek, D. Bushiri, H. Sun, B. Jones, M. Lehmann, J. Levitz, V. Haucke, D. J. Hodson, J. Broichhagen, Sulfonated red and far-red rhodamines to visualize SNAP- and Halo-tagged cell surface proteins. Org. Biomol. Chem. 20, 5967–5980 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.G. Xiang, A. Acosta-Ruiz, A. Radoux-Mergault, M. Kristt, J. Kim, J. D. Moon, J. Broichhagen, A. Inoue, F. S. Lee, M. Stoeber, J. S. Dittman, J. Levitz, Control of Gαq signaling dynamics and GPCR cross-talk by GRKs.Sci. Adv.8, eabq3363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.W. Stauffer, H. Sheng, H. N. Lim, EzColocalization: An ImageJ plugin for visualizing and measuring colocalization in cells and organisms. Sci. Rep. 8, 15764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.M. H. Ulbrich, E. Y. Isacoff, Subunit counting in membrane-bound proteins. Nat. Methods 4, 319–321 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.V. A. Gutzeit, A. Acosta-Ruiz, H. Munguba, S. Häfner, A. Landra-Willm, B. Mathes, J. Mony, D. Yarotski, K. Börjesson, C. Liston, G. Sandoz, J. Levitz, J. Broichhagen, A fine-tuned azobenzene for enhanced photopharmacology in vivo. Chem. Biol. 28, 1648–1663.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.M. Vivaudou, K. W. Chan, J.-L. Sui, L. Y. Jan, E. Reuveny, D. E. Logothetis, Probing the G-protein regulation of GIRK1 and GIRK4, the two subunits of the KACh channel, using functional homomeric mutants. J. Biol. Chem. 272, 31553–31560 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
NMR spectra
LCMS data