Abstract
Helicobacter pylori, a major cause of human gastric disease, is a microaerophilic bacterium that contains neither pyruvate nor 2-oxoglutarate dehydrogenase activity. Previous studies (N. J. Hughes, P. A. Chalk, C. L. Clayton, and D. J. Kelly, J. Bacteriol. 177:3953–3959, 1995) have indicated that the major routes for the generation of acetyl coenzyme A (acetyl-CoA) and succinyl-CoA are via pyruvate:flavodoxin oxidoreductase (POR) and 2-oxoglutarate:acceptor oxidoreductase (OOR), respectively. The purified POR is a heterotetrameric protein, with subunits of 48 (PorA), 36 (PorB), 24 (PorC), and 14 (PorD) kDa. In this study OOR has been purified, and it is similarly composed of polypeptides of 43 (OorA), 33 (OorB), 24 (OorC), and 10 (OorD) kDa. Both POR and OOR are oxygen labile and are likely to be major contributors to the microaerophilic phenotype of H. pylori. Unlike POR, OOR was unable to use a previously identified flavodoxin (FldA) as an electron acceptor. Although the purified enzymes were unable to reduce NAD(P), electrons from both pyruvate and 2-oxoglutarate could reduce NADP in cell extracts, consistent with a role for these oxidoreductases in the provision of NADPH as a respiratory electron donor. The H. pylori por, oor, and fldA genes were cloned and sequenced. The deduced por gene products showed significant sequence similarity to archaeal four-subunit 2-oxoacid:acceptor oxidoreductases. However, the amino acid sequences of OorA and -B were more closely related to that of the two-subunit POR of the aerobic halophile Halobacterium halobium. Both porD and oorD encode integral ferredoxin-like subunits. POR and OOR are probably essential enzymes in H. pylori, as insertion inactivation of porB and oorA appeared to be lethal.
Helicobacter pylori is a curved or spiral gram-negative, microaerophilic bacterium which is now recognized as the major etiological agent in chronic active type B gastritis. H. pylori infection is also strongly associated with gastric and duodenal ulceration (42), and long-term infection may predispose individuals to the development of gastric carcinoma (15, 31). In the western world, about 30 to 50% of individuals may be infected with H. pylori, making it among the most common infections in humans.
The physiology and metabolism of H. pylori are not well understood. The bacterium possesses enzymes of the Entner-Doudoroff and pentose-phosphate pathways (5, 29, 30), but although it is capable of glucose metabolism, respiratory activity is greater with some organic acids, such as pyruvate (6). Recently, it was shown that in contrast to the usual pyruvate dehydrogenase complex employed by aerobes, the main route for pyruvate assimilation in H. pylori is via a pyruvate:flavodoxin oxidoreductase (POR) (EC 1.2.7.1) (20). This enzyme was purified to homogeneity and shown to belong to a newly recognized group of four-subunit PORs. The in vivo electron acceptor of this enzyme is likely to be an endogenous flavodoxin, which was partially purified and shown to be reduced by POR. A 2-oxoglutarate:acceptor oxidoreductase (OOR) activity (EC 1.2.7.3) has also been detected in H. pylori (19, 20), catalyzing the analogous, reversible oxidative decarboxylation of 2-oxoglutarate to form succinyl coenzyme A (succinyl-CoA), a major intermediate of the tricarboxylic acid (TCA) cycle. OOR enzymes have been purified from a number of thermophilic bacteria and archaea. The OOR enzyme of Thermococcus litoralis is composed of four subunits, which have molecular masses comparable to those of four-subunit POR enzymes (25), whereas the OORs of Halobacterium halobium and Hydrogenobacter thermophilus consist of two subunits of 88 and 36 kDa (21) and 70 and 35 kDa (45), respectively. In an analogous fashion to POR, OOR enzymes catalyze the reduction of low-potential electron acceptors, such as ferredoxins (Fd), according to the following reaction: 2-oxoglutarate + CoA + Fdox ↔ succinyl-CoA + CO2 + Fdred
The presence of these 2-oxoacid:acceptor oxidoreductases in H. pylori is of importance for several reasons. First, unlike the 2-oxoacid dehydrogenase multienzyme complexes, these enzymes are highly oxygen labile. This may at least partly explain the microaerophilic nature of H. pylori. Second, POR has been demonstrated to be involved in the reduction of metronidazole (20), a drug commonly used in H. pylori eradication regimes. The reduction of the drug’s nitro group is essential to generate various short-lived derivatives capable of causing DNA damage (see reference 14 for review). Metronidazole resistance in H. pylori isolates is commonly observed (16, 17), but the molecular basis for resistance is unclear. Hoffman et al. (19) have reported complex metabolic changes in metronidazole-resistant mutants of H. pylori, including an apparent repression of POR activity. More recently, Smith and Edwards (38) reported that oxygen scavenging is compromised in resistant strains, as isogenic mutants were found to possess approximately one-third the NADH oxidase activity of wild-type strains. The resulting increase in intracellular oxygen in resistant strains may thus prevent metronidazole activation by increasing futile cycling to the inactive, oxidized form.
In anaerobic bacteria, the generation of low-potential reductants, such as ferredoxin or flavodoxin, by POR or OOR can be utilized to power a number of processes, including hydrogen evolution (2, 11) and nitrogen fixation (4, 34, 35). In this paper, a role in the provision of NADPH for respiratory electron transport in H. pylori is proposed. We also report the purification of the H. pylori OOR enzyme; the cloning and characterization of the por, oor, and fldA genes of H. pylori NCTC 11637; and the expression of POR in Escherichia coli. This study also allows definitive functions to be assigned to several genes identified in the recently released complete genome sequence of H. pylori 26695 (40).
MATERIALS AND METHODS
Enzymes and chemicals.
Restriction endonucleases were purchased from Promega and MBI Fermentas. Recombinant Pyrococcus furiosus DNA polymerase (Pfu) was obtained from Stratagene. Hyperfilm MP and an l-U-14C-amino acid mixture (1.85 to 2.2 GBq mmol−1) were obtained from Amersham. All other chemicals were purchased from Sigma.
Bacterial strains.
H. pylori (NCTC 11637) was obtained from the National Collection of Type Cultures, Colindale, England. Helicobacter cinaedi, Helicobacter nemestrinae, Helicobacter felis, Helicobacter muridarium, Helicobacter acinonyx, Helicobacter mustelae, Campylobacter jejuni, Campylobacter coli, and H. pylori 4187E and 8091 were obtained from A. McClaren, Glaxo-Wellcome Ltd. Expression of H. pylori POR was carried out with the vector pET21a (Novagen), which was propagated in E. coli XL-1 Blue (Stratagene). E. coli BL21(DE3) was used as the host strain for POR expression studies.
Growth of bacterial strains.
All species of Helicobacter were routinely cultivated under microaerobic conditions (5% O2, 10% CO2, 85% N2 [all vol/vol]) at 37°C on Columbia agar supplemented with 5% (vol/vol) chocolated horse blood and 10 μg each of amphotericin B, vancomycin, and polymyxin B ml−1. C. jejuni and C. coli were grown on the same medium except that polymyxin B was omitted. For liquid culture, brain heart infusion (BHI) broth plus 5% (vol/vol) horse or fetal calf serum, supplemented with the antibiotics described above, was used.
POR and OOR assays.
Quantitative measurements of POR activity were carried out with the artificial electron acceptor methyl viologen, using the assay previously described (20). A rapid-screening technique with 96-well microtiter plates was used to detect OOR-containing fractions from purification experiments. Each microtiter assay mixture contained the following: enzyme fraction, 100 to 200 μl; 1 M 2-oxoglutarate, 2 μl; 1 M methyl viologen, 1 μl; and 100 mM CoA, 1 μl. The wells were covered with a layer of mineral oil immediately after the addition of enzyme to aid in the exclusion of oxygen. OOR-containing fractions rapidly turned blue, before being reoxidized. The quantitative assay for OOR was carried out by a method identical to that described for POR (20) but with 5 mM pyruvate replaced by 5 mM 2-oxoglutarate.
Purification of H. pylori POR and partial purification of flavodoxin FldA.
Purification of POR and partial purification of FldA were performed as previously described (20).
Partial purification of OOR.
In the following protocol, the column chromatography was carried out in air, but the exposure of the enzyme to oxygen was reduced by flushing all buffers with N2 prior to use and transferring all purification fractions and enzyme extracts immediately into vessels also flushed with N2. For the small-scale purification of H. pylori OOR, 1.4 g (wet weight) of NCTC 11637 cells, grown in BHI broth under microaerobic conditions, was resuspended in 5 ml of 10 mM Tris-HCl (pH 8.0)–1 mM dithiothreitol. The cells were sonicated (three cycles of 15 s) and centrifuged for 30 min at 100,000 × g at 4°C. The supernatant was loaded onto a MonoQ HR5/5 ion-exchange column (Pharmacia) preequilibrated in 50 mM Tris-HCl (pH 8.0)–1 mM dithiothreitol (buffer A). The column was washed with 10 column volumes of buffer A plus 50 mM KCl. The activity was eluted with an increasing linear gradient of KCl from 50 to 250 mM over 15 column volumes. Active fractions were pooled and brought to 1.5 M ammonium sulfate. The sample was then loaded onto a phenyl-Superose HR5/5 column (Pharmacia) equilibrated with 1.5 M ammonium sulfate in buffer A, and any unbound protein was removed by washing with 5 column volumes. The column was developed with a decreasing linear gradient of ammonium sulfate from 1.5 to 0.75 M over 30 column volumes. Active fractions were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For N-terminal sequencing, proteins were separated on an SDS–12% polyacrylamide gel, electroblotted onto a polyvinylidene difluoride membrane, and stained with amido black, and individual bands were excised for N-terminal sequencing by the automated Edman degradation procedure.
Substrate oxidation by H. pylori sonicated cells and membrane fractions.
For respiration studies, 500 ml of BHI broth was inoculated with H. pylori NCTC 11637 and grown under microaerobic conditions with gentle agitation for 24 h. Cells were harvested at 8,000 × g for 15 min at 4°C, washed once in 20 mM phosphate buffer (pH 7.0), and resuspended in a small volume of the same buffer. Cells were lysed by sonication, and cell debris and unbroken cells were removed by centrifugation at 13,000 × g for 20 min at 4°C. The cell extract was then stored on ice for use the same day. Membranes were prepared by centrifugation of the cell extract (150,000 × g, 2 h, 4°C), and the pelleted membranes were resuspended in phosphate buffer. Substrate oxidation was determined by measuring changes in the dissolved oxygen concentrations in a Clarke-type oxygen electrode calibrated with air-saturated 20 mM potassium phosphate buffer, pH 7.0 (219 nmol of dissolved O2 ml−1 at 37°C). Baselines were determined by reducing all dissolved oxygen with excess sodium dithionite. Cell or membrane suspensions were maintained at 37°C and stirred at a constant rate.
Isolation and manipulation of DNA.
Helicobacter and Campylobacter genomic DNAs were isolated by the method described by Majewski and Goodwin (27). Plasmid DNAs were purified with the Qiagen Plasmid Midi Kit. All standard molecular biology protocols were carried out as described by Sambrook et al. (33).
Detection of heterologous por and oor genes.
Southern blotting was carried out by using digoxigenin labeling and chemiluminescence detection according to the instructions of the manufacturer (Boehringer Mannheim). Genomic DNAs were digested overnight with HindIII, and the fragments were separated on a 0.7% (wt/vol) agarose gel and blotted onto a nylon membrane (Hybond N; Amersham). Hybridization with digoxigenin-labeled porA and oorB probes and high-stringency washes (0.1% [vol/vol] SSC, 0.1% [wt/vol] SDS) were performed at 68°C.
DNA sequencing.
Sequencing was carried out by the Perkin-Elmer Dye Terminator method. Sequence analysis was carried out with the Wisconsin Genetics Computer Group package. Random H. pylori NCTC 11637 sequences were generated from genomic DNA which was sonicated and partially digested with four base cutter restriction enzymes. Fragments were then cloned into pBluescript (Stratagene) prior to sequencing with T7 and T3 primers. The gridded H. pylori NCTC 11637 plasmid library was generated by partial digestion of genomic DNA with Sau3A to generate fragments with an average size of 4 kb. Fragments were cloned into pUC18, and E. coli TG1 was transformed with the library. Six thousand recombinants, corresponding to a 14-fold representation of the genome, were then gridded onto Hybond N (Amersham) membranes in preparation for probing.
Expression of H. pylori POR in E. coli.
Primers were designed to introduce an NdeI restriction site (underlined) at the porC start codon (5′-ACGTCATATGTTTCAAATTAGATGGCATGCA-3′) and a BamHI site in a sequence downstream of the 3′ end of porB (5′-ACGTGGATCCTGGTTTGCTCATTCCATAGGGCTT-3′). PCR was carried out on H. pylori (NCTC 11637) genomic DNA with Pfu DNA polymerase on a Perkin-Elmer DNA Thermal Cycler 480 (25 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 45°C, and extension for 4 min at 72°C). The 3.4-kb PCR product was ligated into NdeI/BamHI-restricted pET21a to generate plasmid pNJH301, which was ultimately transformed into E. coli BL21(DE3). For expression, BL21(DE3, pNJH301) cells were grown aerobically in Luria-Bertani medium with 100 μg of ampicillin ml−1 to mid-exponential phase. The cells were harvested, washed once in M9 medium (33), resuspended in M9 plus 0.2% glucose, and grown aerobically with shaking for 90 min. IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM) was then added, and the culture was incubated for a further 90 min in a microaerobic atmosphere (5% O2, 10% CO2, 85% N2 [vol/vol]), or in air in order to test expression under aerobic conditions. Rifampin was then added to a final concentration of 200 μg ml−1, and the incubation was continued for a further 90 min under microaerobic conditions. The cells were then harvested by centrifugation and resuspended in 2 ml of buffer A. All buffers and vessels used to store the enzyme were flushed with N2 prior to being sealed to reduce exposure of the enzyme to oxygen. The cell suspension was then sonicated (five times for 15 s each), an oxygen-scavenging system (0.1 U of catalase, 3 U of glucose oxidase, and 0.4% [wt/vol] glucose) was added, and after centrifugation (14,000 × g, 10 min, 4°C) the supernatant was collected and stored in screw-cap tubes on ice prior to assaying.
Radiolabeling of proteins expressed from pNJH301.
The cells were propagated as described above, except that aerobic conditions were used throughout. After the incubation in the presence of rifampin, 1 μCi of a 14C-amino acid mixture was added to the cell suspension and incubated aerobically at 37°C for 2 h. Yeast extract was then added to give a final concentration of 0.1% (wt/vol), the cells were harvested, and a cell extract was prepared as described above. Approximately 10 μg of cell extract protein was used for SDS-PAGE analysis. Following electrophoresis, the gel was stained with Coomassie blue, destained and soaked in Amplify (Amersham), and then vacuum dried and exposed to Hyperfilm MP (Amersham) for 72 h at −70°C.
Insertion inactivation of por and oor genes.
Plasmid pAL21 contained a 774-bp insert cloned into pBluescript, of which 753 bp was the 5′ end of the porB gene. A 0.7-kb DNA fragment encoding the chloramphenicol acetyltransferase gene from C. coli (43) was ligated into a unique NcoI site in porB, flanked by 183 and 585 bp of insert DNA on either side, to give pAL21R. The pUC18-based plasmid construct pNJH4B9S contained a 1.35-kb SphI fragment encoding the 3′ end of oorA and the 5′ end of oorB. A 1.1-kb PCR-derived kanamycin resistance cassette containing the aphIII gene from C. coli (41) was ligated into a unique NcoI site in oorA in pNJH4B9S to give pNJH4B9SK. The chloramphenicol acetyltransferase cassette described above was also ligated into a unique MscI site in oorA to give plasmid pNJH4B9SC. The plasmids pAL21R, pNJH4B9SK, and pNJH4B9SC were then introduced into H. pylori NCTC 11637 by natural transformation according to the following protocol. Ten milliliters of BHI broth plus serum and antibiotics was inoculated with H. pylori and grown overnight. The culture was then diluted to give a starting optical density at 710 nm of approximately 0.1 U and allowed to grow for a further 8 h. Plasmid DNA (2 μg) was then added to 1 ml of this culture and incubated overnight with agitation in a microaerobic atmosphere. The cells were then pelleted, resuspended in 100 μl of BHI, and spread onto Columbia agar supplemented with horse blood, the standard antibiotics listed above, and either 30 μg of chloramphenicol ml−1 or 50 μg of kanamycin ml−1 to select for mutants arising by homologous recombination. The resulting agar plates were grown for 1 week and then examined for the appearance of resistant colonies.
Nucleotide sequence accession numbers.
The nucleotide sequences of the H. pylori por, oor, and fldA genes have been submitted to the GenBank/EMBL data bank and assigned accession numbers AF021092, AF021094, and AF021093, respectively.
RESULTS
Partial purification and N-terminal sequencing of the H. pylori OOR.
Cell extracts of H. pylori contain both pyruvate- and 2-oxoglutarate-dependent oxidoreductase activities, which are capable of reducing low-potential artificial electron acceptors (19, 20). As previous work had shown that the purified H. pylori POR enzyme was specific for pyruvate decarboxylation (20), the 2-oxoglutarate activity must be due to a separate enzyme. This activity was partially purified by using a two-step procedure involving ion-exchange chromatography on MonoQ (Pharmacia) and hydrophobic interaction chromatography on phenyl-Superose (Pharmacia). This resulted in a greater-than-60-fold increase in specific OOR activity but yielded less than 5% of the original activity (Table 1). The low yield was most likely due to the inevitable exposure of the enzyme to O2 during purification. With cell extract stored anaerobically on ice for 24 h, the addition of 1 mM thiamine pyrophosphate (TPP) to the standard OOR assay mixture increased OOR activity by 133%. However, the addition of TPP failed to stimulate the POR activity of this extract. Thus, the low yield may also be influenced by the leaching of TPP from the native OOR enzyme complex. OOR activity eluted from the MonoQ column at 0.15 M NaCl, just ahead of POR, which eluted at 0.19 M NaCl. The OOR enzyme was extremely unstable and could not be further purified by gel filtration chromatography without a complete loss of activity. The purified preparation retained OOR activity for only approximately 12 h when stored on ice under anaerobic conditions and was completely inactivated after freezing to −20 or −70°C. The omission of dithiothreitol from chromatography buffers and the exposure of the cell extract to oxygen for extended periods also resulted in inactivation of the enzyme.
TABLE 1.
Purification scheme for H. pylori OORa
| Step | Activity (U)b | Protein (mg) | Sp act (U mg−1) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 0.775 | 51.23 | 0.015 | 100 | 1 |
| MonoQ | 0.071 | 0.454 | 0.156 | 9.1 | 10 |
| Phenyl-Superose | 0.037 | 0.040 | 0.925 | 4.8 | 62 |
Data are for the partial purification of OOR from 1.4 g (wet weight) of H. pylori NCTC 11637 cells.
Units of activity are micromoles of methyl viologen reduced minute−1.
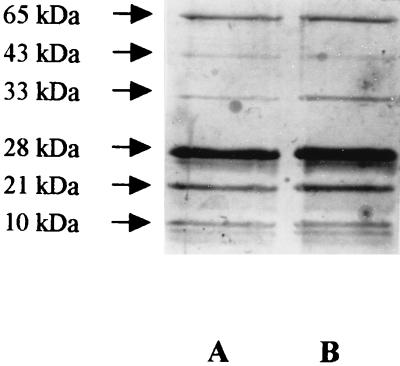
SDS-PAGE analysis revealed the presence of six major polypeptides in the purified OOR preparation, with molecular masses of 65, 43, 33, 28, 21, and 10 kDa (Fig. 1). N-terminal sequences were obtained for all of these proteins (Table 2). The enzyme preparation was found to contain low levels of urease activity, and the presence of the contaminating urease A (65-kDa) and B (28-kDa) subunits was confirmed by comparison of their N-terminal sequences to those previously published for the H. pylori urease enzyme (12). The 43-, 33-, 21-, and 10-kDa polypeptides represented the four subunits of the OOR enzyme (see below) and were designated OorA, OorB, OorC, and OorD respectively.
FIG. 1.
SDS-PAGE analysis of partially purified H. pylori OOR. Lanes A and B show two fractions resulting from the final phenyl-Superose (Pharmacia) chromatography step in the H. pylori OOR purification protocol. The proteins were separated on an SDS–12% polyacrylamide gel and detected by silver staining. Six major bands with molecular masses of 65, 43, 33, 28, 21, and 10 kDa (arrows) were detected. The 28- and 65-kDa bands were determined by N-terminal sequence analysis to be the urease A and B subunits, respectively. The remaining four bands, with molecular masses of 43, 33, 21, and 10 kDa, represent the OorA, -B, -C, and -D subunits, respectively.
TABLE 2.
N-terminal sequence analysis of proteins in the partially purified OOR fractiona
| Subunit | Molecular mass (kDa) | Sequence |
|---|---|---|
| OorA | 43 | M R E I I S D G N E |
| OorB | 33 | A F N Y D E Y L R V |
| OorC | 21 | M E A Q L R F T G V |
| OorD | 10 | X K M S A P D G V ab |
| Urease A | 28 | M K K I S R K E Y V |
| Urease B | 65 | M K L T P K E L D K |
Each of the major polypeptides shown in Fig. 1 was subjected to N-terminal sequence analysis.
The final signal generated by N-terminal sequencing of OorD has been tentatively assigned an alanine; however, DNA sequencing has since identified a threonine at this position. Also, the N terminus of this protein may have been modified (indicated by X).
Analysis of OOR activity and identification of pyruvate- and 2-oxoglutarate-dependent NADP reduction in cell extracts.
The partially purified OOR enzyme was tested for its dependency on various components of the OOR assay mixture. In the presence of 0.1 mM CoA and 5 mM 2-oxoglutarate, the specific activity of the enzyme was 0.925 μmol of methyl viologen reduced min−1 mg of protein−1 (Table 1). The omission of either 2-oxoglutarate or CoA from the reaction mixture, or the replacement of 5 mM 2-oxoglutarate with 5 mM pyruvate, completely prevented the reduction of methyl viologen. Thus, in H. pylori OOR specifically catalyzes the oxidative decarboxylation of 2-oxoglutarate, and POR catalyzes solely the oxidative decarboxylation of pyruvate. As has also been found with other 2-oxoacid oxidoreductases, the partially purified OOR preparation could not utilize NAD or NADP as an electron acceptor when NAD or NADP was added to the complete reaction mixture in place of methyl viologen.
Km values for the substrates of both the POR and OOR activities in cell extracts were determined. The Km values of POR and OOR for pyruvate and 2-oxoglutarate were 0.222 and 0.309 mM, respectively. Interestingly, the affinity of the POR enzyme for CoA was found to be much higher than that of the OOR enzyme, with respective Km values of 1.97 and 13.3 μM.
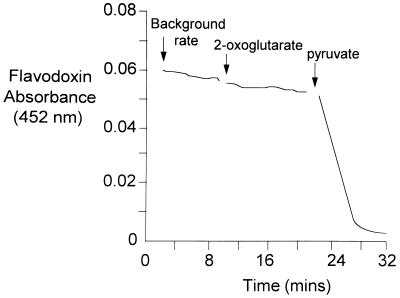
Earlier studies have shown that the in vivo electron acceptor for POR in H. pylori is likely to be a flavodoxin (20). To determine whether this flavodoxin acted as an electron acceptor for the OOR enzyme, a MonoQ fraction containing both POR and OOR activities was incubated anaerobically with CoA and the H. pylori flavodoxin, which was purified as previously described (20). 2-Oxoglutarate was then added, and the reduction of the flavodoxin was monitored at 452 nm (Fig. 2). No reduction was observed in the presence of 2-oxoglutarate. However, following the addition of pyruvate, a rapid quenching of absorption at 452 nm was observed, indicating that only the POR enzyme reduced the flavodoxin. Attempts were made to identify other ferredoxin and flavodoxin proteins by fractionating H. pylori cytoplasmic proteins by MonoQ ion-exchange chromatography. The resulting fractions were then scanned for the characteristic absorption spectra of ferredoxins and flavodoxins. However, we were not able to identify a candidate low-potential electron acceptor for the OOR enzyme.
FIG. 2.
A flavodoxin (FldA) acts as an electron acceptor for the POR enzyme but not for OOR. A MonoQ-purified fraction of H. pylori cell extract, containing both POR and OOR activities, was incubated initially in the presence of sufficient partially purified H. pylori flavodoxin (20) to give a starting absorbance of 0.06 (at 452 nm) and 0.1 mM CoA under anaerobic conditions. The background rate for the reduction of the flavodoxin protein was monitored at 452 nm. 2-Oxoglutarate (5 mM) was then added anaerobically to the cuvette. No increase in the rate of flavodoxin reduction was observed in the presence of this substrate. However, following the addition of 5 mM pyruvate, reduction proceeded rapidly, indicating that the isolated flavodoxin was specifically reduced by electrons derived from the POR reaction.
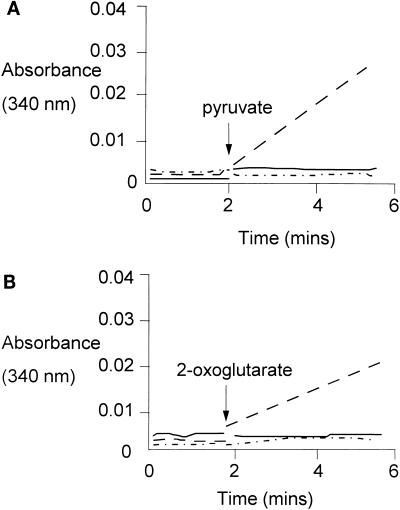
As expected, the purified POR and OOR were unable to utilize NAD or NADP as an electron acceptor, but a CoA- and pyruvate- or 2-oxoglutarate-dependent reduction of NADP could be demonstrated in cell extracts incubated under anaerobic conditions (Fig. 3). No activity was observed with NAD as the electron acceptor, and the NADP reductase activity was completely abolished under aerobic assay conditions, suggesting that pyruvate or 2-oxoglutarate dehydrogenase activities, which are commonly assayed under aerobic conditions, were not active in the cell extract. Further evidence that 2-oxoacid dehydrogenases are not present in H. pylori has been provided by the complete genome sequence of strain 26695, which lacks homologs of these genes (40).
FIG. 3.
Pyruvate- and 2-oxoglutarate-dependent reduction of NADP by H. pylori cell extract. The pyruvate (A)- and 2-oxoglutarate (B)- and CoA-dependent reduction of NADP by H. pylori cell extract, under anaerobic conditions, is shown. The assays were carried out in a 2-ml volume, and the assay mixtures contained 5 mM 2-oxoglutarate or pyruvate, 0.1 mM CoA, 100 μl of H. pylori cell extract, and 1 mM NADP. The point of 2-oxoacid addition is indicated. The effect of pyruvate or 2-oxoglutarate addition on NADP reduction is indicated by the dashed line. The rate of NADP reduction under aerobic conditions is indicated by the solid line, and the rate of NAD reduction, added in place of NADP, is indicated by the dashed and dotted line. Reduction of NADP was observed in the presence of both pyruvate and 2-oxoglutarate and was dependent on anaerobic conditions. Furthermore, NADP could not be replaced by NAD.
NADPH is used in preference to NADH as a respiratory electron donor in H. pylori.
Since the experiments described above showed that electrons derived from the POR and OOR reactions preferentially mediate the reduction of NADP, oxygen uptake by H. pylori in the presence of NADPH and NADH was examined. In most bacteria, NADH acts as the major electron donor for respiration. However, Table 3 shows that membrane preparations of H. pylori, which typically catalyze both succinate and ascorbate/TMPD (tetramethyl-1,4-phenylenediaminedihydrochloride) oxidase activities, did not exhibit detectable rates of oxygen uptake with NADH as the electron donor. However, NADPH-dependent respiration was easily measurable, implying that this is a major respiratory electron donor in H. pylori in vivo. Cell extracts exhibited both activities, presumably due to the presence of additional (soluble) dehydrogenases.
TABLE 3.
Substrate oxidation of cell extracts and membrane fractions of H. pylori NCTC 11637
| Fraction | Substrate (concn) | Respiration rate (nmol of O2 min−1 mg of protein−1)a |
|---|---|---|
| Cell extract | NADPH (0.5 mM) | 4.6 |
| NADH (0.5 mM) | 1.6 | |
| Membrane fraction | Succinate (25 mM) | 0.8 |
| NADPH (250 μM) | 5.1 | |
| NADH (250 μM) | Not detectable | |
| TMPD (1.0 mM) | 15.5 | |
| Ascorbate (1.0%, wt/vol) + TMPD (1.0 mM) | 73.4 |
Data are means from two or more replicate experiments.
Cloning, sequencing, and organization of the H. pylori por and oor genes.
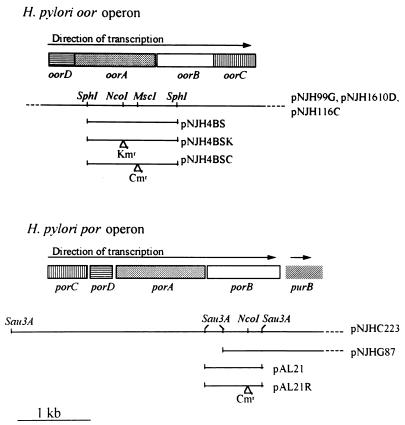
The N-terminal sequences previously published for the four POR subunits (20) and those reported above for OOR were compared with the translations of the random genomic DNA sequences obtained in the Glaxo-Wellcome H. pylori NCTC 11637 sequencing project (8). The 38-residue N-terminal sequence of PorB was identified in the translation of one of the reading frames of the 774-bp insert in plasmid pAL21. The insert in pAL21 was then used as a hybridization probe to screen the H. pylori pUC18 plasmid library in order to obtain clones with larger inserts. This resulted in the identification of two plasmids, pNJHC233 (5.0-kb insert) and pNJHG87 (4.5-kb insert), which were used to sequence a 4,373-bp region containing all four por genes (Fig. 4). A similar approach with the N termini of the OOR subunits resulted in the identification of three plasmids containing the oor genes. The plasmids pNJH1610D (6.0-kb insert), pNJH99G (8.3-kb insert), and pNJH1116C (6.0-kb insert) were used to obtain the complete sequence of a 3,350-bp region which encompassed the oor genes (Fig. 4).
FIG. 4.
Organization of H. pylori por and oor genes. A graphical representation of the oor and por operons of H. pylori NCTC 11637 is shown. Also indicated are the regions covered by plasmids used in this study and the positions of insertion of kanamycin (Kmr) and chloramphenicol (Cmr) resistance cassettes for inactivation studies.
In each of these separate sequenced regions, four adjacent and complete open reading frames (ORFs) were identified which displayed appropriate H. pylori codon usage. The eight ORFs were unequivocally identified as the genes encoding the subunits of POR and OOR by concordance between the N-terminal amino acid sequences determined for each of the POR and OOR subunits and those deduced from the DNA sequence analysis.
The por genes are arranged in the order 5′-porC-porD-porA-porB-3′. A partially sequenced fifth ORF, the product of which was 42% identical over 161 residues with the adenylosuccinate lyase enzyme (PurB) of Bacillus subtilis (13), was located 70 bp downstream of porB and was designated purB. This enzyme plays a role in purine biosynthesis, an activity apparently unrelated to the POR enzyme. Each of the ORFs was preceded by potential Shine-Dalgarno sequences, and the properties of the deduced products are shown in Table 4. The oor genes are arranged in the order 5′-oorD-oorA-oorB-oorC-3′, again preceded by potential Shine-Dalgarno sequences. Translational coupling was observed between oorD and oorA and between oorB and oorC (a TAATG motif in each case). Table 4 also shows the properties of the oor genes and their deduced products.
TABLE 4.
Characteristics of the H. pylori oor and por initiation and termination sites and predicted gene products
| ORF | Proposed Shine-Dalgarno sequencea | No. of amino acid residues | Predicted mass (kDa) | Mass (kDa)b | Predicted isoelectric point |
|---|---|---|---|---|---|
| oorD | aaAGGAgaatgaatg | 113 | 12.43 | 10 | 6.89 |
| oorA | agGGAGaggcaactaatg | 371 | 40.95 | 43 | 6.44 |
| oorB | gaAGGAgctttaaaatg | 274 | 30.64 | 33 | 8.00 |
| oorC | caAGGAaaacaataatg | 184 | 19.99 | 21 | 6.08 |
| porC | taAGGAgacatattaccatg | 172 | 19.27 | 24 | 9.84 |
| porD | taAGGAttatacatg | 131 | 15.11 | 14 | 6.63 |
| porA | taAGGAaaaaatatg | 409 | 44.52 | 47 | 8.06 |
| porB | aaAGGAaatatcatg | 329 | 36.41 | 36 | 6.67 |
| purBc | taAGGAttgtcggtg | 171 | NId | NI | NI |
Predicted Shine Dalgarno sequences were identified by complementarity to the 16S rRNA-binding site of H. pylori. Nucleotides complementing the 16S rRNA-binding site are indicated by capital letters. Initiation codons downstream of Shine-Dalgarno regions are underlined.
Calculated from SDS-PAGE analysis of the purified enzymes.
Incomplete sequence of gene.
NI, not identified.
Recently the complete genome sequence of H. pylori 26695 has become available (40). Examination of this genome has confirmed the presence of homologous porCDAB and oorDABC operons. The HP numbers assigned to each of the individual genes are HP1108 to -1111 for the por operon and HP0588 to -0591 for the oor operon. The nucleotide sequences of the por and oor multigene regions have also been entered under GenBank accession numbers AE000617 and AE000572, respectively. The gene order is identical in each operon, and the purB gene, located downstream of the por genes in H. pylori 11637, is also found downstream of the por genes in strain 26695. The percent identities between the predicted por and oor gene products of strains 11637 and 26695 are as follows (the number of amino acids aligning is given in parentheses): PorD, 100 (129); PorC, 98 (164); PorB, 100 (314); PorA, 95 (408); OorD, 98 (113); OorC, 97 (183); OorB, 97 (274); and OorA, 96 (374).
Identification of por and oor homologs in other Helicobacter strains.
Genomic DNA preparations of bacteria closely related to H. pylori were screened by Southern blotting for hybridization to porA and oorB genes. With the high-stringency conditions described in Materials and Methods, both probes hybridized with DNA from H. acinonyx, H. felis, and H. pylori NCTC 11637, 4187E, and 8091. Under the same conditions, no hybridization was detected with H. muridarum or H. mustelae (results not shown). Although POR activity has been observed in many Campylobacter species, including C. jejuni and C. coli (9), Southern blotting with genomic DNAs from these two species failed to detect homologous sequences under the conditions described (results not shown), indicating a greater divergence in sequence.
Amino acid sequence similarities and identification of potential TPP- and cation-binding sites and iron-sulfur centers in the deduced proteins.
Surprisingly, although POR and OOR of H. pylori are biochemically similar enzymes, each composed of four subunits, BESTFIT alignments of the amino acid sequences of the corresponding polypeptides revealed a fairly low overall level of identity: PorA-OorA, 25.0%; PorB-OorB, 18.9%; PorC-OorC, 23.3%, and PorD-OorD, 20.6%. FASTA database searches clearly showed that POR of H. pylori is most closely related to the recently recognized group of four-subunit PORs from hyperthermophiles (22). Each H. pylori POR subunit sequence displayed the most similarity to its corresponding subunit from the Thermotoga maritima and P. furiosus enzymes (between 38.1 and 44.2% identities). The subunit sequences could also clearly be aligned with corresponding domains in single-subunit PORs, as has previously been reported with the P. furiosus and T. maritima POR sequences and the closely related P. furiosus ketoisovalerate:ferredoxin oxidoreductase enzyme (22).
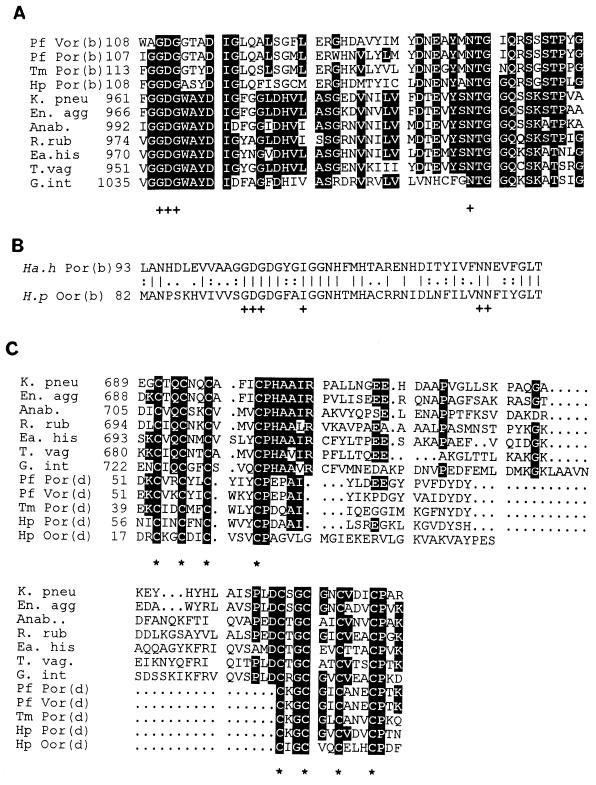
All PORs isolated to date contain the cofactor TPP. A conserved sequence motif (GD/QG-X25–30-NN) has been identified for TPP binding (18). Similar motifs have also been identified in both PorB and OorB (Fig. 5A and B). This motif is part of a highly conserved region in all PORs, but due to insufficient amounts of the purified H. pylori enzymes, we have not been able to perform biochemical analyses to confirm the presence of TPP. PorB and OorB contain vicinal cysteines (C-X2-C), which are also present in other PORs. This motif has been proposed to play a role in heavy metal cation binding (37).
FIG. 5.
Alignments of predicted TPP- and Fe-S-binding sites in H. pylori POR, OOR, and related enzymes. (A and B) PileUp alignments indicating putative conserved regions for TPP binding based on the consensus motif proposed by Hawkins et al. (18) in the H. pylori PorB subunit and related enzymes (A) and in the H. pylori OorB subunit and Halobacterium halobium PorB subunits (B). (C) Regions of conservation-associated Fe-S-binding sites found in the d subunits and regions of 2-oxoacid oxidoreductases. The pattern of cysteine residues, marked with asterisks, is typical of that found in bacterial ferredoxins (39). Residues conserved in >50% of the organisms are indicated by black boxes. The position of the first amino acid in each alignment is also indicated. Abbreviations (accession numbers are given in parentheses: Pf Vor (b) and (d), P. furiosus isoketovalerate:ferredoxin oxidoreductase B and D subunits (X85250); Pf Por (b) and (d), P. furiosus pyruvate:ferredoxin oxidoreductase B and D subunits (X85250); Tm Por (b) and (d), T. maritima pyruvate:ferredoxin oxidoreductase B and D subunits (X85171); Hp Por (b) and (d), H. pylori PorB and PorD (this study); Hp Oor (b) and (d), H. pylori OorB and OorD (this study); Ha.h Por (b), Halobacterium halobium pyruvate:ferredoxin oxidoreductase B subunit (X64521); K. pneu, K. pneumoniae NifJ (X13109); En. agg, Enterobacter agglomerans NifJ (X78558); Anab., Anabeana sp. strain PCC 7120 (L14925); R. rub, Rhodospirillum rubrum pyruvate oxidoreductase (X77515); Ea. his, Entamoeba histolytica pyruvate oxidoreductase (U30149); T. vag, Trichomonas vaginalis pyruvate:ferredoxin oxidoreductase (U16822); G. int, Giardia intestinalis POR (L27221).
Interestingly, the H. pylori OorA and OorB sequences are far more closely related to those of the α and β subunits, respectively, of the two-subunit Halobacterium halobium pyruvate:ferredoxin oxidoreductase (21, 32) than to the PorA and -B subunits of the hyperthermophiles. H. pylori OorA displays 31.8% identity over 385 amino acids to the Halobacterium halobium α subunit, in comparison to only 20.2% over 387 amino acids for the PorA subunit of P. furiosus. H. pylori OorB displays 37.5% amino acid identity to the Halobacterium halobium B subunit over 175 residues. The amino acid sequences of OorC and OorD are most closely related to those of P. furiosus OorC (26% identity over 177 residues) and Anabaena variabilis ferredoxin (35.4% identity over 65 residues). Sequences similar to those of OorD, -A, -B, and -C have also been identified in the genome sequence of Methanococcus jannaschii (accession number U67482). However, the gene order is different in this organism (C-D-A-B), and to the best of our knowledge, this enzyme has not been purified and thus its substrate specificity is unknown.
Most bacterial-type ferredoxins possess two cysteine-rich sequences, each of which conforms to the consensus (-C-X2-C-X2-C-X3-C-) (39). The two units are separated by a connector region of variable length and cooperate to form two [4Fe-4S] centers. This consensus is clearly present in both PorD and OorD and in the D subunits and domains of other POR enzymes (Fig. 5C). The exception to this is the Halobacterium halobium POR enzyme, which contains only one such cysteine-rich unit (32). The iron-sulfur compositions of the H. pylori POR and OOR enzymes could not be determined due to insufficient amounts of purified enzyme. However, from the amino acid sequence it would appear that the enzymes contain at least two [4Fe-4S] clusters per molecule, and this composition has also been found in the four-subunit enzymes of P. furiosus (1), T. maritima (2), and Methanococcus maripaludis (44).
Expression of H. pylori POR in E. coli.
In order to further examine the oxygen lability of the POR enzyme in a heterologous background, the porCDAB operon was PCR amplified with primers designed to introduce an NdeI site into the start codon of porC, and the product was cloned into the translation initiation site of pET21a to give pNJH301. Expression from the plasmid was examined by radiolabeling of the IPTG-induced translation products. Four polypeptides of the correct molecular masses for the porA, -B, -C, and -D gene products were produced after induction of E. coli BL21(DE3, pNJH301) (results not shown). A significant increase in POR activity was also detected in cell extracts (Table 5) prepared from cells which had been induced under microaerobic conditions in M9 medium. No active enzyme was obtained if the cells were induced under aerobic conditions. The background activity observed in the control strain is due to a low endogenous activity of pyruvate oxidoreductase apparently constitutively expressed in E. coli (3).
TABLE 5.
Expression of POR activity in E. coli BL21(DE3, pNJH301)a
| Growth stage | POR activity (nmol of methyl viologen reduced min−1 mg−1)
|
||
|---|---|---|---|
| Microaerobic induction
|
Aerobic induction (pNJH301) | ||
| pET21a control | pNJH301 | ||
| Pre-IPTG addition | 7.5 | 6.2 | 6.2 |
| Post-IPTG addition | 5.8 | 27.4 | 6.0 |
| Post-rifampin addition | 5.9 | 38.4 | 5.8 |
Cell extract was prepared from E. coli BL21(DE3) harboring either pNJH301, for expression of H. pylori POR, or the vector control, pET21a, at various stages during the induction of recombinant POR expression. Samples were taken immediately before the addition of 1 mM IPTG to the growth medium, 90 min after the addition of 1 mM IPTG, and 90 min after the addition of 200 μg of rifampin ml−1 to inhibit host RNA polymerases. Following the addition of IPTG, cells were cultured under either aerobic or microaerobic conditions. A sevenfold overexpression of POR activity was achieved only in cells induced under reduced oxygen concentrations; under aerobic conditions, no increase in POR activity could be detected.
Insertion inactivation of H. pylori porB and oorA.
Plasmids pAL21R, pNJH4B9SK, and pNJH4B9SC were used for natural transformation of H. pylori NCTC 11637 in attempts to obtain chromosomal null mutants of either porB or oorA by allelic exchange. No antibiotic-resistant colonies (<10−9 per CFU) were obtained with any of these constructs. As a control for the insertion inactivation procedure, a plasmid containing a null mutation in a known nonessential chemotaxis gene (cheY) was used. In this plasmid the chloramphenicol resistance gene was flanked by 109 and 953 bp of H. pylori DNA. Transformation with this plasmid yielded chloramphenicol-resistant colonies at a frequency of 2.6 × 10−5 per CFU. Several of these colonies were selected and shown to contain a disrupted chromosomal cheY gene by PCR (data not shown). The lack of colonies with the por and oor constructs therefore suggests that these are essential genes in H. pylori.
Identification of the structural gene (fldA) of the H. pylori POR-specific flavodoxin.
Partial purification of the POR-specific flavodoxin from H. pylori has been reported previously (20). SDS-PAGE analysis indicated that the resulting purification fraction contained a major protein of 17 kDa, a molecular mass similar to those of other flavodoxins (results not shown). The N-terminal sequence for the excised band was determined to be -GKIGIFFGT-. A comparison of this sequence with translations of the Glaxo-Wellcome H. pylori NCTC 11637 random-sequence database (8) identified a plasmid encoding 400 bp of the fldA gene. The insert was used to screen the H. pylori plasmid library to identify a plasmid, pKB1, from which a 1,004-bp region of DNA was sequenced. The region contains the structural gene for this flavodoxin, which was confirmed by concordance between the N-terminal sequence of the isolated protein and the predicted amino acid translation. fldA encodes a protein of 173 amino acids with a predicted molecular mass and pI of 18.37 kDa and 4.46, respectively. The low pI value and the predominance of acidic residues in comparison to basic residues (21 versus 11%) are typical features of other flavodoxins (28). A homologous fldA sequence is also present in the genome sequence of H. pylori 26695 (HP gene number 1161, accession number AE000622) (40). The predicted amino acid sequences of the two flavodoxins are 92% identical over 163 amino acid residues.
DISCUSSION
Previous biochemical studies have emphasized the importance of 2-oxoacid oxidoreductases in the generation of acylthioesters in H. pylori (19, 20). The genome sequence of strain 26695 has supported these assertions, as homologs of pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, and pyruvate:formate lyase genes have not been identified. This paper produces evidence for a second essential acceptor:oxidoreductase in H. pylori, which catalyzes the specific oxidative decarboxylation of 2-oxoglutarate. The end product of this reaction, succinyl-CoA, is a major intermediate of the TCA cycle. However, enzymatic analysis has shown that the cycle is incomplete in H. pylori, and in particular, succinate thiokinase activity has not been detected (9a). Homologs of succinate thiokinase genes also could not be identified in the published genome sequence of strain 26695 (40). So what is the likely fate of succinyl-CoA in H. pylori? Succinyl-CoA is required for lysine biosynthesis in the tetrahydrodipicolinate N-succinyl transferase reaction, and a homolog of this enzyme has been identified in the genome sequence (40). A gene for a putative 3-oxoadipate-CoA transferase has also been identified. This enzyme catalyzes the conversion of succinyl-CoA and 3-oxoadipate to succinate and 3-oxoadipyl-CoA, a reaction of the 3-oxoadipate pathway of aromatic compound degradation. However, the importance of these pathways in the growth of H. pylori is unknown.
Four types of 2-oxoacid oxidoreductases can be distinguished, based on subunit structure and sequence similarities. The most ancient appear to be the four-subunit enzymes typical of a number of thermophilic archaea and bacteria, notably P. furiosus (1), Archaeoglobus fulgidus (24), and T. maritima (2). P. furiosus also possesses a two-subunit indolepyruvate:ferredoxin oxidoreductase (23, 36). The 71-kDa α subunit of this enzyme possesses domains similar to the A, B, and the ferredoxin-like D domains described for four-subunit 2-oxoacid oxidoreductases. Halobacterium halobium also contains a two-subunit POR, which in contrast lacks a distinctive ferredoxin-like subunit or domain (32). Finally, single-subunit enzymes are typified by the NifJ proteins of Klebsiella pneumoniae (4) and Anabaena (35) and consist of four domains which have probably arisen as a result of gene fusion events from an ancestral four-subunit enzyme (22). This study has shown that although they are similar in overall composition and properties, the two H. pylori four-subunit enzymes are not closely related to each other in sequence but instead appear to have evolved independently. The H. pylori POR shows greatest sequence similarity to that of P. furiosus, while the H. pylori OorA and OorB subunits are highly similar in amino acid sequence to the Halobacterium halobium two-subunit POR.
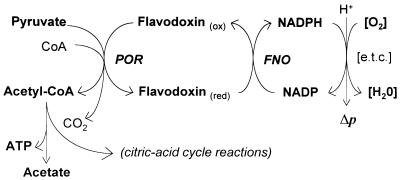
The fact that H. pylori possesses 2-oxoacid oxidoreductases, rather than the more usual dehydrogenase complexes found in aerobes, has a number of consequences for the physiology of this bacterium. First, it is clear that in their purified form they are very oxygen labile, and they are no different in this respect from the corresponding enzymes purified from obligate anaerobes. Expression of the porCDAB genes in E. coli resulted in the formation of an active enzyme only when the cells were induced under reduced oxygen concentrations. The possession of these important enzymes may thus contribute significantly to the observed microaerophilic phenotype of H. pylori. Indeed, the failure of attempts to insertionally inactivate the cognate genes would indicate that both POR and OOR are essential for viability, and thus their inhibition by oxygen could be potentially lethal. However, because the bacterium has a respiratory metabolism, it seems likely that there is some form of protection in vivo which, for example, could maintain a low intracellular oxygen concentration and allow the enzymes to operate satisfactorily in vivo. Second, because the electrons derived from oxidation of the 2-oxoacids are used to reduce a low-potential acceptor, identified as a flavodoxin in the case of POR, these electrons must be removed in order to achieve redox balance and to regenerate the oxidized acceptor. In many anaerobes, which contain ferredoxin-linked 2-oxoacid oxidoreductases, the substrate-derived electrons are disposed of through the evolution of hydrogen gas via the hydrogenase enzyme. H. pylori contains hydrogenase activity (10, 26) which appears to function as an uptake hydrogenase, although its physiological role is unclear. Under aerobic or microaerobic conditions, the evolution of hydrogen would be thermodynamically unfavorable. In this study, we have provided evidence that 2-oxoacid and CoA-dependent NADP reduction can occur in cell extracts, i.e., in the presence of flavodoxin in the case of POR and an as-yet-unidentified electron acceptor in the case of OOR, acting as intermediate electron acceptors. The finding that the overall reaction is specific for NADP rather than NAD is significant in view of the observation that NADPH and not NADH is the most effective electron donor to the respiratory chain, a conclusion which has also been reached in other studies (6, 7). We therefore propose an indirect role for POR and OOR in energy conservation in H. pylori by the provision of NADPH, as illustrated in Fig. 6. We assume that a flavodoxin and/or ferredoxin:NADP oxidoreductase, which links 2-oxoacid oxidation to NADP reduction, is active in H. pylori, but this remains to be identified biochemically. Interestingly, a putative NAD(P)H:flavin oxidoreductase gene has been identified in the genome sequence of H. pylori 26695 (40). Future studies will ascertain the physiological role of this candidate enzyme.
FIG. 6.
Proposed pathway for electron flow to NADP from reduced flavodoxin in H. pylori. The proposed flow of electrons generated in the POR reaction from flavodoxin is shown. The reduced flavodoxin in turn donates electrons to NADP via a hypothetical NADP:flavodoxin oxidoreductase (FNO). NADPH then enters the electron transport chain ([e.t.c.]). The acetyl-CoA generated in the POR reaction may enter the reactions of the incomplete TCA cycle found in H. pylori or may generate ATP by substrate level phosphorylation, yielding acetate.
It should also be noted with regard to energy conservation that acetate is a major product of the aerobic metabolism of pyruvate in H. pylori (5), and thus ATP can also be generated from acetyl-CoA by substrate level phosphorylation. Putative genes for the enzymes responsible for this reaction, phosphotransacetylase and acetate kinase, have been identified in the genome sequence of H. pylori 26695 (40).
Finally, as POR and OOR have been implicated in the bioreduction of nitroimidazole drugs, particularly metronidazole (19, 20), a more detailed knowledge of the electron transport pathways leading from these enzymes will be relevant to an understanding of the mechanisms of metronidazole activation and resistance, which are at present poorly characterized.
ACKNOWLEDGMENTS
This work has been funded by Glaxo-Wellcome Ltd. through a Glaxo scholarship to N.J.H.
We thank N. Crocker for Applied Biosystems nucleotide sequencing, N. Freeman and A Moir for N-terminal sequencing, A. McClaren for provision of bacterial strains, K. Broughton for completion of the flavodoxin sequence, and C. Jackson for the gift of the CheY-chloramphenicol construct. We also thank A. A. Davison for carrying out respiration studies and for access to unpublished data.
REFERENCES
- 1.Blamey J M, Adams M W W. Purification and characterisation of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta. 1993;1161:19–27. doi: 10.1016/0167-4838(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 2.Blamey J M, Adams M W W. Characterisation of an ancestral type of pyruvate ferredoxin oxidoreductase from the hyperthermophilic bacterium, Thermotoga maritima. Biochemistry. 1994;33:1000–1007. doi: 10.1021/bi00170a019. [DOI] [PubMed] [Google Scholar]
- 3.Blaschkowski H P, Neuer G, Ludwig-Festl M, Knappe J. Routes of flavodoxin and ferredoxin reduction in Escherichia coli: CoA acylating pyruvate:flavodoxin and NADPH:flavodoxin oxidoreductases participating in the activation of pyruvate formate lyase. Eur J Biochem. 1982;123:563–569. [PubMed] [Google Scholar]
- 4.Cannon M, Cannon F, Buchanan-Wollaston V, Ally D, Ally A, Beynon J. The nucleotide sequence of the nifJ gene of Klebsiella pneumoniae. Nucleic Acids Res. 1988;16:11379. doi: 10.1093/nar/16.23.11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalk P A, Roberts A D, Blows W M. Metabolism of pyruvate and glucose by intact cells of Helicobacter pylori studied by 13C NMR spectroscopy. Microbiology. 1994;140:2085–2092. doi: 10.1099/13500872-140-8-2085. [DOI] [PubMed] [Google Scholar]
- 6.Chang H T, Marcelli S W, Davison A A, Chalk P A, Poole R K, Miles R J. Kinetics of substrate oxidation by whole cells and cell membranes of Helicobacter pylori. FEMS Microbiol Lett. 1995;129:33–38. doi: 10.1016/0378-1097(95)00130-W. [DOI] [PubMed] [Google Scholar]
- 7.Clayton C L, Hughes N J, Chalk P A. Molecular characterisation of a major NADPH dehydrogenase of Helicobacter pylori. Am J Gastroenterol. 1994;89:1293. [Google Scholar]
- 8.Clayton, C. L., A. Tay, C. O’Donnell, and P. A. Chalk. 1995. Elucidation of Helicobacter pylori metabolism by random genome sequencing. Gut 37(Suppl. 1):A67.
- 9.Daucher J A, Krieg N R. Pyruvate:ferredoxin oxidoreductases in Campylobacter species. Can J Microbiol. 1995;41:198–201. [Google Scholar]
- 9a.Davison, A. A. (University of Sheffield). Personal communication.
- 10.Davison, A. A., D. J. Kelly, P. J. White, P. A. Chalk, and C. L. Clayton. 1995. Sequencing and inactivation of a Helicobacter pylori hydrogenase. Gut 37(Suppl. 1):A18.
- 11.Docampo R, Moreno S N J, Mason R P. Free radical intermediates in the reaction of pyruvate:ferredoxin oxidoreductase in Tritrichomonas foetus hydrogenosomes. J Biol Chem. 1987;262:12417–12420. [PubMed] [Google Scholar]
- 12.Dunn B E, Campbell G P, Prez-Perez G I, Blaser M J. Purification and characterization of urease from Helicobacter pylori. J Biol Chem. 1990;265:9464–9469. [PubMed] [Google Scholar]
- 13.Ebbole D J, Zalkin H. Cloning and characterization of a 12 gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987;262:8274–8287. [PubMed] [Google Scholar]
- 14.Edwards D I. Nitroimidazole drugs—action and resistance mechanisms. I. Mechanisms of action. J Antimicrob Chemother. 1993;31:9–20. doi: 10.1093/jac/31.1.9. [DOI] [PubMed] [Google Scholar]
- 15.Forman D, Newell D G, Fullerton F, Yarnell J W G, Stacey A R, Wald N, Sitas F. Association between Helicobacter pylori infection and risk of gastric cancer: evidence from a prospective investigation. Br Med J. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glupczinski Y. Results of a multicentre European survey in 1991 of metronidazole resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1992;11:777–781. [PubMed] [Google Scholar]
- 17.Goodwin C S, Marshall B J, Blincow E D, Wilson D H, Blackbourn D, Phillips M. Prevention of nitroimidazole resistance in Campylobacter pylori by coadministration of colloidal bismuth subcitrate: clinical and in vitro studies. J Clin Pathol. 1988;41:207–210. doi: 10.1136/jcp.41.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins C F, Borges A, Perham R N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman P S, Goodwin A, Johnsen J, Magee K, Veldhuzyen van Zanten S J O. Metabolic activities of metronidazole-sensitive and resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J Bacteriol. 1996;178:4822–4829. doi: 10.1128/jb.178.16.4822-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes N J, Chalk P A, Clayton C L, Kelly D J. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J Bacteriol. 1995;177:3953–3959. doi: 10.1128/jb.177.14.3953-3959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerscher L, Oesterhelt D. Purification and properties of two 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. Eur J Biochem. 1981;116:587–594. doi: 10.1111/j.1432-1033.1981.tb05376.x. [DOI] [PubMed] [Google Scholar]
- 22.Kletzin A, Adams M W. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate:ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol. 1996;178:248–257. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kletzin A, Adams M W. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic archea. Adv Protein Chem. 1996;48:101–180. doi: 10.1016/s0065-3233(08)60362-9. [DOI] [PubMed] [Google Scholar]
- 24.Kunow J, Linder D, Thauer R K. Pyruvate:ferredoxin oxidoreductase from the sulfate-reducing Archaeoglobus fulgidus: molecular composition, catalytic properties, and sequence alignments. Arch Microbiol. 1995;163:21–28. doi: 10.1007/BF00262199. [DOI] [PubMed] [Google Scholar]
- 25.Mai X H, Adams M W W. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J Bacteriol. 1996;178:5890–5896. doi: 10.1128/jb.178.20.5890-5896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier R J, Fu C, Gilbert J, Moshiri F, Olsen J, Plaut A G. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol Lett. 1996;141:71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- 27.Majewski S I, Goodwin C S. Restriction endonuclease analysis of the genome of Campylobacter pylori with a rapid extraction method: evidence for considerable variation. J Infect Dis. 1988;157:465–471. doi: 10.1093/infdis/157.3.465. [DOI] [PubMed] [Google Scholar]
- 28.Mayhew S G, Ludwig M L. Flavodoxins and electron-transferring flavoproteins. In: Boyer P D, editor. The enzymes. Vol. 12. New York, N.Y: Academic Press; 1975. pp. 57–118. [Google Scholar]
- 29.Mendz G L, Hazell S L. Evidence for a pentose-phosphate pathway in Helicobacter pylori. FEMS Microbiol Lett. 1991;84:331–336. [Google Scholar]
- 30.Mendz G L, Hazell S L, Burns B P. The Entner-Doudoroff pathway in Helicobacter pylori. Arch Biochem Biophys. 1994;312:349–356. doi: 10.1006/abbi.1994.1319. [DOI] [PubMed] [Google Scholar]
- 31.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 32.Plaga W, Lottspeich F, Oesterhelt O. Improved purification, crystallization and primary structure of pyruvate:ferredoxin oxidoreductase from Halobacterium halobium. Eur J Biochem. 1992;205:391–397. doi: 10.1111/j.1432-1033.1992.tb16792.x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbour Press; 1989. [Google Scholar]
- 34.Schmitz O, Kentemich T, Zimmer W, Hundeshagen B, Bothe H. Identification of the nifJ gene coding for pyruvate:ferredoxin oxidoreductase in dinitrogen-fixing cyanobacteria. Arch Microbiol. 1993;160:62–67. [PubMed] [Google Scholar]
- 35.Shah V K, Stacey G, Brill W J. Electron transport to nitrogenase: purification and characterisation of pyruvate:flavodoxin oxidoreductase, the nifJ gene product. J Biol Chem. 1983;258:12064–12068. [PubMed] [Google Scholar]
- 36.Siddiqui M A, Fujiwara S, Imanaka T. Indolepyruvate ferredoxin oxidoreductase from Pyrococcus sp. KOD1 possesses a mosaic structure showing features of various oxidoreductases. Mol Gen Genet. 1997;254:433–439. doi: 10.1007/pl00008607. [DOI] [PubMed] [Google Scholar]
- 37.Silver S, Misra T K. Plasmid-mediated heavy metal resistance. Annu Rev Microbiol. 1988;42:717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- 38.Smith M A, Edwards D I. Oxygen scavenging, NADH oxidase and metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1997;39:347–353. doi: 10.1093/jac/39.3.347. [DOI] [PubMed] [Google Scholar]
- 39.Stigerwald V J, Beckler G S, Reeve J N. Conservation of hydrogenase and polyferredoxin structures in the hyperthermophilic archaebacterium Methanothermus fervidus. J Bacteriol. 1990;172:4715–4718. doi: 10.1128/jb.172.8.4715-4718.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenny K, Fitxegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterbeck T R, Peterson J D, Kelly J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes M S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;387:538–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 41.Trieu-Cuat P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between Gram-positive and Gram-negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tytgat G N J, Lee A, Graham D Y, Dixon M F, Rokkas T. The role of infectious agents in peptic ulcer disease. Gastroenterol Int. 1993;6:76–89. [Google Scholar]
- 43.Wang Y, Taylor D E. Chloramphenicol resistance in Campylobacter coli—nucleotide sequence, expression and cloning vector construct. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y L, Whitman W B. Abstracts of the 94th General Meeting of the American Society for Microbiology 1994. Washington, D.C: American Society for Microbiology; 1994. Purification and characterisation of pyruvate oxidoreductase from Methanococcus maripaludis; p. 287. [Google Scholar]
- 45.Yoon K S, Ishii M, Igarashi Y, Kodama T. Purification and characterization of 2-oxoglutarate ferredoxin oxidoreductase from a thermophilic, obligately chemolithoautotrophic bacterium, Hydrogenobacter thermophilus TK-6. J Bacteriol. 1996;178:3365–3368. doi: 10.1128/jb.178.11.3365-3368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]