Abstract
Goat milk is enriched in fatty acids which are beneficial to human health. Previous research has revealed that 98% of milk fat is composed of triglycerides. However, the mechanisms regulating milk fat composition remain unclear. Forkhead box protein O1 (FoxO1) is a crucial regulatory factor involved in lipid metabolism across various cell types. Chromatin immunoprecipitation sequencing (ChIP)—seq data) and RNA sequencing (RNA-seq) data revealed that have indicated a close association between FoxO1 was closely related to lipid metabolism during lactation in dairy goats. The objective of this study was to investigate the mechanisms by which FoxO1 regulates lipid metabolism in goat mammary epithelial cells (GMECs). FoxO1 knockdown significantly downregulated the expression of adipose triglyceride lipase (ATGL) and suppressed the activity of the ATGL promoter. Consistently, the number of lipid droplets decreased significantly in FoxO1-overexpressing cells and increased in ATGL-knockdown cells. To further verify the effect of FoxO1 on ATGL promoter activity, cells were transfected with four promoter fragments of different lengths. We found that the core region of the ATGL promoter was located between −882 bp and −524 bp, encompassing two FoxO1 binding sites (FKH1 and FKH2). Mutations in the FoxO1 binding sites significantly downregulated ATGL promoter activity in GMECs. Luciferase reporter assays demonstrated that FoxO1 overexpression markedly enhanced ATGL promoter activity. Furthermore, site-directed mutation confirmed that FKH1 and FKH2 sites were simultaneously mutated significantly attenuated the stimulatory effect of FoxO1 on ATGL promoter activities simultaneous mutation of FKH1 and FKH2 sites significantly attenuated the stimulatory effect of FoxO1 on ATGL promoter activity. ChIP assays showed that FoxO1 directly binds to the FKH2 element located in the ATGL promoter in vivo. Finally, immunofluorescence staining revealed that insulin promotes the translocation of FoxO1 from the nucleus to the cytoplasm, thereby attenuating the FoxO1-induced activation of the ATGL promoter. Collectively, these findings uncover a novel pathway where by FoxO1 may regulate lipid metabolism in GMECs specifically by modulating the transcriptional activity of ATGL.
Keywords: adipose triglyceride lipase promoter, forkhead box protein O1, goat mammary epithelial cell, lipolysis, transcriptional regulation
This study uncovered a novel pathway whereby FoxO1 may regulate lipid metabolism in GMECs by controlling ATGL expression, offering fresh insights into the mechanism by which FoxO1 affects lipid metabolism in dairy g oats.
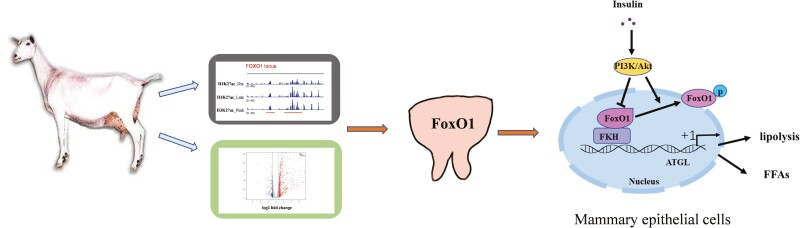
Graphical Abstract
ChIP-seq data from mammary tissue at different lactation periods and RNA-seq data from mammary epithelial cells indicated that FoxO1 is a key transcription factor that regulates lipid metabolism in dairy goats. In addition, FoxO1 promotes the lipolysis by binding to FoxO1 binding sites located in ATGL promoter in an insulin-dependent manner.
Introduction
The composition and content of fatty acids (FAs) are important indices for evaluating the nutritional value and economic output of milk (Park et al., 2007). Goat milk is considered the best substitute for breast milk because of its high content of short- and medium-chain fatty acids and small fat globules, which are more easily absorbed by newborns and the elderly. Studies have shown that most milk fat is secreted in the form of cytoplasmic lipid droplets in mammary epithelial cells (Chong et al., 2011), and 98% of milk fat is composed of triglycerides. These triglycerides are a source of stored energy and can be hydrolyzed into glycerol and free fatty acids to meet the energy needs of animals during periods of negative energy balance (Liu et al., 2020). Therefore, studying the regulatory mechanism of triglyceride synthesis and hydrolysis is helpful in elucidating the metabolic balance between milk fat synthesis and other physiological processes related to lipid metabolism.
Adipose triglyceride lipase (ATGL) catalyzes the crucial initial step in the breakdown of adipose triglycerides, releasing fatty acids (FAs), and diglycerides (DG; Obrowsky et al., 2013; Li et al., 2022b). Previous studies have demonstrated the important role of ATGL in lipid droplet degradation and triglyceride decomposition in tissues with high lipid synthesis, such as adipose tissue, small intestine, and liver (Trites and Clugston, 2019). For instance, ATGL-knockout mice not only exhibited disrupted β-cell function and reduced insulin secretion but also experienced ectopic lipid deposition in various organs. Notably, triglyceride accumulation in the heart leads to heart failure and fatality (Janek et al., 2018; Trites and Clugston, 2019). Recent evidence has suggested that the expression of ATGL is significantly higher during lactation compared to the dry period in dairy goats (Li et al., 2014). Moreover, overexpressing ATGL in goat mammary epithelial cells (GMECs) significantly reduced lipid droplet accumulation and intracellular triglyceride content (Li et al., 2022a).
Forkhead box protein O1 (FoxO1) is a critical regulatory factor primarily involved in transmitting various cytokine signals (Peng et al., 2020; Sabir et al., 2022). In mouse hepatocytes, constitutively active FoxO1 increases fatty acid oxidation while decreasing triglyceride deposition in the liver (Zhang et al., 2016). FoxO1 promotes ATGL transcriptional activity by binding to its promoter, thereby negatively regulating triacylglycerol accumulation in adipocytes (Chakrabarti and Kandror, 2009; Zhang et al., 2016). Additionally, ATGL is activated upon binding to CGI-58 and localizes to the surface of lipid droplets. Conversely, the protein encoded by G0S2 acts as a specific inhibitor of ATGL-mediated lipolysis (Yang et al., 2010). Overexpressing FoxO1 promotes the expression of CGI-58, consequently upregulating ATGL mRNA levels (Miao et al., 2015). Recent studies have suggested that palmitate inhibits the expression of CGI-58 and G0S2 while significantly increasing the expression of ATGL through FoxO1 overexpression in hepatocytes. This results in reduced lipid accumulation and enhanced lipid decomposition (Zhao et al., 2021). However, the interaction between FoxO1 and ATGL in dairy goats remains unclear.
In this study, we aim to explore the regulatory mechanism of FoxO1on ATGL transcription during lactation in dairy goats. We found that FoxO1 functions as a positive regulator of triacylglycerol (TAG) hydrolysis in GMECs. Site-directed mutation and Chromatin immunoprecipitation (ChIP) assays revealed that FoxO1 regulates the transcription of ATGL via binding to the ATGL promoter.
Materials and Methods
Ethics statement
All the experimental procedures were approved by the Animal Care and Use Committee of Northwest A&F University, Yang Ling, China (Approval Number: DK2021054).
Cell culture
GMECs were isolated from three peak-lactating goats. The protocol for purification and cultured procedure as previously described (Wang et al., 2010; Zhu et al., 2015). Briefly, mammary tissue was collected using a surgical approach and washed immediately with D-Hank’s solution. Then the tissue was divided into 1 mm3 pieces and plated in 60 mm dishes and cultured in 5% CO2 at 37 °C until epithelial cells separated from the tissue block. Culture medium was changed every 48 h before the cells were transferred to a new culture dish. Subsequently, cells were digested from the tissue block with 0.25% trypsin-EDTA solution. Then, the fibroblasts were removed by differential adhesion method, which adhered to culture dishes faster than GMECs. The purified GMECs were transferred to a new culture dish and continued incubation. Finally, the GMEC isolated from the three goats were cultured individually to passage six to run the independent experiment. Each experiment was repeated at least three times. The culture medium contained 90% DMEM/F12 (#A4192001, Thermo Fisher Scientific), penicillin/ streptomycin (100 U/mL, #080092569), insulin (5 μg/mL, #I6729, Sigma–Aldrich), hydrocortisone (1 mg/mL, #H0888, Sigma-Aldrich), epidermal growth factor (10 ng/mL, PHG0311, Invitrogen), and 10% fetal bovine serum (10,099 − 141, Invitrogen).
Generation of recombinant adenoviruses
The packaging of adenovirus was according to a previously described (He et al., 2020). Briefly, the coding sequence (CDS) of FoxO1 from dairy goats was cloned into pAdTrack-CMV vector (#16405, Addgene) using KpnI (R3142, NEB, Ipswich, MA, USA) and XhoI (R0146, NEB, Ipswich, MA, USA) restriction sites. After linearization, pAdTrack-CMV-FoxO1 vector or pAdTrack-CMV empty vector was transformed into BJ5183 competent cells for recombination with pAdEasy-1 vector. The recombinant plasmids were linearized with PacI (R0547, NEB, Ipswich, MA, USA) and transfected into 293A cell lines to generate Ad-FoxO1 or Ad-GFP adenoviruses. The clone primers of FoxO1 are as follows: sense: 5ʹ- CGGGATCCATGGCCGAAGCGCCCCAGGT, antisense: 5ʹ- CCGCTCGAGTCAGCCTGACACCCAGCTGT.
Due to the low interference efficiency of the designed small interfering RNA (siRNA) for FoxO1, we utilized FoxO1-specific short hairpin RNA (shRNA) synthesized by Invitrogen. The synthesized shRNA was annealed to generate the double-stranded shRNA, which was cloned into the pAdEasy-U6-CMV-EGFP vector using BamHI (1010, Takara Bio Inc., Otsu, Japan) and EcoRI (1040, Takara Bio Inc., Otsu, Japan) restriction sites. The specific packaging was performed using the procedure described above. The shRNA sequence of FoxO1 as follows: sense: 5ʹ-tcgagGTGTCCGAGATCAGTAACCCTGAGAATTCAAGAGATTCTCAGGGTTACTGATCTCGGACATTTTTTa-3ʹ; antisense: 5ʹ-agcttAAAAAATGTCCGAGATCAGTAACCCTGAGAATCTCTTGAATTCTCAGGGTTACTGATCTCGGACACc-3ʹ.
Small interference RNA for ATGL
The siRNA of ATGL was donated by Jun Li (Li et al., 2014). The siRNA-ATGL was transfected into GMECs at ~ 80% to 90% confluence using Lipofectamine RNAiMAX (Invitrogen Corp., Carlsbad, CA, USA) following the manufacturer’s instructions. Cells were collected and used in experiments after transfected for 48 h. siRNA oligo was followed: 5-cggtcatcgcagctgtcaa-3ʹ.
RNA isolation and high-throughput sequencing
GMECs (1 × 105) were plated into 12 well plates and cultured overnight. Cells were infected with adenovirus vector containing FoxO1 at ~80% to 90% confluence. After being infected for 48 h, the culture medium was removed and cells were washed three times with PBS. Adding 500 μL trizol (Invitrogen, CA, USA) per well to extract total RNA according to the manufacturer. The RNA quality and integrity were determined by spectrophotometer and agarose gel electrophoresis analysis respectively. The ratio of 260/280 was 1.9 to 2.0:1and the ratio of 28S:18S is 2:1, which indicated the total RNA was acceptable. Sequencing library construction and high throughput sequencing were finished by Huada Company. Differentially expressed genes (DEGs) between the control and FoxO1 overexpression group were analyzed by EdgR software with a threshold of P < 0.05 and log2-fold change ≥ 0.75. Gene Ontology Database was used to analyze the GO enrichment of DEGs. KEGG enrichment of DEGs was analyzed by Kyoto Encyclopedia of Genes and Genomes pathway.
Cellular triacylglycerol assay
GMECs (3 × 105) were plated into six well plates overnight and co-transfected with adenovirus vector containing FoxO1 and siRNA-ATGL for 48 h. The TAG was detected using a triacylglycerol assay kit (E1013, Applygen Technologies Inc. Beijing, China) according to the manufacturer. Cells were washed three times with cold PBS, and 200 μL triglyceride lysate was lysed on ice for 10 min. Then, cells were scraped with a cell spatula into a 1.5 mL centrifuge tube together with the lysate. Cell suspension was fragmented by sonication for 5 min, and the supernatant was collected and split into two centrifuge tubes after being centrifuged at 4 °C 15,000 g for 10 min. TAG content was normalized for protein concentration (μg/mg protein).
Oil red O staining
GMECs (1 × 105) were plated into 12 well plates overnight and co-transfected with adenovirus vector containing FoxO1 and siRNA-ATGL for 48 h. The culture medium was removed and cells were washed three times with PBS. Then, cells were fixed with 4% paraformaldehyde for 30 min at room temperature and washed three times with PBS again. Then, the lipid droplet was stained by 0.5% Oil Red O (0.05 g Oil Red O dissolved in 10 mL of isopropanol) at room temperature (RT). Finally, GMECs were observed under a confocal microscope (Leica Geosystems, Co., Ltd, Germany).
Free fatty acids assay
GMECs (1 × 106) were incubated into 60 mm dishes and co-transfected with an adenovirus vector containing FoxO1 and siRNA-ATGL for 48 h. After incubation, the cells were washed three times with PBS and lysed for 10 min in lysis buffer. Subsequently, the cells were scraped using a spatula and transferred to a 1.5 mL centrifuge tube. The supernatant was collected after centrifuged at 4°C 6800 g for 10 min. The content of free fatty acids (FFAs) was determined using the free fatty Acids (FFA) Content Assay Kit (BC0590, Solarbio, Beijing, China).
Protein extraction and western blot
GMECs (1 × 106) were plated into 60 mm dishes overnight before experiments. After treatment with insulin or LY294002 (ab120243, Abcam, Cambridge, USA) for 24 h, an inhibitor of PI3K, cells were lysed for 10 min on ice using RIPA buffer (R0010, Solarbio, Beijing, China) containing 1 × protease inhibitor (04693132001, Roche, Mannheim, Germany). The cells and lysate were transferred to a 1.5 mL centrifuge tube using a cell spatula. The supernatant was collected by centrifugation at 15,000 g for 10 min at 4 °C. The protein concentration was determined using a BCA protein assay kit (#23225, Thermo Fisher Scientific, USA). Subsequently, the protein samples were boiled at 100 °C for 10 minutes and stored at −80 °C.
Western blot procedure follows our previously described (Tian et al., 2018; He et al., 2020). Briefly, 20 μg protein samples for SDS–PAGE with 10% separation glue and transferred into PVDF (#03010040001, Roche, Mannheim, Germany) for 16 V constant pressure with a semi-dry membrane system (Bio-Rad Laboratories Inc., Hercules, CA, USA). Then, PVDF membrane was blocked with 5% skim milk (232100, BD, Franklin Lakes, New Jersey, USA) for 4 h, and incubated with primary antibody at 4 °C overnight. TBST was washed for 30 min, and the secondary antibody was incubated at room temperature for 2 h. Antibodies in this study include anti-FoxO1 (#2880, Cell Signaling Technology, Danvers, USA; 1:1000), anti-AKT (#4691, Cell Signaling Technology, Danvers, USA; 1:1000), anti-pFoxO1 (#9461, Cell Signaling Technology, Danvers, USA; 1:1000), anti-pAKT (#4060, Cell Signaling Technology, Danvers, USA;1:1000), anti-pPI3K (#4228T, Cell Signaling Technology, Danvers, USA;1:1000), anti-β-actin (CW0096, CW Biotech, Beijing, China; 1:1000), and polyclonal goat anti-mouse HRP-conjugated IgG (CW0102, Biotech, Beijing, China; 1:4000). Primary antibodies were diluted with primary antibody dilution buffer (A1810, Solarbio, Beijing, China) and secondary antibody were diluted with 5% skim milk. Signals were detected by ECL exposure system (BioRad Laboratories, Inc., Hercules, CA, USA). Protein abundance was calculated by Image J software.
Immunofluorescence analysis
GMECs (1 × 105) were seeded in 12 well plates overnight and incubated with insulin or without insulin for 24 h. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature after washed three times with PBS. Then, cells were blocked for 2 h with 5% BSA, which following by incubation with primary antibody of pFoxO1 (#9461, Cell signaling Technology, Danvers, USA; 1:200) overnight at 4 °C. Subsequently, cells were incubated with Alexa Fluor488-conjugated goat anti-rabbit IgG (SA00006–2, Proteintech, Chicago, USA; 1:300) at room temperature for 2 h which away from light. The fluorescence was observed under the confocal microscope (Biotek Technologies Inc., USA).
Plasmids construction and bioinformatics analysis
ATGL gene 5ʹ flanking sequence was amplified with PCR using Forword and Reverse primers from goat genome DNA which lab-preserved. PCR products were cloned into the pMD19-T vector and sequenced. Then, using the following website to predict the putative transcription factor binding sites located in ATGL promoter: TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and PATCH public 1.0 (http://www.gene-regulation.com/cgi-bin/pub/programs/patch/bin/patch.cgi).
To construct different deletion fragments, primers were designed at 2024 bp, 1399 bp, 882 bp, and 524 bp upstream of ATGL promoter transcription start site and 216 bp downstream of the transcription start site. Using full-length sequence of ATGL promoter as a template to amplify DNA of different fragment lengths, and subcloned into the pGL3-basic vector which digested with MIU I and BgLII. The constructs of site-directed mutants for FKH1 and FKH2 were generated by overlapping extended PCR as previously described (He et al., 2021). Briefly, for mutant promoters, two DNA fragments containing the designated mutations were generated using PCR. Then, the two DNA fragments are used as templates to generate full-length DNA fragment by overlapping extended PCR. The FKH mutants were constructed using pGL-(−882/+126) constructs as templates. Primers are shown in Table 1.
Table 1.
Primers used for cloning, deletion, site-directed mutagenesis, and ChIP-PCR of ATGL promoter
| Primer name | Primer | Primer sequences2 (5ʹ-3ʹ) | Binding(bp) |
|---|---|---|---|
| Cloning primers | Forward | GGCAGAACTCGCAATCCTA | −2024 |
| Reverse | CACGCCGATATGGTAGAC | +216 | |
| 5ʹ deletion primers | F-2024 F-1399 |
ATTACGCGTGGCAGAACTCGCAATCCTA ATTACGCGTCTCAGTCCTTACTTGAACCT | −2024 −1399 |
| F-882 | ATTACGCGTTCAGAACAGAGACAGGACT | −882 | |
| F-524 | ATTACGCGTGATTCTGTGATGAGTCTCG | −524 | |
| R + 216 | GGAAGATCTCACGCCGATATGGTAGAC | +216 | |
| Site-directed | FKH1 | CGCTGCCTGTGTGTTTACGGGG | −549 |
| ChIP-PCR | FKH –mut | CGCTGCCTGTGCGGATACGGGG | −549 |
| FKH1-anti-mut | CCCCGTATCCGCACAGGCAGCG | −549 | |
| FKH2 | AGACGCCAGTGTTCTCTCTGGGAAT | −585 | |
| FKH 2-mut | AGACGCCAGCGGTATCTCTGGGAAT | −585 | |
| FKH 2-anti-mut | ATTCCCAGAGATACCGCTGGCGTCT | −585 | |
| FKH1-F | GCTGCCTGTGTGTTTACGG | −548 | |
| FKH1-R | CCCAGAGCAGTGCGGAT | −352 | |
| FKH2-F | GTAGACCCACAGCCCAAGGA | −707 | |
| FKH2-R | GTAAACACACAGGCAGCG | −552 |
“+” and “−” represent upstream and downstream from the transcriptional start site; bold represents mutation sites, Italics indicate restriction enzyme sites.
Transfection and luciferase assays
GMECs (8 × 104) were seeded in 48-well plates before experiments. GMECs were co-transferred with pcDNA3.1-FoxO1 overexpression vector and ATGL promoter (300 ng of total DNA per well) according to X-treme GENE HP DNA transfection reagent at ~80% to 90% confluence. Then, cells were cultured with serum-free DMEM/F12 medium for 12 h before treatment with insulin. Renilla luciferase vector (pRL-TK) was used as internal control and the ratio of pGL3-ATGL to pRL-TK was 30:1. Cells were washed three times with PBS and lysed for 40 min in lysis buffer at room temperature. The method of luciferase activity measurement followed by the instructions of Dual-Luciferase Reporter Assay (#0000499188, Promega, USA).
Chromatin immunoprecipitation assays and sequenced
We collected mammary gland tissue samples from six dairy goats at different lactation stages, including peak lactation, mid-lactation, and the dry period. The samples were processed in our laboratory as follows: First, the samples were homogenized in a mortar and the grinding bar in the presence of liquid nitrogen. During the grinding process, any sample sticking to the pestle was scraped into the mortar using a small spoon. Subsequently, the tissue sample and GMECs were fixed with 1% methanol-free formaldehyde dissolved in cold PBS (#28906, Thermo Fisher Scientific, USA) and incubated at room temperature (RT)for 10 min. Add 0.125M glycine to stop fixation and centrifuge at 1000 g for 5 min at 4 °C to obtain the cell pellet. The cells were then resuspended in ice-cold PBS three times and lysed with 300 μL SDS lysis buffer (P0013G, Beyotime, Shanghai, China) containing 1× protease inhibitor. The chromatin was sheared into 100–500 bp fragments using a Bioruptor Sonication system (Bioruptor Pico, Diagenode, Belgium) with the following program: 30 s ON, 30 s OFF, with the number of 10–12 cycles.
For ChIP-seq assay, the chromatin in the immunoprecipitation (IP) group was incubated with 5ul H3K27ac antibody (ab4729, Abcam, Cambridge, USA) or IgG antibody (ab171870, Abcam, Cambridge, USA) at 4 °C overnight. For ChIP-PCR assay, the chromatin in the IP group was incubated with 5 μL FoxO1 antibody (ab39670, Abcam, Cambridge, USA) or IgG antibody at 4 °C overnight. The beads were washed using high salt buffer, low salt buffer, and IP buffer respectively. Protein–DNA complexes were reverse cross-linked at 65 °C for 4 h. Finally, DNA was purified using PCI (phenol–chloroform–isoamyl alcohol 25:24:1) and used for ChIP-qPCR and ChIP-seq. The primers used in the ChIP-PCR are listed in Table 1. The ChIP-seq data were aligned with the genome to produce a bw file. The Integrated Genomics Viewer (IGV) was used to display the H3K27ac patterns in different genes.
Data analysis
The data were analyzed by SPSS 20.0 and presented as mean ± SEM of at least three independent experiments. Each experiment consisted of three biological replicates, and all experiment was individually repeated three times. The results of RT-qPCR were analyzed using the 2−ΔΔCt method, where Ct represents the cycle threshold. Student’s t-test was used to compare the results between the two groups. For multiple comparisons, one-way ANOVA was performed followed by Duncan’s test. Statistically significant differences were defined as P < 0.05 indicated by * and P < 0.01 indicated by **.
Results
FoxO1 screening in dairy goat dairy using ChIP-Seq
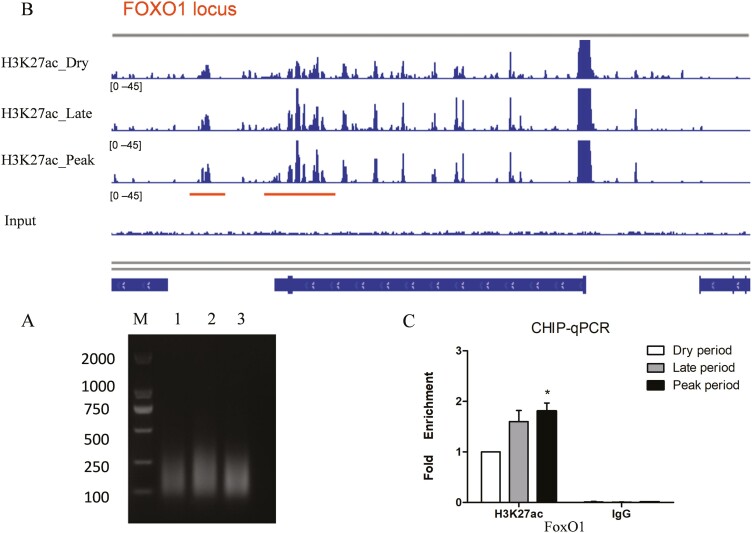
H3K27ac is a chromatin marker that indicates an active regulatory region. To identify and screen key transcriptional regulation factors in mammary gland samples of dairy goats, ChIP-seq assay was performed. As shown in Figure 1A, the DNA fragments were concentrated in the 100 to 500 bp size range, which is suitable for ChIP assays. The enriched DNA was purified and used for sequencing. The ChIP data were aligned to the goat genome, and peak calling was performed using MASC2. Visualization in Integrative Genomics Viewer (IGV) demonstrated higher enrichment of H3K27ac in the FoxO1 enhancer region during the peak lactation stage than compared to the dry period (Figure 1B). ChIP-qPCR confirmed that H3K27ac occupancy on the FoxO1 enhancer region was higher during peak lactation stage (Figure 1C). Therefore, we speculate that FoxO1 is an important transcription factor involved in the lactation process of the mammary gland.
Figure 1.
ChIP-seq of H3K27ac in dairy goat mammary gland. (A) The sonication product of chromosome DNA. Samples of mammary gland tissues at different lactation stages (peak-lactation mid-lactation and dry period; n = 6 goats) were grind until homogenization. The samples were lysed and the chromatin was sheared into 100–500 bp fragments using bioruptor sonication system (B) Distribution of H3K27ac in the FoxO1 loci. IGV was used to display the H3K27Ac pattern in FoxO1 enhancer region. (C) The fold enrichment of H3K27ac in different lactation stages was detected by ChIP-qPCR. Data were presented as mean ± SEM for three individual cultures. *P < 0.05.
FoxO1 regulates fatty acid metabolism via ATGL
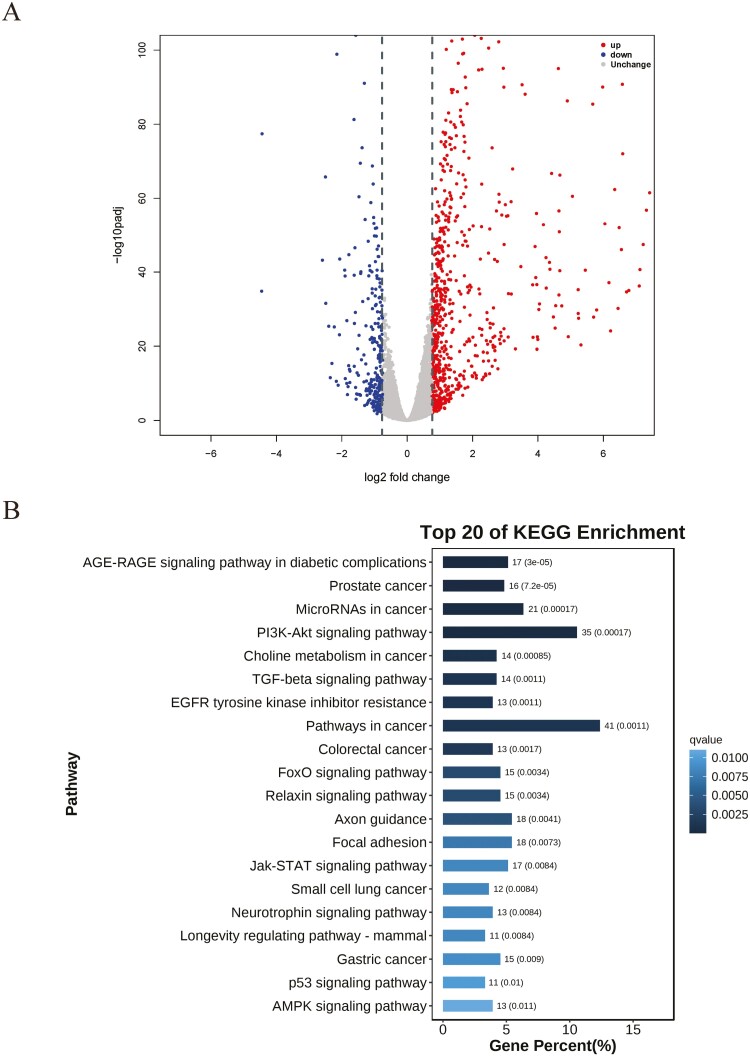
To investigate the function of FoxO1, we first performed RNA-seq in goat mammary epithelial cells (GMECs). We identified a total of 1,084 differentially expressed genes (DEGs) between the FoxO1-overexpressing and control groups. Among these, 806 genes were upregulated, while 278 were downregulated (Figure 2A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed enrichment of PI3K-Akt signaling pathway, TGF-beta signaling pathway, Jak-STAT signaling pathway, p53 signaling pathway, and AMPK signaling pathway (Figure 2B), which are closely related to lipid metabolism (Soukupova et al., 2021; Fang et al., 2022; Savova et al., 2023; Yao et al., 2023). These findings further supported the hypothesis that FoxO1 might play an important role in lipid homeostasis.
Figure 2.
RNA-seq analysis between control and FoxO1 overexpression group. (A) Volcano plot of differentially expressed genes (DEGs) between control and FoxO1 overexpression group. Red dots represent up-regulated genes, and blue dots represent down-regulated genes (B) The top 20 KEGG signaling pathways enriched of DEGs between control and FoxO1 overexpression groups.
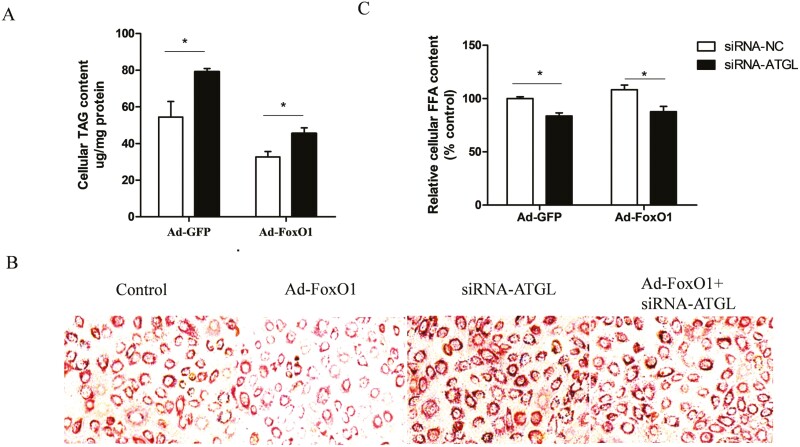
ATGL is the rate-limiting enzyme for triglyceride lipolysis (Zhang et al., 2022). FoxO1 knockdown markedly downregulated ATGL mRNA levels (He et al., 2020). Here, we aimed to explore whether FoxO1 mediates ATGL activation to regulate lipolysis in GMECs. For this purpose, we infected GMECs with an adenoviral vector expressing FoxO1(Ad-FoxO1) or an adenoviral vector containing GFP (Ad-GFP), followed by a 12 h transfection with small interfering RNA (siRNA) targeting ATGL (siRNA- ATGL). We found that FoxO1 overexpression markedly decreased cellular triglyceride (TAG) content compared to the Ad-GFP group in siRNA-NC cells. Conversely, the content of cellular TAG in cells co-transfected with Ad-FoxO1 and siRNA-ATGL was markedly higher than that in cells co-transfected with Ad-FoxO1 and siRNA- NC (Figure 3 A, P < 0.05). These results suggest that the inhibitory effect of FoxO1 on TAG content is attenuated by ATGL knockdown. Oil red O staining confirmed a notable decrease in the number of lipid droplets in FoxO1-overexpressing cells, whereas an increase was observed in the ATGL-knockdown group. Notably, we observed that ATGL interference attenuated the FoxO1-induced inhibition in the lipid droplets formation (Figure 3B). Additionally, we observed a slight increase in the relative content of cellular FFAs in FoxO1-overexpressing cells, while knockdown of ATGL decreased cellular FFAs. However, the overexpression of FoxO1 had no effect on FFA content in siRNA-ATGL cells. Moreover, the cellular FFA content in cells co-transfected with Ad-FoxO1 and siRNA-ATGL was lower than that in cells co-transfected with Ad-FoxO1 and siRNA-NC (Figure 3 C, P < 0.05). Taken together, our findings support the hypothesis that FoxO1 regulates lipid metabolism might by modulating ATGL expression.
Figure 3.
FoxO1 promotes lipolysis by regulating ATGL expression. (A) Effects of FoxO1 overexpression on cellular TAG content in ATGL knockdown cells. Cells were infected with Ad-FoxO1 or Ad-GFP after transfected with siRNA-ATGL or siRNA-NC. (B) Oil red O staining of lipid droplet after overexpression FoxO1, followed by 12 h transfected with siRNA-ATGL. (C) Effects of FoxO1 overexpression on cellular FFA content in ATGL knockdown cells. Ad-FoxO1: Adenoviral vector expressing FoxO1; Ad-GFP: Adenoviral vector expressing GFP; siRNA-ATGL: small interference RNA for ATGL; siRNA-NC: Scramble RNA. one-way ANOVA was used for multiple comparisons followed by Duncan’s test. Data were presented as mean ± SEM for three individual cultures. *P < 0.05.
Cloning and characterization of the goat ATGL promoter
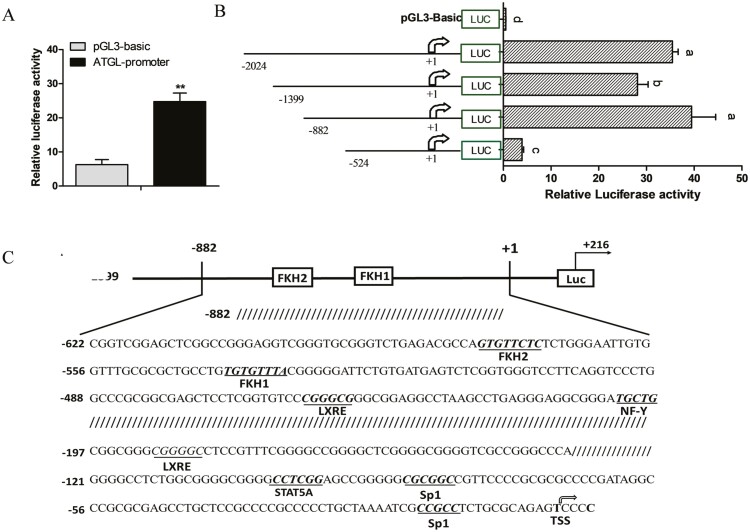
To investigate the mechanism of transcriptional regulation of ATGL by FoxO1, we obtained a 2,240 bp genomic fragment containing the 5ʹ flanking sequence of the ATGL promoter. We then co-transfected GMECs with the pGL3-ATGL and pRL-TK vectors and measured the relative luciferase activity after 48 h. We observed that the ATGL promoter was more active in transfected cells than in control cells (Figure 4A).
Figure 4.
Deletion analysis and characterization of ATGL promoter. (A) Relative luciferase activity of ATGL promoter. GMECs were co-transfected with ATGL promoter and TK-Renilla luciferase vector for 48 h. (B) Relative luciferase activity analysis of deletion fragments of ATGL promoter. GMECs were transfected with different lengths of ATGL promoter (−2024 bp/+216 bp, −1399 bp/+216 bp, −882 bp/+216 bp, −524 bp/+216 bp) for 48 h. Three independent replicates were shown as mean ± SEM. **P < 0.01 vs. control. Different lowercase letters represent significant differences between different groups (P < 0.05); the same lowercase letters mean no significant differences among groups (P > 0.05). (C) Schematic representation of goat ATGL promoter region. The position of transcription factor predicted binding sites was underlined, and the transcription start site was indicated by a black arrow.
To explore the role of the core region of the ATGL promoter, we obtained four promoter fragments of various lengths (−2,024/+216, −1,399/+216, −882/+216, −524/+216) via progressive deletion and subcloned them into luciferase reporter gene vectors. We transfected these recombinant constructs into GMECs to determine the contribution of each region to the activity of the ATGL promoter. We found that deletion of the ATGL promoter region from −2,024 to −1,399 markedly decreased the activity of the promoter (Figure 4B, P < 0.05). Conversely, we found that the activity of the ATGL promoter was significantly increased following the progressive deletion of the region from –1,399 to –882 bp. Importantly, promoter activity was almost abolished upon further deletion of the region from –882 bp to –524 bp (Figure 4B, P < 0.01). These results indicated that the region from –882 bp to –524 bp is important for maintaining the basic transcriptional activity of the ATGL promoter. Additionally, bioinformatics analysis indicated the presence of two FoxO1 binding sites in the ATGL promoter (Figure 4C).
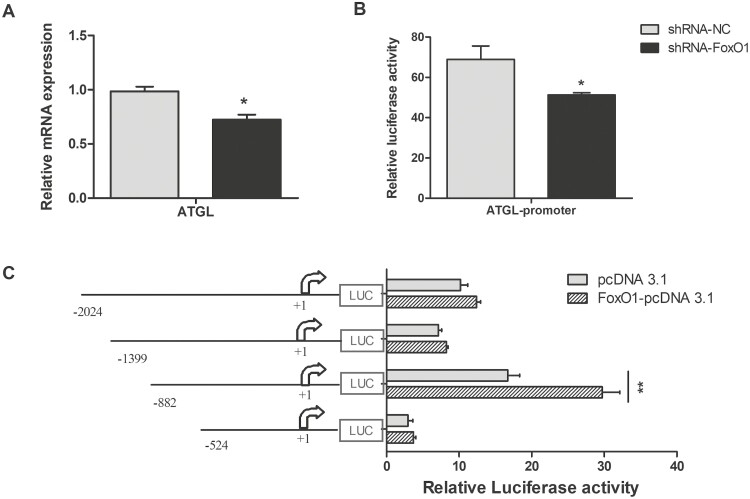
FoxO1 promotes activation of ATGL transcription
Transcription factors, such as SREBP1, LXR, and PPAR, always bind to cis-elements in the sequence of their target genes to regulate downstream gene expression (Shi et al., 2013; Yao et al., 2016; Lyu et al., 2021). Adenovirus containing FoxO1-specific shRNA was generated to evaluate the effect of FoxO1 on the expression of ATGL. We found that FoxO1 knockdown significantly decreased mRNA level (Figure 5A, P < 0.05) and promoter activity of ATGL (Figure 5 B, P < 0.05) compared with control group. To determine the regulatory role of FoxO1 on the ATGL promoter, constructs containing truncated versions ATGL promoter and FoxO1-overexpressing vectors were co-transfected into GMECs. We found that overexpression of FoxO1 significantly upregulated the activity of the ATGL promoter in cells transfected with the −882 bp/216 bp fragment, whereas there was no significant effect on the activity of the ATGL promoter in cells transfected with the −2,024 bp/216 bp, −1,399 bp/216 bp, and −524 bp/216 bp fragments (Figure 5C, P > 0.05). Thus, we speculated the existence of negative regulatory elements upstream of the ATGL promoter and an important FoxO1 positive cis-acting element within the −882 bp/216 bp region of the ATGL promoter.
Figure 5.
Effects of FoxO1 on expression of ATGL and ATGL promoter activity. (A and B) The mRNA levels analysis of ATGL and relative luciferase activity analysis of ATGL promoter in FoxO1 knockdown cells. (C) Relative luciferase activity analysis of different lengths of ATGL promoter in FoxO1 overexpression cells. Different lengths of ATGL promoter (−2024 bp/+216 bp, −1399 bp/+216 bp, −882 bp/+216 bp, −524 bp/+216 bp) and FoxO1-pcDNA3.1 constructs or empty vectors were co-transfected into GMECs and incubate for 48 h, relative luciferase was detected. Data were analyzed with Student’s t-test. Values are shown as mean ± SEM for three biological replicates. *P < 0.05.
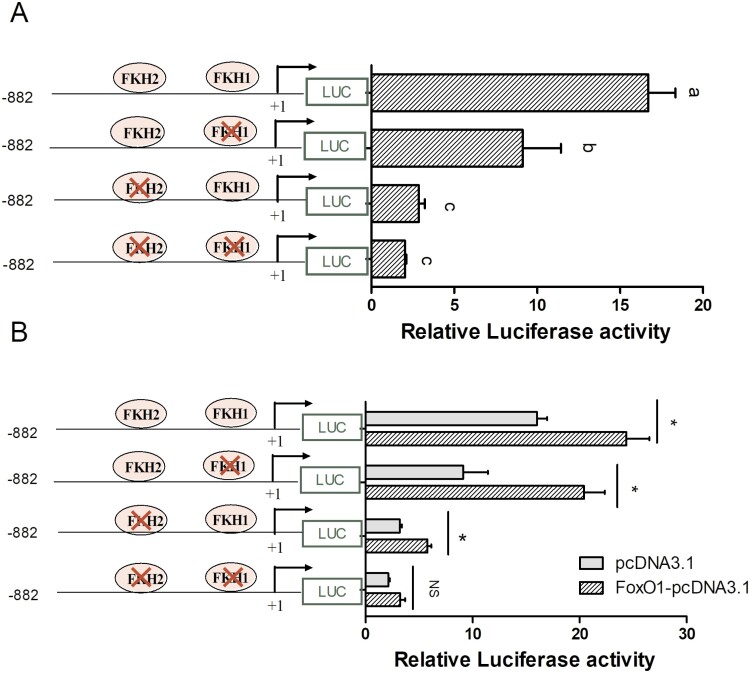
FoxO1 promotes ATGL promoter activity via FoxO1 binding sites
To investigate the mechanism by which FoxO1 regulates ATGL expression, we performed a bioinformatics analysis of the ATGL promoter sequence. We identified two FoxO1 binding sites (FKH) within the −882 bp/+216 fragment of the ATGL promoter. To determine whether the FKH elements influence the ATGL promoter activity, we generated constructs with mutated FKH sites in the –882 bp/+216 bp fragment and transfected them into GMECs. When either FKH1 or FKH2 mutation was mutated, or when both FKH sites were mutated simultaneously, the relative luciferase activity of ATGL promoter significantly decreased compared to the −882 bp/+216 fragment containing two FKH elements (Figure 6A, P < 0.05). These results indicate that FKH elements likely play a crucial role in maintaining the transcriptional activity of the ATGL promoter.
Figure 6.
FoxO1 binding sites are critical for maintaining ATGL promoter activity. (A) Relative luciferase activity analysis of FoxO1 binding sites (FKH) mutated in the region −882 bp/+216 of the ATGL promoter. GMECs were co-transfected with individually or simultaneously mutated FKH site constructs and TK-Renilla luciferase vector for 48 h (n = 3 biological replicates), and relative luciferase was detected. One-way ANOVA was performed with Duncan’s test for multiple comparisons. Different lowercase letters represent significant differences between different groups (P < 0.05), while the same lowercase letters mean no significant differences among groups (P > 0.05). (B) Effects of FoxO1 overexpression on ATGL promoter activity after FKH sites were mutated in the −882 bp/+216 bp region. GMECs were co-transfected with the pcDNA3.1(+) vector or FoxO1-pcDNA3.1 and the wild-type pGL-(−882/+216) or FKH sites were individually or simultaneously mutated constructs. After transfection 48 h, cells were lysed, and luciferase activities were detected (n = three biological replicates). Data were analyzed with Student’s t-test. Values are shown as mean ± SEM three biological replicates. *P < 0.05.
To further confirm the involvement of FKH elements in FoxO1-mediated regulation of the transcription of ATGL, we performed luciferase reporter assays to measure the activity of the ATGL promoter with individually or simultaneously mutated FKH sites. We found that overexpression of FoxO1 significantly upregulated the activity of the wild-type ATGL promoter (Figure 6B, P < 0.01). Additionally, FoxO1 overexpression increased the ATGL promoter activity in groups with single FKH1 or FKH2 mutations compared to the empty vector control group (Figure 6 B, P < 0.05). However, although overexpression of FoxO1 slightly increased ATGL promoter activity when both FKH1 and FKH2 sites were mutated simultaneously, the difference was not statistically significant (P > 0.05). Collectively, we hypothesized that FKH sites may be required for FoxO1-induced promotion of ATGL promoter activity.
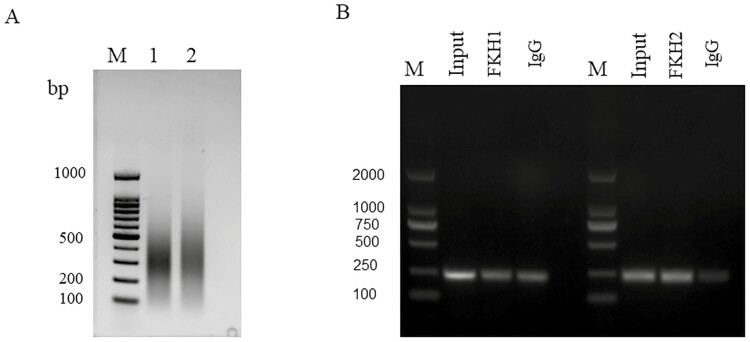
To ascertain whether FoxO1 directly binds to the FKH element to regulate ATGL transcription in vivo, we performed chromatin immunoprecipitation (IP) to investigate the interaction between FoxO1 and ATGL. We found that the DNA fragments were concentrated within the 100 to 500 bp size range, which was suitable for ChIP assays (Figure 7A). ChIP PCR analysis demonstrated the presence of a single band in the input groups, indicating the suitability of PCR amplification primers. In addition, we also amplified single-target bands containing FKH sites in both FKH1 and FKH2 groups (Figure 7B), confirming that FoxO1 binds to the FKH element on the ATGL promoter in GMECs.
Figure 7.
FoxO1 promotes ATGL transcriptional activity via directly binding FKH sites in GMECs. (A) The sonication product of chromosome DNA. M: DNA marker. (B) ChIP-PCR products were analyzed to assess FoxO1 occupancy of ATGL promoters in GMECs. Normal rabbit IgG was used as negative control.
Insulin inhibits FoxO1 transcriptional activity via PI3K/Akt signaling pathway
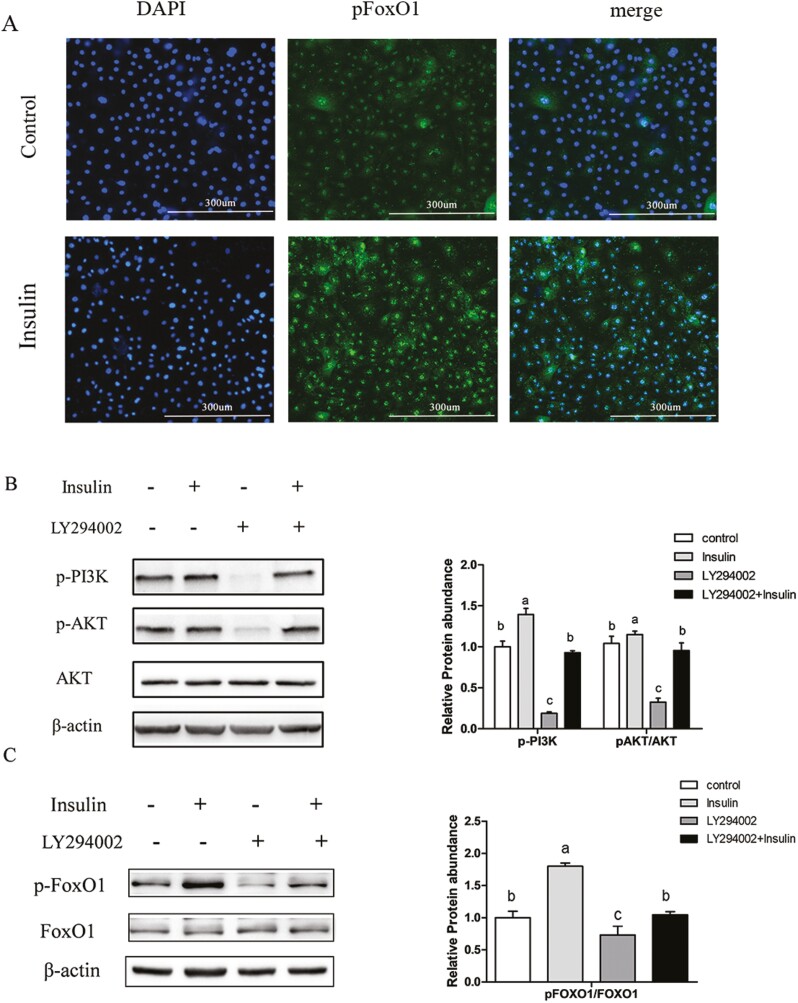
FoxO1 acts as an important mediator of insulin, and its transcriptional activity is reduced upon insulin-mediated phosphorylation (Puigserver et al., 2003). Our previously study illustrated that insulin reduced the levels of nuclear FoxO1 protein and suppressed FoxO1 transcriptional activity in GMECs (He et al., 2020). To explore how insulin affects the transcriptional activity of FoxO1, we initially examined the levels of cytoplasmic phosphorylated FoxO1 (pFoxO1) using immunofluorescence. We found that insulin treatment elevated cytosolic pFoxO1 protein levels compared to control group (Figure 8A). Additionally, insulin treatment increased the protein levels of phosphorylated PI3K (p-PI3K) and phosphorylated Akt (p-Akt), while these effects were reversed following incubation with the PI3K inhibitor LY294002 (Figure 8B). Western blot analysis showed that insulin treatment significantly increased pFoxO1 levels, whereas treatment with LY294002 markedly reduced them compared to control group. Furthermore, insulin-mediated stimulation was almost abolished after incubation with LY294002 (Figure 8C). These results indicate that insulin treatment modulates the PI3K/AKT signaling pathway to phosphorylate FoxO1 in GMECs.
Figure 8.
The transcriptional activity of FoxO1 was regulated by insulin through the PI3K/Akt signaling pathway. (A) Immunofluorescence staining of pFoxO1 in cells incubated with or without insulin. GMECs (1 × 105) were seeded into 12 well plates and incubated overnight. Cells were incubated with serum-free medium for 12 h, followed by incubation with or without insulin for 24 h. The levels of cytosolic pFoxO1 were examined (n = 3). Scale bar = 300 μm. (B) Levels of phosphorylated AKT (pAKT)and PI3K(p-PI3K) proteins determined by western blot in GMECs. Cells were plated into 60 mm dishes and treated with insulin or LY294002 for 24 h, protein abundances of p-PI3K, p-AKT, and AKT were measured. Relative protein abundance was normalized to β-actin or total kinase. (C) Total FoxO1 and pFoxO1 protein levels in GMECs incubation with insulin or LY294002 (panel left); quantitative analysis of pFoxO1 in GMECs (panel right). Protein abundance was calculated by Image J software. One-way ANOVA was used for multiple comparisons followed by Duncan’s test. Data were presented as mean ± SEM for three independent experiments. All experiments were repeated three times. Different lowercase letters represent significant differences between different groups, while the same lowercase letters mean no significant differences among groups.
FoxO1 promotes ATGL promoter activity in an insulin-dependent manner
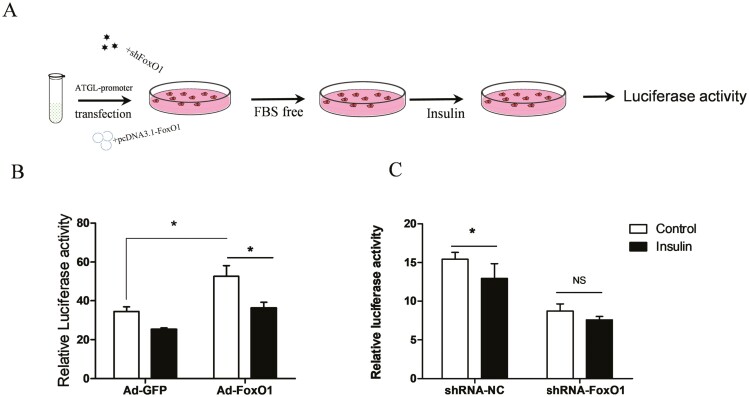
To investigate the effect of FoxO1 on the activity of the ATGL promoter under insulin treatment, we co-transfected GMECs with pGL3-ATGL and FoxO1-overexpressing vectors or adenovirus containing FoxO1-specific shRNA before incubation with insulin (Figure 9A). Overexpression of FoxO1 alone significantly increased the activity of the ATGL promoter compared to the Ad-GFP group in control cells (P < 0.05). Insulin treatment decreased the FoxO1-induced ATGL promoter activity compared to the control group in FoxO1-overexpressing cells (Figure 9B, P < 0.05). To further verify the effect of endogenous FoxO1 on the transcription of ATGL, we transfected GMECs with short hairpin RNA against FoxO1 (shRNA-FoxO1) adenovirus, followed by insulin treatment. We observed that insulin treatment alone significantly reduced the activity of ATGL promoter (Figure 9C, P < 0.05). Nonetheless, insulin treatment has no effect on the activity of the ATGL promoter after FoxO1 knockdown compared to the FoxO1 knockdown alone group (Figure 9C, P > 0.05). Collectively, these results suggest that insulin attenuates FoxO1-induced activation of the ATGL promoter and may affect ATGL transcriptional activity via FoxO1.
Figure 9.
FoxO1 promotes ATGL promoter activity in an insulin-dependent way. (A) Experimental design outlined. GMECs (8 × 104) were seeded in 48-well plates and incubated overnight. Cells were co-transfected with the ATGL promoter or pRL-TK vector and pcDNA3.1(+) vector or FoxO1-pcDNA3.1, or co-transfected with the ATGL promoter or pRL-TK vector and adenovirus containing FoxO1-specific shRNA or adenovirus containing GFP, and incubation for 12 h. Next, cells were incubated with serum-free medium for 12 h, followed by incubation with or without insulin for 24 h. Finally, GMECs were lysed and used for luciferase activity analysis. Three biological replicates were performed for each experiment and all experiment was individually repeated three times. (B) Relative luciferase activity of ATGL promoter after FoxO1 overexpression in GMECs treatment with or without insulin. GMECs were co-transfected with Ad-GFP or Ad-FoxO1 and ATGL promoter vector for 12 h, followed by 12 h incubation with serum-free medium before being treated with insulin for 24 h. GMECs were lysed and used for luciferase activity analysis. (C) Effects of FoxO1 knockdown on ATGL promoter activity in GMECs treatment with or without insulin. Data are shown as mean ± SEM for three independent replicates. “*”P < 0.05; “NS” P > 0.05.
Discussion
The mammary gland is a unique organ responsible for milk synthesis and secretion in preparation for lactation (Jena et al., 2019). Exploring the active transcription factor in the mammary gland is of paramount scientific significance to understanding the lipid metabolism. To our knowledge, this is the first study to perform chromatin immunoprecipitation sequencing (ChIP-seq) assays to screen transcription factors in mammary glands. We found that FoxO1 was actively expressed during the peak lactation, and directly binds to the ATGL promoter to regulate the expression of ATGL. Overall, our results indicate that FoxO1 plays an important role in lipid metabolism in goat mammary epithelial cells (GMECs).
Lipid metabolism is a complex biochemical process that is regulated by multiple transcriptional factors. FoxO1 is an important mediator of insulin involved in lipid metabolism and other biological processes (Ponugoti et al., 2012). Studies have shown that FoxO1 can inhibit SREBP1 activity by binding to the SREBP1 promoter, thereby regulating lipid synthesis in mammals (He et al., 2020). Additionally, FoxO1 decreases adipogenesis by repressing the expression of PPARG or ChREBP (Fajas et al., 2002; Ido-Kitamura et al., 2012), indicating that FoxO1 participates in lipid metabolism through various pathways. Moreover, ATGL is a key enzyme that catalyzes lipolysis and is predominantly responsible for the hydrolysis of TAG (Zimmermann et al., 2004; Schweiger et al., 2006). Studies have shown that ATGL expression is regulated through the PI3K/Akt pathway, where FoxO1 exerts important effects (Li et al., 2017). Furthermore, FoxO1 directly binds to the ATGL promoter to promote ATGL transcription and negatively regulate TAG accumulation in adipocytes (Chakrabarti and Kandror, 2009). In the present study, the KEGG pathway analysis revealed enrichment of the p53 signaling pathway and the AMPK signaling pathway. ATGL expression was directly regulated by FoxO1, and insulin inhibited the expression of ATGL in GMECs by modulating FoxO1 activity, which is consistent with previous studies. Further, Milk fat, milk protein, and lactose are the main components of goat milk, and triglycerides are the predominant form of milk fat present in milk (Haemmerle et al., 2003). It is necessary to study the regulatory mechanisms underlying TAG hydrolysis. Although the KEGG results for FoxO1 also indicate its involvement in some other important pathways, we focused on one specific pathway in this study. The remaining pathways should be explored in the future.
Phosphorylation is an important posttranscriptional modification of FoxO1 that directly alters its subcellular localization and activity (Yu, 2014). Phosphorylated FoxO1 is anchored in the cytoplasm by the 14-3-3 protein and interacts with ubiquitin E3 ligase to induce its degradation (Schall et al., 2015). AKT acts downstream of PI3K in the insulin signaling pathway and the nuclear translocation of FoxO1 depends on insulin (Tzivion et al., 2011). In this study, insulin treatment significantly increased cytosolic pFoxO1 protein levels in GMECs, and this effect was attenuated after treatment with a PI3K/AKT inhibitor, indicating that FoxO1 can be phosphorylated by insulin via the PI3K/AKT signaling pathway. Moreover, insulin treatment inhibited FoxO1-induced activation of the ATGL promoter, which is supported by previous data (Gonzalez et al., 2011).In addition, insulin-like growth factor 1(IGF1) also phosphorylates FoxO1 through the PI3K signaling pathway in several cell types (Yang et al., 2011). Insulin signals via the insulin receptor (IR) to regulate metabolic changes and cellular growth, while IGF1 signals via the IGF1 receptor (IGF1R) to regulate growth and development. However, IR and the highly homologous IGF1R share common downstream signaling pathways that regulate metabolic processes via the PI3K/Akt signaling pathway (Bhardwaj et al., 2021; Lero and Shaw, 2021). We also mainly focused on common downstream signaling pathways in this study.
The luciferase activity of ATGL promoter markedly increased when deleting the region from −1399 to −882, suggesting the presence of negative regulators or silencers in this region. Bioinformatics analysis of the core promoter region of ATGL identified two potential FoxO1-binding sites. Mutating of the FKH sites significantly reduced the activity of the ATGL promoter, consistent with the results of a previous study performed using 3T3-L1 cells (Chakrabarti and Kandror, 2009). Moreover, a single mutation in FKH2 attenuated the promoting effect of FoxO1 on the transcription of ATGL, whereas a mutation of FKH1 slightly enhanced the promoting effect of FoxO1. It is speculated that the FKH1 may act as the binding site for other transcription factors, as well or involved in the formation of a transcription inhibition complex. The ChIP assay showed a clear band in both the FKH1 and IgG groups, suggesting a lack of specificity for FKH1. In addition, we found that a signal brand was detected in both FKH2 and input groups, suggesting that FoxO1 protein directly binds to the FKH2 promoter in vivo. However, the input band is as bright as the FKH2 bands, indicating that ChIP did not achieve much enrichment. Although ChIP is a very versatile tool, the cross-linking time of transcription factors and the purification method of DNA will affect the enrichment efficiency of DNA. We will further optimize the reaction conditions to improve the enrichment efficiency. Furthermore, earlier studies have found that many FoxO1-regulated target gene promoters do not contain FoxO1-binding sites (Dong et al., 2008). Therefore, although we identified binding sites of transcription factors such as LXR and STAT5 in the goat ATGL promoter, whether they regulate the transcriptional activity of ATGL promoter remains to be further studied.
In conclusion, we demonstrated that FoxO1 promoted the degradation of intracellular TAG by upregulating the expression of ATGL. In addition, we confirmed the requirement of a core region of the ATGL promoter for the basal transcription activity of ATGL. We further demonstrated that FoxO1 promotes the transcription of the ATGL gene by directly binding to FoxO1 binding sites in the ATGL promoter. Insulin phosphorylates FoxO1 through the PI3K signaling pathway and inhibits FoxO1-induced ATGL promoter activity. Our study indicated that FoxO1 plays an important role in lipid metabolism by regulating the transcription of ATGL in GMECs.
Glossary
Abbreviations
- ADRP
adipose differentiation-related protein
- ATGL
adipose triglyceride lipase; BTN1A1, butyrophilin subfamily 1 member A1
- ChREBP
Carbohydrate response element binding protein
- ChIP
Chromatin immunoprecipitation; FoxO1, forkhead box protein O1
- DG
diglyceride
- DEG
differentially expressed gene
- FKH
FoxO1 binding site
- FFA
free fatty acid
- GMECs
Goat mammary epithelial cells
- GO
gene ontology
- IR
insulin receptor
- IGF1
insulin-like growth factor 1; IGF1R insulin-like growth factor 1 receptor
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LXR
liver X receptor
- PBS
phosphate buffer saline
- PPAR
peroxisome proliferators-activated receptors; SREBP1, Sterol regulatory element binding protein 1
- STAT5
signal transducer and activator of transcription-5
- TAG
Triacylglycerol
- TBST
tris-buffered saline tween 20
- TSS
transcriptional start site
Contributor Information
Qiuya He, Shaanxi Key Laboratory of Molecular Biology for Agriculture, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China.
Liangjiahui Gao, Shaanxi Key Laboratory of Molecular Biology for Agriculture, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China.
Fuhong Zhang, Shaanxi Key Laboratory of Molecular Biology for Agriculture, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China.
Weiwei Yao, Shaanxi Key Laboratory of Molecular Biology for Agriculture, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China.
Jiao Wu, Shaanxi Key Laboratory of Molecular Biology for Agriculture, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China.
Ning Song, College of Animal Science and Technology, Anhui Province Key Laboratory of Local Livestock and Poultry Genetic Resource Conservation and Bio-breeding, Anhui Agricultural University, Hefei 230036, China.
Jun Luo, Shaanxi Key Laboratory of Molecular Biology for Agriculture, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China.
Yong Zhang, Shaanxi Key Laboratory of Molecular Biology for Agriculture, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China.
Funding
This research was supported by the National Natural Science Foundation of China (Beijing, China; 31772575; 32202643).
Conflict of interest statement
The authors declare that they have no conflict of interest.
References
- Bhardwaj, G., Penniman C. M., Jena J., Suarez Beltran P. A., Foster C., Poro K., Junck T. L., A. O.Hinton, Jr, Souvenir R., Fuqua J. D.,. et al. 2021. Insulin and IGF-1 receptors regulate complex I-dependent mitochondrial bioenergetics and supercomplexes via FoxOs in muscle. J. Clin. Investig. 131:e146415. doi: 10.1172/JCI146415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti, P., and Kandror K. V... 2009. FoxO1 controls insulin-dependent adipose triglyceride lipase (ATGL) expression and lipolysis in adipocytes. J. Biol. Chem. 284:13296–13300. doi: 10.1074/jbc.C800241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, B. M., Reigan P., Mayle-Combs K. D., Orlicky D. J., and McManaman J. L... 2011. Determinants of adipophilin function in milk lipid formation and secretion. Trends Endocrinol. Metab. 22:211–217. doi: 10.1016/j.tem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. C., Copps K. D., Guo S., Li Y., Kollipara R., Depinho R. A., and White M. F... 2008. Inactivsation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 8:65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas, L., Landsberg R. L., Huss-Garcia Y., Sardet C., Lees J. A., and Auwerx J... 2002. E2Fs regulate adipocyte differentiation. Develop. Cell 3:39–49. doi: 10.1016/s1534-5807(02)00190-9. [DOI] [PubMed] [Google Scholar]
- Fang, C., Pan J., Qu N., Lei Y., Han J., Zhang J., and Han D... 2022. The AMPK pathway in fatty liver disease. Front. Physiol. 13:970292. doi: 10.3389/fphys.2022.970292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, E., Flier E., Molle D., Accili D., and Mcgraw T. E... 2011. Hyperinsulinemia leads to uncoupled insulin regulation of the GLUT4 glucose transporter and the FoxO1 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 108:10162–10167. doi: 10.1073/pnas.1019268108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle, G., Zimmermann R., and Zechner R... 2003. Letting lipids go: hormone-sensitive lipase. Curr. Opin. Lipidol. 14:289–297. doi: 10.1097/00041433-200306000-00009. [DOI] [PubMed] [Google Scholar]
- He, Q., Luo J., Wu J., Yao W., Li Z., Wang H., and Xu H... 2020. FoxO1 Knockdown Promotes Fatty Acid Synthesis via Modulating SREBP1 Activities in the Dairy Goat Mammary Epithelial Cells. J. Agric. Food Chem. 68:12067–12078. doi: 10.1021/acs.jafc.0c05237. [DOI] [PubMed] [Google Scholar]
- He, Q., Luo J., Wu J., Li Z., Yao W., Zang S., and Niu H... 2021. ELOVL6 promoter binding sites directly targeted by sterol regulatory element binding protein 1 in fatty acid synthesis of goat mammary epithelial cells. J. Dairy Sci. 104:6253–6266. doi: 10.3168/jds.2020-19292. [DOI] [PubMed] [Google Scholar]
- Ido-Kitamura, Y., Sasaki T., Kobayashi M., Kim H. J., Lee Y. S., Kikuchi O., Yokota-Hashimoto H., Iizuka K., Accili D., and Kitamura T... 2012. Hepatic FoxO1 integrates glucose utilization and lipid synthesis through regulation of Chrebp O-glycosylation. PLoS One 7:e47231. doi: 10.1371/journal.pone.0047231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janek, S., Anna F. L., Kajetan B., Annelie B., Elia S., Zsofia B., Sarah B., Jana G., Niklas B., and Robert K... 2018. Adipose tissue ATGL modifies the cardiac lipidome in pressure-overload-induced left ventricular failure. PLoS Genet. 14:e1007171. doi: 10.1371/journal.pgen.1007171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena, M. K., Jaswal S., Kumar S., and Mohanty A. K... 2019. Molecular mechanism of mammary gland involution: an update. Dev. Biol. 445:145–155. doi: 10.1016/j.ydbio.2018.11.002. [DOI] [PubMed] [Google Scholar]
- Lero, M. W., and Shaw L. M... 2021. Diversity of insulin and IGF signaling in breast cancer: implications for therapy. Mol. Cell. Endocrinol. 527:111213. doi: 10.1016/j.mce.2021.111213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Luo J., Wang H., Shi H., Zhu J., Yao D., Sun Y., and Yu K... 2014. Adipose triglyceride lipase regulates lipid metabolism in dairy goat mammary epithelial cells. Gene 554:125–130. doi: 10.1016/j.gene.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Li, Y., Ma Z., Jiang S., Hub W., Li T., Di S., Wang D., and Yang Y... 2017. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog. Lipid Res. 66:42–49. doi: 10.1016/j.plipres.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Li, J., Wang Y., Yang P., Han H., Zhang G., Xu H., and Quan K... 2022a. Overexpression of ATGL impairs lipid droplet accumulation by accelerating lipolysis in goat mammary epithelial cells. Anim. Biotechnol. 1–9. doi: 10.1080/10495398.2022.2136678. [DOI] [PubMed] [Google Scholar]
- Li, T., Guo W., and Zhou Z... 2022b. Adipose triglyceride lipase in hepatic physiology and pathophysiology. Biomolecules 12:57. doi: 10.3390/biom12010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Li C., Pryce J., and Rochfort S... 2020. Comprehensive characterization of bovine milk lipids: triglycerides. ACS Omega 5:12573–12582. doi: 10.1021/acsomega.0c01841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu, J., Fukunaga K., Imachi H., Sato S., Kobayashi T., Saheki T., Ibata T., Yoshimura T., Iwama H., and Murao K... 2021. Oxidized LDL downregulates ABCA1 expression via MEK/ERK/LXR pathway in INS-1 cells. Nutrients 13:3017. doi: 10.3390/nu13093017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, H., Ou J., Zhang X., Chen Y., and Liang H... 2015. Macrophage CGI-58 deficiency promotes IL-1β transcription by activating the SOCS3-FOXO1 pathway. Clin. Sci. (Colch) 128:493–506. doi: 10.1155/2015/542029. [DOI] [PubMed] [Google Scholar]
- Obrowsky, S., Chandak P. G., Patankar J. V., Povoden S., Schlager S., Kershaw E. E., Bogner-Strauss J. G., Hoefler G., Levak-Frank S., and Kratky D... 2013. Adipose triglyceride lipase is a TG hydrolase of the small intestine and regulates intestinal PPARα signaling. J. Lipid Res. 54:425–435. doi: 10.1194/jlr.M031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y. W., Juárez M., Ramos M., and Haenlein G... 2007. Physico-chemical characteristics of goat and sheep milk. Small Ruminant Res. 68:88–113. doi: 10.1016/j.smallrumres.2006.09.013. [DOI] [Google Scholar]
- Peng, S., Li W., Hou N., and Huang N... 2020. A review of FoxO1-regulated metabolic diseases and related drug discoveries. Cells 9:184. doi: 10.3390/cells9010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponugoti, B., Dong G., and Graves D. T... 2012. Role of forkhead transcription factors in diabetes-induced oxidative stress. Exp. Diabetes Res. 2012:939751. doi: 10.1155/2012/939751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver, P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D.,. et al. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1a interaction. Nature 423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Sabir, U., Irfan H. M., Alamgeer, I.Umer, Z. R. Niazi, and Asjad H. M. M... 2022. Phytochemicals targeting NAFLD through modulating the dual function of forkhead box O1 (FOXO1) transcription factor signaling pathways. Naunyn Schmiedebergs Arch. Pharmacol. 395:741–755. doi: 10.1007/s00210-022-02234-2. [DOI] [PubMed] [Google Scholar]
- Savova, M. S., Mihaylova L. V., Tews D., Wabitsch M., and Georgiev M. I... 2023. Targeting PI3K/AKT signaling pathway in obesity. Int. J. Biol. Sci. 159:114244. doi: 10.1016/j.biopha.2023.114244. [DOI] [PubMed] [Google Scholar]
- Schall, D., Schmitt F., Reis B., Brandt S., and Beer-Hammer S... 2015. SLy1 regulates T-cell proliferation during Listeria monocytogenes infection in a Foxo1-dependent manner. Eur. J. Immunol. 45:3087–3097. doi: 10.1002/eji.201545609. [DOI] [PubMed] [Google Scholar]
- Schweiger, M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., and Zimmermann R... 2006. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- Shi, H. B., Luo J., Yao D. W., Zhu J. J., Xu H. F., Shi H. P., and Loor J. J... 2013. Peroxisome proliferator-activated receptor-γ stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase. J. Dairy Sci. 96:7844–7853. doi: 10.3168/jds.2013-7105. [DOI] [PubMed] [Google Scholar]
- Soukupova, J., Malfettone A., Bertran E., Hernández-Alvarez M. I., Peñuelas-Haro I., Dituri F., Giannelli G., Zorzano A., and Fabregat I... 2021. Epithelial-mesenchymal transition (EMT) induced by TGF-β in hepatocellular carcinoma cells reprograms lipid metabolism. Int. J. Mol. Sci. 22:5543. doi: 10.3390/ijms22115543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H., Luo J., Zhang Z., Wu J., Zhang T., and Bionaz M... 2018. CRISPR/Cas9-mediated stearoyl-CoA desaturase 1 (SCD1) deficiency affects fatty acid metabolism in goat mammary epithelial cells. J. Agric. Food Chem. 66:10041–10052. doi: 10.1021/acs.jafc.8b03545. [DOI] [PubMed] [Google Scholar]
- Trites, M. J., and Clugston R. D... 2019. The role of adipose triglyceride lipase in lipid and glucose homeostasis: lessons from transgenic mice. Lipids Health Dis. 18:204. doi: 10.1186/s12944-019-1151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion, G., Dobson M., and Ramakrishnan G... 2011. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim. Biophys. Acta 1813:1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Luo J., Wang W., Zhao W., and Lin X... 2010. Characterization and culture of isolated primary dairy goat mammary gland epithelial cells. Chin. J. Biotechnol. 26:1123–1127. CNKI:SUN:SHWU.0.2010-08-017. [PubMed] [Google Scholar]
- Yang, X., Lu X., Lombès M., Rha G. B., Chi Y. I., Guerin T. M., Smart E. J., and Liu J... 2010. The G0/G1 switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Xu H., Yu S., Cao H., Fan J., Ge C., Fransceschi R. T., Dong H. H., and Xiao G... 2011. Foxo1 mediates insulin-like growth factor 1 (IGF1)/insulin regulation of osteocalcin expression by antagonizing Runx2 in osteoblasts. J. Biol. Chem. 286:19149–19158. doi: 10.1074/jbc.M110.197905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, D., Luo J., He Q., Shi H., Li J., Wang H., Xu H., Chen Z., Yi Y., and Loor J. J... 2016. SCD1 alters long‐chain fatty acid (LCFA) composition and its expression is directly regulated by SREBP‐1 and PPARγ 1 in dairy goat mammary cells. J. Cell. Physiol. 232:635–649. doi: 10.1002/jcp.25469. [DOI] [PubMed] [Google Scholar]
- Yao, Y., Luo Z. P., Li H. W., Wang S. X., Wu Y. C., Hu Y., Hu S., Yang C. C., Yang J. F., Wang J. P.,. et al. 2023. P38γ modulates the lipid metabolism in non-alcoholic fatty liver disease by regulating the JAK-STAT signaling pathway. FASEB J. 37:e22716. doi: 10.1096/fj.202200939RR. [DOI] [PubMed] [Google Scholar]
- Ju, Y., Xu T., Zhang H., Yu. A.. 2014. FOXO1-dependent DNA damage repair is regulated by JNK in lung cancer cells. Int. J. Oncol. 44:1284–1292. doi: 10.3892/ijo.2014. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Bu S. Y., Mashek M. T., O-Sullivan I., Sibai Z., Khan S. A., Ilkayeva O., Newgard C. B., Mashek D. G., and Unterman T. G... 2016. Integrated regulation of hepatic lipid and glucose metabolism by adipose triacylglycerol lipase and FoxO proteins. Cell Rep. 15:349–359. doi: 10.1016/j.celrep.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R., Meng J., Yang S., Liu W., Shi L., Zeng J., Chang J., Liang B., Liu N., and Xing D... 2022. Recent advances on the role of ATGL in cancer. Front. Oncol. 12:944025. doi: 10.3389/fonc.2022.944025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, N., Tan H., Wang L., Han L., and Liu X... 2021. Palmitate induces fat accumulation via repressing FoxO1-mediated ATGL-dependent lipolysis in HepG2 hepatocytes. PLoS One 16:e0243938. doi: 10.1371/journal.pone.0243938. eCollection 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Sun Y., Luo J., Wu M., Li J., and Cao Y... 2015. Specificity protein 1 regulates gene expression related to fatty acid metabolism in goat mammary epithelial cells. Int. J. Mol. Sci. 16:1806–1820. doi: 10.3390/ijms16011806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A.,. et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]