Abstract
Objective:
Chromosome 8q arm (chr8q) is the most amplified chromosomal segment in advanced metastatic castration-resistant prostate cancer after chXq12. These regions harbor important oncogenes driving prostate cancer progression, including MYC that plays a role in various hallmarks of cancer, including cell cycle progression and immune surveillance and has been well established in prostate cancer progression. Herein we characterize the co-expression patterns of chr8q genes and their clinical utility in more than 7,000 radical prostatectomy (RP) samples.
Materials and Methods:
Copy Number (CN) alterations of 336 genes on chr8q21 to chr8q24 were extracted from two primary prostate cancer cohorts (TCGA, n=492; MSK-primary, n=856) and three metastatic prostate cancer cohorts (MSK-met, N=432; MSK-mCSPC, N=424; SU2CPNAS , n=444) from cBioPortal. Expression data for the 336 genes was extracted from 6,135 RP samples from Decipher GRID registry. For survival analysis, patients were grouped into top 10% and top 25% by band expression vs remaining patients. Hazard ratios were calculated using Cox proportional hazards models.
Results:
Genes on chr8q were highly co-amplified and co-expressed. Copy number alterations and overexpression of chr8q genes in primary disease were associated with higher Gleason scores, increased risk of metastases, and increased prostate cancer specific mortality (PCSM). Additionally, our data demonstrated high expression of MYC alone was not associated with differences in metastases free survival (MFS) while high expression of other chr8q bands were associated with decreased MFS. By combining chr8q data with an established genomic classifier like Decipher, we were able to develop a new model that was better at predicting metastases than Decipher alone.
Conclusions:
Our findings highlight the clinical utility of chr8q data, which can be used to improve prognostication and risk prediction in localized prostate cancer.
Keywords: Genomics, Risk Stratification, Radical Prostatectomy, Chromosomal Amplification, Prognostication
1. INTRODUCTION
Chromosome 8q arm (chr8q) is the most amplified chromosomal segment in advanced metastatic castration-resistant prostate cancer after chXq12.1 These regions harbor important oncogenes driving prostate cancer progression. Ch8q24.2 contains MYC while chXq12 contains androgen receptor (AR). MYC is amplified in about 20% in mCRPC without neuroendocrine phenotype while it is amplified in 40% in neuroendocrine mCRPC.1–3
Early studies suggest involvement of gene amplification in human prostate cancers and, more specifically, the amplification of the 8q21.1–24.2 band region. More recent work suggests that amplification and over-expression of the 8q24 area from increased enhancer activity can lead to increased risk of prostate cancer.4–6 Furthermore, earlier studies suggest that the c-myc oncogene is a primary driver of oncogenesis associated with the 8q24 locus.7–12 These studies suggest that overexpression is a common predictor of biochemical failure in prostate cancer patients.
While these earlier studies are helpful in establishing the context behind the impact of myc as an individual gene involved in prostate cancer, many of these scientific articles are over a decade old, underscoring the importance of newer data that afford a more nuanced understanding of 8q arm overexpression and the association with the c-myc oncogene.11,13–15 Herein we characterize the co-expression patterns of chr8q genes and their clinical utility in more than 7,000 modern RP samples.
2. MATERIALS & METHODS
2.1. Data source and study populations
Genes on chr8q arm that have high amplification rate (>10%) in mCRPC (SU2CPNAS cohort) were identified. Copy Number (CN) alterations of 336 genes on chr8q21 to chr8q24 (Supplementary Table S1) were extracted from two primary prostate cancer cohorts (TCGA, n=492; MSK, n=85616) and three metastatic prostate cancer cohorts (MSK,n=60816; MSK-mCSPC,n=42417; SU2CPNAS , n=44418). Sample processing is described in greater detail in the source references.16–18 All five cohorts were publicly available on cBioPortal (https://www.cbioportal.org/). TCGA and SU2CPNAS has whole-exome sequencing data, while the others were run on MSK-IMPACT which is a targeted sequencing assay.16–18 Only 6 genes out of the 336 were covered by MSK-IMPACT including MYC. Additionally, CN alterations of AR and the other 6 genes on chrX12q, where AR is located, were extracted from TCGA and SU2CPNAS, and only AR was covered by MSK-IMPACT.
Expression data for the 336 genes was extracted from 6,135 RP samples from the Decipher GRID, which is a 22-gene RNA-based genomic Classifier that independently predicts risks of metastases.19 1135 samples were from three retrospective cohort (MCI: n=545, MCII: n=235, JHMI: n=355) with long-term outcome follow up, and 5,000 are prospective RP with known Gleason scores and Decipher risk groups. These cohorts were used to assess the overall correlation and expression patterns of genes on chr8q arm, and their associations with clinical and outcome variables. Genes on chr8q were grouped into 10 bands(q21_1:3, q22_1:3,q23, q24_1:3).
2.2. Statistical analysis
To calculate overall band expression, average expression of genes within bands was calculated as representative score for the band. For survival analysis, patients were grouped into top 10% (patients with band score above 90th percentile) and remaining patients or top 25% vs remaining patients.
For survival analysis, the hazard ratio (HR) of the top10% or top 25% compared to remaining patients were calculated using cox-proportional hazard model. HR was calculated with and without adjusting for high-risk Decipher group in MCII and JHMI cohorts. MCI was excluded from survival analyses as Decipher was built using that cohort. We finally incorporated the bands with Decipher score to build a new signature. A GLMNET model was used to build a signature (DecipherPLUSchr8) using Mayo cohort I that was used originally to build Decipher scores and validated in two independent cohorts.
3. RESULTS
MYC and chr8q arm amplification rates across several cohorts with different disease stages, both primary and metastatic, are shown in Table 1. MYC and chr8q were more amplified in the primary setting compared to AR, but AR was more amplified in mCRPC. When looking at the amplification profile of all genes and chr8q bands in mCRPC cohort (SU2CPNAS , n=444)(Figure 1, Supplementary Table S2), we found different amplification patterns across bands. Genes on chr8q24_3 had different amplification pattern compared to remaining bands. Most genes within chr8q arm bands are highly co-amplified (Supplementary Table S3), but have different amplification patterns compared to AR. About 50–60% of samples with AR amplification have chr8q amplification, and about 70–90% of samples with MYC amplification have chr8q amplification. When doing similar analysis using gene expression data from mCRPC (Figure S1), we found similar trends of co-overexpression among genes within bands. These two results suggest that chr8q bands may harbor independent genomic information that might have different clinical impact.
Table 1:
Summary of copy number amplification of AR, MYC and chr8qarm across multiple prostate cancer cohorts with different disease stages
| Primary | Metastatic | ||||
|---|---|---|---|---|---|
| Study | TCGA | MSK-IMPACT (Eur Urol 2020) | MSK-IMPACT (CCR2020) | MSK-IMPACT (Eur Urol 2020) | SU2C_(PNAS 2019) |
| Tissue type | Primary | Primary | mCSPC | metastatic | mCRPC |
| Sequencing tech | Whole Exom | Targeted-MSKIMPACT | Targeted-MSKIMPACT | Targeted-MSKIMPACT | Whole Exom |
| Number of samples | 492 | 856 | 424 | 432 | 444 |
| AR-AMP | 1.2% | 1.1% | 4.0% | 39.0% | 49.0% |
| MYC-AMP | 8.0% | 2.9% | 7.0% | 12.0% | 24.0% |
| chr8q-AMP (mean) | 6.5% | 2.0% | 4.9% | 7.3% | 20.7% |
Figure 1:
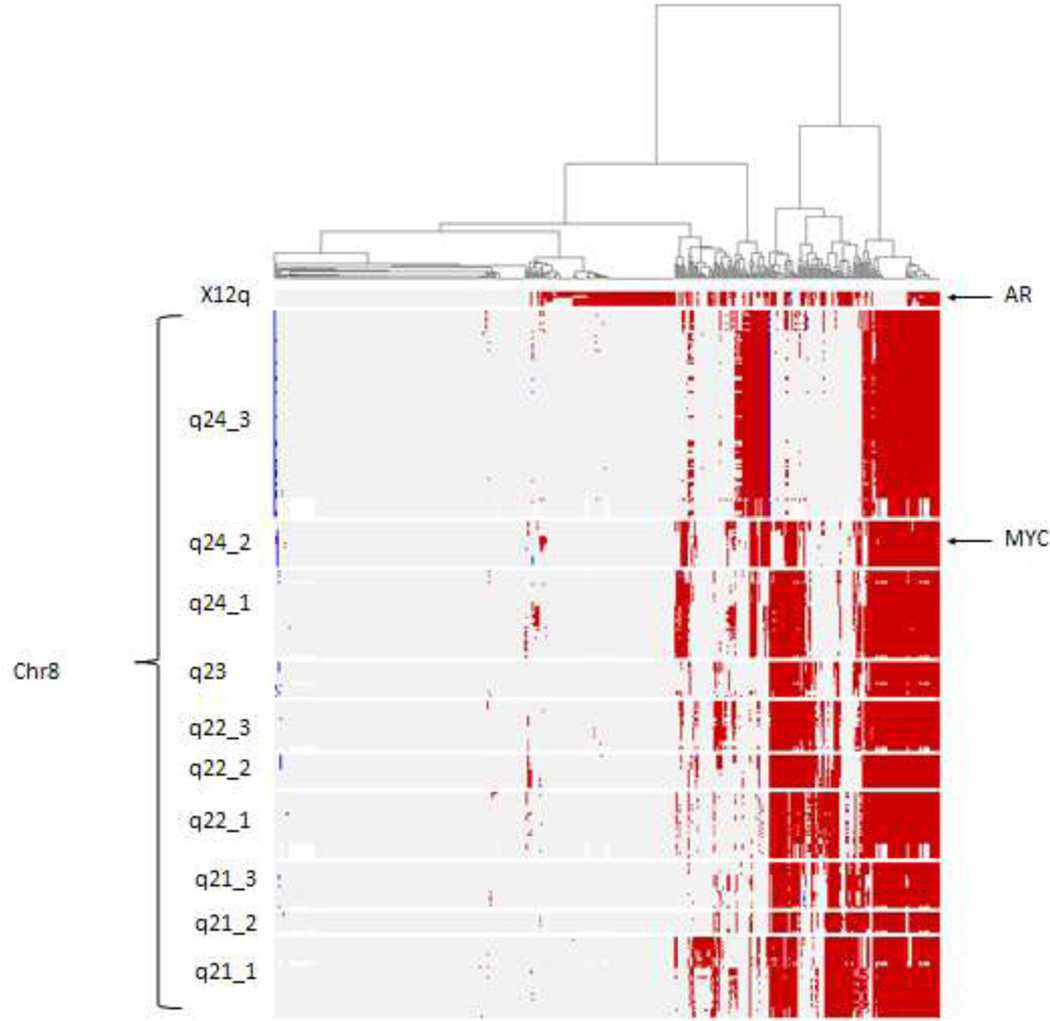
Heatmap of copy number alterations (amplification:red, deletion:blue) of genes in X12q arm hosting AR and chr8qarm in mCRPC cohort (SU2C-PNAS 2019). Heatmap shows coamplification of most genes in chr8q arm.
When analyzing amplification patterns in primary prostate cancer (TCGA-PRAD cohort, Supplementary Table S4), we observed a similar chr8q co-amplification pattern (Figure S2). Patients with co amplified bands were associated with greater risk of disease progression.
To further characterize the clinical implications of chr8q over-expression, we next studied the expression patterns of chr8q genes in a cohort of 5000 RP samples from Decipher GRID where pathological variables and Decipher risk groups were available. Heatmap (Figure 2) showed coexpression of bands. Patients with overexpression of the bands were associated with high Decipher scores and high Gleason scores (GS8–10). When evaluating by mean expression of genes within bands, most of the bands were highly correlated with each other, but some of them were correlated with Decipher scores (Figure S3).
Figure 2:
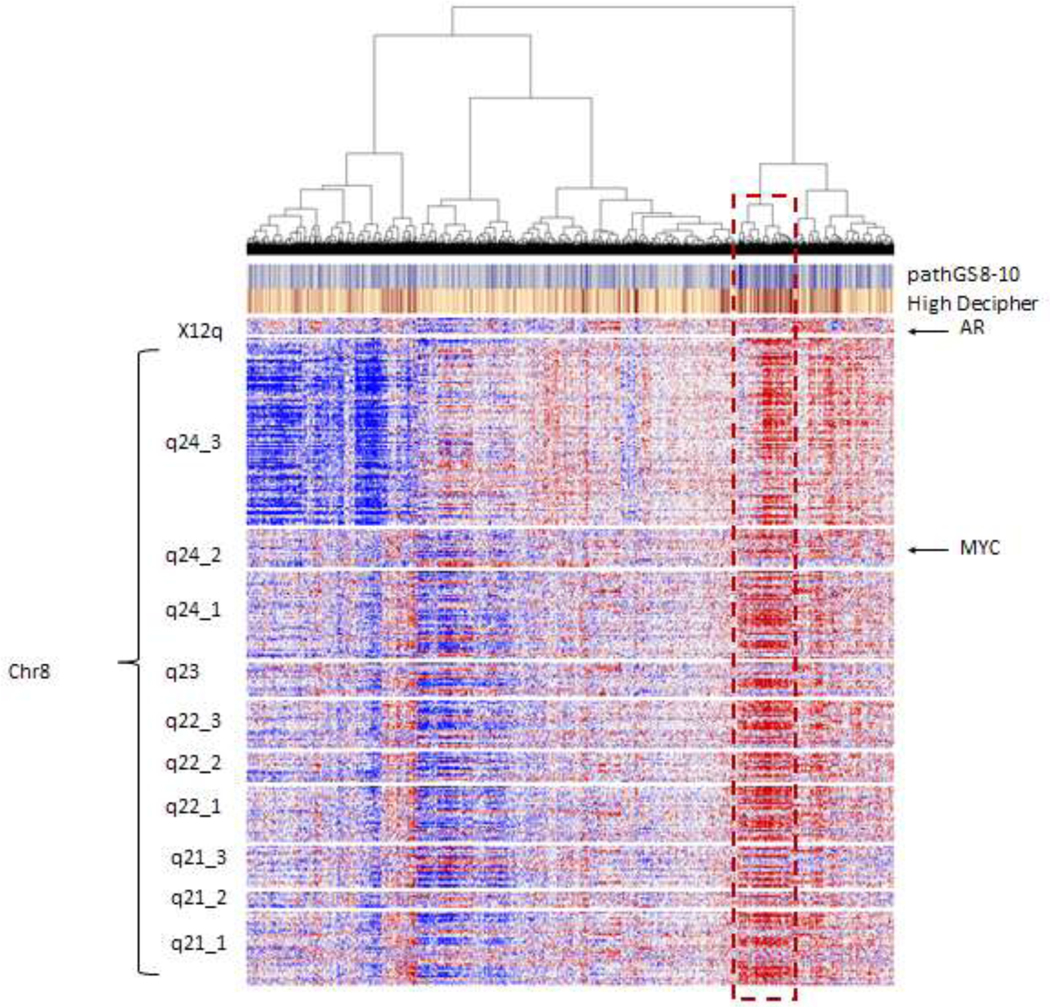
Heatmap of gene expression of genes in X12q arm hosting AR and chr8qarm in 5000 primary RP samples from Decipher GRID cohort . Heatmap shows co-expression of most genes in chr8q arm. Samples with high expression are associated with high Decipher scores defining a an aggressive subgroup of potential metastatic outcome.
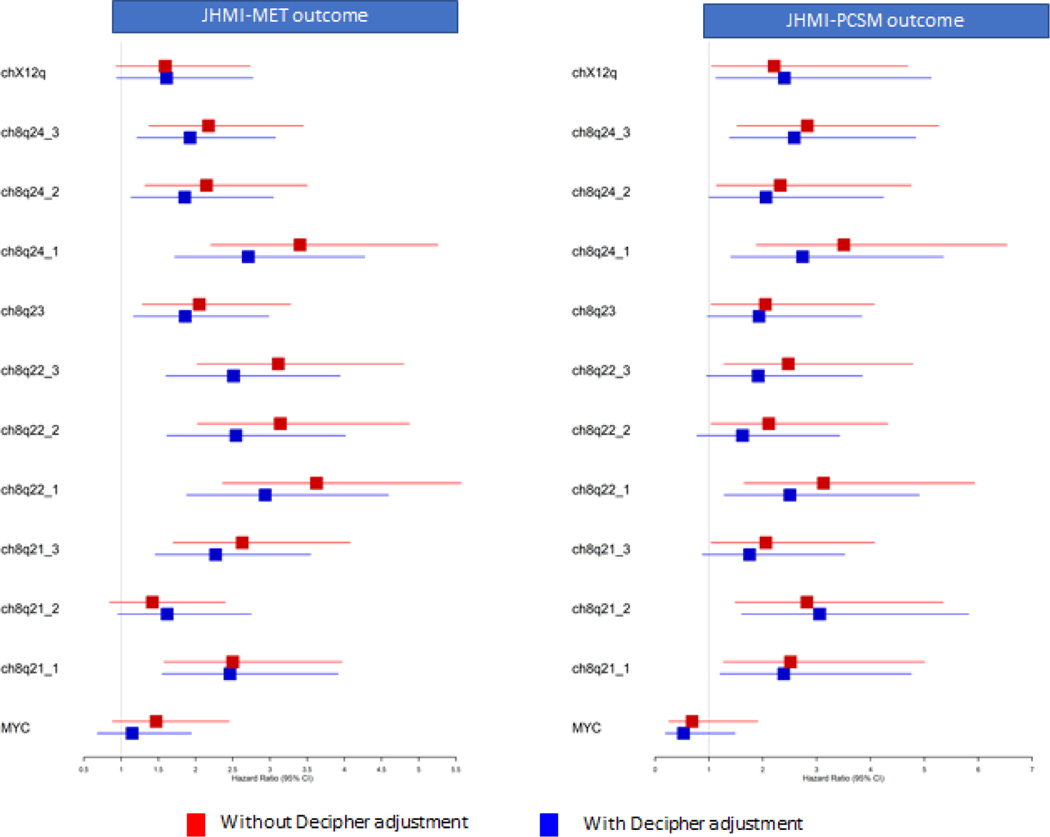
We performed a similar analysis in a retrospective natural history cohort (JHMI) (Figure S4) where patients did not receive any treatment from RP till onset of metastasis. Overexpression of bands was associated with metastatic outcome. To further investigate the associations with metastatic outcome, we averaged the expression of genes within bands and then split patients into top 10% versus others. Across most bands, high expression of bands was associated with metastatic outcome (Figure 3, Figure S5), but MYC alone and chrXq12 were not associated with metastatic outcome. Most of the bands were also associated with PCSM outcome (Figure 3). Even when adjusting for high Decipher score, most bands remained independently prognostic, indicating that these bands harbor independent prognostic power compared to Decipher (Supplementary Table S5). Similar results were observed in an independent case cohort from Mayo Clinic II (MCII, n=235) (Figure S6).
Figure 3:
Hazard ratios of chr8q bands for metastasis (MET) and prostate cancer specific mortality (PCSM) in localized natural history cohort without (red) and with adjusting for high Decipher. Most chr8q bands are associated with poor outcome and remained significant after adjusting for high Decipher.
We further split samples into top 25% vs others to show the robustness of the prognostic power of chr8q arm. Patients with top 25% expression of chr8q bands remained independently associated with metastatic outcome and PCSM (Supplementary Table S6).
We finally incorporated the average expression of chr8q bands with Decipher to build a new signature. A GLMNET model was used to build a signature (DecipherPLUSchr8) using Mayo cohort I that was used originally to build Decipher and validated in two independent cohorts. In the training cohort, ch8q22_3 and ch8q22_1 have higher weights than Decipher (7.12 , 5.52 vs 4.73). Compared to Decipher, DecipherPLUSchr8 showed improvement in AUC of predicting metastasis and high DecipherPLUSchr8 (>0.6) was more associated with metastatic outcome in Mayo cohort II (HR 6.63 vs 4.63 Decipher) and JHMI (HR 3.54 vs 3.0 Decipher).
4. DISCUSSION
In this large study using 5,000 RP samples, we found that increased copy number alterations and overexpression of chr8q genes in primary disease were associated with higher Decipher and Gleason scores, increased risk of metastases, and increased PCSM. By combining chr8q data with an established genomic classifier like Decipher, we were able to develop a new model that was better at predicting metastases than Decipher alone. Our findings highlight the clinical utility of chr8q data, which can be used to improve prognostication and risk prediction in localized prostate cancer.
Amongst the genes of interest located on chr8q includes MYC, which has been heavily studied in its role in prostate cancer progression.3,9–11,20 MYC has been suggested to play a role in driving metastasis and castration resistance.12 One study has found that overexpression of MYC is an independent predictor for biochemical recurrence.11 We found that high expression of MYC alone was not associated with metastatic outcome. However, overexpression of most other bands on chr8q was independently prognostic for metastasis and PCSM, even after adjusting for high Decipher. Additionally, our data demonstrated high expression of MYC was not associated with differences in MFS while high expression of other chr8q bands were associated with decreased MFS.
Our findings are in accordance with previous studies that utilized either FISH or CGH data and found that increased amplification of chr8q arm to be a marker of aggressiveness in prostate cancer.4,5,21 This study underscores the potential clinical significance of whole arm 8q gains in localized disease to improve risk prediction in prostate cancer. Recent studies have also shown that Black men have a higher proportion of chr8q gains compared to other races and also have more biologically aggressive cancer.22–25 These findings underscore the importance of improving equity in access to genomic testing and clinical trials, particularly for populations that may benefit from therapeutic targets for chr8q. In summary, whole arm chr8q gain and overexpression are associated with metastatic potential in localized disease, and therapeutic targets for that chromosomal segment should focus beyond MYC alone.
Limitations with our study include use of a retrospective cohort. Additionally, our data uses mRNA expression data, which only identifies bands of interest on chr8q. However, further studies are needed to identify specific genes and gene products. Finally, further studies with long term clinical follow-up are needed to determine prognostic implications of chr8q.
5. CONCLUSIONS
In summary, our study shows whole arm chr8q gain and overexpression are associated with metastatic potential in localized disease. However, high expression of MYC alone was not associated with metastatic outcome. Our findings highlight that chr8q expression data holds prognostic information for risk stratification in localized prostate cancer. Thus, development of therapeutic targets for chr8q should focus beyond MYC. Finally, further studies with long term clinical follow-up are needed to determine prognostic implications of chr8q.
Supplementary Material
Highlights:
Whole arm chr8q amplification and overexpression is associated with metastatic potential in localized disease
Chr8q expression data, and not only MYC, holds prognostic information for risk stratification in localized prostate cancer
Funding:
Dr. Mahal is funded by the Prostate Cancer Foundation and (PCF), the American Society for Radiation Oncology (ASTRO), the Department of Defense, and the Sylvester Comprehensive Cancer Center. Dr. Dee is funded in part through the Cancer Center Support Grant from the National Cancer Institute (P30 CA008748).
Footnotes
Concept and Design: All authors
Acquisition, analysis, interpretation of data: All authors
Drafting of the manuscript: Dr. Alshalalfa, Dr. Nguyen, Dr. Dee, Dr. Mahal Critical revision of the manuscript for important intellectual content: All authors Statistical analysis: Dr. Alshalalfa
Conflicts of Interest: No conflicts of interest relevant to this submission reported.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran H, Rickman DS, Park K, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–495. doi: 10.1158/2159-8290.CD-11-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin Y, Xu L, Chang Y, et al. N-Myc promotes therapeutic resistance development of neuroendocrine prostate cancer by differentially regulating miR-421/ATM pathway. Mol Cancer. 2019;18(1):11. doi: 10.1186/s12943-019-0941-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Gammal AT, Brüchmann M, Zustin J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16(1):56–64. doi: 10.1158/1078-0432.CCR-09-1423 [DOI] [PubMed] [Google Scholar]
- 5.Steiner T, Junker K, Burkhardt F, Braunsdorf A, Janitzky V, Schubert J. Gain in chromosome 8q correlates with early progression in hormonal treated prostate cancer. Eur Urol. 2002;41(2):167–171. doi: 10.1016/s0302-2838(01)00030-6 [DOI] [PubMed] [Google Scholar]
- 6.Sato K, Qian J, Slezak JM, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst. 1999;91(18):1574–1580. doi: 10.1093/jnci/91.18.1574 [DOI] [PubMed] [Google Scholar]
- 7.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4(6):a014241. doi: 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Targeting MYC, the Emperor of Oncoproteins | Prostate Cancer Foundation. Accessed August 22, 2022. https://www.pcf.org/scientific-retreat/video/targeting-myc/
- 9.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and Prostate Cancer. Genes Cancer. 2010;1(6):617–628. doi: 10.1177/1947601910379132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH. Myc confers androgen-independent prostate cancer cell growth. J Clin Invest. 2003;112(11):1724–1731. doi: 10.1172/JCI19035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawksworth D, Ravindranath L, Chen Y, et al. Overexpression of C-MYC oncogene in prostate cancer predicts biochemical recurrence. Prostate Cancer Prostatic Dis. 2010;13(4):311–315. doi: 10.1038/pcan.2010.31 [DOI] [PubMed] [Google Scholar]
- 12.Arriaga JM, Panja S, Alshalalfa M, et al. A MYC and RAS co-activation signature in localized prostate cancer drives bone metastasis and castration resistance. Nat Cancer. 2020;1(11):1082–1096. doi: 10.1038/s43018-020-00125-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Den Berg C, Guan XY, Von Hoff D, et al. DNA sequence amplification in human prostate cancer identified by chromosome microdissection: potential prognostic implications. Clin Cancer Res. 1995;1(1):11–18. [PubMed] [Google Scholar]
- 14.Sotelo J, Esposito D, Duhagon MA, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107(7):3001–3005. doi: 10.1073/pnas.0906067107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia L, Landan G, Pomerantz M, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5(8):e1000597. doi: 10.1371/journal.pgen.1000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen B, Mota JM, Nandakumar S, et al. Pan-cancer Analysis of CDK12 Alterations Identifies a Subset of Prostate Cancers with Distinct Genomic and Clinical Characteristics. Eur Urol. 2020;78(5):671–679. doi: 10.1016/j.eururo.2020.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stopsack KH, Nandakumar S, Wibmer AG, et al. Oncogenic Genomic Alterations, Clinical Phenotypes, and Outcomes in Metastatic Castration-Sensitive Prostate Cancer. Clin Cancer Res. 2020;26(13):3230–3238. doi: 10.1158/1078-0432.CCR-20-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.W A JC, G H, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(23). doi: 10.1073/pnas.1902651116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spratt DE, Dai DLY, Den RB, et al. Performance of a Prostate Cancer Genomic Classifier in Predicting Metastasis in Men with Prostate-specific Antigen Persistence Postprostatectomy. Eur Urol. 2018;74(1):107–114. doi: 10.1016/j.eururo.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JK, Phillips JW, Smith BA, et al. N-Myc Drives Neuroendocrine Prostate Cancer Initiated from Human Prostate Epithelial Cells. Cancer Cell. 2016;29(4):536–547. doi: 10.1016/j.ccell.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stopsack KH, Whittaker CA, Gerke TA, et al. Aneuploidy drives lethal progression in prostate cancer. Proc Natl Acad Sci U S A. 2019;116(23):11390–11395. doi: 10.1073/pnas.1902645116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stopsack KH, Nandakumar S, Arora K, et al. Differences in Prostate Cancer Genomes by Self-reported Race: Contributions of Genetic Ancestry, Modifiable Cancer Risk Factors, and Clinical Factors. Clin Cancer Res. 2022;28(2):318–326. doi: 10.1158/1078-0432.CCR-21-2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dess RT, Hartman HE, Mahal BA, et al. Association of Black Race With Prostate Cancer-Specific and Other-Cause Mortality. JAMA Oncol. 2019;5(7):975–983. doi: 10.1001/jamaoncol.2019.0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahal BA, Alshalalfa M, Kensler KH, et al. Racial Differences in Genomic Profiling of Prostate Cancer. N Engl J Med. 2020;383(11):1083–1085. doi: 10.1056/NEJMc2000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahal BA, Gerke T, Awasthi S, et al. Prostate Cancer Racial Disparities: A Systematic Review by the Prostate Cancer Foundation Panel. Eur Urol Oncol. 2022;5(1):18–29. doi: 10.1016/j.euo.2021.07.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.