Abstract
Native mass spectrometry (nMS) has emerged as an important tool in studying the structure and function of macromolecules and their complexes in the gas phase. In this review, we cover recent advances in nMS and related techniques including sample preparation, instrumentation, activation methods, and data analysis software. These advances have enabled nMS-based techniques to address a variety of challenging questions in structural biology. The second half of this review highlights recent applications of these technologies and surveys the classes of complexes that can be studied with nMS. Complementarity of nMS to existing structural biology techniques and current challenges in nMS are also addressed.
Keywords: native mass spectrometry, ion mobility, protein complexes, structural biology
INTRODUCTION
In this brief review, we first describe significant technological and methodological developments that have improved data quality and interpretation or enabled new measurements in native mass spectrometry (nMS) (Figure 1). The establishment of nMS as a powerful structural biology tool for biomedical research is then illustrated with several examples in the second half of this review. Limits on text length and reference numbers prevent us from comprehensively covering all contributions to this growing field; the reader is referred to the cited reviews for more detailed coverage.
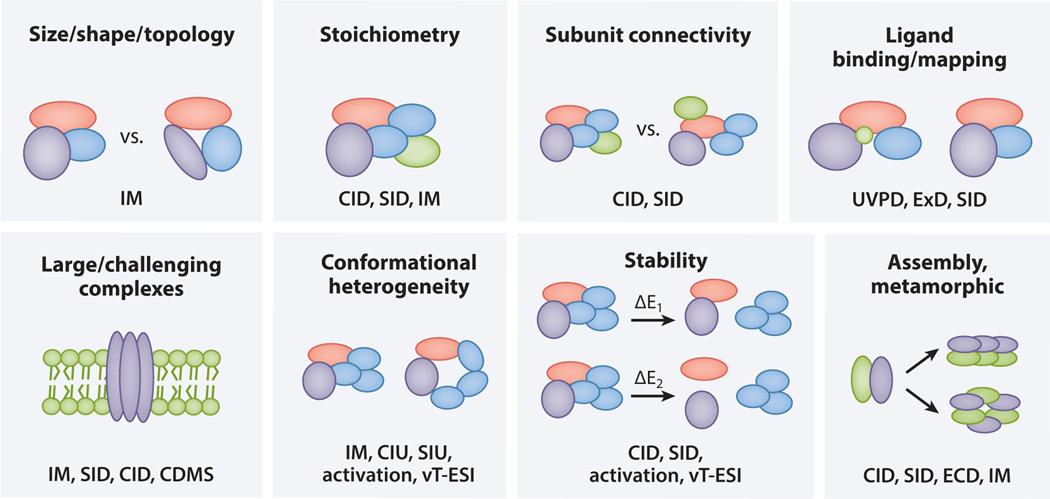
Figure 1.
Diverse applications of nMS. Each box includes an application of nMS depicted with a schematic, and the nMS-based techniques used to address that application are listed in the lower portion of the box. Abbreviations: CDMS, charge detection mass spectrometry; CID, collision-induced dissociation; CIU, collision-induced unfolding; ECD, electron capture dissociation; ExD, electron transfer dissociation (or electron capture dissociation); IM, ion mobility; nMS, native mass spectrometry; SID, surface-induced dissociation; SIU, surface-induced unfolding; UVPD, ultraviolet photodissociation; vT-ESI, variable temperature electrospray ionization.
NATIVE MASS SPECTROMETRY TECHNOLOGIES
Sample Preparation and Separations
A critical step in an nMS experiment is the preparation or transfer of samples into an MS-compatible solution, which typically involves exchange into a solution of a volatile salt such as ammonium acetate. However, low concentrations of additional components can be retained if required for stability or activity (102). Traditionally, sample preparation is done offline, via buffer exchange spin columns, diafiltration, or dialysis. In recent years, however, more online sample preparation and separation methods have been developed, allowing for increasing throughput and sample complexity. A recent report demonstrated that rapid online buffer exchange (OBE) can be performed directly prior to nMS analysis using short size-exclusion chromatography columns, enabling the separation of proteins and protein complexes from nonvolatile buffer components (158). OBE enables higher throughput than offline buffer exchange and allows the sample to be kept in its preferred buffer until immediately before nMS analysis. More recently, OBE has been coupled with immobilized metal affinity chromatography OBE (IMAC-OBE) to screen overexpressed His-tagged proteins from cell lysates (16). Alternatively, for abundant overexpressed proteins, if lysis is performed in an MS-compatible buffer, then samples can be centrifuged and the supernatant directly used for MS analysis, reducing the sample purification steps required (161, 162).
Recently, a novel method for protein desalting was developed utilizing nanoscale nanoelectrospray ionization (nESI) emitters,which reduce initial droplet size and therefore the number of salt ions in each droplet, leaving fewer adducts on proteins after solvent evaporation (74). Nanoscale emitters permit analysis of protein complexes (153,154) and even membrane proteins (152) over a wide variety of buffer,salt,and detergent conditions and reduce sample preparation requirements.
The coupling of longer size exclusion chromatography (SEC) columns with nMS has also been demonstrated for the separation of protein mixtures directly prior to MS analysis, reducing signal suppression caused by differential ionization efficiencies in direct infusion experiments (32, 39). Online SEC-nMS has been used to study protein mixtures, thermally stressed biopharmaceuticals, and protein–ligand interactions (including nucleic acids) (32). SEC-nMS has the potential to not only increase sample throughput, but also widen the range of sample types that are amenable to nMS. Ion exchange has also been coupled with nMS, using a salt or pH gradient, and has been used to study antibody charge variants and a range of designed heterodimers of similar size but differing pIs (4, 23). Hydrophobic interaction chromatography (HIC) can have high separation power but has been challenging to couple with nMS, as it typically requires buffers with high salt concentrations. Even so, the coupling of HIC with nMS for antibody drug conjugates has recently been demonstrated via the incorporation of an SEC step after HIC separation to allow for salt removal (38). More recently, HIC was directly coupled with MS using MS-compatible buffers (165). As online separation approaches advance, enabling higher sample throughput and improving nMS’s amenability to more complex samples, we anticipate widespread adoption of nMS.
Ion Sources for Native Mass Spectrometry
Retaining native protein structure during the transfer of analytes from solution to the gas phase is a key stage in then MS workflow and is typically accomplished by nESI (47, 166). Unfortunately, ammonium acetate and residual nonvolatile solution components can adduct with proteins, causing peak broadening and reducing apparent mass resolution. Several strategies have been developed to ameliorate this issue. Collisional activation is effective and widely used for removing adducts after the ionization stage but risks gas-phase restructuring (33, 48, 51, 169).
Aside from influencing gas-phase adduction, variations in source design and solution conditions can manipulate ion charge states, which can alter fragmentation pathways and/or gas-phase conformation (151). Charge-reducing reagents such as triethylammonium acetate (TEAA), ethylene diamine diacetate (EDDA), and trimethylamine N-oxide (TMAO) (83, 126) are commonly used to reduce ion charge state, resulting in more native-like fragmentation. In the past several years, many alternative reagents have been described, from imidazole derivatives (159) to alkali metal acetate salts (127) and polyamines (106). For membrane proteins in particular, charge-reducing detergents such as tetraethylene glycol monooctyl ether (C8E4) (90), n-dodecyl-N,N-dimethylamine-N-oxide (LDAO) (126), and oligoglycerol detergents (OGDs) (160) can be used, as well as reagents including TMAO (126), and polyamines (106).
Structural features of protein complexes can also be probed using newly developed variable temperature electrospray ionization (vT-ESI) sources, and readers are encouraged to consult Russell and coworkers (89) in this volume.
While nESI is the most commonly used method of ionization for nMS studies, desorption ESI (DESI) has also been shown to be a promising technique in such studies. Recently, the Robinson group demonstrated the utility of DESI in studying lipid and drug binding to a membrane protein [G protein–coupled receptor (GPCR)]. Analyzing membrane samples from a surface allows for the possibility of high-throughput screening and measurement of selectivity of agonists, and enables the formation of a more planar and spatially heterogeneous lipid distribution that is more representative of the cellular surface compared to solubilized membrane proteins (3). In addition, the Cooper lab has demonstrated the power of nano-DESI for nMS imaging of proteins and their complexes from tissue samples. This provides the exciting opportunity to study proteins from their native environment. Furthermore, Hale & Cooper (65) showed that it was possible to perform location-targeted top-down sequencing experiments with this setup.
New Paradigms in Mass Analysis
nMS has benefited from the widespread adoption of high-resolution, high-accuracy MS platforms. The challenges of transmitting, detecting, and resolving charge states and proteoforms of large, heterogeneous macromolecular assemblies have also prompted the development of novel measurement strategies, including collision cross-section (CCS) and charge detection mass spectrometry (CDMS) measurements in Orbitraps (83, 114, 128), electrostatic linear ion trap (42, 86) analyzers, and mass analysis of large complexes by mass photometry (146, 147).
CDMS in linear ion traps and Orbitraps has enabled ultrahigh mass analysis because it permits the independent measurement of ion mass and charge simultaneously (86).To this end,the Jarrold group (69) and Williams group (41, 42) have independently developed two distinct CDMS systems based around the electrostatic linear ion trap geometry with reflectron mirrors on either side of detection tubes. The charge accuracy of these devices is high compared to that of Orbitraps; the Jarrold group has demonstrated accuracies as low as 0.20e (87, 125) and detection limits of approximately 7e.
Because the Orbitrap uses image charge detection,CDMS is also possible on this platform (82, 168). The Orbitrap CDMS technique involves the collection of hundreds to thousands of low-intensity scans (time-domain transients corresponding to approximately single-ion events) and specialized data processing (81) to produce a mass spectrum. By allowing determination of charge states and reducing ion–ion interactions in the Orbitrap cell, CDMS has enabled ultrasensitive analysis of heterogeneous protein assemblies such as ribosomes and adeno-associated viruses (82, 168) and improved mass spectral resolution (83, 115).
Ion Mobility
Ion mobility (IM) enables separation of ions in the gas phase based on their rotationally averaged CCS and can provide a wealth of information on oligomeric state and conformation (8, 55). IM is often coupled with prior collisional activation [collision-induced unfolding (CIU) or surface-induced dissociation (SID)] to investigate the conformational space of proteins and their fragments.The main IM techniques employed for nMS are drift tube IM (DT-IM),traveling wave IM (TWIM), and trapped IM spectrometry (TIMS).
In DT-IM, analyte ions are propelled through a pressurized drift tube under a uniform weak electric field. DT-IM enables direct measurement of an ion’s CCS from first principles and therefore does not require calibrants (112). Typically, DT-IM is coupled to time-of-flight (ToF) mass analyzers, as the IM separation is slow (milliseconds) compared to ToF spectrum acquisition (microseconds); this coupling enables the collection of mass measurements across the full arrival time distribution (128, 148). Conversely, higher-resolution trapping MS instruments (i.e., Orbitraps) experience an inherent duty cycle mismatch, as IM separation and mass measurement scans occur on approximately the same timescale (76, 113). Recently, the Russell group developed a new IM-Orbitrap platform that overcomes this duty cycle mismatch. The group designed a reverse-entry ion source (REIS) and a periodic focusing DT-IM analyzer that retains the ability to measure CCS on first principles while also enabling high-resolution mass measurements on the Orbitrap (126, 127). This instrument was able to successfully measure CCS values of protein complexes from 8.6 to 810 kDa, demonstrating the power of this new platform to combine first-principles CCS measurements with high-mass-accuracy Orbitrap analyzers (114).
In TWIM experiments, an oscillating electric field is used to produce a traveling voltage wave that pushes ions through a drift gas. TWIM enables enhanced separation and the use of longer path lengths without significant ion loss compared to DT-IM but cannot directly measure CCS (60). Instead, calibrants consisting of ions of known CCS and charge states with similar properties (i.e., shape, mass, and charge) are used to determine the CCS of the analytes of interest (54, 149). Recently, Waters Corporation introduced the SELECT SERIES Cyclic IMS instrument containing a 98-cm path length traveling wave cyclic IM (cIM) device that enables single or multipass separations to increase resolving power. This new instrument has been used to measure conformations and unfolding pathways of monomeric cytochrome C and multimeric Concanavalin A proteins with considerably higher resolution than linear IM instruments (R up to approximately 750 for the reverse peptides SDGRG and GRGDS after 100 passes versus R approximately 45 on a Synapt G2) (40, 61, 62).
TIMS represents one of the newest IM methods and was recently commercialized by Bruker Daltonics. In TIMS, ions are exposed to a parallel gas flow that pushes them toward the detector, but their motion is opposed by an electric field. The field strength is slowly decreased, allowing ions to eject in order of decreasing mobility. Generally, CCS values are obtained based on a calibration curve (20, 130). TIMS has been used to study native protein complexes, protein– nucleic acid complexes, and protein–ligand complexes (17, 77, 78, 100, 123).
Activation Methods
The activation and subsequent dissociation of native-like ions provides important insight into their interactions, organization, inter subunit connectivity, and ligand binding sites. Several activation methods yield fragmentation pathways that correlate with secondary, tertiary, and quaternary structural features (66, 107).
In collision-induced dissociation (CID), ions are activated through a series of low-energy collisions with gas molecules. CID results in slow accumulation of ion internal energy that generally causes dissociation through the unfolding or elongation and subsequent ejection of a highly charged monomer (asymmetric charge partitioning) (46, 80). CID can also cause structural rearrangement without dissociation and should be used with appropriate caution when attempting to derive structural features of complexes (137). CID as part of a CIU experiment, however, has been used extensively to study protein unfolding and changes in protein stability (33).
Electron- and photon-based activation methods, including electron transfer dissociation (ETD), electron capture dissociation (ECD), ultraviolet photodissociation (UVPD), and infrared multiphoton dissociation (IRMPD),have been shown to preserve noncovalent interactions and/or produce backbone fragments that retain bound ligands. Protein backbone cleavages induced by these methods occur in regions of greater structural flexibility and surface exposure, providing insights into protein conformation, sequence, and ligand binding sites (178).
SID, i.e., collisional activation with a surface, has emerged as a promising activation method, as it has been shown to produce compact, charge-symmetric fragments that are reflective of the quaternary structure while also generally preserving bound ligands for a range of complex types (170). SID devices have been installed on a range of instrument platforms, and one is now commercially available on the Waters SELECT SERIES Cyclic IMS platform (144, 159). SID has been used to study the stability, structure, ligand binding, and assembly pathways of a range of protein complexes, such as membrane proteins, designed heterocomplexes, and large complexes such as the 20S proteasome (150).
Gas-Phase Chemistry
Once noncovalent complexes are in the gas phase, a wide variety of controlled chemical manipulations can be applied to probe them through ion–ion and ion–molecule reactions (49). Recent developments have focused on reduction of charge state by proton transfer charge reduction (PTCR) (75), cation to anion proton transfer reactions (CAPTR), electron transfer or capture, analysis of protein conformation by gas-phase hydrogen/deuterium exchange (HDX) (120), and other manipulations such as covalent cross-linking and multiply charged ion attachment (49).
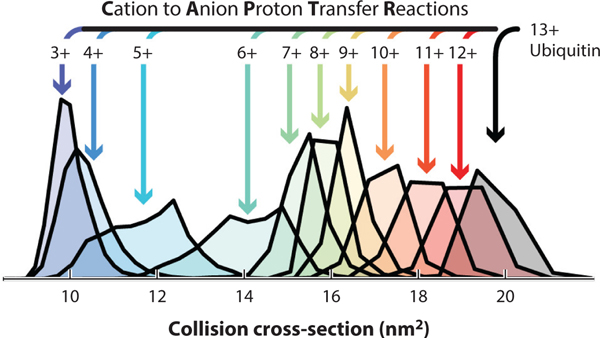
PTCR is a tool for reducing spectral congestion by decreasing the charge states of ions at an analyte-dependent rate using ion–ion proton transfer to negatively charged reagent ions such as fluoranthene or perfluoroperhydrophenanthrene (71, 75), although it has been used only sparingly with nMS (6). A similar technique, CAPTR, involves a reaction between cation analytes (proteins) and reagent anions [e.g., perfluoro-1,3-dimethylcyclohexane (PDCH)] to reduce the analyte charge state (95). The Bush lab has published several studies using CAPTR to resolve mixtures of native proteins and complexes (e.g., 95). The Bush lab also showed that ubiquitin ions of various charge states generated from CAPTR of the 13+ precursor exhibited a range of CCS,despite originating from the same precursor structure (Figure 2), suggesting the occurrence of protein folding or restructuring in the gas phase. For the larger multidomain serum albumin (66 kDa), the final CAPTR CCS depended on starting solution conditions (57), supporting the kinetic trapping hypothesis that has allowed nMS to flourish.
Figure 2.
Collision cross-sections of ubiquitin ions generated from the cation to anion proton transfer reaction of the 13+ charge state. Figure adapted with permission from Reference 96; copyright 2016 American Chemical Society.
Electron transfer and electron capture are equally viable methods for manipulating charge states of native protein complexes. Lermyte and coworkers (98, 99) demonstrated extensive electron transfer without dissociation (ETnoD) charge reduction for several proteins, to as low as singly or doubly charged species, on Synapt G2 and G2-S platforms using 1,4-dicyanobenzene and p-nitrotoluene as reagent anions. The Barran lab investigated conformations of cytochrome c and myoglobin, observing depletion of compact conformers after charge reduction and, as in CAPTR, changes in protein folding in the gas phase upon charge reduction (79). The Koltashov lab has used electron transfer charge reduction to determine the binding ratio of heparin of varying chain lengths with antithrombin-III (175) and has combined solution-phase supercharging and gas-phase charge reduction to investigate heterogeneous hemoglobin and haptoglobin samples (172).
Software Tools for Native Mass Spectrometry
Data analysis is often a bottleneck in nMS studies, but fortunately multiple tools have emerged in recent years to aid data interpretation. Software can be divided into two broad categories: tools to aid deconvolution (converting from m/z to mass domain) and tools for IM data interpretation. Data deconvolution can be performed using both commercial tools [e.g., Intact Mass from Protein Metrics (10) or BioPharma Finder from Thermo Scientific] and noncommercial options such as UniDec (111, 129) and iFAMS (27). Intact Mass (10) and MetaUnidec (129) nMS software have enabled batch deconvolution and reporting of data files, increasing data analysis throughput. Batch deconvolution coupled with automated sample running has the potential to transform nMS into a truly high-throughput method.
As discussed above, rotationally averaged CCS can be determined by IM. Combining experimental CCS with computationally determined theoretical CCS can provide additional information, allowing the comparison of models and proposed structures to experimental values (67, 114). Several approaches have been previously reported for CCS determination (108, 117, 140). The simplest approach, projection approximation (PA), models ions using a collection of overlapping hard spheres, with radii equal to the hard sphere collision distances (108). PA often leads to underestimation of the CCS, but it is a less computationally demanding method and is often used with an empirically derived correction factor (7). The PA model has been implemented in many forms, including in a tool called Ion Mobility Projection Approximation Calculation Tool (IMPACT), which can determine CCS from X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, small-angle X-ray scattering (SAXS), and electron microscopy (EM) data (31, 110). An alternative method is the projection super approximation (PSA), which determines CCS in a similar fashion to PA but also accounts for collective shape and size effects, which can increase accuracy (11). The trajectory method is computationally more expensive but considers the ion as a collection of atoms and accounts for long-range interactions, collisions between the ion and buffer gas, and multiple collisions (117). A version of the trajectory method has recently been implemented into a package known as Collidoscope (44), which offers faster analysis than previous versions. Specialized tools have also been developed to analyze IM-based studies on the stability, unfolding, and interactions of proteins and complexes (2, 43, 118). Recent progress on computational tools for top-down MS and nMS can be found at http://nativems.osu.edu/training.
DRIVING BIOMEDICAL RESEARCH THROUGH NATIVE MASS SPECTROMETRY
As the rapid advances in nMS technology enable more in-depth analysis of a broader range of macromolecular complexes in the gas phase, the technique is gaining in popularity for driving biomedical research through collaborative structural biology. This section highlights selected achievements in which nMS provided critical structural information to complement other structural biology approaches.
Protein–Protein Complexes
Most proteins interact with other biomolecules to form assemblies essential for their biological function. nMS has emerged as a leading tool to characterize various properties of complexes, including their individual subunits, stoichiometries, relative binding affinities, and architecture (177). Ahnert and colleagues (1) used nMS and large-scale analysis of existing protein structures to elucidate some guiding principles of protein assembly and topology that can be accessed in the gas phase.
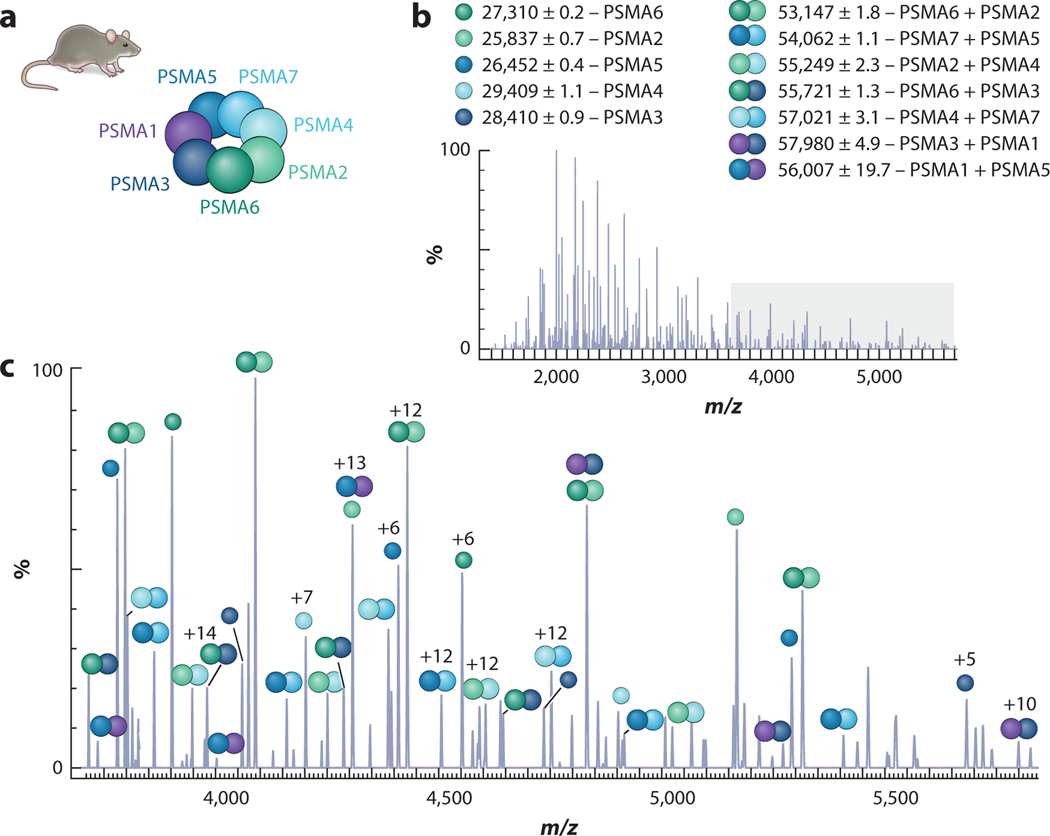
Recently, Vimer and colleagues (160) integrated several nMS techniques to study structural features among 20S proteasome orthologs from five species (Figure 3). Each proteasome adopts a cylindrical structure composed of four hetero-heptameric rings (approximately 700 kDa), but these structures vary in complexity. Subunit connectivity was determined using SID and CID, while rough architecture was determined from CCS values calculated from IM. The kinetic stabilities of each complex were measured using CIU, and top-down MS3 was used for sequence analysis. The combined results of these experiments demonstrated that these complexes vary in their size, kinetic stability, and subunit variants. The nMS results were corroborated by the solving of the rat 20S proteasome structure by cryo-EM, demonstrating how nMS approaches can reliably guide structural studies, even for complexes that lack high-resolution structures (160).
Figure 3.
MS3 analysis of rat 20S proteasome α-ring containing seven different α subunits (PSMA1–7). Samples were subjected to HCD, and fragments were identified based on mass. (a) Deduced structural organization of the α-ring based on identified subcomplexes labeled in panels b and c. Panel c represents an enlarged view of the gray-shaded area in panel b. Figure adapted with permission from Reference 160; copyright 2020 American Chemical Society. Abbreviations: HCD, higher-energy collision dissociation; MS, mass spectrometry.
Many protein–protein interactions are mediated by specific inter- and intramolecular interactions. Double-mutant cycle analysis (DMC) is a strategy to measure the energetic coupling between specific amino acids at a binding interface or within proteins (122, 145). In DMC, two residues are mutated individually and in combination, and the effects on binding or folding are measured. If the residues interact directly or indirectly, then the effect of the double mutant will differ from the sum of the two individual mutations. DMC has been used to identify the binding location of capsaicin in the ion channel TRPV1 (171) and protein–peptide interactions (84) and has been combined with nMS to study protein–protein interactions in Escherichia coli lysates (30) and interprotein contacts in the gas phase (145).
nMS has also been used to probe endogenous protein–protein interactions in tissues and cells (9, 58, 121, 133). For example, Skinner et al. (141) used nMS and multistage tandem MS to identify and characterize 125 intact endogenous complexes and 217 distinct proteoforms from mouse heart and human cancer cell lines, providing insight into how protein complexes exist in the cell, including preservation of endogenous interactions, modifications, and ligands. The Sun group has developed and utilized native capillary zone electrophoresis (CZE) to separate heterogeneous protein complexes from ribosomal isolates and cell lysates (139). Further work with native CZE has focused on characterization of antibodies, as well as structural and conformational analysis of large protein complexes (21).
Protein–Ligand Complexes
Protein function is often regulated by conformational changes imparted by ligand interaction. For protein systems with multiple ligands, the binding cooperativity is also critical. Therefore, deciphering allosteric and cooperative properties upon ligand binding provides mechanistic insight into protein complex function. nMS has emerged as a leading tool to probe the stoichiometry, binding constants,and allosteric properties of ligand binding, as well as their effects on overall complex stability (63, 138).
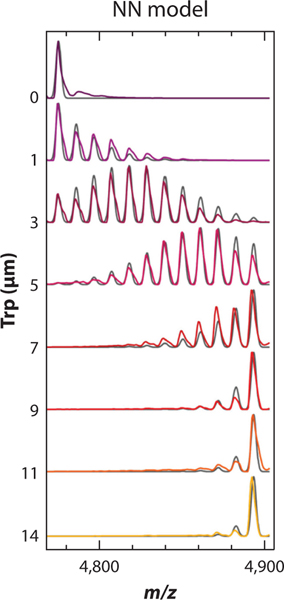
For example, Holmquist and colleagues (72) used nMS to measure the cooperativity of tryptophan (Trp)–trp RNA-binding attenuation protein (TRAP), a ring-shaped homo-oligomeric complex composed of 11 subunits and 11 Trp binding sites located at the subunit interface. A Trp titration experiment and thermodynamic modeling revealed that Trp binds to TRAP according to a nearest-neighbor cooperative model whereby binding of Trp to one subunit modestly enhances Trp binding to immediately adjacent subunits (Figure 4). Some other notable examples of probing ligand cooperativity and allostery using nMS include studies of lipid binding to membrane proteins (28, 101, 124) and nucleotide and substrate binding to chaperones (37, 92).
Figure 4.
Population distributions of tryptophan (Trp)–trp RNA-binding attenuation protein (TRAP) complexes reveal homotropic cooperativity in Trp binding to TRAP. The mass spectrum for the 19+ charge state of 1 μM TRAP incubated with the indicated concentration of Trp is displayed (color), overlaid with simulated populations computed from fits to the nearest-neighbor (NN) cooperative model (gray). Figure adapted with permission from Reference 72; copyright 2020 American Chemical Society.
Ligand binding sites can also be mapped using nMS coupled to single- or multistep activation techniques. In general, CID causes ligand loss and thus is not often used for these purposes. However, CIU can provide information on the overall conformational landscape and stability of a protein upon ligand binding (33, 45). Electron- and photon-based activation methods (e.g., ETD, ECD, UVPD) and SID generally preserve bound ligand, and the covalent fragmentation of the protein backbone in ETD, ECD, and UVPD can enable localization of ligand binding sites based on which fragments retain the ligand (178).
Nucleoprotein Complexes
Nucleoprotein complexes present some additional challenges in nMS analysis: (a) Nonvolatile cations such as Mg2+ are sometimes required for assembly; (b) ionization suppression from metal ions and/or free RNA or DNA can result in weak signal intensity; (c) adduction of nonvolatile cations may reduce apparent mass resolution and accuracy; and (d) charge–charge interactions (common in nucleoproteins) are difficult to dissociate by CID, although they can be dissociated by SID in some cases (56, 109). Despite these challenges, nMS has been used to rigorously study many nucleoprotein complexes, including their stoichiometries, that have evaded analysis by complementary techniques due to their size, heterogeneity, and/or stability (22, 94, 134). nMS has also recently been used to measure the interaction between the SARS-CoV-2 nucleocapsid protein and a range of ligands including RNA, antibodies, and cyclophilin A (103).
When combined with IM and molecular modeling, nMS has also measured the stability, assembly pathways, and/or structure of nucleoprotein complexes, including DNA Pol III subcomplexes (109),nucleosomes (132), topoisomerase (77), aminoacyl-tRNA synthetase complexes (22), HIV-1 interaction with Gag protein (134), viral matrix proteins (94), and RNase P (93, 106). For example, nMS has been used to study the assembly pathways of Redβ oligomerization on DNA and has provided mechanistic insights into DNA repair processes in bacteriophage λ (18).
Top-down nMS has also been used to map binding locations of nucleic acids on nucleoprotein complexes and ligand binding on nucleic acids (135, 136, 164, 174). For example, Schneeberger and colleagues (136) used nMS and CID to locate the binding site of rev response element (RRE) RNA on rev protein complexes, an important step in HIV-1 virus assembly. The results demonstrate that rev protein initially binds to the upper stem of RRE IIB RNA but is then relocated to a binding site on RRE that enables rev protein dimerization, highlighting the utility of nMS techniques in probing the assembly pathways, stoichiometry, and binding interfaces of nucleoprotein complexes.
Membrane Proteins
Due to their low expression yields, heterogeneity, and requirement for solubilization in a membrane mimetic, membrane proteins have proven challenging to structurally characterize. nMS’s low sample requirements and ability to handle heterogeneous samples have brought it to the forefront in membrane protein studies. An initial challenge in such studies was how to solubilize the proteins under nMS-friendly conditions (5). In recent years, multiple membrane mimetics have been utilized, including detergent micelles (91), nanodiscs (85), bicelles (73), amiphols (97), and styrene maleic acid lipid particles (SMALPs) (68, 70), and analysis has even been conducted directly from membranes and in destabilized lipid vesicles (25, 26). nMS reports on individual states, as opposed to a bulk ensemble measurement, and thus has revealed the specificity of lipid interactions, stability imparted upon lipid binding, and even the thermodynamics of binding (2, 12, 29, 90). In addition, nMS has been used to study nucleotide and drug binding to GPCRs, a particularly challenging class of proteins due to their low yield and instability post–membrane extraction (173). IM-MS has also been used to study the ATP-binding cassette transporter P-glycoprotein, demonstrating that the protein exists in an equilibrium between different conformational states that can be readily interconverted upon ligand and lipid binding (109).
nMS has recently been exploited to identify ligands and endogenous lipids bound to membrane proteins, which often appear as unassigned or poorly resolved density in solved structures (59). One such approach has identified endogenous lipids by combining nMS, controlled delipidation, and solution-phase lipid profiling techniques (64). Recently, a multistage MS approach (MSn) was also presented in which membrane proteins are introduced into the MS within detergent micelles. In the first stage of activation, the protein complex is released from the micelle (MS2); the assembly is then isolated and dissociated to release proteins or ligands (MS3) for further fragmentation (MS4) for proteoform sequencing or ligand identification. Using this approach, Gault & coworkers (59) were able to observe lipid binding and identify the endogenous lipid species to the outer mitochondrial membrane translocator protein (TSPO), which could then be fit into the previously solvedX-ray structure. Multiple lipids could often be modelled into poorly resolved maps, and thus, defining the lipid headgroup, side chain asymmetry, and chain length distribution can improve phospholipid modeling (59).
Antibodies and Glycoproteins
nMS, often coupled with liquid-phase separation techniques, has been applied to the study of antibody–drug conjugates, antibody–antigen complexes, and bispecific antibodies (19). While the intact mass alone can be informative (e.g., providing information on the extent of glycosylation or insight into antibody–antigen complexes), coupling with complementary methods such as activation can often provide more information. For example, the Ruotolo group has shown that intentionally unfolding antibodies with gas-phase collisions (known as CIU) and monitoring the unfolding with IM can distinguish among antibody isoforms containing different numbers and patterns of disulfide bonds and differing extents of glycosylation (155, 156). Glycosylation is the most complex protein modification and is not only essential for many cellular functions, but also often present on biotherapeutic proteins, influencing their efficacy and safety. The characterization of glycosylation is therefore of great interest, and a recent review has summarized the role of nMS in studies of glycosylation (151). The heterogeneity and flexibility of oligosaccharides often pose a problem for traditional, higher-resolution structural biology techniques, frequently resulting in proteins being deglycosylated before analysis, which has motivated the development of alternative methods. High-resolution nMS is proving useful in glycoprotein analysis, enabling the identification of previously unreported glycosylation sites on human C9 and C8 proteins (52, 53). High-resolution nMS can be coupled with complementary techniques such as IM, to obtain conformational information, or 193-nm UVPD, to map binding epitopes (100, 116).
Computationally Designed Proteins
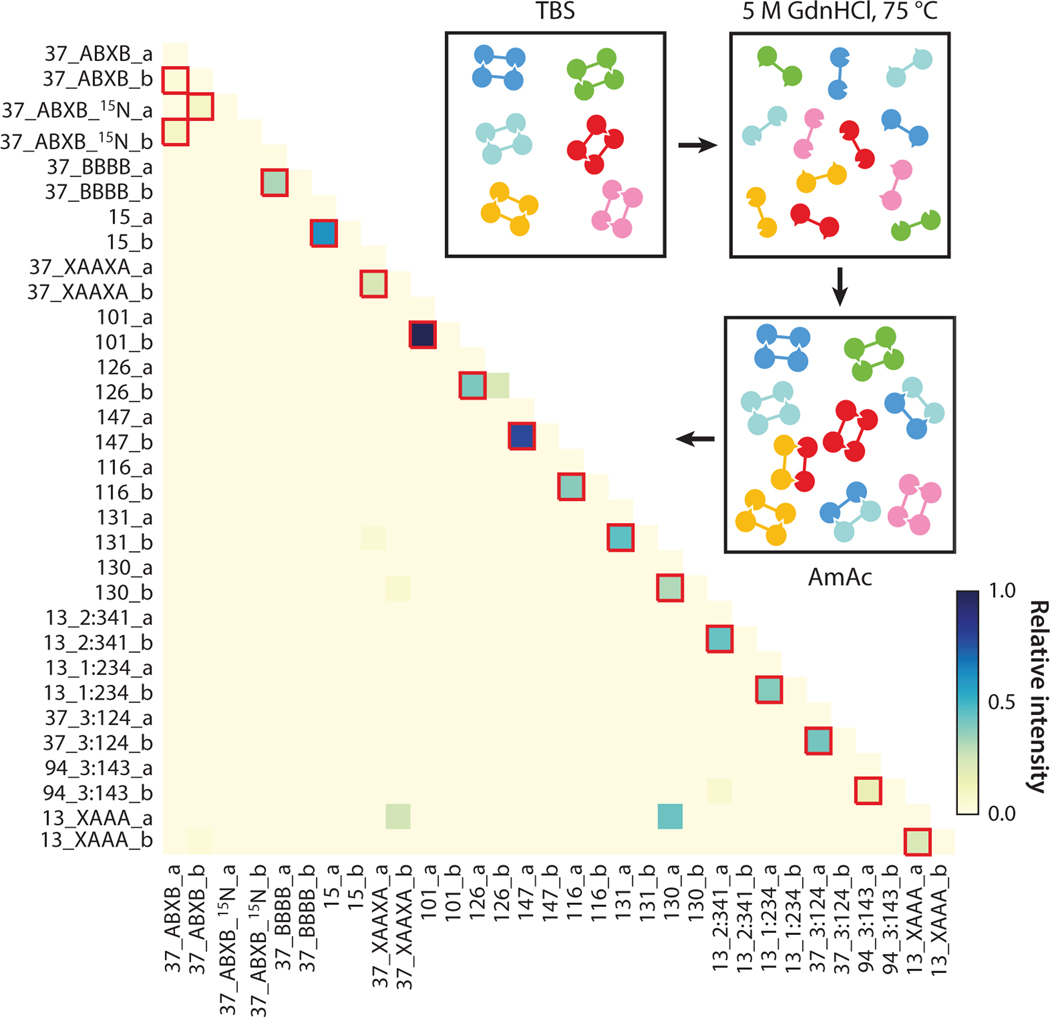
Protein re-engineering and de novo design have great potential in the generation of novel materials for chemical and medical applications. Characterizing designs using traditional structural biology techniques can be time consuming and sample intensive. nMS has shown promise as a rapid,high-throughput method of screening different designs to confirm complex formation (158) and of studying pH-driven conformational changes (14), co-operativity of designed protein-logic gates (24), and transmembrane β-barrels (163). When coupled with SID, nMS has also been used to confirm subunit arrangement (23, 131). The study of designed protein complexes has also been aided by online native separations coupled with nMS (23, 158). In one such study, to test the interaction specificity of 16 heterodimer designs, the dimers were mixed, denatured, reannealed by dialysis, and then characterized using ion exchange chromatography coupled online with nMS. Significantly, the mixing experiments highlighted the specificity of the designs, with all 16 designed pairs recovered and only a low number of off-target dimers observed (23), as shown in Figure 5. The advent of CDMS has also aided in the characterization of designed multimeric complexes, particularly hexamers designed to form asymmetric virus capsids (176).
Figure 5.
All-against-all orthogonality of 16 pairs of heterodimers assessed by online ion exchange chromatography coupled with native mass spectrometry. Red boxes indicate designed cognate pairs. Exchange of unlabeled and partially 15N-labeled DHD37_ABXB results in a distribution of overlapping species. Figure adapted with permission from Reference 23; copyright 2018 Springer Nature Limited. Abbreviations: AmAc, ammonium acetate; TBS, Tris-buffered saline.
Measuring Large Molecules
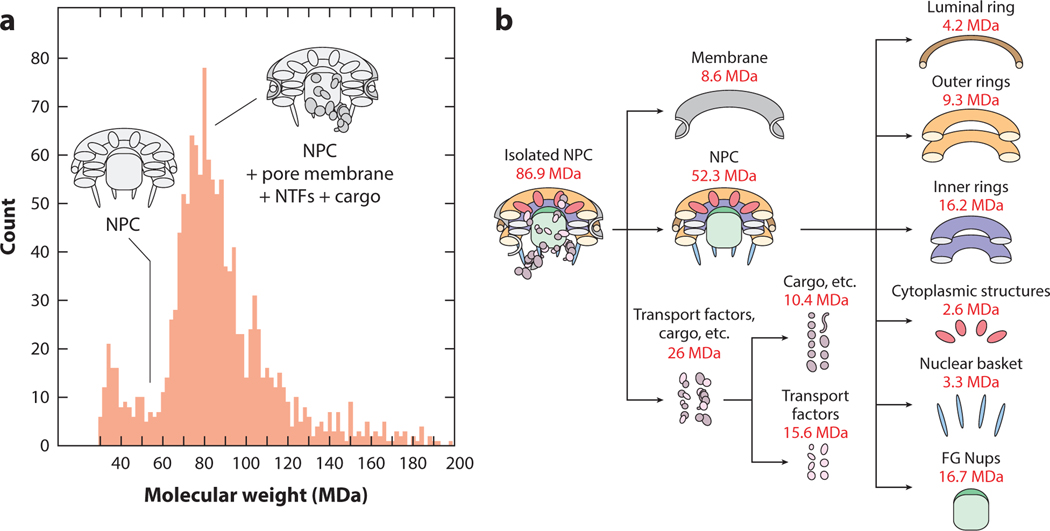
While the mass range and transmission efficiency of conventional Orbitrap, FT-ICR, and ToF mass spectrometers are continuing to increase, unconventional means of mass analysis still lie at the forefront for measuring MDa-size particles (86). As described above, CDMS is pushing the limit of what can be measured, including intact virus particles (13, 35, 36, 105), exosomes (15), lipoproteins (104), amyloid fibers (34), and a 552-protein nuclear pore complex (NPC) from Saccharomyces cerevisiae (88) (Figure 6). The NPC gates RNA and proteins between cytoplasm and nucleoplasm (Figure 6b) and is amenable to measurement by CDMS (Figure 6a). The measured mass of approximately 80 MDa implies that the large complex and its many substituents remain intact and measurable by CDMS in the gas phase. A variety of complementary tools (e.g., cross-linking, cryo-electron tomography, etc.) were used in this study to determine the masses and orientations of the individual subunits in the complex. In another recent study, the SARSCoV-2 spike protein was characterized, better defining the heterogeneous glycosylation profile and average glycan mass (119).
Figure 6.
(a) Charge detection mass spectrum of a 552-protein NPC and (b) breakdown of its composition. Figure adapted with permission from Reference 88; copyright 2018 Macmillan Publishers Limited, part of Springer Nature. Abbreviations: FG Nups, phenylalanine-glycine nucleoporins; NPC, nuclear pore complex; NTF, nuclear transport factor.
Orbitrap mass spectrometry has also demonstrated the ability to analyze MDa-size particles, including viruses (50, 51, 82, 143, 157, 167, 168) and bacteria (142, 143). In one study, the Heck lab used an Exactive Plus Orbitrap to study the stoichiometry of viral and bacterial nanoparticles up to approximately 4.5 MDa, which was possible due to the reduction of rf frequencies throughout the instrument (143). Both conventional ensemble measurements and CDMS have been applied in these studies (82). For example, Kafader et al. (82) compared spectra of 27 nm (3.2 MDa) MS2 virus-like particles using standard ensemble MS measurements on an Orbitrap with CDMS measurements. While the charge states remain unresolved by standard MS, the latter approach has allowed measurement of m/z and charge and thus determination of mass. CDMS clearly has advantages over conventional MS in these cases, where high sample heterogeneity or limited instrument resolution hinders mass determination, and is likely to expand its presence in nMS in the near future.
CONCLUSIONS AND FUTURE DIRECTIONS
nMS is one important tool in a suite of complementary structural biology tools that are used in parallel to solve structural questions. nMS is complementary to cryo-EM, for example, in (a) identification of multiple proteoforms that make high-resolution structural characterization difficult; (b) identification of the ligand corresponding to missing density in an X-ray or cryo-EM structure; (c) definition of stoichiometry, subunit connectivity, and CCS for a complex for which no or only low-resolution structures can be observed using cryo-EM; and (d) identification of subunit mixing in cases where partners of a complex are too similar to be distinguished via their cryoEM structure. Innovations in nMS technology and techniques have led to improved separations; sample introduction; ionization; activation; and mass, m/z, and shape measurement, enabling increasingly robust structural and dynamic analysis of macromolecular complexes. Complementary advances in computational frameworks will increasingly allow the integration of nMS data into structural modeling, for example, in cryo-EM 3D reconstructions. All of these nMS advancements have enabled applications to increasingly complex sample systems and illustrate the emerging role of nMS as an important member of an integrated suite of structural biology tools.
Despite these exciting advances in nMS, some challenges remain to enabling faster and more rigorous analysis on a broader swath of sample types. For example, existing platforms are often unable to resolve heterogeneous protein or nucleoprotein complex mixtures containing small mass differences (e.g., addition of small ligands or post-translational modifications), although charge changing experiments can sometimes solve that problem. Improvements to single molecule or charge detection MS, including improvements to throughput, would also solve this problem. Another challenge is the need to resolve and desolvate complexes that are sprayed in high concentrations of additives such as metal ions, small molecules (e.g., ATP), specific and nonspecific lipids, or nMS-incompatible buffer components that may be required for maintaining the integrity of the complex. Improving gas-phase desalting and adduct removal techniques, or removal of membrane mimics, while also being gentle enough to prevent restructuring of the complex would also aid in improved identification and resolution. Further improvements in the resolution of IM would enable better separation and characterization of different conformers of a protein or nucleoprotein complex or subcomplex and aid in the identification of species that overlap in m/z space. Coupling IM techniques with different analyzer types that are useful in nMS (e.g.,Orbitrap), and developing platforms where IM can be placed in different positions within the instrument without worries about pressure, will increase the versatility and utility of IM techniques in nMS. However, increasing IM resolution without sacrificing ion transmission or increasing the instrument footprint is an important challenge. Another remaining challenge is increasing the throughput and ease of nMS analysis, for example, through the streamlined integration of software, development of online separation techniques to enable screening, and integration of multiple MS/MS and IM approaches on a single instrument platform. We are especially excited about the future possibility of integrating nMS and other structural biology tools into a single platform using the same sample for MS- and non-MS-based measurements.
ACKNOWLEDGMENTS
We acknowledge the support of native mass spectrometry in our laboratory through National Institutes of Health grant P41 GM128577. We gratefully acknowledge the contributions of the large number of talented investigators throughout the world who have contributed to this topic, even though we could not cite all of the work in this brief review.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ahnert SE, Marsh JA, Hernández H, Robinson CV, Teichmann SA. 2015. Principles of assembly reveal a periodic table of protein complexes. Science 350(6266):aaa2245 [DOI] [PubMed] [Google Scholar]
- 2.Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV. 2015. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat. Commun 6:8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrose S, Housden NG, Gupta K, Fan J, White P, et al. 2017. Native desorption electrospray ionization liberates soluble and membrane protein complexes from surfaces. Angew. Chem 129(46):14655–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey AO,Han G,Phung W,Gazis P,Sutton J,et al.2018.Chargevariantnativemassspectrometrybenefits mass precision and dynamic range of monoclonal antibody intact mass analysis.mAbs 10(8):1214–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrera NP, Bartolo ND, Booth PJ, Robinson CV. 2008. Micelles protect membrane complexes from solution to vacuum. Science 321(5886):243–46 [DOI] [PubMed] [Google Scholar]
- 6.Bashyal A, Sanders JD, Holden DD, Brodbelt JS. 2019. Top-down analysis of proteins in low charge states. J. Am. Soc. Mass Spectrom 30(4):704–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benesch JL, Ruotolo BT. 2011. Mass spectrometry: come of age for structural and dynamical biology.Curr. Opin. Struct. Biol 21(5):641–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Nissan G, Sharon M. 2018. The application of ion-mobility mass spectrometry for structure/function investigation of protein complexes. Curr. Opin. Chem. Biol 42:25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Nissan G, Vimer S, Warszawski S, Katz A, Yona M, et al. 2018. Rapid characterization of secreted recombinant proteins by native mass spectrometry. Commun. Biol 1:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bern M, Caval T, Kil YJ, Tang W, Becker C, et al. 2018. Parsimonious charge deconvolution for native mass spectrometry. J. Proteome Res 17(3):1216–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleiholder C, Wyttenbach T, Bowers MT. 2011. A novel projection approximation algorithm for the fast and accurate computation of molecular collision cross sections (I). Method. Int. J. Mass Spectrom 308(1):1–10 [Google Scholar]
- 12.Bolla JR, Agasid MT, Mehmood S, Robinson CV. 2019. Membrane protein-lipid interactions probed using mass spectrometry. Annu. Rev. Biochem 88:85–111 [DOI] [PubMed] [Google Scholar]
- 13.Bond KM, Lyktey NA, Tsvetkova IB, Dragnea B, Jarrold MF. 2020. Disassembly intermediates of the brome mosaic virus identified by charge detection mass spectrometry. J. Phys. Chem. B 124(11):2124–31 [DOI] [PubMed] [Google Scholar]
- 14.Boyken SE, Benhaim MA, Busch F, Jia M, Bick MJ, et al. 2019. De novo design of tunable, pH-drivenconformational changes. Science 364(6441):658–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown BA, Zeng X, Todd AR, Barnes LF, Winstone JMA, et al. 2020. Charge detection mass spectrometry measurements of exosomes and other extracellular particles enriched from bovine milk. Anal. Chem 92(4):3285–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busch F, VanAernum ZL, Lai SM, Gopalan V, Wysocki VH. 2021. Analysis of tagged proteins using tandem affinity-buffer exchange chromatography online with native mass spectrometry. Biochemistry 60(24):1876–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler KE, Takinami Y, Rainczuk A, Baker ES, Roberts BR.2021.Utilizing ion mobility-mass spectrometry to investigate the unfolding pathway of Cu/Zn superoxide dismutase. Front. Chem 9:614595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldwell BJ, Norris A, Zakharova E, Smith CE, Wheat CT, et al.2021.Oligomeric complexes formed by Redβ single strand annealing protein in its different DNA bound states. Nucleic Acids Res. 49(6):3441–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campuzano IDG, Sandoval W. 2021. Denaturing and native mass spectrometric analytics for biotherapeutic drug discovery research: historical, current, and future personal perspectives. J. Am. Soc. Mass Spectrom 32(8):1861–85 [DOI] [PubMed] [Google Scholar]
- 20.Chai M, Young MN, Liu FC, Bleiholder C. 2018. A transferable, sample-independent calibration procedure for trapped ion mobility spectrometry (TIMS). Anal. Chem 90(15):9040–47 [DOI] [PubMed] [Google Scholar]
- 21.Chen D, McCool EN, Yang Z, Shen X, Lubeckyj RA, et al. 2021. Recent advances (2019–2021) of capillary electrophoresis-mass spectrometry for multilevel proteomics. Mass Spectrom. Rev In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Tanimoto A, So BR, Bakhtina M, Magliery TJ, et al. 2019. Stoichiometry of triple-sieve tRNA editing complex ensures fidelity of aminoacyl-tRNA formation. Nucleic Acids Res. 47(2):929–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Boyken SE, Jia M, Busch F, Flores-Solis D, et al. 2019. Programmable design of orthogonal protein heterodimers. Nature 565(7737):106–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Kibler RD, Hunt A, Busch F, Pearl J, et al. 2020. De novo design of protein logic gates. Science 368(6486):78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chorev DS, Baker LA, Wu D, Beilsten-Edmands V, Rouse SL, et al. 2018. Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science 362(6416):829–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chorev DS, Tang H, Rouse SL, Bolla JR, von Kügelgen A, et al.2020.The use of sonicated lipid vesicles for mass spectrometry of membrane protein complexes. Nat. Protoc 15(5):1690–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleary SP, Thompson AM, Prell JS. 2016. Fourier analysis method for analyzing highly congested mass spectra of ion populations with repeated subunits. Anal. Chem 88(12):6205–13 [DOI] [PubMed] [Google Scholar]
- 28.Cong X, Liu Y, Liu W, Liang X, Laganowsky A. 2017. Allosteric modulation of protein-protein interactions by individual lipid binding events. Nat. Commun 8:2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong X, Liu Y, Liu W, Liang X, Russell DH, Laganowsky A. 2016. Determining membrane protein-lipid binding thermodynamics using native mass spectrometry. J. Am. Chem. Soc 138(13):4346–49 [DOI] [PubMed] [Google Scholar]
- 30.Cveticanin J, Netzer R, Arkind G, Fleishman SJ, Horovitz A, Sharon M. 2018. Estimating inter-protein pairwise interaction energies in cell lysates from a single native mass spectrum. Anal. Chem 90(17):10090–94 [DOI] [PubMed] [Google Scholar]
- 31.Degiacomi MT, Benesch JLP. 2016. EMIM: software for relating ion mobility mass spectrometry and electron microscopy data. Analyst 141(1):70–75 [DOI] [PubMed] [Google Scholar]
- 32.Deslignière E, Ley M, Bourguet M, Ehkirch A, Botzanowski T, et al. 2021. Pushing the limits of native MS: online SEC-native MS for structural biology applications. Int. J. Mass Spectrom 461:116502 [Google Scholar]
- 33.Dixit SM, Polasky DA, Ruotolo BT. 2018. Collision induced unfolding of isolated proteins in the gas phase: past, present, and future. Curr. Opin. Chem. Biol 42:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doussineau T, Mathevon C, Altamura L, Vendrely C, Dugourd P, et al. 2016. Mass determination of entire amyloid fibrils by using mass spectrometry. Angew. Chem 128(7):2386–90 [DOI] [PubMed] [Google Scholar]
- 35.Dunbar CA, Callaway HM, Parrish CR, Jarrold MF. 2018. Probing antibody binding to canine parvovirus with charge detection mass spectrometry. J. Am. Chem. Soc 140(46):15701–11 [DOI] [PubMed] [Google Scholar]
- 36.Dunbar CA, Rayaprolu V, Wang JC-Y, Brown CJ, Leishman E, et al. 2019. Dissecting the components of Sindbis virus from arthropod and vertebrate hosts: implications for infectivity differences. ACS Infect. Dis 5(6):892–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyachenko A, Gruber R, Shimon L, Horovitz A, Sharon M. 2013. Allosteric mechanisms can be distinguished using structural mass spectrometry. PNAS 110(18):7235–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehkirch A, D’Atri V, Rouviere F, Hernandez-Alba O, Goyon A, et al. 2018. An online four-dimensional HIC×SEC-IM×MS methodology for proof-of-concept characterization of antibody drug conjugates. Anal. Chem 90(3):1578–86 [DOI] [PubMed] [Google Scholar]
- 39.Ehkirch A, Hernandez-Alba O, Colas O, Beck A, Guillarme D, Cianférani S. 2018. Hyphenation of size exclusion chromatography to native ion mobility mass spectrometry for the analytical characterization of therapeutic antibodies and related products. J. Chromatogr. B 1086:176–83 [DOI] [PubMed] [Google Scholar]
- 40.Eldrid C,Ujma J,Kalfas S,Tomczyk N,Giles K,et al.2019.Gas phase stability of protein ions in a cyclic ion mobility spectrometry traveling wave device. Anal. Chem 91(12):7554–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott AG, Harper CC, Lin H-W, Williams ER. 2017. Mass, mobility and MSn measurements of single ions using charge detection mass spectrometry. Analyst 142(15):2760–69 [DOI] [PubMed] [Google Scholar]
- 42.Elliott AG,Merenbloom SI,Chakrabarty S,Williams ER.2017.Single particle analyzer of mass: a charge detection mass spectrometer with a multi-detector electrostatic ion trap.Int.J.Mass Spectrom 414:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eschweiler JD, Rabuck-Gibbons JN, Tian Y, Ruotolo BT. 2015. CIUSuite: a quantitative analysis package for collision induced unfolding measurements of gas-phase protein ions. Anal. Chem 87(22):11516–22 [DOI] [PubMed] [Google Scholar]
- 44.Ewing SA, Donor MT, Wilson JW, Prell JS. 2017. Collidoscope: an improved tool for computing collisional cross-sections with the trajectory method. J. Am. Soc. Mass Spectrom 28(4):587–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fantin SM, Parson KF, Niu S, Liu J, Polasky DA, et al. 2019. Collision induced unfolding classifies ligands bound to the integral membrane translocator protein. Anal. Chem 91(24):15469–76 [DOI] [PubMed] [Google Scholar]
- 46.Felitsyn N, Kitova EN, Klassen JS. 2001. Thermal decomposition of a gaseous multiprotein complex studied by blackbody infrared radiative dissociation: investigating the origin of the asymmetric dissociation behavior. Anal. Chem 73(19):4647–61 [DOI] [PubMed] [Google Scholar]
- 47.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science 246(4926):64–71 [DOI] [PubMed] [Google Scholar]
- 48.Flick TG, Cassou CA, Chang TM, Williams ER. 2012. Solution additives that desalt protein ions in native mass spectrometry. Anal. Chem 84(17):7511–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foreman DJ, McLuckey SA. 2020. Recent developments in gas-phase ion/ion reactions for analytical mass spectrometry. Anal. Chem 92(1):252–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forsberg E, Fang M, Siuzdak G. 2017. Staying alive: measuring intact viable microbes with electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom 28(1):14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fort KL, van de Waterbeemd M, Boll D, Reinhardt-Szyba M, Belov ME, et al. 2018. Expanding the structural analysis capabilities on an Orbitrap-based mass spectrometer for large macromolecular complexes. Analyst 143(1):100–5 [DOI] [PubMed] [Google Scholar]
- 52.Franc V, Yang Y, Heck AJR. 2017. Proteoform profile mapping of the human serum complement component C9 revealing unexpected new features of N-,O-,and C-glycosylation.Anal.Chem 89(6):3483–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franc V, Zhu J, Heck AJR. 2018. Comprehensive proteoform characterization of plasma complement component C8αβγ by hybrid mass spectrometry approaches. J. Am. Soc. Mass Spectrom 29(6):1099–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.France AP, Migas LG, Sinclair E, Bellina B, Barran PE. 2020. Using collision cross section distributions to assess the distribution of collision cross section values. Anal. Chem 92(6):4340–48 [DOI] [PubMed] [Google Scholar]
- 55.Gabelica V, Marklund E. 2018. Fundamentals of ion mobility spectrometry. Curr. Opin. Chem. Biol 42:51–59 [DOI] [PubMed] [Google Scholar]
- 56.Gabelica V, Vreuls C, Filée P, Duval V, Joris B, Pauw ED.2002.Advantages and drawbacks of nanospray for studying noncovalent protein-DNA complexes by mass spectrometry. Rapid Commun. Mass Spectrom 16(18):1723–28 [DOI] [PubMed] [Google Scholar]
- 57.Gadzuk-Shea MM, Bush MF.2018.Effects of charge state on the structures of serum albumin ions in the gas phase: insights from cation-to-anion proton-transfer reactions, ion mobility, and mass spectrometry. J. Phys. Chem. B 122(43):9947–55 [DOI] [PubMed] [Google Scholar]
- 58.Gan J, Ben-Nissan G, Arkind G, Tarnavsky M, Trudeau D, et al. 2017. Native mass spectrometry of recombinant proteins from crude cell lysates. Anal. Chem 89(8):4398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gault J, Liko I, Landreh M, Shutin D, Bolla JR, et al. 2020. Combining native and “omics” mass spectrometry to identify endogenous ligands bound to membrane proteins. Nat. Methods 17(5):505–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. 2004. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom 18(20):2401–14 [DOI] [PubMed] [Google Scholar]
- 61.Giles K, Ujma J, Wildgoose J, Pringle S, Richardson K, et al. 2019. A cyclic ion mobility-mass spectrometry system. Anal. Chem 91(13):8564–73 [DOI] [PubMed] [Google Scholar]
- 62.Giles K, Williams JP, Campuzano I. 2011. Enhancements in travelling wave ion mobility resolution. Rapid Commun. Mass Spectrom 25(11):1559–66 [DOI] [PubMed] [Google Scholar]
- 63.Gruber R, Horovitz A. 2018. Unpicking allosteric mechanisms of homo-oligomeric proteins by determining their successive ligand binding constants. Philos. Trans. R. Soc. B 373(1749):20170176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta K,Li J,Liko I,Gault J,Bechara C,et al.2018.Identifying key membrane protein lipid interactions using mass spectrometry. Nat. Protoc 13(5):1106–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hale OJ, Cooper HJ. 2021. Native mass spectrometry imaging of proteins and protein complexes by nano-DESI. Anal. Chem 93(10):4619–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. 2012. Charge-state dependent compaction and dissociation of protein complexes: insights from ion mobility and molecular dynamics. J. Am. Chem. Soc 134(7):3429–38 [DOI] [PubMed] [Google Scholar]
- 67.Harvey SR, Seffernick JT, Quintyn RS, Song Y, Ju Y, et al. 2019. Relative interfacial cleavage energetics of protein complexes revealed by surface collisions. PNAS 116(17):8143–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hellwig N, Peetz O, Ahdash Z, Tascón I, Booth PJ, et al. 2018. Native mass spectrometry goes more native: investigation of membrane protein complexes directly from SMALPs. Chem. Commun 54(97):13702–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hogan JA, Jarrold MF. 2018. Optimized electrostatic linear ion trap for charge detection mass spectrometry. J. Am. Soc. Mass Spectrom 29(10):2086–95 [DOI] [PubMed] [Google Scholar]
- 70.Hoi KK, Bada Juarez JF, Judge PJ, Yen H-Y, Wu D, et al.2021.Detergent-free lipodisq nanoparticles facilitate high-resolution mass spectrometry of folded integral membrane proteins.Nano Lett.21(7):2824–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holden DD, Brodbelt JS. 2016. Ultraviolet photodissociation of native proteins following proton transfer reactions in the gas phase. Anal. Chem 88(24):12354–62 [DOI] [PubMed] [Google Scholar]
- 72.Holmquist ML, Ihms EC, Gollnick P, Wysocki VH, Foster MP. 2020. Population distributions from native mass spectrometry titrations reveal nearest-neighbor cooperativity in the ring-shaped oligomeric protein TRAP. Biochemistry 59(27):2518–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hopper JTS, Yu YT-C, Li D, Raymond A, Bostock M, et al. 2013. Detergent-free mass spectrometry of membrane protein complexes. Nat. Methods 10(12):1206–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu J, Guan Q-Y, Wang J, Jiang X-X, Wu Z-Q, et al. 2017. Effect of nanoemitters on suppressing the formation of metal adduct ions in electrospray ionization mass spectrometry.Anal.Chem 89(3):1838–45 [DOI] [PubMed] [Google Scholar]
- 75.Huguet R, Mullen C, Srzentic K, Greer JB, Fellers RT, et al. 2019. Proton transfer charge reduction enables high-throughput top-down analysis of large proteoforms. Anal. Chem 91(24):15732–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ibrahim YM, Garimella SVB, Prost SA, Wojcik R, Norheim RV, et al. 2016. Development of an ion mobility spectrometry-Orbitrap mass spectrometer platform. Anal. Chem 88(24):12152–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeanne Dit Fouque K, Garabedian A, Leng F, Tse-Dinh Y-C, Fernandez-Lima F. 2019. Microheterogeneity of topoisomerase IA/IB and their DNA-bound states. ACS Omega 4(2):3619–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeanne Dit Fouque K, Garabedian A, Leng F, Tse-Dinh Y-C, Ridgeway ME, et al. 2021. Trapped ion mobility spectrometry of native macromolecular assemblies. Anal. Chem 93(5):2933–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jhingree JR, Beveridge R, Dickinson ER, Williams JP, Brown JM, et al. 2017. Electron transfer with no dissociation ion mobility-mass spectrometry (ETnoD IM-MS): the effect of charge reduction on protein conformation. Int. J. Mass Spectrom 413:43–51 [Google Scholar]
- 80.Jurchen JC, Williams ER. 2003. Origin of asymmetric charge partitioning in the dissociation of gas-phase protein homodimers. J. Am. Chem. Soc 125(9):2817–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kafader JO, Beu SC, Early BP, Melani RD, Durbin KR, et al. 2019. STORI plots enable accurate tracking of individual ion signals. J. Am. Soc. Mass Spectrom 30(11):2200–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kafader JO, Melani RD, Durbin KR, Ikwuagwu B, Early BP, et al. 2020.Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes. Nat. Methods 17(4):391–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kafader JO, Melani RD, Senko MW, Makarov AA, Kelleher NL, Compton PD. 2019. Measurement of individual ions sharply increases the resolution of Orbitrap mass spectra of proteins. Anal. Chem 91(4):2776–83 [DOI] [PubMed] [Google Scholar]
- 84.Karlsson OA, Sundell GN, Andersson E, Ivarsson Y, Jemth P. 2016. Improved affinity at the cost of decreased specificity: a recurring theme in PDZ-peptide interactions. Sci. Rep 6:34269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keener JE, Zambrano DE, Zhang G, Zak CK, Reid DJ, et al. 2019. Chemical additives enable native mass spectrometry measurement of membrane protein oligomeric state within intact nanodiscs. J. Am. Chem. Soc 141(2):1054–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keifer DZ, Pierson EE, Jarrold MF.2017.Charge detection mass spectrometry: weighing heavier things. Analyst 142(10):1654–71 [DOI] [PubMed] [Google Scholar]
- 87.Keifer DZ, Shinholt DL, Jarrold MF. 2015. Charge detection mass spectrometry with almost perfect charge accuracy. Anal. Chem 87(20):10330–37 [DOI] [PubMed] [Google Scholar]
- 88.Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, et al. 2018. Integrative structure and functional anatomy of a nuclear pore complex. Nature 555(7697):475–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laganowsky A, Clemmer DE, Russell DH. 2022. Variable-temperature native mass spectrometry for studies of protein folding, stabilities, assembly, and molecular interactions. Annu. Rev. Biophys 51:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, et al. 2014. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510(7503):172–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laganowsky A, Reading E, Hopper JTS, Robinson CV. 2013. Mass spectrometry of intact membrane protein complexes. Nat. Protoc 8(4):639–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lai AL, Clerico EM, Blackburn ME, Patel NA, Robinson CV, et al. 2017. Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron electron resonance. J. Biol. Chem 292(21):8773–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lai LB, Tanimoto A, Lai SM, Chen W-Y, Marathe IA, et al. 2017. A novel double kink-turn module in euryarchaeal RNase P RNAs. Nucleic Acids Res. 45(12):7432–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Landeras-Bueno S, Wasserman H, Oliveira G, VanAernum ZL, Busch F, et al. 2021. Cellular mRNA triggers structural transformation of Ebola virus matrix protein VP40 to its essential regulatory form. Cell Rep. 35(2):108986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laszlo KJ, Bush MF. 2015. Analysis of native-like proteins and protein complexes using cation to anion proton transfer reactions (CAPTR). J. Am. Soc. Mass Spectrom 26(12):2152–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Laszlo KJ, Munger EB, Bush MF. 2016. Folding of protein ions in the gas phase after cation-to-anion proton-transfer reactions. J. Am. Chem. Soc 138(30):9581–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leney AC, McMorran LM, Radford SE, Ashcroft AE. 2012. Amphipathic polymers enable the study of functional membrane proteins in the gas phase. Anal. Chem 84(22):9841–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lermyte F, Łącki MK, Valkenborg D, Gambin A, Sobott F. 2017. Conformational space and stability of ETD charge reduction products of ubiquitin. J. Am. Soc. Mass Spectrom 28(1):69–76 [DOI] [PubMed] [Google Scholar]
- 99.Lermyte F, Williams JP, Brown JM, Martin EM, Sobott F. 2015. Extensive charge reduction and dissociation of intact protein complexes following electron transfer on a quadrupole-ion mobility-time-of-flight MS. J. Am. Soc. Mass Spectrom 26(7):1068–76 [DOI] [PubMed] [Google Scholar]
- 100.Liu FC, Cropley TC, Ridgeway ME, Park MA, Bleiholder C. 2020. Structural analysis of the glycoprotein complex avidin by tandem-trapped ion mobility spectrometry-mass spectrometry (tandemTIMS/MS). Anal. Chem 92(6):4459–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu Y, LoCaste CE, Liu W, Poltash ML, Russell DH, Laganowsky A. 2019. Selective binding of a toxin and phosphatidylinositides to a mammalian potassium channel. Nat. Commun 10:1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lorenzen K, van Duijn E. 2010. Native mass spectrometry as a tool in structural biology. Curr. Protoc. Protein Sci 62(1):17.12 [DOI] [PubMed] [Google Scholar]
- 103.Lutomski CA, El-Baba TJ, Bolla JR, Robinson CV. 2021. Multiple roles of SARS-CoV-2 N protein facilitated by proteoform-specific interactions with RNA, host proteins, and convalescent antibodies. JACS Au 1(8):1147–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lutomski CA, Gordon SM, Remaley AT, Jarrold MF. 2018. Resolution of lipoprotein subclasses by charge detection mass spectrometry. Anal. Chem 90(11):6353–56 [DOI] [PubMed] [Google Scholar]
- 105.Lutomski CA, Lyktey NA, Zhao Z, Pierson EE, Zlotnick A, Jarrold MF. 2017. Hepatitis B virus capsid completion occurs through error correction. J. Am. Chem. Soc 139(46):16932–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma X, Lai LB, Lai SM, Tanimoto A, Foster MP, et al. 2014. Uncovering the stoichiometry of Pyrococcus furiosus RNase P, a multi-subunit catalytic ribonucleoprotein complex, by surface-induced dissociation and ion mobility mass spectrometry. Angew. Chem. Int. Ed 53(43):11483–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Macias LA, Santos IC, Brodbelt JS. 2020. Ion activation methods for peptides and proteins. Anal. Chem 92(1):227–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mack E. 1925. Average cross-sectional areas of molecules by gaseous diffusion methods. J. Am. Chem. Soc 47(10):2468–82 [Google Scholar]
- 109.Marcoux J, Wang SC, Politis A, Reading E, Ma, et al. 2013. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. PNAS 110(24):9704–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marklund EG, Degiacomi MT, Robinson CV, Baldwin AJ, Benesch JLP. 2015. Collision cross sections for structural proteomics. Structure 23(4):791–99 [DOI] [PubMed] [Google Scholar]
- 111.Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV. 2015. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem 87(8):4370–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mason EA, Schamp HW. 1958. Mobility of gaseous ions in weak electric fields. Ann. Phys 4(3):233–70 [Google Scholar]
- 113.McCabe JW, Hebert MJ, Shirzadeh M, Mallis CS, Denton JK, et al. 2021. The IMS paradox: a perspective on structural ion mobility-mass spectrometry. Mass Spectrom. Rev 40(3):280–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McCabe JW, Mallis CS, Kocurek KI, Poltash ML, Shirzadeh M, et al. 2020. First-principles collision cross section measurements of large proteins and protein complexes. Anal. Chem 92(16):11155–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McGee JP, Melani RD, Yip PF, Senko MW, Compton PD, et al. 2021. Isotopic resolution of protein complexes up to 466 kDa using individual ion mass spectrometry. Anal. Chem 93(5):2723–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mehaffey MR, Lee J, Jung J, Lanzillotti MB, Escobar EE, et al. 2020. Mapping a conformational epitope of hemagglutinin A using native mass spectrometry and ultraviolet photodissociation. Anal. Chem 92(17):11869–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. 1996. Structural information from ion mobility measurements: effects of the long-range potential. J. Phys. Chem 100(40):16082–86 [Google Scholar]
- 118.Migas LG,France AP,Bellina B,Barran PE.2018.ORIGAMI: a software suite for activated ion mobility mass spectrometry (aIM-MS) applied to multimeric protein assemblies. Int. J. Mass Spectrom 427:20–28 [Google Scholar]
- 119.Miller LM,Barnes LF,Raab SA,Draper BE,El-Baba TJ,et al.2021.Heterogeneity of glycan processing on trimeric SARS-CoV-2 spike protein revealed by charge detection mass spectrometry. J. Am. Chem. Soc 143(10):3959–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mistarz UH,Chandler SA,Brown JM,Benesch JLP,Rand KD.2019.Probing the dissociation of protein complexes by means of gas-phase H/D exchange mass spectrometry.J.Am.Soc.Mass Spectrom 30(1):45–57 [DOI] [PubMed] [Google Scholar]
- 121.Olinares PDB, Dunn AD, Padovan JC, Fernandez-Martinez J, Rout MP, Chait BT. 2016. A robust workflow for native mass spectrometric analysis of affinity-isolated endogenous protein assemblies. Anal. Chem 88(5):2799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pagano L, Toto A, Malagrinò F, Visconti L, Jemth P, Gianni S. 2021. Double mutant cycles as a tool to address folding, binding, and allostery. Int. J. Mol. Sci 22(2):828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Panczyk EM, Snyder DT, Ridgeway ME, Somogyi Á, Park MA, Wysocki VH. 2021. Surface-induced dissociation of protein complexes selected by trapped ion mobility spectrometry. Anal. Chem 93(13):5513–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Patrick JW, Boone CD, Liu W, Conover GM, Liu Y, et al. 2018. Allostery revealed within lipid binding events to membrane proteins. PNAS 115(12):2976–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pierson EE, Contino NC, Keifer DZ, Jarrold MF. 2015. Charge detection mass spectrometry for single ions with an uncertainty in the charge measurement of 0.65 e. J. Am. Soc. Mass Spectrom 26(7):1213–20 [DOI] [PubMed] [Google Scholar]
- 126.Poltash ML, McCabe JW, Patrick JW, Laganowsky A, Russell DH. 2019. Development and evaluation of a reverse-entry ion source Orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom 30(1):192–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Poltash ML, McCabe JW, Shirzadeh M, Laganowsky A, Clowers BH, Russell DH. 2018. Fourier transform-ion mobility-Orbitrap mass spectrometer: a next-generation instrument for native mass spectrometry. Anal. Chem 90(17):10472–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Poltash ML, McCabe JW, Shirzadeh M, Laganowsky A, Russell DH. 2020. Native IM-Orbitrap MS: resolving what was hidden. Trends Anal. Chem 124:115533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reid DJ, Diesing JM, Miller MA, Perry SM, Wales JA, et al. 2019. MetaUniDec: high-throughput deconvolution of native mass spectra. J. Am. Soc. Mass Spectrom 30(1):118–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ridgeway ME, Lubeck M, Jordens J, Mann M, Park MA. 2018. Trapped ion mobility spectrometry: a short review. Int. J. Mass Spectrom 425:22–35 [Google Scholar]
- 131.Sahasrabuddhe A, Hsia Y, Busch F, Sheffler W, King NP, et al. 2018. Confirmation of intersubunit connectivity and topology of designed protein complexes by native MS. PNAS 115(6):1268–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Saikusa K, Osakabe A, Kato D, Fuchigami S, Nagadoi A, et al. 2018. Structural diversity of nucleosomes characterized by native mass spectrometry. Anal. Chem 90(13):8217–26 [DOI] [PubMed] [Google Scholar]
- 133.Sakamoto W, Azegami N, Konuma T, Akashi S. 2021. Single-cell native mass spectrometry of human erythrocytes. Anal. Chem 93(17):6583–88 [DOI] [PubMed] [Google Scholar]
- 134.Sarni S, Biswas B, Liu S, Olson ED, Kitzrow JP, et al. 2020. HIV-1 Gag protein with or without p6 specifically dimerizes on the viral RNA packaging signal. J. Biol. Chem 295(42):14391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schneeberger EM-, Breuker K. 2017. Native top-down mass spectrometry of TAR RNA in complexes with a wild-type tat peptide for binding site mapping. Angew. Chem 129(5):1274–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schneeberger E-M, Halper M, Palasser M, Heel SV, Vušurović J, et al. 2020. Native mass spectrometry reveals the initial binding events of HIV-1 rev to RRE stem II RNA. Nat. Commun 11:5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sever AIM, Yin V, Konermann L. 2021. Interrogating the quaternary structure of noncanonical hemoglobin complexes by electrospray mass spectrometry and collision-induced dissociation. J. Am. Soc. Mass Spectrom 32(1):270–80 [DOI] [PubMed] [Google Scholar]
- 138.Sharon M, Horovitz A.2015.Probing allosteric mechanisms using native mass spectrometry. Curr.Opin. Struct. Biol 34:7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shen X, Kou Q, Guo R, Yang Z, Chen D, et al. 2018. Native proteomics in discovery mode using size-exclusion chromatography-capillary zone electrophoresis-tandem mass spectrometry. Anal. Chem 90(17):10095–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shvartsburg AA, Jarrold MF. 1996. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem. Phys. Lett 261(1):86–91 [Google Scholar]
- 141.Skinner OS, Haverland NA, Fornelli L, Melani RD, Do Vale LHF, et al. 2018. Top-down characterization of endogenous protein complexes with native proteomics. Nat. Chem. Biol 14(1):36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Snijder J, Kononova O, Barbu IM, Uetrecht C, Rurup WF, et al. 2016. Assembly and mechanical properties of the cargo-free and cargo-loaded bacterial nano compartment encapsulin. Biomacromolecules 17(8):2522–29 [DOI] [PubMed] [Google Scholar]
- 143.Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, et al. 2014. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. J. Am. Chem. Soc 136(20):7295–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Snyder DT, Panczyk EM, Somogyi A, Kaplan DA, Wysocki V.2020.Simple and minimally invasive SID devices for native mass spectrometry. Anal. Chem 92(16):11195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sokolovski M, Cveticanin J, Hayoun D, Korobko I, Sharon M, Horovitz A. 2017. Measuring inter-protein pairwise interaction energies from a single native mass spectrum by double-mutant cycle analysis. Nat. Commun 8:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Soltermann F, Foley EDB, Pagnoni V, Galpin M, Benesch JLP, et al.2020.Quantifying protein-protein interactions by molecular counting with mass photometry. Angew. Chem. Int. Ed 59(27):10774–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sonn-Segev A, Belacic K, Bodrug T, Young G, VanderLinden RT, et al. 2020. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun 11:1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Srebalus CA, Li J, Marshall WS, Clemmer DE. 1999. Gas-phase separations of electrosprayed peptide libraries. Anal. Chem 71(18):3918–27 [DOI] [PubMed] [Google Scholar]
- 149.Stiving AQ, Jones BJ, Ujma J, Giles K, Wysocki VH. 2020. Collision cross sections of charge-reduced proteins and protein complexes: a database for collision cross section calibration. Anal. Chem 92(6):4475–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stiving AQ, VanAernum ZL, Busch F, Harvey SR, Sarni SH, Wysocki VH.2019.Surface-induced dissociation: an effective method for characterization of protein quaternary structure. Anal. Chem 91(1):190–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Struwe WB, Robinson CV. 2019. Relating glycoprotein structural heterogeneity to function—insights from native mass spectrometry. Curr. Opin. Struct. Biol 58:241–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Susa AC, Lippens JL, Xia Z, Loo JA, Campuzano IDG, Williams ER. 2018. Submicrometer emitter ESI tips for native mass spectrometry of membrane proteins in ionic and nonionic detergents. J. Am. Soc. Mass Spectrom 29(1):203–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Susa AC, Xia Z, Williams ER. 2017. Native mass spectrometry from common buffers with salts that mimic the extracellular environment. Angew. Chem. Int. Ed 56(27):7912–15 [DOI] [PubMed] [Google Scholar]
- 154.Susa AC, Xia Z, Williams ER. 2017. Small emitter tips for native mass spectrometry of proteins and protein complexes from nonvolatile buffers that mimic the intracellular environment. Anal. Chem 89(5):3116–22 [DOI] [PubMed] [Google Scholar]
- 155.Tian Y, Han L, Buckner AC, Ruotolo BT. 2015. Collision induced unfolding of intact antibodies: rapid characterization of disulfide bonding patterns, glycosylation, and structures. Anal. Chem 87(22):11509–15 [DOI] [PubMed] [Google Scholar]
- 156.Tian Y, Ruotolo BT. 2018. Collision induced unfolding detects subtle differences in intact antibody glycoforms and associated fragments. Int. J. Mass Spectrom 425:1–9 [Google Scholar]
- 157.van de Waterbeemd M, Snijder J, Tsvetkova IB, Dragnea BG, Cornelissen JJ, Heck AJR. 2016. Examining the heterogeneous genome content of multipartite viruses BMV and CCMV by native mass spectrometry. J. Am. Soc. Mass Spectrom 27(6):1000–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.VanAernum ZL, Busch F, Jones BJ, Jia M, Chen Z, et al. 2020. Rapid online buffer exchange for screening of proteins, protein complexes and cell lysates by native mass spectrometry. Nat. Protoc 15(3):1132–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.VanAernum ZL, Gilbert JD, Belov ME, Makarov AA, Horning SR, Wysocki VH. 2019. Surface-induced dissociation of noncovalent protein complexes in an extended mass range Orbitrap mass spectrometer. Anal. Chem 91(5):3611–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vimer S, Ben-Nissan G, Morgenstern D, Kumar-Deshmukh F, Polkinghorn C, et al.2020.Comparative structural analysis of 20S proteasome ortholog protein complexes by native mass spectrometry. ACS Cent. Sci 6(4):573–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vimer S, Ben-Nissan G, Sharon M. 2020. Direct characterization of overproduced proteins by native mass spectrometry. Nat. Protoc 15(2):236–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vimer S, Ben-Nissan G, Sharon M. 2020. Mass spectrometry analysis of intact proteins from crude samples. Anal. Chem 92(19):12741–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vorobieva AA, White P, Liang B, Horne JE, Bera AK, et al. 2021. De novo design of transmembrane β barrels. Science 371(6531):eabc8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vušurović J, Breuker K. 2019. Relative strength of noncovalent interactions and covalent backbone bonds in gaseous RNA-peptide complexes. Anal. Chem 91(2):1659–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wei B, Han G, Tang J, Sandoval W, Zhang YT. 2019. Native hydrophobic interaction chromatography hyphenated to mass spectrometry for characterization of monoclonal antibody minor variants. Anal. Chem 91(24):15360–64 [DOI] [PubMed] [Google Scholar]
- 166.Wilm M,Mann M.1996.Analytical properties of the nanoelectrospray ion source.Anal.Chem 68(1):1–8 [DOI] [PubMed] [Google Scholar]
- 167.Wörner TP, Bennett A, Habka S, Snijder J, Friese O,et al.2021. Adeno-associated virus capsid assemblyis divergent and stochastic. Nat. Commun 12:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wörner TP, Snijder J, Bennett A, Agbandje-McKenna M,Makarov AA,Heck AJR.2020.Resolving heterogeneous macromolecular assemblies by Orbitrap-based single-particle charge detection mass spectrometry. Nat. Methods 17(4):395–98 [DOI] [PubMed] [Google Scholar]
- 169.Wysocki VH, Jones CM, Galhena AS, Blackwell AE.2008. Surface-induced dissociation shows potential to be more informative than collision-induced dissociation for structural studies of large systems. J. Am. Soc. Mass Spectrom 19(7):903–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wysocki VH, Joyce KE, Jones CM, Beardsley RL. 2008. Surface-induced dissociation of small molecules, peptides, and non-covalent protein complexes. J. Am. Soc. Mass Spectrom 19(2):190–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yang F,Xiao X,Cheng W,Yang W,Yu P,et al.2015. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol 11(7):518–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang Y, Niu C, Bobst CE, Kaltashov IA.2021.Charge manipulation using solution and gas-phase chemistry to facilitate analysis of highly heterogeneous protein complexes in native mass spectrometry. Anal. Chem 93(7):3337–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yen H-Y, Hopper JTS, Liko I, Allison TM, Zhu Y, et al. 2017. Ligand binding to a G protein-coupled receptor captured in a mass spectrometer. Sci. Adv 3(6):e1701016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang J, Loo RRO, Loo JA. 2017. Structural characterization of a thrombin-aptamer complex by high resolution native top-down mass spectrometry. J. Am. Soc. Mass Spectrom 28(9):1815–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhao Y, Abzalimov RR, Kaltashov IA. 2016. Interactions of intact unfractionated heparin with its client proteins can be probed directly using native electrospray ionization mass spectrometry. Anal. Chem 88(3):1711–18 [DOI] [PubMed] [Google Scholar]
- 176.Zhao Z, Wang JC-Y, Zhang M, Lyktey NA, Jarrold MF, et al. 2021. Asymmetrizing an icosahedral virus capsid by hierarchical assembly of subunits with designed asymmetry. Nat. Commun 12:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhou M, Lantz C, Brown KA, Ge Y, Paša-Tolić L, et al. 2020. Higher-order structural characterisation of native proteins and complexes by top-down mass spectrometry. Chem. Sci 11(48):12918–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou M,Liu W,Shaw JB.2020.Charge movement and structural changes in the gas-phase unfolding of multimeric protein complexes captured by native top-down mass spectrometry. Anal. Chem 92(2):1788–95 [DOI] [PubMed] [Google Scholar]