Abstract
We studied the ancestry of virulence-associated genes in Escherichia coli by examining chromosomal regions specific to pathogenic isolates. The four virulence determinants examined were the alpha-hemolysin (hly) loci hlyI and hlyII, the type II capsule gene cluster kps, and the P (pap) and S (sfa) fimbria gene clusters. All four loci were shown previously to be associated with pathogenicity islands of uropathogenic E. coli isolates. The hly, kps, sfa, and pap regions each have an unexpected clustered distribution among the E. coli collection of reference (ECOR) strains, but all these regions were absent from a collection of diarrheagenic E. coli isolates. Strains in the ECOR subgroup B2 typically had a combination of at least three of the four loci, and all strains in subgroup D had a copy of the kps and pap clusters. In contrast, only four strains in subgroup A had either hly, kps, sfa, or pap, and no subgroup A strains had all four together. Strains of subgroup B1 were devoid of all four virulence regions, with the exception of one isolate that had a copy of the sfa gene cluster. This phylogenetic distribution of strain-specific sequences corresponds to the ECOR groups with the largest genome size, namely, B2 and D. We propose that the pathogenicity islands are ancestral to subgroups B2 and D and were acquired after speciation, with subsequent horizontal transfer into some group A, B1, and E lineages. These results suggest that the hly, kps, sfa, and pap pathogenicity determinants may play a role in the evolution of enteric bacteria quite apart from, and perhaps with precedence over, their ability to cause disease.
Escherichia coli is a genetically diverse species, the majority of isolates of which are commensal organisms of the intestinal tract. However, some isolates are opportunistic pathogens causing intestinal and extraintestinal infections in a range of hosts. For example, the enteropathogenic E. coli (EPEC) is the leading cause of severe infantile diarrhea in the developing world, and enterohemorrhagic E. coli (EHEC) O157:H7 has recently emerged as the cause of bloody diarrhea and hemolytic-uremic syndrome in major food-borne outbreaks in the United States, Europe, and Asia (33, 34).
Pathogenicity islands (PAIs) encompass large segments of sometimes unstable chromosomal DNA (5 to 200 kb) containing virulence gene clusters (11, 14, 19) that are often flanked by insertion sequence elements or tRNA genes (2, 6, 14). Five such islands have previously been identified in pathogenic E. coli isolates (10, 14). PAIs I, II, IV, and V carry a number of virulence gene clusters, among them the P (pap) and P-related (prf) fimbria gene clusters and two alpha-hemolysin loci, hlyI and hlyII (2, 3, 16, 21). The locus LEE (locus of enterocyte effacement) was identified in an EPEC strain (23); LEE carries a different set of virulence genes than those found in PAI I, II, IV, or V but is inserted in the same chromosomal site as PAI I, at 82 min at the selenocysteine-specific tRNA (2, 23).
We examined the occurrence, phylogenetic distribution, and genetic diversity of the four virulence determinants hly, kps, pap, and sfa among E. coli natural isolates. The purpose was to test the conventional view that chromosomal virulence determinants are temporary insertions into the bacterial chromosome which come and go as the lineage undergoes expansion and diversification (2, 3, 11, 14, 19). The hly locus contains the gene for alpha-hemolysin production, whereas the kps gene cluster contains genes for type II capsule (4, 8, 21, 26). The pap and sfa gene clusters encode P and S fimbriae that are associated mostly with uropathogenic E. coli strains (3, 9, 12, 13, 17). The distribution of the four virulence determinants was confined predominately to two lineages of the E. coli reference collection (ECOR) strains, subgroups B2 and D, with sporadic occurrences in subgroups A, B1, and E.
Does the observed phylogenetic clustering we have found reflect common descent of the pathogenicity determinants in the subgroup B2 and D lineages? If it does, then this would imply that these particular pathogenicity determinants have a long-term persistence in the genomes of these particular bacterial lineages. Did horizontal transfer of PAIs play an important role during the evolution of E. coli, and what role, if any, do PAIs play in genome size evolution? To address these issues we also analyzed nucleotide sequence variation at the three loci hlyA, kpsD, and papH to determine levels of genetic diversity compared with the housekeeping gene mdh for malate dehydrogenase. These data were analyzed for evidence of horizontal transfer among ECOR subgroups and long-term persistence within subgroups.
MATERIALS AND METHODS
Bacterial strains.
Two E. coli reference collections of natural isolates were examined: the ECOR collection and the diarrheagenic E. coli (DEC) collection (25, 34). The ECOR collection consists of 72 strains, 62 of which were recovered from healthy humans and animals, and 10 from women with urinary tract infections. The ECOR collection encompasses much of the total variation found within this species, and the major lineages are divided into five groups: A, B1, B2, D, and E. The DEC collection is made up of 15 strains recovered from patients infected with an organism from one of three enteric diseae categories: EPEC, EHEC, and enterotoxigenic E. coli (ETEC). Genomic DNA was extracted with the G-Nome DNA isolation kit from Bio 101 (Vista, Calif.).
PCR and DNA probe construction.
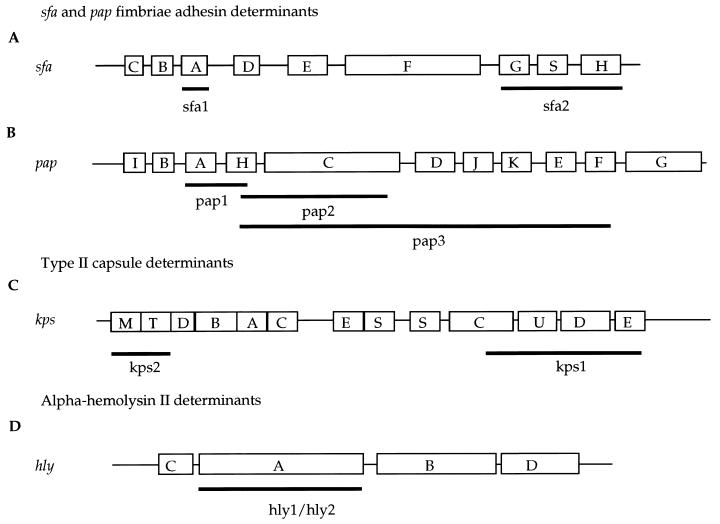
Nine sets of primers for amplification of sequences of the hly, kps, pap, and sfa gene clusters were used to obtain probes to screen strains for the presence of these virulence determinants (Table 1). In the 7.9-kb sfa operon there are nine genes, and two probes were designed for this region: sfa1 from the 5′ end of the gene cluster and sfa2 from the 3′ region (Table 1 and Fig. 1A). We applied long-range PCR (7) to obtain three probes, pap1, pap2, and pap3, from the 9-kb pap gene cluster (Table 1 and Fig. 1B). Probe kps1 represents kps region 1, and probe kps2 represents kps region 3 (Table 1 and Fig. 1C). Two alpha-hemolysin gene probes, hly1 and hly2, were made from a hemolysin gene cluster in an EHEC strain and a uropathogenic E. coli strain, J96, respectively (Table 1 and Fig. 1D). Following amplification, the PCR products were purified with the Qiaquick PCR purification kit (Qiagen, Inc., Chatsworth, Calif.). All probes were prepared with DNA from ECOR 52 as a template. The probes were labeled with fluorescein-conjugated nucleotides according to the manufacturer’s instructions (Amersham, Arlington Heights, Ill.).
TABLE 1.
Forward and reverse primers used in construction of probes
| Gene | Forward and reverse primer | PCR product size (bp) | Probe | Reference |
|---|---|---|---|---|
| sfaA1 | 5′ CGG TGT GCG TAG TTC AAT 3′ | 32 | ||
| sfaA2 | 5′ CAC CCG CAT GGA TAA AAA 3′ | 641 | sfa1 | 32 |
| sfaG1 | 5′ CAT TAA CTC CCG AAA CTT 3′ | 29 | ||
| sfaH2 | 5′ CAT TAC CGC CAC AAC TGC 3′ | 1,862 | sfa2 | 29 |
| papA1 | 5′ TTA AAG GTA ATC GGG TCA T 3′ | 22 | ||
| papA2 | 5′ GGA ATC AGA GAA AAG GTT 3′ | 856 | pap1 | 22 |
| papA1 | 5′ TTA AAG GTA ATC GGG TCA T 3′ | 22 | ||
| papC2 | 5′ CGC TTC AGG TCA ACA GAG G 3′ | 3,512 | pap2 | 22 |
| papH1 | 5′ AAT ACT GGG GAG AAG AGC AC 3′ | 22 | ||
| papF2 | 5′ ATT ACG AAA GGG CAC TGA AG 3′ | 6,086 | pap3 | 22 |
| kpsE1 | 5′ AAA AGA CCC GTG TAG AAG C 3′ | 26 | ||
| kpsC2 | 5′ CAA AAG GTC AGA GCC AAG T 3′ | 3,852 | kps1 | 26 |
| kpsD2 | 5′ CGTTACGGG AAT GCT GCT T 3′ | 26 | ||
| kpsM1 | 5′ GTC CAG AAA GTC ACC GTA GA 3′ | X53819a | ||
| kpsT2 | 5′ CAA TCG CCA CAT CAC AAA AC 3′ | 1,335 | kps2 | X53819a |
| hly1 | 5′ TCA GTC CTC TTT CTT TCC T 3′ | U12572a | ||
| hly2 | 5′ CTC TGG CAA CGG TCT CTC C 3′ | 2,899 | hly1 | U12572a |
| hlyA1 | 5′ GAC AAA GCA CGA AAG ATG 3′ | 8 | ||
| hlyA2 | 5′ CAA CTG CAA TAA AGA AGC 3′ | 2,930 | hly2 | 8 |
GenBank accession number.
FIG. 1.
Gene organization of kps, sfa, pap, and hly gene clusters. (A and B) Organization of pap and sfa gene clusters. Boxes indicate genes, and uppercase letters indicate gene designation(s). (C) The kps genes in region 1 (five genes [kpsSCUDE]) and region 3 (two genes [kpsMT]) are conserved among E. coli isolates that synthesize serologically distinct capsules. The number of genes in the central region 2 (kpsDBACES) reflects the size and complexity of the polysaccharide repeating unit. (D) The alpha-hemolysin determinant from E. coli J96 is represented; it consists of four genes. Black bars below gene clusters indicate the position of DNA fragments used as probes.
DNA hybridization.
Bacterial genomic DNA was digested with EcoRI, and the digests were separated by electrophoresis in 0.6% agarose gels in 1× Tris-borate-EDTA. DNA fragments were transferred by alkaline blotting to Hybond-N+ membranes (Amersham). Membranes were prehybridized for 30 min at 65°C in a solution of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, and 5% dextran sulfate. Hybridizations to labeled probes were carried out overnight at 65°C. Hybridized fragments were detected with the enhanced chemiluminescence system (Amersham).
Nucleotide sequencing.
PCR products from three genes, hlyA, kpsD, and papH, were sequenced with an Applied Biosystems model 373A automated DNA sequencing system with a DyeDeoxy terminator cycle sequencing kit. For all strains, both strands were sequenced. From eight ECOR isolates, representing three ECOR subgroups, A, B2, and D, 570 bp of the hlyA gene was sequenced. A 729-bp region of the kpsD gene was sequenced from each of nine ECOR strains from subgroups A, B2, and D. A 462-bp portion of the papH gene was sequenced from six ECOR isolates.
Statistical analysis.
To assay possible intragenic recombination events the Stephens test was used (31). The Stephens test identifies nonrandom clustering of polymorphic sites that may reflect the results of recombination events. The Stephens method examines the distribution of polymorphic sites relative to the phylogenetic partitions they support. Polymorphic sites supporting a particular phylogenetic partition are expected to be randomly distributed along the sequence if there is no recombination. Nucleotide sequences were analyzed by use of the computer program MEGA (20). Phylogenetic trees were constructed from synonymous site variation by the neighbor-joining method (28).
Nucleotide sequence accession numbers.
The nucleotide sequences of the genes described in this paper have been deposited in the GenBank database under the accession no. AF037572 to AF037588.
RESULTS
Distribution of virulence determinants associated with PAIs.
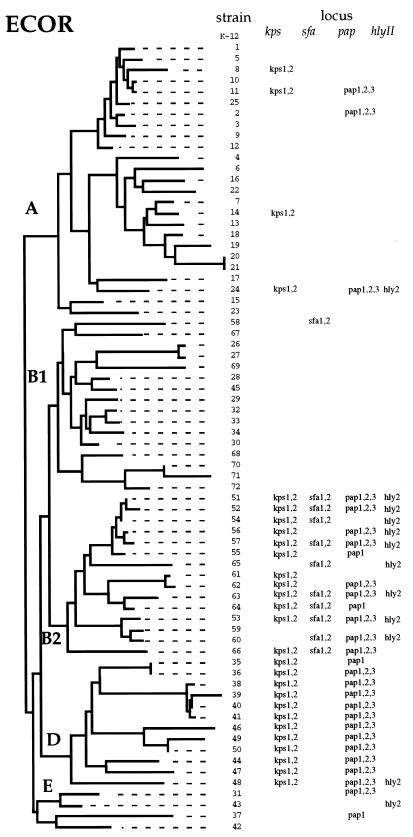
We determined the distribution of four virulence-associated regions in the 72 isolates of the ECOR reference collection and the 15 isolates of the DEC reference collection. The loci assayed were the two adhesin gene clusters sfa and pap; the kps capsule operon; and the alpha-hemolysin loci hlyI and hlyII. The distribution of the four virulence determinants was confined predominately to two lineages of the ECOR strains, subgroup B2 and D (Fig. 2).
FIG. 2.
Distribution of the kps, sfa, pap, and hly complexes among the ECOR collection of natural isolates. The tree is based on electrophoretic variation among 38 enzymes (15). Only for the pap operon did the three probes used give differing results.
The sfa locus was found almost exclusively in ECOR group B2 strains (10 of 15) and one isolate of subgroup B1 (ECOR 58); moreover, this was the only one of the assigned regions detected in subgroup B1. The pap gene cluster was detected in all subgroup D strains and most subgroup B2 strains (11 of 15), as well as in three group A and two group E isolates. In two subgroup B2 strains (ECOR 55 and ECOR 64), one subgroup D isolate (ECOR 35), and one subgroup E isolate (ECOR 37), the majority of the pap cluster was apparently deleted, as only the pap1 probe gave a positive hybridization signal. Four strains had multiple copies of sfa (ECOR 52, ECOR 54, ECOR 63, and ECOR 60) and papA (ECOR 11, ECOR 24, ECOR 49, and ECOR 50).
The kps operon was present in all subgroup D isolates, the majority of B2 isolates (12 of 15), and four isolates of group A (Fig. 2). The hly1 probe from an EHEC hly sequence did not hybridize with any of the ECOR strains; however, the hly2 probe from the uropathogenic E. coli strain J96 hybridized with ECOR subgroup B2 strains (9 of 16) and one strain each of subgroups A, D, and E.
Nucleotide sequence variation.
The single genes hlyA (hlyII), kpsD (kps), and papH (pap) from each of the three virulence gene clusters were sequenced from representative isolates of the ECOR collection.
Within the hlyA alleles from nine E. coli strains, there were only six polymorphic sites in the 570-bp segment sequenced. ECOR 24 and ECOR 43 had virtually identical sequences and are distinct at five of the six polymorphic sites detected in the other ECOR strains examined (Fig. 3). The hlyA locus has a GC content of 40%, which is atypical for genes of E. coli, and a codon adaptation index (CAI) of 0.228.
FIG. 3.
Distribution of polymorphic nucleotide and amino acid sites along the kspD, papH, and hlyA genes among ECOR isolates. Vertical numbers indicate the nucleotide position along the gene. Dots indicate nucleotide identity.
At the papH locus there were 22 polymorphic sites, which resulted in nine amino acid replacements, six of which were found in ECOR 49 (Fig. 4). The papH locus had a typical E. coli GC content of 50%.
FIG. 4.
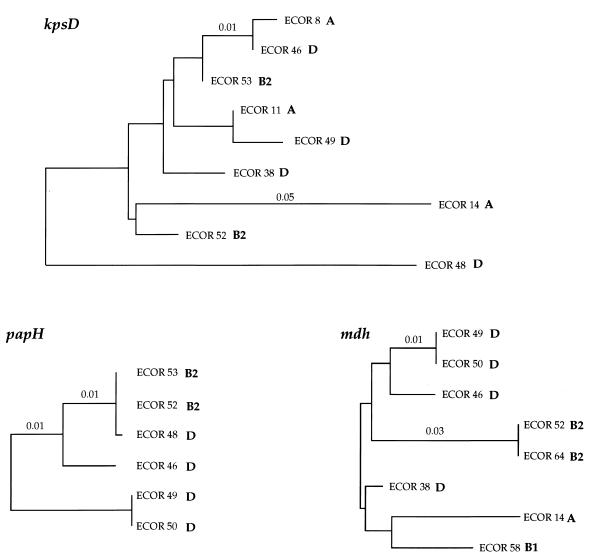
Evolutionary relationships among three chromosomal loci. Pairwise genetic distances were estimated from the numbers of substitutions (18, 24). Letters indicate subgroups of the ECOR strains, and numbers indicate genetic distance.
There were 48 polymorphic sites in the 729-bp region of kpsD sequenced in nine ECOR isolates. ECOR 14 and ECOR 48 were the most divergent in nucleotide sequence and accounted for much of the variation among the strains (Fig. 4). The GC content of kpsD is 52%, and this coding region has a CAI of 0.331 (30).
Rates of synonymous and nonsynonymous substitution.
For hlyA we estimated the numbers of synonymous (silent) substitutions per 100 synonymous sites (kS) and nonsynonymous (replacement) substitutions per 100 nonsynonymous sites (kN) (24). There were reduced levels of synonymous and nonsynonymous site variation at the hlyA locus relative to the gene mdh, coding for malate dehydrogenase; however, the kN/kS ratio was high because three of six polymorphic sites encoded amino acid replacements. The kN/kS ratio indicates the relative degree of functional constraints experienced by an evolving protein. Examination of the genes in Table 2 indicates that both mdh and kpsD have an increased level of functional constraint relative to hlyA and papH. At the kps and papH loci, the levels of variation at synonymous sites were similar to that of the housekeeping gene mdh (Table 2). However, at both of these loci there were elevated levels of nonsynonymous site variation; in the case of papH, most of the variation was accounted for by polymorphism contributed by the ECOR 49 papH sequence (Fig. 2).
TABLE 2.
Nucleotide sequence polymorphism among three virulence genes and one housekeeping gene from natural isolates of E. coli
| Gene | No. of isolates | Length (bp) | No. of polymorphic:
|
kS (102) | kN (102) | kN/kS ratio | CAI (30) | GC content | |
|---|---|---|---|---|---|---|---|---|---|
| Nucleotides | Amino acids | ||||||||
| hlyA | 8 | 570 | 6 | 3 | 0.89 ± 0.53 | 0.27 ± 0.16 | 0.33 | 0.23 | 40 |
| kpsD | 8 | 729 | 48 | 10 | 5.84 ± 0.99 | 0.52 ± 0.16 | 0.09 | 0.33 | 52 |
| papH | 6 | 462 | 22 | 9 | 5.29 ± 1.54 | 1.07 ± 0.35 | 0.21 | 0.24 | 50 |
| mdh | 8 | 864 | 20 | 2 | 3.10 ± 0.80 | 0.14 ± 0.10 | 0.05 | 0.58 | 52 |
Distribution of polymorphic nucleotide sites.
To test for nonrandom clustering of polymorphic nucleotide sites, a pattern that may be indicative of intragenic recombination, we used the Stephens test (31). Among the six papH sequences, there were 22 polymorphic sites in the 462 bp sequenced, and the Stephens test identified two statistically significant partitions. The first identified a 177-bp segment of invariant sites (P < 0.006) in ECOR 49, and the second identified a 156-bp region of invariant sites shared between ECOR 49 and ECOR 50.
For the 48 polymorphic sites among the nine kpsD sequences (shown in Fig. 3), six partitions were found for which there were statistically significant P values for either clustered polymorphic sites or segments composed of invariant sites. The first partition grouped ECOR 8 and ECOR 46 kpsD sequences and was based on only three polymorphic sites (sites 2, 9, and 25) and a 159-bp segment of consecutive nonpolymorphic sites (P < 0.006). The second partition identified a cluster of nine polymorphic sites (P < 0.002) in ECOR 14, eight of which were found at the 5′ end of the kpsD gene. The third partition involved only three clustered sites in ECOR 8 with a 324-bp segment of consecutive nonpolymorphic sites (P < 0.006). The fourth partition identified 15 clustered polymorphic sites (P < 0.000), the majority of which were in the 3′ end of the kps gene and separated ECOR 48 from the rest. Two sites grouped strains ECOR 14 and ECOR 49, and the probability of two polymorphic sites as close as or closer than 2 bp is 0.009. The final statistically significant kps partition identified grouped kps sequences from ECOR 11 and ECOR 49 and is based on a run of 275 bp of consecutive nonpolymorphic sites (P < 0.006).
Evolutionary trees for the kpsD and papH sequences.
For purposes of comparison, the housekeeping gene mdh from eight ECOR strains (5) analyzed at the kpsD and papH loci was used to construct an mdh gene tree (Fig. 4). Previous analysis has shown that the mdh gene tree is congruent with phylogenetic relationships based on multilocus enzyme electrophoresis and DNA hybridization analysis (5), thus providing a reliable measure of overall chromosomal relationships. No phylogenetic tree was constructed for hlyA, as there was not sufficient nucleotide sequence polymorphism to determine relationships among strains. Among all trees there are three isolates in common: ECOR 46, ECOR 49, and ECOR 52. Relationships among the ECOR strains were not congruent among the kpsD, papH, and mdh trees, particularly at the kps locus, at which phylogenetic comparisons with the mdh gene tree indicate several possible horizontal transfer events. The long branch length found in the ECOR 14 and ECOR 48 kpsD sequences can be accounted for by possible intragenic recombination events from an unknown source, identified by the Stephens test, involving the 5′ region in ECOR 14 and the 3′ region in ECOR 48.
DISCUSSION
Pathogenic isolates of E. coli are known to carry large chromosomal regions required for virulence, which are termed PAIs. Natural isolates are polymorphic for the presence and copy number of these DNA sequences. For example, the pathogenicity islands PAI I and PAI II of uropathogenic strains (2, 3), which are 70 and 190 kb in length, as well as the class II capsule synthesis operon (kps), are absent from E. coli K-12 (4). The prevailing view of E. coli as a relatively benign organism obtaining novel sequences from an outside source and thereby becoming pathogenic or switching disease syndrome or host has been supported by many authors (2, 3, 11, 14, 19). For example, Whittam and colleagues have proposed that the O157:H7 clone emerged when an EPEC O55:H7-like progenitor was lysogenized by a bacteriophage containing Shiga-like toxin genes (34).
Recently, Pupo and colleagues (27) have described the relationship between pathogenic and nonpathogenic isolates of E. coli. The pathogenic isolates included EPEC, EHEC, ETEC, enteroinvasive E. coli, and urinary tract infection strains, and the nonpathogenic isolates were represented by the ECOR strains. The authors concluded that pathogenic strains arising from E. coli do not have a single evolutionary origin within E. coli but have arisen many times, with the exception of Shigella and the EHEC clones. The Shigella isolates examined were all closely related to each other, clustering within ECOR subgroup A strains, confirming earlier conclusions that Shigella species are a clonal lineage of E. coli (35). Further, the results verified earlier findings of Whittam et al. (34) that strains causing the same disease do not form a monophyletic group. Wieler et al. (36) recently examined the insertion sites of the LEE locus in DEC strains and showed that the LEE insertion site differs in relation to the clonal lineage of the strains, which they conclude is due to multiple insertions during the evolution of these pathogens.
As shown by the phylogenetic distribution in Fig. 2, the majority of ECOR B2 and D isolates contain at least two of the four regions examined. Most isolates of subgroup B2 and all isolates of subgroup D have a copy of kps and pap; in addition, subgroup B2 strains have a copy of sfa. Among the other ECOR lineages there is only a sporadic occurrence of these sequences. In particular, among subgroup A strains (which is the largest set of lineages represented in the ECOR collection [25 of 72]), only five strains show hybridization to the probes for hly, kps, and pap, and only two strains (ECOR 11 and ECOR 24) contain sequences with homology to both the kps and pap regions.
The sporadic occurrence of hly, kps, pap, and sfa among ECOR group A isolates could have resulted from either widespread loss from strains within this group, which is unlikely, or transfer from perhaps a group B2 or D strain. The hlyA locus has been detected on an E. coli plasmid (8), which suggests a mechanism of horizontal transfer. Given the low GC content of the hlyA gene, 40% (compared to an average of 52% for most E. coli genes), and the limited nucleotide sequence polymorphism, this region may have been recently acquired from an unknown source. Alternatively, the highly conserved nucleotide sequence at hlyA may be due to selective constraints at the protein level. These conflicting views may be addressed with additional nucleotide sequence information from this gene cluster. Among the kpsD sequences examined, those of subgroup A strains ECOR 8 and ECOR 11 showed identity with those of subgroup B2, which is most easily explained by recent horizontal transfer between these subgroups. Marklund and coworkers (22) have proposed that E. coli acquired the pap locus after the speciation of E. coli and suggest that the different pap genes could have been acquired by horizontal gene transfer. Moreover, they proposed that the recent genetic exchanges involving the entire pilin gene clusters have occurred in response to selection pressures exerted by the host. Among the papH sequences studied, only that of ECOR 49 is radically different, and all of the amino acid substitutions are encoded in the first hundred bases of this gene; based on additional nucleotide sequence analysis, it appears that this diversity resulted from horizontal transfer at the papA locus, 63 bp upstream of papH, and that the variation in ECOR 49 is a result of hitchhiking (5a).
The stratification in the distribution of these virulence regions may be a consequence of the fact that the majority of the isolates of groups B2 and D were recovered from primates. For example, among isolates with sequences that show hybridization to the kps probe, all but two were isolated from primates; likewise all pap- and sfa-positive strains, with one exception, are all from primates. In contrast, ECOR isolates of subgroup B1 were predominantly isolated from nonprimate hosts (Table 3).
TABLE 3.
Sources of E. coli reference collection strainsa
| Host | Strains having indicated virulence-associated region
|
|||
|---|---|---|---|---|
| hlyII | kps | pap | sfa | |
| Human | 24, 43, 48, 51, 54, 56, 60, 63 | 8, 11, 14, 24, 35, 36, 38, 39, 40, 41, 47, 48, 49, 50, 51, 53, 55, 56, 61, 62, 63, 64 | 2, 11, 24, 35, 36, 38, 39, 40, 41, 48, 49, 50, 51, 53, 54, 55, 56, 59, 60, 62, 63, 64 | 51, 53, 54, 60, 63, 64 |
| Other primates | 52, 57 | 46, 52, 57, 66 | 37, 46, 52, 57 | 52, 57, 65, 66 |
| Nonprimate | 44, 47 | 31, 44 | 58 | |
Numbers are ECOR strain numbers (25).
Natural isolates of E. coli may vary in genome size by up to 1 Mb (1, 25a). The mechanism(s) of chromosome size variation in natural isolates of E. coli remains largely unknown. It may be relevant to these observations that isolates belonging to ECOR subgroup A have a significantly smaller genome size than those of either subgroup B2 or D, which have the largest genome size among natural isolates of E. coli (1). Comparisons of genome size with the presence of regions of interest in ECOR strains indicate that ECOR 14, which has a putative PAI present, has a genome size of 5,000 kb and differs in size by more than 300 kb from a closely related lineage, ECOR 13, where we found none of the four regions present. Isolates of group B2 and D (ECOR 51, 62, 63, 38, and 40) for which genome size is available range in size from 4,952 to 5,302 kb, and all isolates have two or more of the regions under study associated with them.
To determine the relationship between uropathogenic E. coli and DEC, isolates from the DEC collection were examined (34). The DEC collection comprises 15 clones representing E. coli isolates recovered from patients with enteric disease in one of three disease categories, EPEC, EHEC, and ETEC. Among the 15 DEC isolates examined, none had sequences which hybridized with the hlyII, pap, sfa, and kps probes. This result confirms earlier findings that DEC strains require a very different set of virulence determinants (the LEE locus) than those of uropathogenic E. coli isolates. The result also suggests that isolates may not typically carry PAIs for multiple disease categories. Further, our data support the view of long-term persistence of PAIs within E. coli lineages and indicate that whereas some pathogenicity determinants may be genetically labile, others are maintained for long periods of evolutionary time. Currently we are investigating whether hly, kps, pap, and sfa are confined to a single PAI. Future studies will include analysis of the insertion sites of putative PAIs in subgroup B2 and D strains.
ACKNOWLEDGMENT
This research was supported by grant GM322 from the National Institutes of Health.
REFERENCES
- 1.Bergthorsson U, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum G, Falbo V, Caprioli A, Hacker J. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. 1995;126:189–195. doi: 10.1111/j.1574-6968.1995.tb07415.x. [DOI] [PubMed] [Google Scholar]
- 4.Boulnois G J, Roberts I S. Genetics of capsular polysaccharide production in bacteria. Curr Top Microbiol Immunol. 1990;150:1–18. doi: 10.1007/978-3-642-74694-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Boyd E F, Nelson K, Wang F-S, Whittam T S, Selander R K. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Boyd, E. F., and D. L. Hartl. Unpublished data.
- 6.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 7.Cheng S, Fockler C, Barnes W M, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felmlee T, Pellett S, Welch R A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163:94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaastra W, Svennerholm A M. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4:444–452. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 10.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 11.Hacker J, Bender L, Ott M, Wiingender J, Lund B, Marre R, Goebel W. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- 12.Hacker J. Role of fimbrial in the pathogenesis of Escherichia coli infections. Can J Microbiol. 1992;38:720–727. doi: 10.1139/m92-118. [DOI] [PubMed] [Google Scholar]
- 13.Hacker J, Kestler H, Hoschützky H, Jann K, Lottspeich F, Korhonen T K. Cloning and characterization of the S fimbrial adhesin II complex of an Escherichia coli O18:K1 meningitis isolate. Infect Immun. 1993;61:544–550. doi: 10.1128/iai.61.2.544-550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker J, Oehler-Blum G, Mühlodorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function, and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 15.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hull S I, Hull R A, Minshew B H, Falkow S. Genetics of hemolysin of Escherichia coli. J Bacteriol. 1982;151:1006–1012. doi: 10.1128/jb.151.2.1006-1012.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hultgren S J, Jones C H, Normark S. Bacterial adhesins and their assembly. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2730–2756. [Google Scholar]
- 18.Jukes T H, Cantor C R. Evolution of protein molecules. New York, N.Y: Academic Press; 1969. [Google Scholar]
- 19.Knapp S, Hacker J, Jarchau T, Goebel W. Large, unstable inserts in the chromosome affect virulence properties of uropathogenic Escherichia coli O6 strain 536. J Bacteriol. 1986;168:22–30. doi: 10.1128/jb.168.1.22-30.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetics analysis, version 1. The Pennsylvania State University, University Park.
- 21.Low D, David V, Lark D, Schoolnik G, Falkow S. Gene clusters governing the production of hemolysin and mannose-resistant hemagglutination are closely linked in Escherichia coli serotype O4 and O6 isolates from urinary tract infections. Infect Immun. 1984;43:353–358. doi: 10.1128/iai.43.1.353-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marklund B I, Tennent J M, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 23.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 25.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Ochman, H. Personal communication.
- 26.Pazzani C, Rosenow C, Boulnois G J, Bronner D, Jann K, Roberts I S. Molecular analysis of region 1 of the Escherichia coli K5 antigen gene cluster: a region encoding proteins involved in cell surface expression of capsular polysaccharide. J Bacteriol. 1993;175:5978–5983. doi: 10.1128/jb.175.18.5978-5983.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Schmoll T, Hoschutzky H, Morschhauser J, Lottspeich F, Jann K, Hacker J. Analysis of genes coding for the sialic acid-binding adhesin and two other minor fimbrial subunits of the S-fimbrial adhesin determinant of Escherichia coli. Mol Microbiol. 1989;3:1735–1744. doi: 10.1111/j.1365-2958.1989.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 30.Sharp P, Li W H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens J C. Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol Biol Evol. 1985;2:539–556. doi: 10.1093/oxfordjournals.molbev.a040371. [DOI] [PubMed] [Google Scholar]
- 32.van Die I, Geffen B, Hoekstra W, Bergmans H E N. Type 1C fimbriae of a uropathogenic Escherichia coli strain: cloning and characterization of the gene involved in the expression of the 1C antigen and nucleotide sequence of the subunit gene. Gene. 1984;34:187–196. doi: 10.1016/0378-1119(85)90127-1. [DOI] [PubMed] [Google Scholar]
- 33.Whittam T S. Genetic variation and evolutionary processes in natural populations of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2708–2720. [Google Scholar]
- 34.Whittam T S, Wolfe M L, Wachsmuth I K, Ørskov F, Ørskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whittam T S, Ochman H, Selander R K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci USA. 1983;80:1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieler L H, McDaniel T K, Whittam T S, Kaper J B. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol Lett. 1997;156:49–53. doi: 10.1111/j.1574-6968.1997.tb12704.x. [DOI] [PubMed] [Google Scholar]