Understanding the pathophysiology of stress-related disorders, like post-traumatic stress disorder (PTSD), involves dissecting the molecular mechanisms regulated by genetic and environmental factors leading to individual differences in the traumatic stress response. Genome-wide association studies (GWASs) of PTSD have revealed heritability, genetic correlations with other psychiatric conditions such as major depressive disorder (MDD), and genomic risk loci enriched in (sub)cortical brain regions and their cell types (Fig. 1A) [1]. Crh17q21.31 is among the PTSD-GWAS loci, and its transcriptomic-based fine mapping has prioritized, among other genes, CRHR1 (corticotropin-releasing hormone receptor 1), a major regulator of the stress response and the resultant glucocorticoid (GC) secretion.
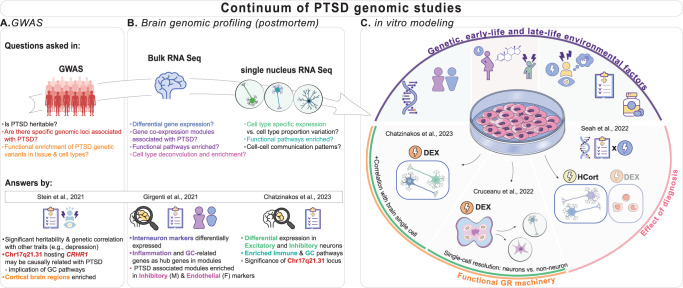
Fig. 1. Continuum of genomic studies of PTSD: from GWAS to brain and cell studies.
A, B With genome-wide association studies (GWASs; A) and postmortem brain genomic studies (B) providing new insights into PTSD, different aspects of PTSD molecular pathophysiology have been unraveled. Genetic and transcriptomic studies have complemented each other, consistently highlighting the involvement of the neuron-based glucocorticoid (GC) pathway in cortical regions. For instance, Stein et al. showed that the 17q21.31 chromosomal region of corticotropin-releasing hormone receptor 1 (CRHR1) gene could be causally implicated with PTSD. Girgenti et al. reported that interneuron markers and GC-related genes are key genes in cortical gene co-expression modules associated with PTSD. Chatzinakos et al. demonstrated that excitatory and inhibitory cortical neurons from individuals with PTSD show dysregulated GC pathways with top differentially expressed genes in 17q21.31 locus. C In vitro cell studies facilitate modeling of genetic and environmental contributions to PTSD pathogenesis. In this context, iPSC-derived cultures preserve the genetic background of the donor and allow studying the biological effects of environmental factors like stress/trauma exposure in utero, early or late life, primary diagnoses, comorbidities, and medications. The evidence of GC signaling dysregulation in PTSD motivated a series of in vitro studies focusing on modeling the effect of stress exposure mediated by GC signaling. These studies used two main GC agents: dexamethasone (DEX), a potent agonist of the glucocorticoid receptor (GR), and hydrocortisone (HCort), which is a less potent agonist of both the GR and mineralocorticoid receptor (MR). Chatzinakos et al. stimulated iPSC-derived mature neurons from neurotypical controls with DEX and showed that the transcriptomic signals correlate with postmortem PTSD brain neuronal transcriptomic signatures (from panel A), making DEX-exposed iPSC-derived neurons a good model for PTSD’s GC signaling, and a system that can differentiate PTSD from depression. Cruceanu et al. examined the effect of DEX exposure on brain organoids derived from neurotypical controls, and the differential response of the consisting cell types, from neural progenitors to mature neurons. Note both studies also demonstrated functional GR machinery. Finally, Seah et al. assessed gene-environment interactions in PTSD in iPSC-derived neurons and peripheral blood mononuclear cells (PBMC) from individuals with PTSD and neurotypical controls exposed to HCort and DEX, respectively. The transcriptomic responses to GCs were heightened in the cell lines derived from the individuals with PTSD, validating the involvement of a dysregulated GC system in PTSD.
Brain genomic profiling, e.g., a bulk transcriptomic study on postmortem cortical tissue [2], uncovered global and sex-specific molecular signatures of PTSD including differentially expressed interneuron markers (ELFN1) and inflammation- and GC-related genes as key drivers of disease-associated networks (Fig. 1B) with moderate to low overlap with MDD. However, cell-type specificity in these analyses relied on inference, underlining the need for single-cell genomic approaches. The first single-nucleus RNA-sequencing study in PTSD showed excitatory and inhibitory neurons’ transcriptomic changes in the dorsolateral prefrontal cortex associated with PTSD, involving dysregulation of immune, mitochondrial and GC-signaling pathways (Fig. 1B) [3]. Genes in the 17q21.31 locus (ARL17B, LINC02210-CRHR1, LRRC37A2), and other GC-regulated genes (SLC16A6, TAF1C) showed prominent cell-type-specific expression alterations. PTSD and MDD findings were more distinct at the cell type compared to the bulk-tissue level, while PTSD results showed the highest replication rate. While valuable for understanding PTSD’s molecular pathophysiology, these studies are limited to describing the “end-stage” of the disease as the temporal trajectories and relative contributions of genetic and environmental effects (stress exposures, clinical dimensions, comorbidities, medications) cannot be statistically deconvoluted.
IPSC-derived cells exposed to stress agents can model “stress in a dish”, capturing both genetic and environmental contributors of the maladaptive stress response (Fig. 1C). Activating the GC-receptor in iPSC-derived neurons using a specific GC-agonist (dexamethasone) induced transcriptomic changes that positively correlated with the neuronal PTSD-signatures and negatively with the MDD-signatures [3]. This showed the relevance of the in vitro stress paradigm for both diseases while confirming the directional differences in their GC regulation. In more complex cellular systems, e.g., iPSC-derived cerebral organoids, cell-type-specific responses to GCs have been also characterized. While transcriptomic changes were observed across cell types, only the neuronal signatures were relevant to neuropsychiatric risk, supporting the significant impact of GC-dysregulation on neuronal-related events [4]. Finally, the preserved PTSD-related genetic background of iPSC-derived lines (proxied by diagnosis) heightened the transcriptomic response to GCs and the alterations overlapped with developmental and adult psychiatric disorders (incl. PTSD)-associated genes [5].
The next steps to revolutionize PTSD genomics involve connecting PTSD-associated genetic and epigenetic factors with their functional consequences on cell-type-specific stress responses. High-throughput cell-type-specific approaches like “cell village” platforms [6], CRISPR-Cas9 screens and massive parallel reporter assays study the impact of disease-associated loci and regulatory regions under basal and stress conditions and, thus, bringing us closer to attributing inherited or acquired molecular signatures to a specific stress-related disease. Such platforms would validate putative causal brain quantitative trait loci and unravel the intricate stress-dependent molecular architecture underlying PTSD stress responses.
Author contributions
AI and NPD drafted and edited the hot topic commentary, prepared the figure, and approved of the final version.
Funding
This work was supported by NIMH (grants P50-MH115874, R01-MH117292, R01 R01-MH10659, and R21-MH121909). NPD was supported by 2015 and 2018 NARSAD Young Investigator grants from the Brain and Behavior Research Foundation, a Jonathan Edward Brooking mental health research fellowship from McLean Hospital, and a KL2 award from Harvard Catalyst/Harvard Clinical and Translational Science Center (NCATS KL2TR002542, UL1TR002541).
Competing interests
NPD has served on scientific advisory boards for BioVie Pharma, Circular Genomics and Sentio Solutions for unrelated work. All authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stein MB, Levey DF, Cheng Z, Wendt FR, Harrington K, Pathak GA, et al. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nat Genet. 2021;53:174–84. doi: 10.1038/s41588-020-00767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girgenti MJ, Wang J, Ji D, Cruz DA, Traumatic Stress Brain Research G. Stein MB, et al. Transcriptomic organization of the human brain in post-traumatic stress disorder. Nat Neurosci. 2021;24:24–33. doi: 10.1038/s41593-020-00748-7. [DOI] [PubMed] [Google Scholar]
- 3.Chatzinakos C, Pernia CD, Morrison FG, Iatrou A, McCullough KM, Schuler H, et al. Single-nucleus transcriptome profiling of dorsolateral prefrontal cortex: mechanistic roles for neuronal gene expression, including the 17q21.31 locus, in PTSD stress response. Am J Psychiatry. 2023. 10.1176/appi.ajp.20220478. [DOI] [PMC free article] [PubMed]
- 4.Cruceanu C, Dony L, Krontira AC, Fischer DS, Roeh S, Di Giaimo R, et al. Cell-type-specific impact of glucocorticoid receptor activation on the developing brain: a cerebral organoid study. Am J Psychiatry. 2022;179:375–87. doi: 10.1176/appi.ajp.2021.21010095. [DOI] [PubMed] [Google Scholar]
- 5.Seah C, Breen MS, Rusielewicz T, Bader HN, Xu C, Hunter CJ, et al. Modeling gene x environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression. Nat Neurosci. 2022;25:1434–45. doi: 10.1038/s41593-022-01161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells MF, Nemesh J, Ghosh S, Mitchell JM, Salick MR, Mello CJ, et al. Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages. Cell Stem Cell. 2023;30:312–32.e13. doi: 10.1016/j.stem.2023.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]