Abstract
Complex I (EC 1.6.99.3) of the bacterium Escherichia coli is considered to be the minimal form of the type I NADH dehydrogenase, the first enzyme complex in the respiratory chain. Because of its small size and relative simplicity, the E. coli enzyme has become a model used to identify and characterize the mechanism(s) by which cells regulate the synthesis and assembly of this large respiratory complex. To begin dissecting the processes by which E. coli cells regulate the expression of nuo and the assembly of complex I, we undertook a genetic analysis of the nuo locus, which encodes the 14 Nuo subunits comprising E. coli complex I. Here we present the results of studies, performed on an isogenic collection of nuo mutants, that focus on the physiological, biochemical, and molecular consequences caused by the lack of or defects in several Nuo subunits. In particular, we present evidence that NuoG, a peripheral subunit, is essential for complex I function and that it plays a role in the regulation of nuo expression and/or the assembly of complex I.
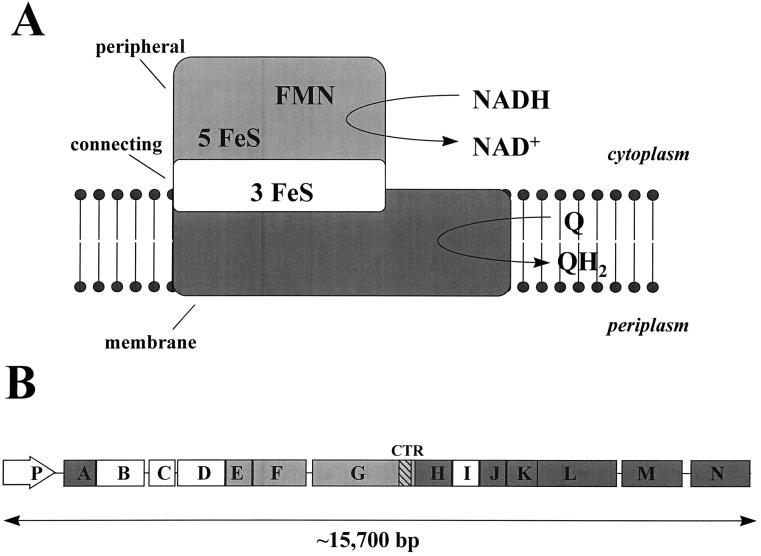
Complex I (NADH:ubiquinone oxidoreductase; EC 1.6.99.3), a type I NADH dehydrogenase that couples the oxidation of NADH to the generation of a proton motive force, is the first enzyme complex of the respiratory chain (2, 35, 47). The Escherichia coli enzyme, considered to be the minimal form of complex I, consists of 14 subunits instead of the 40 to 50 subunits associated with the homologous eukaryotic mitochondrial enzyme (17, 29, 30, 48–50). E. coli also possesses a second NADH dehydrogenase, NDH-II, which does not generate a proton motive force (31). E. coli complex I resembles eukaryotic complex I in many ways (16, 17, 30, 49): it performs the same enzymatic reaction and is sensitive to a number of the same inhibitors, it consists of subunits homologous to those found in all proton-translocating NADH:ubiquinone oxidoreductases studied thus far, it is comprised of a large number of subunits relative to the number that comprise other respiratory enzymes, and it contains flavin mononucleotide and FeS center prosthetic groups. Additionally, it possesses an L-shaped topology (14, 22) like that of its Neurospora crassa homolog (27), and it consists of distinct fragments or subcomplexes. Whereas eukaryotic complex I can be dissected into a peripheral arm and a membrane arm, the E. coli enzyme consists of three subcomplexes referred to as the peripheral, connecting, and membrane fragments (29) (Fig. 1A). The subunit composition of these three fragments correlates approximately with the organization of the 14 structural genes (nuoA to nuoN) (49) of the nuo (for NADH:ubiquinone oxidoreductase) locus (Fig. 1B), an organization that is conserved in several other bacteria, including Salmonella typhimurium (3), Paracoccus denitrificans (53), Rhodobacter capsulatus (12), and Thermus thermophilus (54). The 5′ half of the locus contains a promoter (nuoP), previously identified and located upstream of nuoA (8, 49), and the majority of genes that encode subunits homologous to the nucleus-encoded subunits of eukaryotic complex I and to subunits of the Alcaligenes eutrophus NAD-reducing hydrogenase (17, 29, 30, 49). In contrast, the 3′ half contains the majority of the genes that encode subunits homologous to the mitochondrion-encoded subunits of eukaryotic complex I and to subunits of the E. coli formate-hydrogen lyase complex (17, 29, 30, 49). Whereas the nuclear homologs NuoE, NuoF, and NuoG constitute the peripheral fragment (also referred to as the NADH dehydrogenase fragment [NDF]), the nuclear homologs NuoB, NuoC, NuoD, and NuoI constitute the connecting fragment. The mitochondrial homologs NuoA, NuoH, NuoJ, NuoK, NuoL, NuoM, and NuoN constitute the membrane fragment (29). E. coli complex I likely evolved by fusion of preexisting protein assemblies constituting modules for electron transfer and proton translocation (17–19, 30).
FIG. 1.
Schematic of E. coli complex I and the nuo locus. Adapted with permission of the publisher (17, 29, 30, 49). (A) E. coli complex I is comprised of three distinct fragments: the peripheral (light gray), connecting (white), and membrane (dark gray) fragments (17, 29). The peripheral fragment (NDF) is comprised of the nuclear homologs NuoE, -F, and -G and exhibits NADH dehydrogenase activity that oxidizes NADH to NAD+; the connecting fragment is comprised of the nuclear homologs NuoB, -C, -D, and -I; and the membrane fragment is comprised of the mitochondrial homologs NuoA, -H, and -J to -N and catalyzes ubiquinone (Q) to its reduced form (QH2). FMN, flavin mononucleotide. (B) The E. coli nuo locus encodes the 14 Nuo subunits that constitute complex I. The 5′ half of the locus contains a previously identified promoter (nuoP) and the majority of genes that encode the peripheral and connecting subunits (light gray and white, respectively). The 3′ half of the locus contains the majority of the genes encoding the membrane subunits (dark gray). The 3′ end of nuoG encodes a C-Terminal region (CTR) of the NuoG subunit (hatched).
Because of its smaller size and relative simplicity, researchers recently have begun to utilize complex I of E. coli, and that of its close relative S. typhimurium, to identify and characterize the mechanism(s) by which cells regulate the synthesis and assembly of this large respiratory complex (3, 8, 46) and to investigate the diverse physiological consequences caused by defects in this enzyme (4, 6, 10, 40, 59). Such defects affect the ability of cells to perform chemotaxis (40), to grow on certain carbon sources (4, 6, 10, 40, 57), to survive stationary phase (59), to perform energy-dependent proteolysis (4), to regulate the expression of at least one gene (32), and to maintain virulence (5).
To begin dissecting the processes by which E. coli cells regulate the expression of nuo and the assembly of complex I, we undertook a genetic analysis of the nuo locus. Here, we present the results of studies, performed on an isogenic collection of nuo mutants, that focus on the physiological, biochemical, and molecular consequences caused by the lack of or defects in several Nuo subunits. In particular, we present evidence that NuoG, a peripheral subunit, is essential for complex I function and that it plays a role in the regulation of nuo expression and/or the assembly of complex I.
MATERIALS AND METHODS
Chemicals.
Restriction enzymes were purchased from Promega Corporation (Madison, Wis.), Gibco-BRL Life Technologies (Gaithersburg, Md.), or New England BioLabs (Beverly, Mass.). Enzymes and substrates were obtained from Boehringer Mannheim (Indianapolis, Ind.) or Sigma Chemical Company (St. Louis, Mo.). Radiolabeled materials were purchased from Amersham (Arlington Heights, Ill.), and the bicinchoninic acid (BCA) protein assay reagent was obtained from Pierce Biochemicals (Rockford, Ill.).
DNA manipulations.
Plasmid preparations were performed by the alkaline lysis procedure (42), using the Promega Wizard Miniprep Purification System. Restriction enzyme digestions, ligations, and plasmid transformations were performed as described previously (42). Chromosomal DNA was prepared as described previously (36). Chromosomal transformations were performed with chromosomal DNA (∼0.1 to 1.0 μg) from nuo mutant cells, which was transformed into competent cells prepared as described previously (42).
Bacterial strains and mutant alleles.
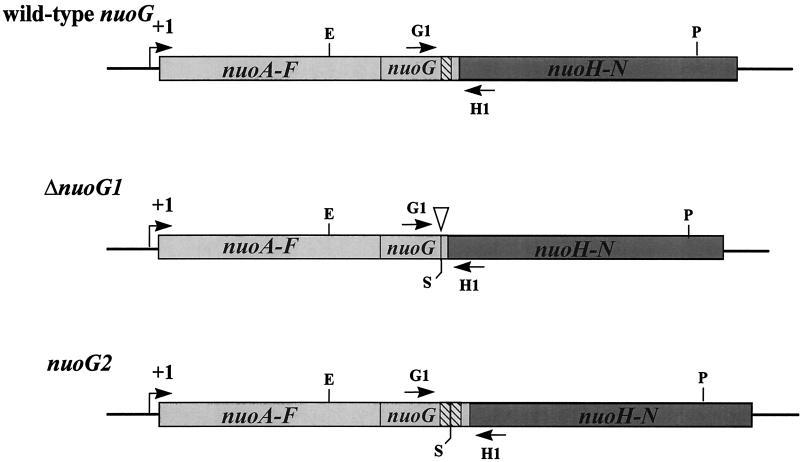
All strains are derivatives of E. coli K-12 and are listed in Table 1. The ΔnuoG1 and nuoG2 mutant alleles (Fig. 2) were constructed by first subcloning a 7.0-kb EcoRI-PstI nuo fragment from pAJW105ΔPstI, a plasmid that encompasses the 3′ end of nuoF to the 3′ end of nuoL, into the site-directed mutagenesis vector pALTER (Promega Corporation). Then, by site-directed mutagenesis, two unique SalI sites flanking the 3′ region of nuoG that encodes a C-terminal region (CTR) of the NuoG subunit were constructed. Next, these sites were used to construct one plasmid (pHF17) that harbored a 235-bp deletion of the CTR (allele ΔnuoG1) and one plasmid (pHF18) that harbored a 235-bp tandem duplication of the CTR (allele nuoG2). HindIII fragments from pHF17 and pHF18 were subsequently subcloned into the temperature-sensitive suicide vector pMAK705 (24) to yield plasmids pHF68 and pHF69, respectively. Finally, alleles ΔnuoG1 and nuoG2 were introduced into the chromosome by means of homologous recombination following transformation with (i) plasmids pHF17 and pHF18, respectively, into the Nuo+ PolA(Ts) host strain CP366 (23) or (ii) plasmids pHF68 and pHF69, respectively, into the closely related Nuo+ strain CP875 (24). The resultant recombinants were screened for the ΔnuoG1 deletion or the nuoG2 duplication by whole-cell PCR (41). One CP366 ΔnuoG1 recombinant (designated strain AJW931), one CP366 nuoG2 recombinant (AJW1470), one CP875 ΔnuoG1 recombinant (AJW1516), and one CP875 nuoG2 recombinant (AJW1517) were selected for further study. The nuo mutant strains CP910 (allele nuoG::Tn10-1) and CP938 [Δ(nuoF-L)-1] are derivatives of the wild-type strain CP875, while CP932 (nuoG::Tn10-1) is a derivative of the wild-type strain CP366 (40). To add to this isogenic set of nuo mutants, generalized transduction with the phage P1kc (44) or transformation with chromosomal DNA was used to transfer mutant nuo alleles from a variety of genetic backgrounds (7, 21, 59) into strain CP875. The location of the nuoB::Km mutation (strain AJW844) relative to that of mutations nuoB-C::Cm and nuoF::miniTn10Cm was verified genetically (97.4 and 70.8% linkages, respectively) and confirmed by Southern blot analysis (data not shown).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| CP366 | polA(Ts) thi-1 thr(Am)-1 leuB6 his-4 rpsL136a lacY1 xyl-5 ara-14 tonA31 tsx-78 rha zig::Tn10a Ace− | 23 |
| CP875 | thi-1 thr(Am)-1 leuB6 his-4 rpsL136 lacY1 ΔlacX74 λlacY | 40 |
| AJW844 | CP875 nuoB::Km | MCN021 × CP875→Kmr Nuo− |
| AJW853 | CP875 nuoB-C::Cmb | MWC230 × CP875→Cmr Nuo− |
| AJW851 | CP875 nuoF::miniTn10Cmc | ZK1363 × CP875→Cmr Nuo− |
| CP938 | CP875 Δ(nuoF-L)-1 | 40 |
| CP910 | CP875 nuoG::Tn10-1a | 40 |
| CP932 | CP366 nuoG::Tn10-1 | 40 |
| AJW931 | CP366 ΔnuoG1d | This study |
| AJW1516 | CP875 ΔnuoG1 | This study |
| AJW1470 | CP366 nuoG2e | This study |
| AJW1517 | CP875 nuoG2 | This study |
| AJW845 | CP875 nuoH::Km | ND1-KanR × CP875→Kmr Nuo− |
| AJW846 | CP875 nuoI::Km | MCN091 × CP875→Kmr Nuo− |
| AJW852 | CP875 nuoM::miniTn10Cmc | ZK1362 × CP875→Cmr Nuo− |
| AJW847 | CP875 nuoN::Km | ANN141 × CP875→Kmr Nuo− |
| AJW932 | CP366 nuoΩ(pHF17)f | This study |
| AJW1459 | AJW931 nuoΩ(pAJW105ΔPstI)g | This study |
| AJW1472 | AJW1470 nuoΩ(pAJW105ΔPstI) | This study |
| AJW1582 | CP875 ΔnuoG1 nuoH::Km | This study |
| AJW1583 | CP875 ΔnuoG1 nuoI::Km | This study |
| AJW1584 | CP875 nuoG2 nuoH::Km | This study |
| MCN021 | nuoB::Km | 7 |
| MWC230 | nuoB-C::Cm | R. Gennis |
| ZK1363 | nuoF::miniTn10Cm | 59 |
| ND1-KanR | nuoH::Km | 7 |
| MCN091 | nuoI::Km | 7 |
| ZK1362 | nuoM::miniTn10Cm | 59 |
| ANN141 | nuoN::Km | 7 |
nuoG::Tn10-1 and zig::Tn10 confer Tcr; rpsL136 confers Strr.
The Cm cassette is located in the intergenic region between nuoB and nuoC.
This mutation has been renamed to reflect its location within the nuo locus.
The ΔnuoG1 mutation is a 235-bp deletion of a 3′ region of nuoG.
The nuoG2 mutation is a 235-bp tandem duplication of a 3′ region of nuoG.
This integration results in a duplication of the region nuoF-L and introduces the ΔnuoG1 allele (see Fig. 3). pHF17 confers Apr.
This integration results in a duplication of the region nuoF-L and introduces the wild-type nuoG allele (see Fig. 3). pAJW105ΔPstI confers Apr.
FIG. 2.
Wild-type nuoG and mutant ΔnuoG1 and nuoG2 alleles. +1, nuo transcription initiation site (49). Restriction enzyme sites: E, EcoRI; P, PstI; S, SalI derived by site-directed mutagenesis to facilitate construction of mutant alleles. Hatched bars, CTR, a 235-bp 3′ region of nuoG present in the wild-type nuoG, deleted in the ΔnuoG1 allele, and duplicated in tandem in the nuoG2 allele. Inverted triangle, location of the CTR deletion. G1 and H1, flanking primers used to amplify the CTR.
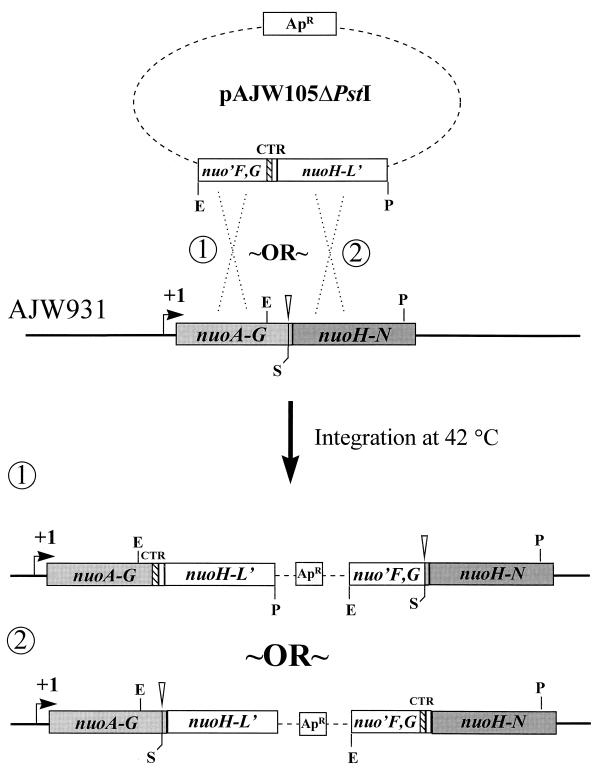
A strain carrying both the wild-type and ΔnuoG1 alleles on its chromosome was isolated following an integrative, homologous recombination event by the Campbell-type mechanism (58) between pAJW105ΔPstI, which carries the wild-type nuoG allele, and the ΔnuoG1 polA(Ts) strain AJW931. Following transformation and a shift in temperature from 32 to 42°C, a single, nonreciprocal, homologous recombination event between the cloned insert in pAJW105ΔPstI and the nuo locus in AJW931 (Fig. 3) resulted in a partial, nontandem duplication of the nuo locus at either end of the vector sequence (23, 38). The resultant strain was designated AJW1459. A similar strain (AJW932) was constructed when the plasmid pHF17, which carries the mutant ΔnuoG1 allele, was integrated into the Nuo+ PolA(Ts) strain CP366 in the same manner. Likewise, a strain (AJW1472) carrying both the wild-type nuoG and mutant nuoG2 alleles on its chromosome was constructed when the plasmid pAJW105ΔPstI was integrated into the nuoG2 polA(Ts) strain AJW1470. Maintenance of integration was verified following each experiment by confirming the vector-encoded ampicillin resistance of each strain at 42°C (23) and the concurrent presence of both the wild-type nuoG and mutant ΔnuoG1 or nuoG2 alleles within the chromosome by PCR.
FIG. 3.
Construction of strain AJW1459 by integrational, homologous recombination. White bars, vector-derived nuo sequence; gray bars, chromosome-derived nuo sequence; dotted lines, vector sequence; ApR, ampicillin resistance. Other designations are as described in the Fig. 2 legend. The plasmid pAJW105ΔPstI encompasses the 3′ end of nuoF to the 3′ end of nuoL and carries the wild-type nuoG allele. This plasmid was transformed into competent AJW931 [ΔnuoG1 polA(Ts)] cells. Cells in which the plasmid had integrated into the chromosome by homologous recombination were identified by selection of ampicillin resistance at the restrictive temperature, 42°C. Strain AJW932 was constructed similarly (not shown), except that the plasmid pHF17, which carries the ΔnuoG1 allele, was integrated into the Nuo+ PolA(Ts) strain CP366. Similarly, strain AJW1472 was constructed by integrating pAJW105ΔPstI into strain AJW1470 [nuoG2 polA(Ts)]. The simultaneous presence of both the wild-type and mutant nuoG alleles in each of these strains was verified by PCR analysis with primers G1 and H1.
Media and growth conditions.
Cells were grown with aeration in tryptone broth (TB) (1% [wt/vol] tryptone and 0.5% [wt/vol] sodium chloride) or in Luria-Bertani medium (TB and 0.5% [wt/vol] yeast extract) (34). When necessary, 100 μg of ampicillin per ml, 15 μg of tetracycline per ml, 34 μg of chloramphenicol per ml, or 40 μg of kanamycin per ml was added. Cells were grown at 32°C, unless otherwise stated.
To obtain growth curves, cells were grown in Luria-Bertani medium (supplemented with the appropriate antibiotics) to mid-exponential phase (optical density at 610 nm [OD610], 0.35 to 0.4), diluted 10−2 into fresh TB (without antibiotics), and aerated until they reached stationary phase.
To test for the ability to use acetate as the sole carbon source, cells were grown in TB to mid-exponential phase prior to harvesting and resuspension at 10−5 (volume of 100 μl). Fifty microliters of the diluted culture was spread on M63 minimal medium plates (40, 44) supplemented with 25 mM sodium acetate (pH 7.0). Each plate was incubated for 55.5 h.
When whole-cell lysates were required, cells were harvested by centrifugation at 3,500 × g for 10 min at 4°C, washed once with phosphate-buffered saline, and lysed by sonication. One hundred microliters of the whole-cell lysate was used either to determine the total protein concentration of each sample by the BCA reagent method with bovine serum albumin as a standard (Pierce) or to perform β-galactosidase assays.
Swarm assays.
Cells were aerated in TB (supplemented with appropriate antibiotics) to mid-exponential phase and inoculated at the center of TB swarm plates (0.25% agar) as described previously (40). Following incubation, the plates were examined for the presence of the inner, aspartate ring (51). The absence of this inner ring was taken as indicative of the Nuo− phenotype, i.e., the lack of functional complex I (40).
Reporter fusion construction and β-galactosidase assays.
A multicopy nuoPA′::lacZYA reporter fusion was constructed by subcloning a 443-bp fragment of plasmid pNUO2.3 (49) that encompasses the nuo promoter (nuoP) and the proximal third of the nuoA gene into pRS415, a transcriptional (operon) fusion vector (45). The resultant plasmid, pHF9, was transformed into both wild-type and nuo mutant cells, and the β-galactosidase activities of the transformants were quantified as a measure of nuo promoter activity (45). β-Galactosidase activity was determined according to the procedure of Miller (34) and Sigma Chemical Co., except that the cells were disrupted by sonication and centrifuged at 16,000 × g for 5 min, and the initial rate of reaction was measured. The protein concentration was determined by the BCA reagent method. β-Galactosidase activity was expressed in terms of the specific activity (units per milligram), where 1 U = 1 μmol of o-nitrophenol formed/min. Determinations of both protein concentration and β-galactosidase activity were performed with a DU640 spectrophotometer (Beckman, Fullerton, Calif.).
RNA extraction and dot blot analysis.
Total cellular RNA was extracted from cells grown in TB to mid-exponential phase by using RNeasy mini-prep kit columns (Qiagen, Santa Clarita, Calif.). RNA samples were treated with DNase I (RQ1 RNase-free DNase I; Promega) and repurified. RNA (0.5 μg) from each strain was directly transferred to multiple GeneScreen Plus nitrocellulose membranes (NEN Life Science Products, Boston, Mass.), using the Schleicher and Schuell (Keene, N.H.) dot blot apparatus, according to the manufacturers’ instructions. The resultant RNA dot blots were hybridized according to the instructions of the manufacturer (NEN Life Science Products) with DNA probes labeled with [α-32P]dCTP by using the RTS RadPrime DNA labeling system (Gibco BRL). Prehybridization was performed at 42°C for 2 h and was followed by hybridization for 20 h at 42°C. The membranes were washed as per the manufacturer’s instructions before autoradiography with XAR film (Kodak, New Haven, Conn.) at −70°C for 8 days.
SDS-PAGE and immunoblot analysis.
Total whole-cell lysate protein (100 μg) and/or purified complex I (a gift from T. Friedrich) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7.5% polyacrylamide gels) as described previously (28). The proteins were transferred electrophoretically overnight to 0.45-μm-pore-size nitrocellulose membranes by using a Trans-Blot Cell apparatus (Bio-Rad Laboratories, Richmond, Calif.). The membranes were blocked with 5% (wt/vol) nonfat dried milk in TBST (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, and 0.1% Tween 20). They were washed in TBST before being subjected to sequential incubation with rabbit anti-E. coli complex I polyclonal antibody 2409 (also a gift from T. Friedrich) for 2 h at room temperature and with goat anti-rabbit immunoglobulin G (heavy- and light-chain specific) alkaline phosphatase-conjugated antibody at appropriate dilutions for 2 h at room temperature. Color development was achieved with nitroblue tetrazolium and 5-bromo-4-chloro-3-indoylphosphate as described previously (25).
Biochemical techniques.
NADH oxidase activity was measured as described previously (15) by monitoring the reduction of ferricyanide by NADH in an assay mixture containing 50 mM Tris-HCl (pH 7.5), 0.1% Triton X-100, 1 mM potassium ferricyanide, and 0.1 mM NADH or 0.1 mM deamino-NADH (d-NADH). Electron paramagnetic resonance (EPR) spectroscopy was performed as described previously (29).
Computer analysis.
Protein sequence analysis was performed with the BestFit, Gap, Publish, and PeptideSort programs within the Wisconsin Package software (version 8.1) of the Genetics Computer Group (20). Autoradiographs of the RNA dot blots were scanned by using DeskScan II (26), and the images were compiled in PowerPoint (33).
RESULTS
Polarity of nuo mutants.
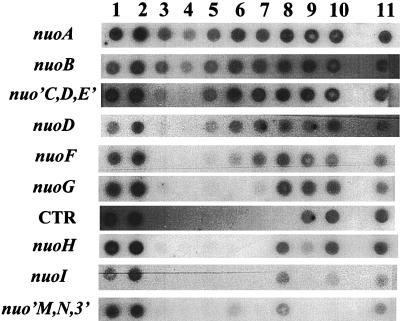
To verify the construction of each mutation and to determine whether that mutation exerts a polar effect upon transcription of downstream genes, we performed RNA dot blot analysis. From cells carrying either the wild-type nuo locus or the mutant allele nuoB::Km, nuoB-C::Cm, nuoF::miniTn10Cm, Δ(nuoF-L)-1, nuoG::Tn10-1, ΔnuoG1, nuoG2, nuoH::Km, nuoI::Km, nuoM::miniTn10Cm, or nuoN::Km, we purified total cellular RNA, transferred that RNA directly to multiple nitrocellulose membranes, and then hybridized each membrane with 1 of 10 nuo probes (Fig. 4). Each probe hybridized to RNA extracted from wild-type cells (Fig. 4, lanes 1 and 2). Similarly, all probes complementary to sequences located upstream of the reported location of each insertion hybridized to RNA extracted from mutant cells. In contrast, probes complementary to sequences downstream of each insertion hybridized poorly, if at all. For example, the upstream probes nuoA, nuoB, nuo′CDE′, nuoD, and nuoF, but not the downstream probes CTR, nuoH, nuoI, and nuo′MN3′, hybridized with RNA from the nuoG::Tn10-1 strain (Fig. 4, lane 7). The faint signal observed with the nuoG probe likely resulted from its hybridization with RNA encoded by the portion of nuoG located upstream of the insertion. We observed similar hybridization patterns with RNA extracted from every nuo mutant constructed by insertion (Fig. 4, lanes 4, 5, 9, 10, and 11), with one exception. RNA from the nuoB::Km mutant (Fig. 4, lane 3) hybridized with the downstream probe nuo′CDE′. In contrast, nuo mutants carrying the deletion allele Δ(nuoF-L)-1 (Fig. 4, lane 6) or ΔnuoG1 (lane 8) or nuo mutants carrying the tandem duplication nuoG2 (AJW1470 and AJW1517) (data not shown) exhibited different patterns. With these three mutations, probes complementary to sequences both upstream and downstream of the respective mutations hybridized. The CTR, nuoG, nuoH, and nuoI probes did not hybridize with RNA isolated from the Δ(nuoF-L)-1 strain (Fig. 4, lane 6), presumably because those genes had been deleted from the chromosome. Likewise, the CTR probe did not hybridize with RNA isolated from the ΔnuoG1 strain (Fig. 4, lane 8). On the basis of these observations, we conclude that each insertion used to construct a nuo mutant exerted a polar effect upon downstream transcription (with the exception of the nuoB::Km mutant). In contrast, the two deletions and the tandem duplication each exerted no polar effect.
FIG. 4.
Autoradiograph of RNA dot blots. RNA was purified from wild-type or nuo mutant cells, transferred directly to multiple nitrocellulose membrane, and probed with the nuo genes or fragments as indicated on the left. Lanes: 1, wild type (strain CP875); 2, wild type (CP366); 3, nuoB::Km mutant (AJW844); 4, nuoB-C::Cm mutant (AJW853); 5, nuoF::miniTn10Cm mutant (AJW851); 6, Δ(nuoF-L)-1 mutant (CP938); 7, nuoG::Tn10-1 mutant (CP910); 8, ΔnuoG1 mutant (AJW931); 9, nuoH::Km mutant (AJW845); 10, nuoI::Km mutant (AJW846); 11, nuoN::Km mutant (AJW847). The hybridization pattern for the nuoM::miniTn10Cm strain (AJW852) (data not shown) was identical to that for the nuoN::Km strain (AJW847) (lane 11). The hybridization pattern for AJW1516 was identical to that for AJW931 (data not shown).
Phenotypes of nuo mutants.
We characterized this isogenic collection of mutants by subjecting them to a series of tests, each shown previously to distinguish wild-type cells from those of nuo mutants.
First, we examined cells for their ability to form chemotactic rings on swarm plates (40). We inoculated cells that were either wild type or mutant for nuo at the center of TB swarm plates. We incubated those plates at 32°C until the outer, serine ring had reached the edge and then examined them for the presence or absence of the inner, aspartate ring. Wild-type cells formed both the serine and aspartate rings. In contrast, all nuo mutants, whether polar or nonpolar, failed to form the inner, aspartate ring (Table 2).
TABLE 2.
Summary of Nuo phenotypes
| Strain | nuo mutation(s) | Swarm plate resulta | TB growth curveb | Growth on M63 + 25 mM acetatec | NADH/d-NADH FeCN activitiesd | EPR spectroscopy resulte |
|---|---|---|---|---|---|---|
| CP875 | None (wild type) | + | + | + | + | + |
| CP366 | None (wild type) | + | + | NDf,g | + | + |
| AJW844 | nuoB::Km | − | − | − | −h | −h |
| AJW853 | nuoB-C::Cm | − | − | − | ND | ND |
| AJW851 | nuoF::miniTn10Cm | − | − | − | −h | ND |
| CP938 | Δ(nuoF-L)-1 | − | − | − | − | − |
| CP910 | nuoG::Tn10-1 | − | − | − | − | − |
| CP932 | nuoG::Tn10-1 | − | − | NDg | ND | ND |
| AJW931 | ΔnuoG1 | − | − | NDg | − | − |
| AJW1516 | ΔnuoG1 | − | − | − | ND | ND |
| AJW1470 | nuoG2 | − | − | NDg | ND | ND |
| AJW1517 | nuoG2 | − | − | − | ND | ND |
| AJW845 | nuoH::Km | − | − | − | −h | −h |
| AJW846 | nuoI::Km | − | − | − | −h | −h |
| AJW852 | nuoM::miniTn10Cm | − | − | − | ND | ND |
| AJW847 | nuoN::Km | − | − | − | −h | +h |
| AJW932 | CP366 nuoΩ(pHF17) | + | + | NDg | ND | ND |
| AJW1459 | AJW931 nuoΩ(pAJW105ΔPstI) | + | + | NDg | ND | ND |
| AJW1472 | AJW1470 nuoΩ(pAJW105ΔPstI) | + | + | NDg | ND | ND |
| AJW1582 | ΔnuoG1 nuoH::Km | − | − | − | ND | ND |
| AJW1583 | ΔnuoG1 nuoI::Km | − | − | − | ND | ND |
| AJW1584 | nuoG2 nuoH::Km | − | − | − | ND | ND |
+, presence of an inner, aspartate ring; −, absence of the inner ring.
+, no growth defect; −, growth defect.
+, good growth (≥1.0-mm colonies); −, poor growth (≤0.5-mm colonies).
+, wild-type levels of both activities in membrane fractions; −, less than 20% of wild-type activities.
+, at least three FeS centers detected; −, no detectable FeS centers.
ND, not done for this study.
The strain background was Ace− prior to introduction of the nuo mutation(s).
The mutation was analyzed in a different, but related, background.
Second, we examined the growth rates of wild-type and mutant cells in TB (40). We grew the cells with aeration in TB and monitored their growth as a function of OD. Prior to mid-exponential phase, the wild-type and mutant cells grew at similar rates. Whereas wild-type cells continued relatively rapid growth, at an OD610 of ca. 0.3 mutant cells suddenly reduced their growth rate. The mutant cells continued their slower growth as they entered stationary phase. Even after 24 h, the mutants’ ODs did not reach that of wild-type cells. Regardless of genetic background, all nuo mutants exhibited similar behavior (Table 2).
Third, to test the cells for their ability to use acetate as a sole carbon source (40), we inoculated M63 minimal plates supplemented with 25 mM acetate or 0.25% glucose with wild-type or mutant cells and incubated those plates at 32°C for 55.5 h before scoring colony size. Relative to wild-type cells, mutant cells formed very small colonies on M63 acetate plates (≤0.5 mm, compared to ≥1.0 mm for wild-type cells). In contrast, all cells formed colonies of equal size (∼1.0 mm) on M63 glucose plates. All nuo mutants tested exhibited similar behavior (Table 2).
Fourth, complex I activity was examined in selected mutants by measuring the NADH and d-NADH ferricyanide activities in their membrane fractions (15). For wild-type strains, the NADH ferricyanide activity was between 1.5 and 2.0 μM NADH/min · mg−1. In general, the d-NADH activity was 0.3 to 0.5 μM less than the NADH activity for wild-type strains. Although the exact values often differed from each other by a factor of two, all nuo mutants exhibited less than 20% of the corresponding wild-type activities (7) (Table 2). The activities were not completely abolished in the nuo mutants, because the second NADH dehydrogenase, NDH-II, also reacts with both substrates (16).
Finally, we used EPR spectroscopy analysis to detect the FeS centers of complex I. Leif et al. had previously identified, by EPR spectroscopy, two binuclear FeS clusters (N1b and N1c) and three tetranuclear FeS clusters (N2, N3, and N4) in isolated wild-type complex I (29). Similar EPR analyses revealed no detectable amounts of complex I or subcomplexes of complex I in the membrane fractions of any of the nuo mutants tested (Table 2). Specifically, the tetranuclear N2, N3, and N4 clusters were not detected in the membranes of nuo mutants, although they were readily detected in wild-type membranes (we did not test for the N1b and N1c clusters). Intriguingly, EPR analysis detected FeS centers (clusters N1b, N1c, N3, and N4) in the cytoplasm of the nuoN mutant (7). The presence of FeS centers in the cytoplasm of nuoN mutants was confirmed by the purification of the peripheral fragment (NDF) from those cells (7).
On the basis of these five phenotypic analyses, we conclude that all of the nuo mutants tested exhibit the pleiotropic Nuo− phenotype, demonstrating the lack of a functional complex I in these cells. Also, from the EPR spectroscopy analysis, it appears that the nuoN mutant possesses a cytoplasmic peripheral fragment (NDF).
Complementation of the ΔnuoG1 and nuoG2 alleles.
We tested for complementation in cis (13) with multiple, independent isolates of strains that carry both the wild-type nuoG and the mutant ΔnuoG1 alleles (strains AJW932 and AJW1459) or that carry both the wild-type nuoG and the mutant nuoG2 alleles (AJW1472) on their chromosomes by scoring each strain for its ability to produce the inner ring in swarm assays (Table 2). Multiple, independent isolates of each strain formed an inner, aspartate ring despite the fact that vector sequence interrupts the partially duplicated nuo locus in these strains (Fig. 3). Additionally, AJW932, AJW1459, and AJW1472 did not exhibit the TB growth defect (Table 2). Since these strains exhibited these two Nuo+ phenotypes, we conclude that the ΔnuoG1 and nuoG2 alleles can be complemented and that both alleles are recessive.
Translational analysis.
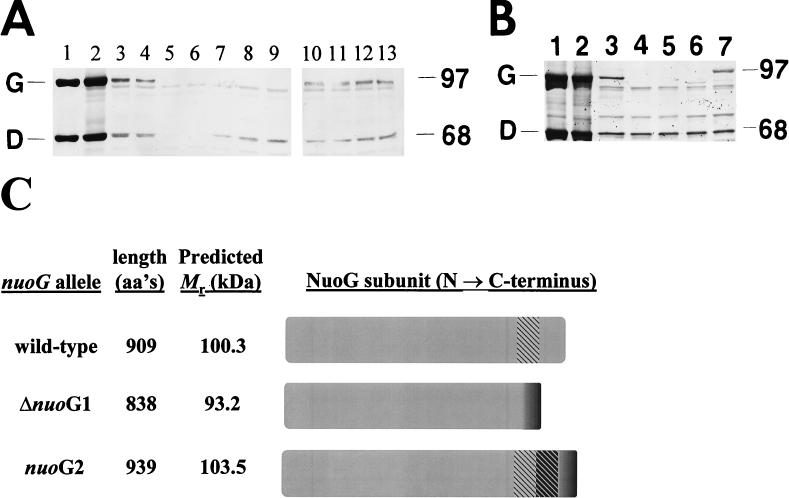
We examined the ability of wild-type and nuo mutant cells to synthesize the NuoCD and NuoG subunits, using a polyclonal antibody (no. 2409) directed against purified complex I. The NuoCD subunit has been identified as a fusion protein in E. coli (14), a finding consistent with that for the bacterium Buchnera aphidicola, which possesses a protein homologous to NuoC at its N terminus and homologous to NuoD at its C terminus (11). We identified the bands that correspond to NuoCD and NuoG by comparing the banding pattern of purified complex I (29) (Fig. 5A and B, lanes 1), that of a mixture of purified complex I and whole-cell lysate from wild-type cells (Fig. 5A and B, lanes 2), and those of whole-cell lysates from wild-type cells alone, strains CP366 (Fig. 5A and B, lanes 3) and CP875 (Fig. 5A, lane 4). We failed to detect NuoCD only in strains that carry an insertion mutation upstream of nuoD (Fig. 5A, lane 6), but we detected NuoCD in all other strains (Fig. 5A, lanes 3, 4, and 7 to 13, and B, lanes 3 to 7); we observed barely detectable levels of NuoCD in the nuoB::Km mutant (Fig. 5A, lane 5). Similarly, we failed to detect NuoG in strains that either lack nuoG or harbor an insertion mutation upstream of or within nuoG (Fig. 5A, lanes 5 to 9, and B, lanes 4 and 5), but we detected NuoG in all other strains (Fig. 5A, lanes 3, 4, and 10 to 13, and B, lane 3). Cells that carry the allele ΔnuoG1 (Fig. 5B, lane 6) or nuoG2 (Fig. 5B, lane 7) synthesized proteins that exhibited faster and slower mobilities, respectively, than the wild-type NuoG. The apparent molecular mass of each variant roughly corresponded to the predicted product of its respective allele (20) (Fig. 5C). In addition, the steady-state level of the smaller variant produced by ΔnuoG1 cells (Fig. 5B, lane 6) seemed to be significantly less than that of the full-length NuoG protein produced by the wild-type cells (Fig. 5B, lane 3). In contrast, the amounts of NuoCD protein produced by the two strains seemed to be roughly equivalent. On the basis of these observations, we conclude that the ΔnuoG1 and nuoG2 alleles result in the synthesis of altered forms of the NuoG subunit. Because NuoG is subject to proteolytic digestion in disrupted cells (14), it is likely that the truncated NuoG variant expressed by ΔnuoG1 cells is less abundant due to increased sensitivity to such digestion.
FIG. 5.
(A and B) Immunoblot analysis with E. coli anti-complex I polyclonal antibody following SDS-PAGE (7.5% polyacrylamide) and transfer to nitrocellulose. Sizes of molecular mass markers (in kilodaltons) are indicated on the right; the NuoG (G) and NuoCD (D) subunits are identified on the left. (A) Analysis of whole-cell lysates prepared from wild-type or nuo polar mutant cells. Lanes: 1, purified complex I; 2, purified complex I mixed with wild-type lysate (strain CP366); 3, wild type (CP366); 4, wild type (CP875); 5, nuoB::Km mutant (AJW844); 6, nuoB-C::Cm mutant (AJW853); 7, nuoF::miniTn10Cm mutant (AJW851); 8, Δ(nuoF-L)-1 mutant (CP938); 9, nuoG::Tn10-1 mutant (CP910); 10, nuoH::Km mutant (AJW845); 11, nuoI::Km mutant (AJW846); 12, nuoM::miniTn10Cm mutant (AJW852); 13, nuoN::Km mutant (AJW847). Cells were grown with aeration in TB at 32°C until cultures reached mid-exponential phase (OD610, 0.35 to 0.4). One hundred micrograms of whole-cell lysate was loaded in each lane. (B) Analysis of whole-cell lysates prepared from wild-type or nuoG mutant cells. Lanes: 1, purified complex I; 2, purified complex I mixed with wild-type whole-cell lysate (CP366); 3, wild type (CP366); 4, Δ(nuoF-L)-1 mutant (CP938); 5, nuoG::Tn10-1 mutant (CP910); 6, ΔnuoG1 mutant (AJW931); 7, nuoG2 mutant (AJW1470). Cells were grown as described for panel A. One hundred micrograms of whole-cell lysate was loaded in each lane. The profile for AJW1516 was identical to that for AJW931, and the profile for AJW1517 was identical to that for AJW1470 (data not shown). (C) Putative NuoG peptides. The lengths and molecular masses (Mr) of translation products of the nuoG alleles as predicted by PeptideSort (20) are shown. aa, amino acid. The CTR of NuoG is hatched in gray. The deletion in ΔnuoG1 shifts the remainder of the NuoG C terminus out of frame (dark gray). The duplication in nuoG2 shifts the duplicated CTR (hatched in black) and the remainder of the NuoG C terminus (dark gray) out of frame.
Effect of nuo mutations on nuo promoter activity.
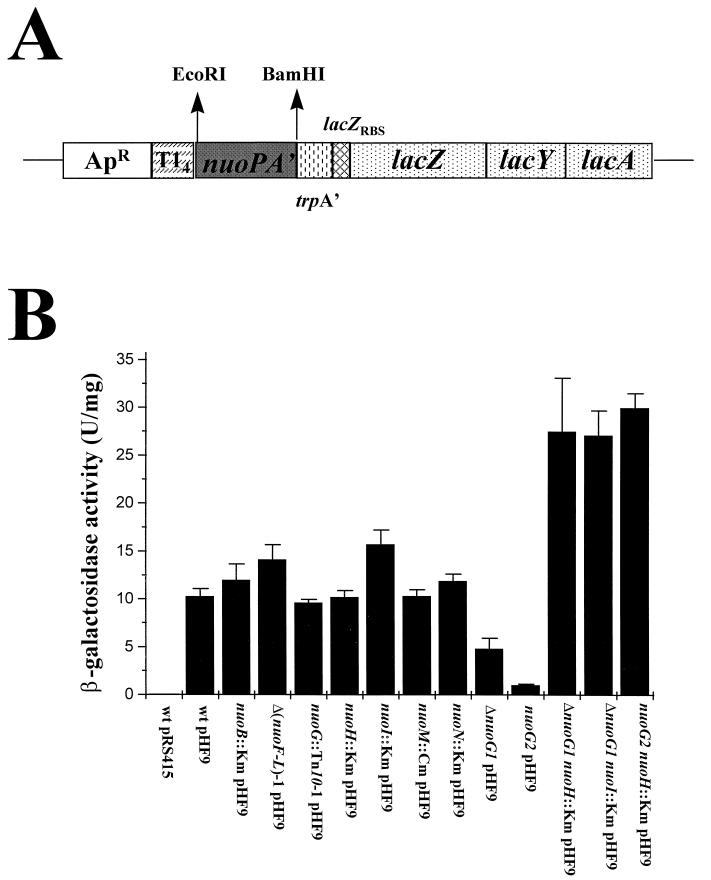
To examine the effect that nuo mutations exert upon nuo promoter activity, we monitored β-galactosidase activity from a nuoPA′::lacZYA transcriptional (operon) fusion. We transformed cells that carry the wild-type nuo locus or the mutant allele nuoB::Km, Δ(nuoF-L)-1, nuoG::Tn10-1, ΔnuoG1, nuoG2, nuoH::Km, nuoI::Km, nuoM::Km, nuoN::Km, ΔnuoG1 nuoH::Km (AJW1582), ΔnuoG1 nuoI::Km (AJW1583), or nuoG2 nuoH::Km (AJW1584) with the multicopy nuoPA′::lacZYA reporter fusion plasmid, pHF9 (Fig. 6A), or with its parental vector, pRS415. We grew the resultant transformants in TB at 32°C, harvested and lysed them as the cultures reached mid-exponential phase, and then measured their β-galactosidase activities (Fig. 6B). Wild-type cells transformed with the vector control pRS415 displayed almost no β-galactosidase activity (specific activity, 0.02 ± ≤0.01). Relative to pHF9 transformants of wild-type cells (specific activity, 10.29 ± 0.76), those of the nonpolar ΔnuoG1 and nuoG2 mutants exhibited reduced β-galactosidase activity (specific activity, 4.83 ± 1.06 and 1.04 ± 0.06, respectively). In contrast, pHF9 transformants of the ΔnuoG1 nuoH::Km, ΔnuoG1 nuoI::Km, and nuoG2 nuoH::Km double mutant strains exhibited significantly higher activities (specific activity, 27.5 ± 5.60, 27.1 ± 2.55, and 30.0 ± 1.49, respectively) than did the respective ΔnuoG1, nuoG2, and wild-type transformants. This increase in activity cannot be due to the presence of the nuoH::Km or nuoI::Km mutation alone, since transformants of those single mutants exhibited activities similar to that of the wild-type transformants (specific activity, 10.20 ± 0.68 and 15.74 ± 1.47, respectively). In fact, transformants of all of the single mutants tested (except for the ΔnuoG1 and nuoG2 mutants) displayed activities similar to that of the wild-type transformant, including the nuoG::Tn10-1 transformant (specific activity, 9.60 ± 0.36). On the basis of these data, we propose that NuoG participates in the regulation of nuo transcription and therefore possibly plays a role in the successful assembly of complex I.
FIG. 6.
Effect of nuo mutations on nuo promoter activity. (A) The multicopy nuoPA′::lacZYA transcriptional (operon) reporter fusion, pHF9, constructed from pRS415 (45). The 443-bp nuo insert consists of the nuo promoter (49) and the proximal third of nuoA. ApR, ampicillin resistance; T14, transcriptional terminator; trpA′, trp operon sequence; lacZYA, lac operon including its translational machinery but missing the lac promoter. (B) β-Galactosidase activity assay. Cells were grown in TB at 32°C to mid-exponential phase (OD610, 0.35 to 0.4), harvested, lysed by sonication, and centrifuged to separate the cytoplasmic fraction from the membrane fraction. The β-galactosidase activity of the cytoplasmic fractions was quantified as units per milligram of protein, where 1 U = 1 μmol of o-nitrophenol formed/min. Results are from at least six independent experiments. Error bars indicate standard errors of the mean. Strains: wt, CP875, nuoB::Km, AJW844; Δ(nuoF-L)-1, CP938; nuoG::Tn10-1, CP910; nuoH::Km, AJW845; nuoI::Km, AJW846; nuoM::miniTn10Cm, AJW852; nuoN::Km, AJW847; ΔnuoG1, AJW1516; nuoG2, AJW1517; ΔnuoG1 nuoH::Km, AJW1582; ΔnuoG1 nuoI::Km, AJW1583; nuoG2 nuoH::Km, AJW1584.
DISCUSSION
We performed a genetic analysis of nuo, the E. coli locus that encodes the proton-translocating NADH dehydrogenase, complex I. Examining physiological, biochemical, and molecular properties of mutants defective in 9 of the 14 nuo genes, we demonstrated that a mutation in any one of the nuo genes tested causes a complex I deficiency as measured by the inability of the mutant cells (i) to form a inner, aspartate ring on chemotaxis swarm plates, (ii) to grow rapidly in TB beyond mid-exponential phase, (iii) to use acetate effectively as a sole carbon source, (iv) to exhibit membrane-associated NADH/ d-NADH FeCN activities, and (v) to detect membrane-associated FeS centers. Since each nuo mutant exhibited all five Nuo− phenotypes, we used the swarm assay as a simple and reliable Nuo− screen. The mechanism for this phenotypic defect remains unclear; however, the absence of the inner ring on swarm plates may result from the cells’ inability to respire aerobically: wild-type cells grown anaerobically in the presence or absence of an electron acceptor also do not form the inner ring (31a).
All of the nuo insertion mutations tested (nuoB-C::Cm, nuoF::miniTn10Cm, nuoG::Tn10-1, nuoH::Km, nuoI::Km, nuoM::miniTn10Cm, and nuoN::Km) prevented transcription of downstream genes, as judged by RNA dot blot analyses, with the exception of the nuoB::Km mutation. The nuoB::Km mutant synthesized barely detectable levels of the NuoCD subunit but did not synthesize any NuoG subunit, as demonstrated by immunoblot analysis. We do not understand the apparent incomplete polarity exhibited by the mutation. These analyses also revealed that both deletion mutations [Δ(nuoF-L)-1 and ΔnuoG1] and the duplication mutation (nuoG2) do not exert polar effects upon the transcription of downstream genes. In addition, complementation analyses showed that both the ΔnuoG1 and nuoG2 mutant alleles are recessive: strains that carry both the wild-type nuoG allele and either of these mutant alleles on their chromosomes formed inner rings on swarm plates and did not exhibit the TB growth defect. Since these strains exhibited these wild-type behaviors, we conclude that the two halves of the nuo locus can be regulated independently despite their separation by vector sequence. Expression of the downstream half of the locus in these strains may result from a promoter located within the vector.
Since all 14 genes appear to be transcribed by ΔnuoG1 and nuoG2 cells, we assume that these mutant cells synthesize all 14 Nuo subunits, including their respective mutant variants of NuoG. We base this assumption, in part, on our ability to detect by immunoblot analysis both wild-type NuoCD and altered NuoG subunits. Thus, ΔnuoG1 and nuoG2 represent the first recessive, nonpolar mutations located within a single nuo gene.
By analyzing selected members of our mutant collection, we demonstrated that the anti-complex I antibody 2409 recognizes both the NuoCD and NuoG subunits. As predicted, the ΔnuoG1 and nuoG2 mutants synthesized smaller and larger variants of the NuoG subunit, respectively. Since these cells presumably synthesized wild-type versions of all the other Nuo subunits, we conclude that functional complex I requires NuoG. EPR spectroscopy detected cytoplasmic FeS centers in the nuoN mutant. The detection of these centers in the cytoplasm instead of associated with the membrane supports the hypothesis that NuoE, NuoF, and NuoG subunits can form the peripheral NDF in the absence of the membrane fragment. Furthermore, Braun and colleagues (9) have demonstrated recently that the NDF is properly assembled in E. coli in the absence of the membrane fragment as long as some subunits of the connecting fragment are present.
On the basis of the following evidence, we hypothesize that NuoG and at least one other downstream subunit affect nuo promoter activity. First, of all the mutants tested, only those carrying the nonpolar mutation ΔnuoG1 or nuoG2 exhibited promoter activity markedly reduced from that exhibited by wild-type cells when transformed with the multicopy nuoPA′::lacZYA operon fusion. This effect was not observed in transformants carrying any of the polar mutations or the large deletion mutation, e.g., nuoG::Tn10 or Δ(nuoF-L)-1 transformants, suggesting that it is not the lack a functional NuoG alone that causes an inhibitory effect at the nuo promoter. Second, the inhibitory effect we observed in the ΔnuoG1 or nuoG2 transformants was alleviated by the additional presence of the polar nuoH::Km or nuoI::Km mutation; in fact, the addition of either of these mutations resulted in a significant increase in promoter activity. This increase cannot be due solely to the presence of either the nuoH::Km or nuoI::Km mutation, since each of those single mutants displayed β-galactosidase activities similar to those of wild-type transformants. These data provide evidence for some kind of feedback regulatory mechanism dependent on both NuoG and at least one downstream subunit (i.e., NuoH to NuoN inclusive). However, we do not know whether the effect of NuoG and the downstream subunit(s) on the promoter is direct.
We have demonstrated here that the C-terminal defects caused by deletion or duplication of the CTR in the NuoG subunit prevent complex I from functioning properly. A role in complex stability and cofactor incorporation has been described for the NuoG homolog in P. denitrificans, NQO3 (55). When the P. denitrificans NuoE and NuoF homologs, NQO2 and NQO1, respectively, were coexpressed in trans, the two subunits formed a subcomplex with 1:1 stoichiometry containing one binuclear [2Fe-2S] cluster. The flavin mononucleotide and the tetranuclear [4Fe-4S] cluster, however, were not incorporated into the subcomplex in situ (although the prosthetic groups could be partially reconstituted in vitro) (55). The authors suggested that interaction with neighboring subunits, such as NQO3, may be required for proper cofactor incorporation and to form a more stable subcomplex. NuoG and its P. denitrificans homolog NQO3 are 24% identical, and both contain at least one binuclear and one tetranuclear FeS cluster, each of which serves as a cofactor (29, 49, 52, 56). The NuoG CTR does not contain any cofactors; however, the deletion or duplication of the CTR may result in improper folding of some other portion(s) of NuoG, thus preventing incorporation of cofactors. In turn, the altered NuoG subunits may not incorporate properly into the NDF or, if incorporated, may prevent the further assembly of other subunits. Another study, using cytoskeletal preparations from bovine cardiac muscle, proposes that the bovine homolog to NuoG, the 75(IP) subunit (originally called the mitoskelin protein), may serve as a structural linker between the surfaces of the mitochondria and the cytoskeleton (39). The authors described the subunit’s ability to self-polymerize into 10-nm-wide filaments in vitro, suggesting that it may function as a cytoskeletal protein. If a similar function exists for the NuoG subunit in E. coli, then it is possible that the CTR deletion or duplication affects its ability to self-assemble or assemble with other Nuo subunits, such as the membrane subunits.
The organization of the nuo locus, the detection of a subassembled NDF in the nuoN mutant (7), the observation that the NDF is properly assembled in the absence of the membrane fragment (9), and the evolution of complex I (17–19, 30) are consistent with the hypothesis that E. coli complex I assembly proceeds first by construction of independently assembled subcomplexes (17, 30), as is the case for N. crassa complex I (15, 43). Perhaps, throughout the evolution of complex I, regulatory components of each protein assembly remained within the nuo locus to coordinate regulation of its two halves, ensuring proper assembly of a complete and functional complex I. On the basis of our findings, we hypothesize that E. coli cells can sense whether all of the Nuo subunits have been synthesized and assembled. If they do not, cells are able to utilize some form of feedback mechanism to regulate the expression of nuo. Similar mechanisms are used by a number of other large protein complexes, e.g., the flagellar apparatus (1) and ribosomes (37), to regulate their expression and assembly.
ACKNOWLEDGMENTS
We thank T. Friedrich for graciously performing NADH/d-NADH ferricyanide activity assays and EPR spectroscopy analyses, for purified complex I and anti-complex I antibody, for providing data prior to publication, and for critical reading of the manuscript. We thank R. Gennis for strains and critical reading of the manuscript and R. Kolter for strains. Finally, we thank J. Nelms, S. Kumari, D. Ellefson, B. McNamara, and C. Beatty for valuable discussions.
This work was supported in part by Public Health Service grant GM46221 from the National Institute of General Medical Sciences. H. Falk-Krzesinski was supported in part by the Eloise Gerry Fellowship Fund of Sigma Delta Epsilon/Graduate Women in Science, Inc.
REFERENCES
- 1.Aizawa S. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 2.Anraku Y, Gennis R B. The aerobic respiratory chain of Escherichia coli. Trends Biochem Sci. 1987;12:262–266. [Google Scholar]
- 3.Archer C D, Elliott T. Transcriptional control of the nuo operon which encodes the energy conserving NADH dehydrogenase of Salmonella typhimurium. J Bacteriol. 1995;177:2335–2342. doi: 10.1128/jb.177.9.2335-2342.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer C D, Wang X, Elliott T. Mutants defective in the energy-conserving NADH dehydrogenase of Salmonella typhimurium identified by a decrease in energy-dependent proteolysis after carbon starvation. Proc Natl Acad Sci USA. 1993;90:9877–9881. doi: 10.1073/pnas.90.21.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrow, P. A. 1996. Personal communication.
- 6.Becker S, Holighaus G, Gabrielczyk T, Unden G. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J Bacteriol. 1996;178:4515–4521. doi: 10.1128/jb.178.15.4515-4521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, A., V. Spehr, and T. Friedrich. 1997. Unpublished data.
- 8.Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-nuoN) of Escherichia coli by electron acceptors, electron donors, and gene regulators. Mol Microbiol. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 9.Braun, M., S. Bungert, and T. Friedrich. Characterization of the overproduced NADH dehydrogenase fragment of the NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochemistry, in press. [DOI] [PubMed]
- 10.Calhoun M, Gennis R B. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J Bacteriol. 1993;175:3013–3019. doi: 10.1128/jb.175.10.3013-3019.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark M A, Baumann L, Baumann P. Buchnera aphidicola (endosymbiont of aphids) contains nuoC(D) genes that encode subunits of NADH dehydrogenase. Curr Microbiol. 1997;35:122–123. [PubMed] [Google Scholar]
- 12.Dupuis A, Peinnequin A, Chevallet M, Lunardi J, Darrouzet E, Pierrard B, Procaccio V, Issartel J. Identification of five Rhodobacter capsulatus genes encoding the equivalent of ND subunits of the mitochondrial NADH-ubiquinone oxidoreductase. Gene. 1995;167:99–104. doi: 10.1016/0378-1119(95)00693-1. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari E, Hoch J A. A single copy, transducible system for complementation and dominance analysis in Bacillus subtilis. Mol Gen Genet. 1983;189:321–325. [Google Scholar]
- 14.Friedrich, T. 1997. Personal communication.
- 15.Friedrich T, Hofhaus G, Ise W, Nehls U, Schmitz B, Weiss H. A small isoform of NADH:ubiquinone oxidoreductase (complex I) without mitochondrially encoded subunits is made in chloramphenicol-treated Neurospora crassa. Eur J Biochem. 1989;180:173–180. doi: 10.1111/j.1432-1033.1989.tb14629.x. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich T, van Heek P, Leif H, Ohnishi T, Forche E, Kunze B, Jansen R, Trowitzsch-Kienast W, Hofle G, Reichenbach H, Weiss H. Two binding sites of inhibitors in NADH:ubiquinone oxidoreductase (complex I) Eur J Biochem. 1994;219:691–698. doi: 10.1111/j.1432-1033.1994.tb19985.x. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich T, Weidner U, Nehls U, Fecke W, Schneider R, Weiss H. Attempts to define distinct parts of NADH:ubiquinone oxidoreductase (complex I) J Bioenerg Biomembr. 1993;25:331–337. doi: 10.1007/BF00762458. [DOI] [PubMed] [Google Scholar]
- 18.Friedrich T, Weiss H. Origin and evolution of the proton-pumping NADH:ubiquinone oxidoreductase (complex I) In: Baltscheffsky H, editor. Origin and evolution of biological energy conversion. New York, N.Y: VCH Publishers; 1996. pp. 205–220. [Google Scholar]
- 19.Friedrich T, Weiss H. Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J Theor Biol. 1997;187:529–541. doi: 10.1006/jtbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 20.Genetics Computer Group, Inc. Wisconsin Package, version 8.0. Madison, Wis: Genetics Computer Group, Inc.; 1994. [Google Scholar]
- 21.Gennis, R. 1995. Personal communication.
- 22.Guenebaut, V., A. Schlitt, K. Leonard, H. Weiss, and T. Friedrich. Consistent structure between bacterial and mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol., in press. [DOI] [PubMed]
- 23.Gutterson N I, Koshland D E. Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci USA. 1983;80:4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 563–566. [Google Scholar]
- 26.Hewlett-Packard Corp. DeskScan II. Palo Alto, Calif: Hewlett-Packard Corp.; 1991. [Google Scholar]
- 27.Hofhaus G, Weiss H, Leonard K. Electron microscopic analysis of the peripheral and membrane parts of mitochondrial NADH dehydrogenase (complex I) J Mol Biol. 1991;221:1027–1043. doi: 10.1016/0022-2836(91)80190-6. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Leif H, Sled V D, Ohnishi T, Weiss H, Friedrich T. Isolation and characterization of the proton-translocating NADH:ubiquinone oxidoreductase from Escherichia coli. Eur J Biochem. 1995;230:538–548. doi: 10.1111/j.1432-1033.1995.tb20594.x. [DOI] [PubMed] [Google Scholar]
- 30.Leif H, Weidner U, Berger A, Spehr V, Braun M, van Heek P, Friedrich T, Ohnishi T, Weiss H. Escherichia coli NADH dehydrogenase I, a minimal form of the mitochondrial complex I. Biochem Soc Trans. 1993;21:998–1001. doi: 10.1042/bst0210998. [DOI] [PubMed] [Google Scholar]
- 31.Matsushita K, Ohnishi T, Kaback H R. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry. 1987;26:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- 31a.McNamara, B. P., and A. J. Wolfe. Unpublished observation.
- 32.Meissler S, Langhammer R, Quinones A. Abstracts of Sonderausgabe zur Frühjahrstagung of the Vereinigung für Allegemeine und Angewandte Mikrobiologie 1997. Hamburg, Germany: Vereinigung für Allegemeine und Angewandte Mikrobiologie; 1997. A Tn5 insertion in the nuo operon of Escherichia coli causes an enhanced dnaN expression and an increase in the frequency of frameshift and deletion mutations, abstr. no. PF107; p. 120. [Google Scholar]
- 33.Microsoft Corp. PowerPoint, version 4.0. Seattle, Wash: Microsoft Corp.; 1994. [Google Scholar]
- 34.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 35.Neijessel O M, Teixeira de Mattos M J. The energetics of bacterial growth: a reassessment. Mol Microbiol. 1994;13:179–182. doi: 10.1111/j.1365-2958.1994.tb00413.x. [DOI] [PubMed] [Google Scholar]
- 36.Neumann B, Pospiech A, Schairer H U. Rapid isolation of genomic DNA from gram-negative bacteria. Trends Genet. 1992;8:332–333. doi: 10.1016/0168-9525(92)90269-a. [DOI] [PubMed] [Google Scholar]
- 37.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 38.Park C P, Hazelbauer G L. Mutations specifically affecting ligand interaction of the TRG chemosensory transducer. J Bacteriol. 1986;167:101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price M G, Gomer R H. Mitoskelin: a mitochondrial protein found in cytoskeletal preparations. Cell Mot Cytoskel. 1989;13:274–287. doi: 10.1002/cm.970130406. [DOI] [PubMed] [Google Scholar]
- 40.Prüß B M, Nelms J M, Park C, Wolfe A J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saiki R K. Amplification of genomic DNA. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 13–20. [Google Scholar]
- 42.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Schmidt M, Friedrich T, Wallrath J, Ohnishi T, Weiss H. Accumulation of the preassembled membrane arm of NADH:ubiquinone oxidoreductase in mitochondria of manganese-limited grown Neurospora crassa. FEBS Lett. 1992;313:8–11. doi: 10.1016/0014-5793(92)81172-i. [DOI] [PubMed] [Google Scholar]
- 44.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 45.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 46.Tran Q H, Bongaerts J, Vlad D, Unden G. Requirement for the proton-pumping NADH dehydrogenase I of Escherichia coli in respiration of NADH to fumarate and its bioenergetic implications. Eur J Biochem. 1997;244:155–160. doi: 10.1111/j.1432-1033.1997.00155.x. [DOI] [PubMed] [Google Scholar]
- 47.Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 48.Walker J E. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 49.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 50.Weidner U, Nehls U, Schneider R, Fecke W, Leif H, Schmiede A, Friedrich T, Zensen R, Schulte U, Ohnishi T, Weiss H. Molecular genetic studies of complex I in Neurospora crassa, Aspergillus niger, and Escherichia coli. Biochim Biophys Acta. 1992;1101:177–180. [PubMed] [Google Scholar]
- 51.Wolfe A J, Conley M P, Kramer T J, Berg H C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu X, Matsuno-Yagi A, Yagi T. Structural features of the 66-kDa subunit of the energy-transducing NADH-ubiquinone oxidoreductase (NDH-1) of Paracoccus denitrificans. Arch Biochem Biophys. 1992;296:40–48. doi: 10.1016/0003-9861(92)90542-5. [DOI] [PubMed] [Google Scholar]
- 53.Xu X, Matsuno-Yagi A, Yagi T. DNA sequencing of the seven remaining structural genes of the gene cluster encoding the energy-transducing NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochemistry. 1993;32:968–981. doi: 10.1021/bi00054a030. [DOI] [PubMed] [Google Scholar]
- 54.Yano T, Chu S S, Sled V D, Ohnishi T, Yagi T. The proton-translocating NADH-quinone oxidoreductase (NDH-1) of thermophilic bacterium Thermus thermophilus HB-8. J Biol Chem. 1997;272:4201–4211. doi: 10.1074/jbc.272.7.4201. [DOI] [PubMed] [Google Scholar]
- 55.Yano T, Sled V D, Ohnishi T, Yagi T. Expression and characterization of the flavoprotein subcomplex composed of 50-kDa (NQO1) and 25-kDa (NQO2) subunits of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. J Biol Chem. 1996;271:5907–5913. doi: 10.1074/jbc.271.10.5907. [DOI] [PubMed] [Google Scholar]
- 56.Yano T, Yagi T, Sled V D, Ohnishi T. Expression and characterization of the 66-kilodalton (NQO3) iron-sulfur subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. J Biol Chem. 1995;270:18264–18270. doi: 10.1074/jbc.270.31.18264. [DOI] [PubMed] [Google Scholar]
- 57.Young I G, Wallace B J. Mutations affecting the reduced nicotinamide adenine dinucleotide dehydrogenase complex of Escherichia coli. Biochim Biophys Acta. 1976;449:376–385. doi: 10.1016/0005-2728(76)90149-3. [DOI] [PubMed] [Google Scholar]
- 58.Youngman P. Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus species. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. West Sussex, England: John Wiley and Sons; 1990. pp. 221–257. [Google Scholar]
- 59.Zambrano M M, Kolter R. Escherichia coli mutants lacking the NADH dehydrogenase I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]