Abstract
Quorum sensing is a phenomenon in which bacteria sense and respond to their own population density by releasing and sensing pheromones. In gram-negative bacteria, quorum sensing is often performed by the LuxR family of transcriptional regulators, which affect phenotypes as diverse as conjugation, bioluminescence, and virulence gene expression. The gene encoding one LuxR family member, named sdiA (suppressor of cell division inhibition), is present in the Escherichia coli genome. In this report, we have cloned the Salmonella typhimurium homolog of SdiA and performed a systematic screen for sdiA-regulated genes. A 4.4-kb fragment encoding the S. typhimurium sdiA gene was sequenced and found to encode the 3′ end of YecC (homologous to amino acid transporters of the ABC family), all of SdiA and SirA (Salmonella invasion regulator), and the 5′ end of UvrC. This gene organization is conserved between E. coli and S. typhimurium. We determined that the S. typhimurium sdiA gene was able to weakly complement the E. coli sdiA gene for activation of ftsQAZ at promoter 2 and for suppression of filamentation caused by an ftsZ(Ts) allele. To better understand the function of sdiA in S. typhimurium, we screened 10,000 random lacZY transcriptional fusions (MudJ transposon mutations) for regulation by sdiA. Ten positively regulated fusions were isolated. Seven of the fusions were within an apparent operon containing ORF8, ORF9, rck (resistance to complement killing), and ORF11 of the S. typhimurium virulence plasmid. The three ORFs have now been named srgA, srgB, and srgC (for sdiA-regulated gene), respectively. The DNA sequence adjacent to the remaining three fusions shared no similarity with previously described genes.
Many species of bacteria sense and respond to their own population density by a phenomenon known as quorum sensing. Each bacterium in a population produces and releases a pheromone that can be detected by its neighbors. As the population density increases, so does the concentration of pheromone, allowing bacteria to gauge their own population density and possibly those of other species. Many of these sensory systems contain a positive feedback loop in which the synthesis of pheromone is increased as more pheromone is detected. The population density required to initiate the positive feedback loop defines a bacterial quorum (for reviews, see references 13, 50, 54, and 58).
Gram-negative bacteria primarily use a variety of N-acylhomoserine lactones as pheromones, while gram-positive bacteria use a variety of peptide pheromones (28). The gram-negative pheromones are referred to as autoinducers, and the corresponding gene regulation is referred to as autoinduction. The paradigm for gram-negative quorum sensing is the regulation of luminescence genes in Vibrio fischeri. When large numbers of bacteria colonize the squid light organ, autoinducer concentration rises to a level that induces the bacterial luminescence genes. Autoinducer binds to LuxR, and the complex acts as a transcriptional activator for the structural luminescence genes. The LuxR-autoinducer complex also activates luxI, which encodes an autoinducer synthase, presumably activating a positive feedback loop (13, 54).
Many pathogenic bacteria utilize quorum sensing to regulate expression of virulence genes. In the gram-negative pathogens, several systems homologous to the V. fischeri lux system have been identified. In Pseudomonas aeruginosa, the secreted virulence factors elastase (encoded by lasB), LasA protease (encoded by lasA), and alkaline protease (encoded by apr) are under control of multiple LuxR/LuxI homologs (7, 29, 41–43, 63). In addition, the plant pathogen Agrobacterium tumefaciens regulates conjugal transfer of the Ti plasmid by using LuxR/LuxI homologs (14, 23, 24, 36). In another plant pathogen, Erwinia carotovora, the antibiotic carbapenem and several exoenzymes that degrade plant cell walls are regulated by LuxR/LuxI homologs (27, 34, 44). One possible explanation for why pathogens use quorum sensing to regulate virulence genes involves the timing of host-bacterium interactions. For example, subsequent to E. carotovora degradation of plant cell walls, the cell wall fragments alert the plant to initiate a defense response against the pathogen. Therefore, to ensure a successful infection, it may be advantageous for an individual bacterium to wait until a large number of bacteria are present before initiating production of exoenzymes (50).
The pathogen described in this report, Salmonella typhimurium, causes millions of cases of gastroenteritis in humans each year and causes a typhoid-like disease in mice. Virulence genes of S. typhimurium are controlled by complex regulatory networks, with many of the genes being regulated to some degree by growth phase and/or nutrient limitation. For instance, SPI1 (Salmonella pathogenicity island 1) and the spv locus (Salmonella plasmid virulence) are two virulence gene clusters that are affected by growth phase (10, 17, 31). These observations led us to hypothesize that population density may be another regulatory input to virulence gene expression in S. typhimurium. To test this hypothesis, we determined that S. typhimurium encodes a LuxR homolog (SdiA) and then performed a screen for genes regulated by sdiA.
MATERIALS AND METHODS
Bacterial strains and media.
Bacteria were grown in Luria-Bertani (LB) broth (Difco) unless otherwise indicated. M9 minimal medium contained (per liter) 11 g of Na2HPO4 · 7H2O, 3 g of KH2PO4, 0.5 g of NaCl, and 1 g of NH4Cl. The pH was adjusted to 7.4, the solution was autoclaved, and then MgSO4 was added to 2 mM, CaCl2 was added to 0.1 mM, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to 40 μg/ml, and glucose or arabinose was added to 0.2%. Where indicated, Casamino Acids (Difco) were added to the M9 medium to 0.2%. Dulbecco modified Eagle medium (DMEM) was purchased from Gibco BRL, and when indicated, normal human serum (Sigma) was added to 10%. LB and M9 plates contained 1.5% agar (Difco).
Strain constructions and lineages are described in Table 1. Transductions were performed with phage P22HTint, followed by streaking to isolation in the presence of 10 mM EGTA and confirmation of smooth lipopolysaccharide (LPS) and lack of pseudolysogeny by cross-streaking transductants against P22vir on Evan’s Blue-Uranine plates (33). Plasmids were introduced by electroporation with a BTX electro cell manipulator 600.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype | Construction or reference |
|---|---|---|
| WX2 | E. coli Δlac sdiA::Kanr | 62 |
| AW40 | E. coli ftsZ84(Ts) | 62 |
| 14028 | Wild-type S. typhimurium | ATCCa |
| IR715 | 14028 Nalr | 56 |
| BA612 | 14028 sdiA::mTn3 (Carbr) | See Materials and Methods |
| TT10288 | S. typhimurium LT2 hisD9953::MudJ hisA9944::MudI | 22 |
| BA1100 series of mutants | 14028 srg::MudJ | This study |
| BA1200 series of mutants | BA612 srg::MudJ/pJVR2 (Camr) | This study |
| BA1300 series of mutants | BA612 srg::MudJ | This study |
| BA1400 series of mutants | BA612 srg::MudJ/pBAD33 (Camr) | This study |
| BA1101 | 14028 srg-5::MudJ | This study |
| BA1102 | 14028 srgB2::MudJ | This study |
| BA1103 | 14028 srgA1::MudJ | This study |
| BA1104 | 14028 rck2::MudJ | This study |
| BA1105 | 14028 rck3::MudJ | This study |
| BA1107 | 14028 srgB1::MudJ | This study |
| BA1109 | 14028 srgC1::MudJ | This study |
| BA1110 | 14028 srg-6::MudJ | This study |
| BA1111 | 14028 rck1::MudJ | This study |
| BA1112 | 14028 srg-7::MudJ | This study |
| pGB2 | pSC101 cloning vector (Specr) | 62 |
| pWSK29 | pSC101 cloning vector (Carbr) | 61 |
| pWSK129 | pSC101 cloning vector (Kanr) | 61 |
| Cosmid 6-10 | sdiA+ cosmid from pLAFR2 library of 14028 chromosomal DNA | See Materials and Methods |
| pBA301 | pWSK29 S. typhimurium sdiA+ sirA+ (Carbr) | 4.4-kb EcoRI fragment of cosmid 6-10 in EcoRI site of pWSK29 |
| pBA302 | pWSK129 S. typhimurium sdiA+ sirA+ (Kanr) | 4.4-kb EcoRI fragment of cosmid 6-10 in EcoRI site of pWSK129 |
| pBA306 | pWSK29 S. typhimurium sdiA+ (Carbr) | 1.4-kb PstI fragment of pBA301 in PstI site of pWSK29 |
| pCX16 | pGB2 carrying E. coli sdiA | 62 |
| pCX39 | Mini-F′ carrying ftsQAZ promoter 2 (sdiA-sensitive) fusion to galK′-lacZYA | 62 |
| pCX40 | Mini-F′ carrying ftsQAZ promoter 1 (sdiA-insensitive) fusion to galK′-lacZYA | 62 |
| pBAD33 | pACYC vector for arabinose-conditional expression | 18 |
| pJVR2 | pBAD33 sdiA+ | S. typhimurium sdiA PCR product in SmaI site of pBAD33 |
ATCC, American Type Culture Collection.
Antibiotics were used at the following concentrations: kanamycin, 60 μg/ml; tetracycline, 20 μg/ml; carbenicillin, 100 μg/ml (chromosomal) and 200 μg/ml (episomal); chloramphenicol, 30 μg/ml; streptomycin, 100 μg/ml; spectinomycin, 100 μg/ml; and naladixic acid, 50 μg/ml.
Molecular biology techniques.
Plasmid DNA was isolated by using ion-exchange columns from Qiagen. Standard methods were used for restriction endonuclease digestion, ligation, and electroporation of plasmid DNA (2). All restriction endonucleases and T4 DNA ligase were purchased from Gibco BRL or Boehringer Mannheim. PCR was performed with Pfu DNA polymerase (Stratagene) according to the instructions of the manufacturer. Southern hybridizations and probes were produced with the Genius nonradioactive nucleic acid detection kit and positively charged nylon membrane from Boehringer Mannheim. DNA probes were labeled with digoxigenin-dUTP (DIG) and detected with anti-DIG-alkaline phosphatase conjugate Fab fragments and the chemiluminescent substrate Lumi-Phos 530.
DNA sequencing was performed with a Pharmacia automated laser fluorescence and an Applied Biosystems 377 fluorescent DNA sequencer at our departmental core facility. Oligonucleotide synthesis was performed by Applied Biosystems automated solid-phase synthesis with standard chemistry at our departmental core facility. Sequence analysis was performed with the following programs: AssemblyLign 1.0.7 (Eastman Kodak), MacVector 6.0 (Oxford Molecular Group), the BLAST programs from the National Center for Biotechnology Information (1), and the Genetics Computer Group (package version 9.0 from the University of Wisconsin).
β-Galactosidase assays.
Expression of lacZ fusions was assessed by using β-galactosidase assays as described by Miller (37).
Cloning of the S. typhimurium sdiA homolog.
An E. coli luxR homolog (sdiA) was identified by performing database searches with the Vibrio fischeri LuxR amino acid sequence and the program TBlastN. sdiA-specific oligonucleotides (BA134 and BA135 [Table 2]) were synthesized, and the E. coli sdiA gene was amplified by PCR. The 654-bp PCR product was DIG labeled and used as a probe to clone the S. typhimurium homolog.
TABLE 2.
Oligonucleotides used
| Name | Sequence | Description |
|---|---|---|
| Universal-PCR | GTAAAACGACGGCCAGTGAGCGCGC | Binds the KpnI side of the pCR-Script multiple cloning site (Stratagene) |
| Reverse-PCR | GGAAACAGCTATGACCATGATTACG | Binds the SacI side of the pCR-Script multiple cloning site (Stratagene) |
| Tn3-bla | GACAGATCGCTGAGATAGGTGCCTC | Binds the end of mTn3 (oriented outward) within the β-lactamase gene |
| BA134 | TGCGTTTTCAGAGGATGGAGAC | Binds nucleotides 501 to 522 of E. coli sdiA (accession no. X03691) |
| BA135 | AGGCAACCTGGGTCTTATTTGG | Binds nucleotides 1154 to 1133 of E. coli sdiA (accession no. X03691) |
| BA174 | CAACAAGAAGAACGTTGATCAAAGG | Binds nucleotides 1970 to 1946 of the sequence reported here (accession no. U88651) |
| BA176 | CGAGTGAAATACGCGTAATAAAC | Binds nucleotides 1678 to 1656 of the sequence reported here (accession no. U88651) |
| BA178 | AATGATTATCAATATCAAAGGCGTG | Binds nucleotides 437 to 461 of the sequence reported here (accession no. U88651) |
| MudOut | CCGAATAATCCAATGTCCTCCCGGT | Binds nucleotides 54 to 30 of the Mu left end (accession no. M64097) |
| MudTaq | AGTGCGCAATAACTTGCTCTCGTTC | Binds nucleotides 701 to 725 of the Mu left end (accession no. M64097) |
| MudAlu | CGAAAAACAAAAACACTGCAAATCATTTCAATAAC | Binds nucleotides 167 to 201 of the Mu left end (accession no. M64097) |
A genomic library of S. typhimurium 14028 prepared in the cosmid vector pLAFR2 has been previously described (32). Individual cosmids from this library were grouped into 30 pools of 30 cosmids each. The 30 pools were digested with HaeIII, separated by agarose gel electrophoresis, and transferred to HybondN+ (Amersham). The blot was probed with the DIG-labeled E. coli sdiA PCR product under low-stringency conditions (hybridization overnight at 65°C followed by four washes in 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% sodium dodecyl sulfate at room temperature). One pool gave a positive signal so the 30 cosmids from this pool were individually digested with EcoRI, and a second Southern blot which identified the positive cosmid, now named cosmid 6-10, was performed.
Plasmid constructions and DNA sequencing.
All plasmid constructions except pJVR2 are described in Table 1. pJVR2 is the S. typhimurium sdiA gene that has been amplified with the high-fidelity DNA polymerase Pfu (Stratagene) and cloned into the SmaI site of pBAD33. The PCR primers (BA176 and BA178 [Table 2]) were designed to amplify sdiA without its promoter, in order to allow control exclusively by the arabinose promoter of pBAD33 (18). Orientation of the insert was confirmed by restriction analysis, and the 5′ and 3′ junctions were confirmed by sequence analysis with oligonucleotides specific to pBAD33 (18).
The 4.4-kb insert of pBA301 was sequenced on both strands by several strategies. Subclones were sequenced with Universal-PCR and Reverse-PCR primers to sequence into the inserts of pBA301, pBA306, and a collection of Sau3A subclones. Successive deletions into the insert of pBA301 were generated by exonuclease III and nuclease S1 digestion followed by religation (2). The resulting plasmids were sequenced with Universal-PCR and Reverse-PCR primers. The DNAs adjacent to a collection of mTn3 insertions were sequenced with primer Tn3-bla. Finally, several oligonucleotides were synthesized to sequence gaps.
Transposon mutagenesis of pBA302 and construction of a chromosomal sdiA::mTn3.
mTn3 mutagenesis was performed as described previously (52). Briefly, Escherichia coli RDP146 (carrying an F′::mTn3 and plasmid pLB101 encoding the Tn3 transposase) was transformed with pBA302. Transformants, now carrying cointegrates between pBA302 and the F′::mTn3, were mated with NS2114Sm. In this strain, the cointegrates are resolved by Cre recombinase. Transpositions within the insert of pBA302 were identified by restriction mapping. Fusion junctions were then sequenced with primer Tn3-bla.
One pBA302::mTn3 isolate, pBA366, had an mTn3 inserted between nucleotides 716 and 717, which is within the sdiA coding sequence. PCR was used to amplify the sdiA::mTn3 from pBA366 with oligonucleotides Universal-PCR and BA174. The ≈3.3-kb PCR product was cloned into the SrfI site of pCR-Script (Stratagene), excised with KpnI, and cloned into the KpnI site of the suicide vector pGP705 (60), resulting in pPV383. SM10λpir + pPV383 was mated with IR715 (S. typhimurium 14028 [Nalr]), selecting carbenicillin and naladixic acid. Double-crossover events were distinguished from single-crossover events by patching the resulting colonies for loss of vector-encoded tetracycline resistance. The mTn3 from several tetracycline-sensitive colonies was transduced into 14028 with P22HTint. The presence of the chromosomal sdiA::mTn3 of these transductants was confirmed by Southern hybridization of PstI-digested genomic DNA probed with the EcoRI fragment from pBA301. One isolate demonstrating the correct hybridization pattern was named BA612 and used for further analysis.
Inverse PCR.
DNA flanking the left end of each MudJ mutation was isolated by inverse PCR as described elsewhere (40, 59), with the following modifications. Chromosomal DNA was isolated according to standard procedures (2), and roughly 1.0 μg was digested with either TaqI or AluI in a 30-μl digestion mixture. One microliter of this digested DNA was self-ligated in a 20-μl T4 DNA ligase reaction mixture. One microliter of this ligation reaction mixture was used as the template in a 100-μl PCR reaction mixture with Pfu polymerase (Stratagene). DNA that had been digested with TaqI was amplified with primers MudOut and MudTaq. DNA that had been digested with AluI was amplified with primers MudOut and MudAlu. The resulting PCR products were gel purified with Qiagen gel extraction spin columns and cloned into the SrfI site of pCR-Script (Stratagene). Clones were sequenced on both strands with Universal-PCR and Reverse-PCR primers.
Nucleotide sequence accession number.
The complete sequence of the 4.4-kb insert of pBA301 was submitted to the GenBank database under accession no. U88651.
RESULTS
Cloning of the S. typhimurium sdiA homolog.
Database searches indicated that the E. coli sdiA gene was the only luxR family member that has been sequenced to date in E. coli or S. typhimurium. To determine whether S. typhimurium encodes an sdiA homolog, we probed an S. typhimurium cosmid bank with DNA encoding the E. coli sdiA gene (see Materials and Methods). A single positive cosmid was isolated and named cosmid 6-10. A 4.4-kb EcoRI fragment responsible for the positive hybridization signal was cloned into the EcoRI sites of pWSK29 and pWSK129 (61) to give pBA301 and pBA302, respectively (Fig. 1).
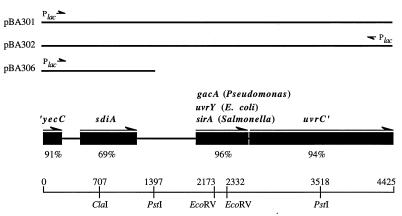
FIG. 1.
Restriction, ORF, and plasmid map of the sdiA region of S. typhimurium. Black boxes, ORFs; arrows, orientation. The percentages of amino acid identity between the S. typhimurium and E. coli homologs are indicated below each ORF. Plasmids discussed in the text are diagrammed above the ORFs, with black lines indicating the DNA region carried by each plasmid. The orientation of the insert with respect to the vector Plac promoter is indicated.
Sequence analysis of the S. typhimurium sdiA region.
The 4.4-kb insert of pBA301 was sequenced on both strands. Four open reading frames (ORFs) were identified, with the first and last being truncated (Fig. 1). The four ORFs share the same spacing and orientation as the homologous region in E. coli. The 3′ end of the first ORF extends from nucleotides 1 to 237 and shares 91% amino acid identity with the homologous ORF in E. coli, which is named yecC. Database searches suggest that yecC encodes an amino acid transport protein of the ABC (ATP-binding cassette) family. YecC is similar to HisP, the ATP-binding component of the Salmonella histidine transporter. Interestingly, YecC is also very similar to the opaline and nopaline transporters in A. tumefaciens (60% amino acid identity in certain regions). These transporters are located on the Ti plasmid, which is known to use quorum sensing to regulate conjugation (14, 23, 24).
The next open reading frame is sdiA, which extends from nucleotides 472 to 1191. SdiA is 69% identical at the amino acid level to its E. coli homolog, while it is 26% identical and 47% similar to LuxR of V. fischeri (Fig. 2). The ORF downstream of sdiA is known as sirA in S. typhimurium, uvrY or ORF2 in E. coli, and gacA in Pseudomonas species. The S. typhimurium sirA gene was recently identified genetically as a regulator of SPI1 gene expression (26). In Pseudomonas fluorescens and Pseudomonas syringae, the gacA gene is required for secretion of several secondary metabolites including antibiotics, HCN, a metalloprotease, and a phospholipase C (15, 30, 47, 49). Pseudomonas aeruginosa requires gacA for infection of both plant and animal hosts (46). As with gacA of the Pseudomonas species and uvrY of E. coli, sirA of S. typhimurium is directly followed by the uvrC gene. uvrC encodes a subunit of the UvrABC DNA excision repair enzyme (39, 51). Assuming that S. typhimurium uvrC is similar in size to the E. coli uvrC gene, all but the last two amino acids of UvrC are encoded by pBA301. The E. coli and S. typhimurium coding regions share 94% amino acid identity.
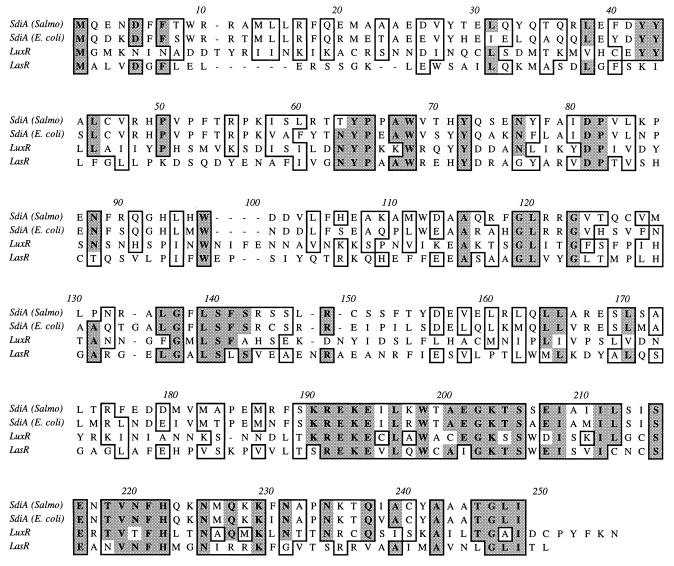
FIG. 2.
Sequence alignment of S. typhimurium SdiA, E. coli SdiA, V. fischeri LuxR, and P. aeruginosa LasR. Putative functional domains have been reviewed elsewhere (13). They include an autoinducer binding domain from residues 79 to 127, a helix-turn-helix DNA binding motif from residues 180 to 230, and a transcriptional activation domain from residues 211 to 250. Similar residues are boxed, while identical residues are boxed and shaded. Alignment was generated by using MacVector 6.0 and the ClustalW algorithm. Salmo, S. typhimurium.
Construction of an S. typhimurium sdiA mutation and an arabinose-inducible sdiA gene.
To identify lacZY fusions that are regulated by sdiA, we constructed a strain in which sdiA was under control of the tightly regulated arabinose-inducible PBAD promoter (18). First, the chromosomal copy of sdiA was disrupted in S. typhimurium 14028 to create BA612. Disruption of sdiA was accomplished by mutagenizing pBA302 with mTn3 as described in Materials and Methods (52). One mutant clone, pBA366, contained an mTn3 between nucleotides 716 and 717 of the sequence reported here, which corresponds to codon 82 of the sdiA ORF. This plasmid-encoded sdiA::mTn3 was used to disrupt the chromosomal sdiA of S. typhimurium by allelic exchange as described in Materials and Methods, resulting in strain BA612.
The second step was to create a system for controlling expression of sdiA. Oligonucleotides were designed to amplify the sdiA gene, including its putative ribosome binding site, but lacking any upstream promoter sequences. This PCR product was cloned into pBAD33, which placed the sdiA gene under control of the PBAD arabinose-inducible promoter (see Materials and Methods). The resulting plasmid, pJVR2, was then transformed into BA612 (14028 sdiA::mTn3), yielding an S. typhimurium strain in which sdiA expression was dependent on the presence of arabinose (18).
S. typhimurium sdiA weakly complements E. coli sdiA.
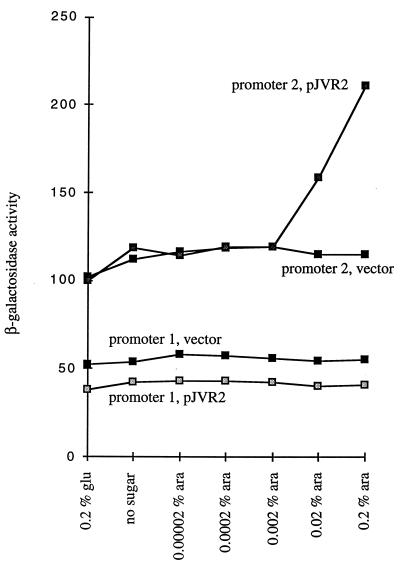
In E. coli, when sdiA is expressed from multicopy plasmids, it suppresses filamentation of an ftsZ(Ts) mutant grown at the nonpermissive temperature (62). The mechanism for this suppression is increased expression from promoter 2 of the ftsQAZ operon by sdiA. Presumably, a fraction of wild-type FtsZ activity is present at the nonpermissive temperature; thus, increasing expression of the ftsZ(Ts) allele raises the concentration of active FtsZ to a level capable of supporting cell division (62). To determine if S. typhimurium sdiA can also activate the E. coli ftsQAZ operon, pJVR2 was transformed into E. coli WX2/pCX39 (sdiA mutant E. coli carrying a lacZ fusion to promoter 2 of ftsQAZ). Expression of β-galactosidase activities from cultures grown in various concentrations of glucose or arabinose were determined (Fig. 3). pJVR2, but not the vector pBAD33, activated promoter 2 of ftsQAZ in the presence of arabinose. This demonstrates that (i) S. typhimurium sdiA is expressed by pJVR2 in the presence of arabinose and (ii) S. typhimurium sdiA can at least partially complement E. coli sdiA for regulation of ftsQAZ promoter 2. Promoter 1 of ftsQAZ is known to be insensitive to E. coli sdiA, and in agreement with this, pJVR2 had no effect on this promoter (WX2/pCX40 [Fig. 3]).
FIG. 3.
Activation of E. coli ftsQAZ promoter 2 by S. typhimurium sdiA. E. coli sdiA mutant carrying either pCX39 or pCX40 (encoding ftsQAZ promoter 2 or promoter 1 lacZ fusions, respectively) was transformed with either pJVR2 (S. typhimurium sdiA under arabinose control) or the vector alone (pBAD33). These strains were grown overnight in 5 ml of buffered LB containing the appropriate antibiotics (LB plus 0.1 M Tris-maleate [pH 8.0] in tubes [18 by 150 mm] with shaking at 37°C). On the following morning, the optical densities at 600 nm of the cultures were equalized to the least dense culture, followed by subculturing (at a dilution of 1:2) into buffered LB containing various arabinose (ara) or glucose (glu) concentrations. After 1 h of agitation at 37°C, the β-galactosidase activity of each culture was determined.
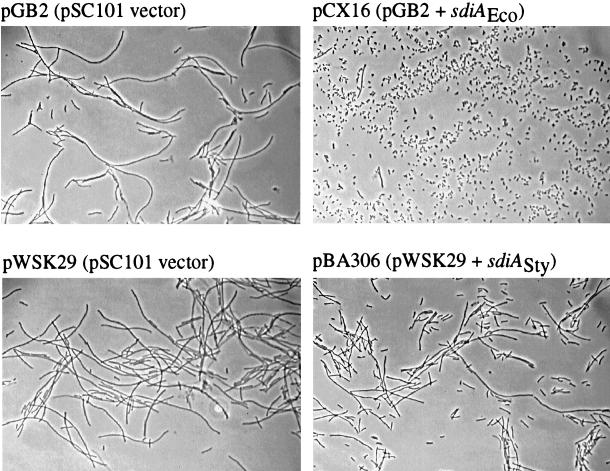
To further assess the ability of S. typhimurium sdiA to complement the filamentation phenotype of an E. coli ftsZ(Ts) mutant, filamentation assays were performed. E. coli sdiA and S. typhimurium sdiA were both expressed from low-copy-number pSC101 replicons and compared directly for their abilities to complement the filamentation phenotype of an E. coli ftsZ(Ts) mutant grown at the nonpermissive temperature. Figure 4 demonstrates that S. typhimurium sdiA does indeed complement the filamentation phenotype, but the complementation is weak compared to that for E. coli sdiA.
FIG. 4.
The sdiA gene from S. typhimurium (Sty) can partially replace the native E. coli (Eco) sdiA gene for suppression of filamentation of an E. coli ftsZ(Ts) strain grown at 42°C. AW40 cells carrying the plasmids indicated above each panel were grown overnight in 5 ml of LB at the permissive temperature of 30°C (in tubes [18 by 150 mm] with shaking). The strains were subcultured at a dilution of 1:100 into LB containing the appropriate antibiotics and shaken at 42°C for 3 h. The cells were fixed in a final concentration of 2% paraformaldehyde and were examined by phase-contrast microscopy. Magnification, ×600.
Isolation of sdiA-regulated genes.
To address the role of sdiA in S. typhimurium, we performed a systematic screen for genes regulated by sdiA using a strategy recently used to identify rpoS-regulated genes (11). BA612/pJVR2 (S. typhimurium containing sdiA under arabinose control) was mutagenized with MudJ as previously described (22) and plated on M9 minimal medium containing glucose as the carbon source (M9 glucose). A total of 10,000 random mutants from these plates were patched on M9 glucose versus M9 arabinose, with all plates containing the colorimetric β-galactosidase substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Mutants with altered β-galactosidase activities on glucose versus arabinose were streaked to isolation, and the differences in color were confirmed.
The next step was to transduce each MudJ fusion into four distinct genetic backgrounds with phage P22HTint. These constructions and backgrounds are listed in Table 1. The first background was wild-type S. typhimurium 14028 (resulting in the BA1100 series of mutants). The second background was a simple retransduction into BA612/pJVR2 (the BA1200 series of mutants) to eliminate the possibility of secondary mutations contributing to the expression phenotype. The BA1300 series of mutants were the result of each MudJ fusion being transduced into the sdiA mutant (BA612) background. The fourth background, BA612/pBAD33 (the BA1400 series of mutants), was an isogenic vector control to show that these fusions were not regulated by arabinose or the pBAD33 vector.
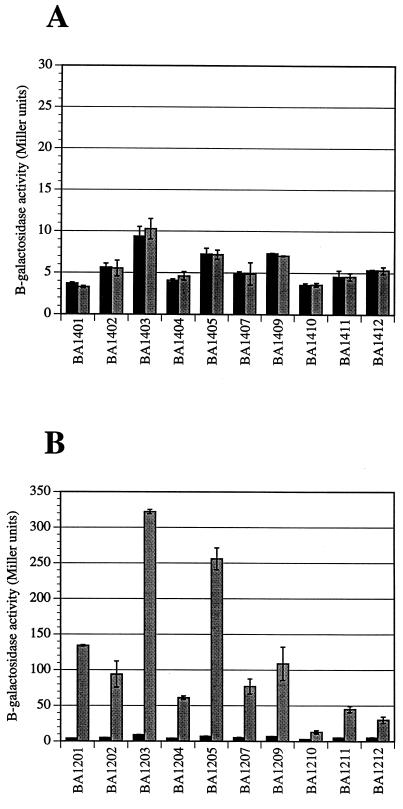
As seen in Fig. 5, the 10 fusions identified in this study are expressed only in the presence of arabinose and only in the arabinose-conditional sdiA background, not in the isogenic control background BA612/pBAD33. This demonstrates that expression of these fusions is sdiA specific and is not due to arabinose or vector sequences. Of course, fusions that failed these tests were isolated and discarded as arabinose-regulated genes.
FIG. 5.
β-Galactosidase activities of sdiA-regulated lacZ fusions after growth in the presence of glucose (dark shaded bars) or arabinose (light shaded bars). Strains were grown overnight in LB (96-well plates at 37°C with no shaking) followed by subculture at a dilution of 1:2 into either LB plus 0.4% glucose or LB plus 0.4% arabinose (resulting in a final sugar concentration of 0.2% [also in 96-well plates]). β-Galactosidase activities were measured after 1 h of growth (37°C, no shaking). These data demonstrate that the fusions are not regulated by arabinose alone (BA612/pBAD33 background) (A) but are regulated by the arabinose-inducible sdiA gene present in BA612/pJVR2 (B). Each strain was assayed at least three times independently. The results shown are from a single representative experiment performed with duplicate cultures (error bars indicate standard deviations).
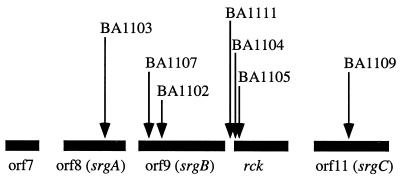
Inverse PCR was used to clone and sequence a small portion of genome flanking each sdiA-regulated fusion as described in Materials and Methods. Database searches were performed, and the results are listed in Table 3. Seven of the MudJ mutations were located within four ORFs on the S. typhimurium virulence plasmid: ORF8, ORF9, rck (resistance to complement killing), and ORF11 (Fig. 6). These ORFs are located at the 3′ terminus of the previously identified pef region (5, 6, 12). We have renamed ORF8, ORF9, and ORF11 srgA, srgB, and srgC, respectively (sdiA regulated gene). Three other positively regulated fusions were also identified. These three fusions shared no similarity with current GenBank entries and are not present in the E. coli genome.
TABLE 3.
sdiA-regulated lacZY fusions
| Strain | Homology/amino acid identity (%) | No. of nucleotides sequenceda | Insertion siteb |
|---|---|---|---|
| BA1101 | No homology | 463 | NA |
| BA1102 | S. typhimurium ORF9 (srgB)/100 | 112 | 10,603 |
| BA1103 | S. typhimurium ORF8 (srgA)/100 | 111 | 10,114 |
| BA1104 | S. typhimurium rck/100 | 252 | 11,412 |
| BA1105 | S. typhimurium rck/100 | 234 | 11,430 |
| BA1107 | S. typhimurium ORF9 (srgB)/100 | 195 | 10,519 |
| BA1109 | S. typhimurium ORF11 (srgC)/100 | 136 | 12,604 |
| BA1110 | No homology | 699 | NA |
| BA1111 | S. typhimurium rck/100 | 315 | 11,350 |
| BA1112 | No homology | 477 | NA |
Size of genomic DNA portion of inverse PCR product.
Nucleotide preceding MudJ insertion site according to the sequence reported in GenBank accession no. L08613. NA, not applicable.
FIG. 6.
ORF and mutation map of the 3′ end of the pef region (12). Black bars, ORFs (orientation is left to right for all genes); arrows, sdiA-regulated MudJ insertions.
The expression of each fusion in the wild-type and sdiA mutant backgrounds was also assessed (BA1100 and BA1300 series mutants). None of the fusions demonstrated any significant activity in either background (data not shown). This experiment was performed with stationary-phase cultures grown in LB or M9 minimal medium with no activity in either case (always less than 10 U). Since 7 of 10 fusions are within a gene cluster containing rck (resistance to complement killing), we assayed the β-galactosidase activities of the rck fusion in both sdiA+ and sdiA mutant backgrounds (BA1105 and BA1305) in M9 glucose, M9 glucose plus Casamino Acids, LB, DMEM, and DMEM containing human serum. β-Galactosidase activities at time points throughout the growth curve for all cultures were assayed. In every case, less than 10 U of activity was obtained, and the residual activity was not dependent on sdiA (data not shown).
DISCUSSION
Pathogenic bacteria sense and respond to a wide variety of environmental signals during the transition from a free-living state to infection of a suitable host. Oxygen tension, osmolarity, iron availability, pH, nutrient limitation, temperature, and even specific bacterium-host physical interactions regulate expression of virulence genes (see reference 3 and references therein). Population density is emerging as yet another environmental factor that is sensed by bacterial pathogens. Most gram-negative bacteria, including pathogens, sense population density by using members of the LuxR-LuxI family of quorum-sensing components in which LuxR is the autoinducer sensor and transcriptional regulator and LuxI is the autoinducer synthase (for reviews, see references 13, 50, 54, and 58). A second type of autoinducer synthase, AinS, has also been identified (16).
E. coli encodes a LuxR homolog named SdiA. In E. coli, sdiA weakly activates ftsQAZ in response to V. fischeri and Vibrio harveyi autoinducer and more dramatically in response to spent E. coli culture supernatant (53). These findings suggest that the native E. coli autoinducer is different in some way from the Vibrio autoinducers. Interestingly, upon completion of the E. coli genome sequencing project, we searched again for the quorum-sensing components LuxR, LuxI, and AinS. SdiA remains the only LuxR homolog, and there is no autoinducer synthase of any known family (LuxI or AinS). This leaves open the nature of the autoinducer (if any) produced by E. coli.
To determine whether S. typhimurium encodes a homolog of SdiA, we probed a cosmid library of the S. typhimurium genome with the E. coli sdiA gene. A 4.4-kb fragment encoding the S. typhimurium sdiA gene was cloned and sequenced. The order, spacing, and orientation of the genes adjacent to sdiA (yecC, sirA, and uvrC) are conserved between E. coli and S. typhimurium. This gene organization may be conserved in Pseudomonas species as well, but to date only gacA (sirA) and uvrC have been sequenced (30, 47). Interestingly, the S. typhimurium yecC, sirA, and uvrC genes all encode products that are at least 90% identical at the amino acid level to the corresponding E. coli homologs. However, sdiA shares only 69% amino acid identity with its E. coli counterpart (Fig. 1 and 2). The function of sdiA may differ in the two organisms, resulting in rapid sequence divergence. This selective pressure could affect either autoinducer specificity or DNA recognition specificity and raised the possibility that the S. typhimurium sdiA gene and the E. coli sdiA gene are not completely interchangeable. To determine whether S. typhimurium sdiA could complement E. coli sdiA, we first tested the ability of S. typhimurium sdiA to activate promoter 2 of E. coli ftsQAZ. We found that S. typhimurium sdiA under control of an arabinose promoter was able to activate E. coli ftsQAZ by 2-fold (Fig. 3). Using a different expression system, Wang et al. found that E. coli sdiA activated ftsQAZ by 5- to 13-fold (62). Second, we tested the ability of S. typhimurium sdiA to suppress the filamentation phenotype of an E. coli ftsZ(Ts) strain grown at nonpermissive temperature. S. typhimurium sdiA was capable of partially suppressing filamentation of the E. coli strain (Fig. 4). Both assays taken together demonstrate that S. typhimurium sdiA can only partially complement E. coli sdiA.
To isolate genes regulated by sdiA in S. typhimurium, we screened 10,000 random lacZY fusions for regulation by S. typhimurium sdiA. Ten sdiA-regulated fusions were isolated. Three genes were not homologous to any sequences present in GenBank, including the recently completed E. coli genome. The remaining seven fusions were located within a four-gene cluster on the S. typhimurium virulence plasmid. The four ORFs ORF8, ORF9, rck, and ORF11 were adjacent to each other at the 3′ end of the previously identified pef region (12). We have assigned the names srgA, srgB, and srgC to ORF8, ORF9, and ORF11 (sdiA-regulated gene), respectively. The clustering of the fusions suggests that these four genes are transcribed independently of the other genes in the pef region. Interestingly, S. typhi, the causative agent of human typhoid fever, lacks a virulence plasmid but has retained homologs of four plasmid-encoded genes in its chromosome: pefI, ORF7, ORF8, and ORF9 (48). The identification of genes not present in E. coli confirms the hypothesis that sdiA of S. typhimurium may not have precisely the same functions as sdiA of E. coli, possibly explaining the relatively large degree of sequence divergence.
The four-plasmid-encoded ORFs regulated by sdiA begin with srgA (ORF8), which encodes a dsbA homolog. DsbA is a disulfide bond isomerase involved with periplasmic protein folding (4). Several bacterial pathogens have fimbriae that require dsbA for proper fimbrial subunit folding and assembly (25, 55, 64). It would be reasonable to speculate that srgA plays some role in Pef assembly; however, mutations in srgA have no apparent effect on fimbrial biogenesis (12).
srgB contains a putative lipoprotein signal sequence but otherwise is not homologous to any characterized genes in the nucleotide databases (12). However, there is an uncharacterized ORF (yjiK) in the chromosome of E. coli that shares 32% amino acid identity.
Rck is an outer membrane protein that is known to confer two phenotypes when expressed in E. coli: adhesion to eukaryotic cells and resistance to human serum (9, 19–21). During formation of the membrane attack complex on a bacterial membrane, complement component C9 is able to attach to the O antigen of LPS, but Rck prevents C9 polymerization into a complete membrane attack complex (20). Other homologs of this family include ail of Yersinia enterocolitica, pagC of S. typhimurium, ompX of E. coli and Enterobacter cloacae, and lom of bacteriophage lambda (35, 38, 45, 57).
The final gene product, SrgC (ORF11), is homologous to several regulatory proteins of the AraC family including EnvY, AppY, and Rns (from E. coli) and VirF (from Y. enterocolitica) (12). However, a function for srgC in S. typhimurium has not been reported.
The environmental signal that SdiA recognizes is unknown, although sequence homology and evidence in E. coli (53) suggest that SdiA is a quorum sensor. The fact that E. coli does not encode a known family of autoinducer synthase makes the E. coli quorum-sensing system unique among gram-negative bacteria. It is possible that S. typhimurium also lacks an autoinducer synthase. In agreement with this, no complementation of N-acylhomoserine lactone biosensors has been seen in dichloromethane-extracted culture supernatants of E. coli and S. typhimurium (57a). One possible explanation for this is that E. coli and S. typhimurium simply produce a novel type of autoinducer. Another possibility is that E. coli and S. typhimurium sense autoinducers produced only by other species of bacteria.
We recently performed Northern blot experiments and gene fusion studies of S. typhimurium sdiA, and we know that it is expressed from the chromosome under laboratory conditions (unpublished data). So why can SdiA expressed from the chromosome not activate the genes identified in this study? One possibility is that the genes regulated by SdiA are also under the control of another regulatory protein(s). In the absence of the correct conditions for expression by these other proteins, chromosomally encoded SdiA may not be able to activate the promoters, while overexpressed SdiA is able to effectively compete with these other influences and activate transcription. Another possibility is that chromosomally encoded SdiA was unable to activate the genes because an autoinducer was not present in the assays. SdiA expressed under arabinose control may have bypassed a signal requirement by overexpression. Precedence for this hypothesis is provided by the fact that overexpression of V. fischeri LuxR in E. coli results in activation of luminescence genes in the absence of autoinducer (8).
While our hypothesis is speculative, E. coli and S. typhimurium may use quorum sensing to detect the transition from a free-living state to the intestinal environment. If this is true, detection of heterologous bacterial pheromones in the intestine would seem sufficient, and E. coli and S. typhimurium would have no need to produce their own autoinducers. The activation of rck in the intestinal environment could presumably confer adhesion to intestinal epithelium and/or prepare the bacterium for complement attack during subsequent steps of infection. We hope to use the fusions identified in this study to screen heterologous bacterial pheromones for sdiA-dependent activation of these genes.
ACKNOWLEDGMENTS
This work was supported by a grant from the N. L. Tartar Research fund to B.M.M.A. and NIH grant AI22933-11 to F.H.
We thank Alex Merz for help with photography of the filamentation strains. We are grateful to Lawrence Rothfield for providing bacterial strains and to Don Guiney, Simon Swift, Susanne Lindgren, Cindy Arvidson, Linda Kenney, Alex Merz, and Lawrence Rothfield for helpful discussions or critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 3.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell J C, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 5.Baumler A J, Tsolis R M, Bowe F A, Kusters J G, Hoffmann S, Heffron F. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect Immun. 1996;64:61–68. doi: 10.1128/iai.64.1.61-68.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi S H, Greenberg E P. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol. 1992;174:4064–4069. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo D M, Heffernan E J, Wu L, Harwood J, Fierer J, Guiney D G. Identification of a domain in Rck, a product of the Salmonella typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect Immun. 1996;64:2019–2023. doi: 10.1128/iai.64.6.2019-2023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang F C, Chen C Y, Guiney D G, Xu Y. Identification of sigma S-regulated genes in Salmonella typhimurium: complementary regulatory interactions between sigma S and cyclic AMP receptor protein. J Bacteriol. 1996;178:5112–5120. doi: 10.1128/jb.178.17.5112-5120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaffney T D, Lam S T, Ligon J, Gates K, Frazelle A, Di M J, Hill S, Goodwin S, Torkewitz N, Allshouse A M, et al. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol Plant-Microbe Interact. 1994;7:455–463. doi: 10.1094/mpmi-7-0455. [DOI] [PubMed] [Google Scholar]
- 16.Gilson L, Kuo A, Dunlap P V. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guiney D G, Libby S, Fang F C, Krause M, Fierer J. Growth-phase regulation of plasmid virulence genes in Salmonella. Trends Microbiol. 1995;3:275–279. doi: 10.1016/s0966-842x(00)88944-1. [DOI] [PubMed] [Google Scholar]
- 18.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heffernan E J, Harwood J, Fierer J, Guiney D. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol. 1992;174:84–91. doi: 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heffernan E J, Reed S, Hackett J, Fierer J, Roudier C, Guiney D. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J Clin Invest. 1992;90:953–964. doi: 10.1172/JCI115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heffernan E J, Wu L, Louie J, Okamoto S, Fierer J, Guiney D G. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect Immun. 1994;62:5183–5186. doi: 10.1128/iai.62.11.5183-5186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes K T, Roth J R. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988;119:9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang I, Cook D M, Farrand S K. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J Bacteriol. 1995;177:449–458. doi: 10.1128/jb.177.2.449-458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang I, Li P L, Zhang L, Piper K R, Cook D M, Tate M E, Farrand S K. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacob D F, Pinkner J, Xu Z, Striker R, Padmanhaban A, Hultgren S J. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc Natl Acad Sci USA. 1994;91:11552–11556. doi: 10.1073/pnas.91.24.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston C, Pegues D A, Hueck C J, Lee C A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 27.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J, Golby P, Reeves P J, Stephens S, et al. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleerebezem M, Quadri L E N, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 29.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 30.Laville J, Voisard C, Keel C, Maurhofer M, Defago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 34.McGowan S, Sebaihia M, Jones S, Yu B, Bainton N, Chan P F, Bycroft B, Stewart G S, Williams P, Salmond G P. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology. 1995;141:541–550. doi: 10.1099/13500872-141-3-541. [DOI] [PubMed] [Google Scholar]
- 35.Mecsas J, Welch R, Erickson J W, Gross C A. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol. 1995;177:799–804. doi: 10.1128/jb.177.3.799-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mel S F, Mekalanos J J. Modulation of horizontal gene transfer in pathogenic bacteria by in vivo signals. Cell. 1996;87:795–798. doi: 10.1016/s0092-8674(00)81986-8. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 38.Miller V L, Bliska J B, Falkow S. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J Bacteriol. 1990;172:1062–1069. doi: 10.1128/jb.172.2.1062-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moolenaar G F, van Sluis C A, Backendorf C, van der Putte P. Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrC structural gene and the region coding for a 24 kD protein. Nucleic Acids Res. 1987;15:4273–4289. doi: 10.1093/nar/15.10.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochman H, Ayala F J, Hartl D L. Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol. 1993;218:309–321. doi: 10.1016/0076-6879(93)18023-6. [DOI] [PubMed] [Google Scholar]
- 41.Ochsner U A, Koch A K, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pulkkinen W S, Miller S I. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J Bacteriol. 1991;173:86–93. doi: 10.1128/jb.173.1.86-93.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 47.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez P J, Alvarez I, Ibanez M, Rotger R. Homologous regions of the Salmonella enteritidis virulence plasmid and the chromosome of Salmonella typhi encode thiol:disulphide oxidoreductases belonging to the DsbA thioredoxin family. Microbiology. 1997;143:1405–1413. doi: 10.1099/00221287-143-4-1405. [DOI] [PubMed] [Google Scholar]
- 49.Sacherer P, Defago G, Haas D. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol Lett. 1994;116:155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 50.Salmond G P, Bycroft B W, Stewart G S, Williams P. The bacterial ‘enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 51.Sancar G B, Sancar A, Rupp W D. Sequences of the E. coli uvrC gene and protein. Nucleic Acids Res. 1984;12:4593–4608. doi: 10.1093/nar/12.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seifert H S, Chen E Y, So M, Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sitnikov D M, Schineller J B, Baldwin T O. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 55.Sone M, Akiyama Y, Ito K. Differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J Biol Chem. 1997;272:10349–10352. doi: 10.1074/jbc.272.16.10349. [DOI] [PubMed] [Google Scholar]
- 56.Stojiljkovic I, Baumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoorvogel J, van Bussel M J, Tommassen J, van der Klundert J A M. Molecular characterization of an Enterobacter cloacae outer membrane protein (OmpX) J Bacteriol. 1991;173:156–160. doi: 10.1128/jb.173.1.156-160.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.Swift, S. Personal communication.
- 58.Swift S, Throup J P, Williams P, Salmond G P, Stewart G S. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends. 1996;21:214–219. [PubMed] [Google Scholar]
- 59.Tsolis R M, Baumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valentine P J, Meyer K, Rivera M M, Lipps C, Pauza D, Maziarz R T, So M, Heffron F. Induction of SIV capsid-specific CTL and mucosal sIgA in mice immunized with a recombinant S. typhimurium aroA mutant. Vaccine. 1996;14:138–146. doi: 10.1016/0264-410x(95)00130-s. [DOI] [PubMed] [Google Scholar]
- 61.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 62.Wang X D, de Boer P A J, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P, Bycroft B W, et al. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H Z, Donnenberg M S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]