Abstract
A paradox in evolutionary biology is how supergenes can maintain high fitness despite reduced effective population size, the suppression of recombination, and the expected accumulation of mutational load. The ruff supergene involves 2 rare inversion haplotypes (satellite and faeder). These are recessive lethals but with dominant effects on male mating strategies, plumage, and body size. Sequence divergence to the wild-type (independent) haplotype indicates that the inversion could be as old as 4 million years. Here, we have constructed a highly contiguous genome assembly of the inversion region for both the independent and satellite haplotypes. Based on the new data, we estimate that the recombination event(s) creating the satellite haplotype occurred only about 70,000 yr ago. Contrary to expectations for supergenes, we find no substantial expansion of repeats and only a modest mutation load on the satellite and faeder haplotypes despite high sequence divergence to the non-inverted haplotype (1.46%). The essential centromere protein N (CENPN) gene is disrupted by the inversion and is as well conserved on the inversion haplotypes as on the noninversion haplotype. These results suggest that the inversion may be much younger than previously thought. The low mutation load, despite recessive lethality, may be explained by the introgression of the inversion from a now extinct lineage.
Keywords: ruff, supergene, genetic load, mating strategies, natural selection
Introduction
Supergenes are defined as a cluster of genes controlling complex phenotypic traits and recombination within supergenes is often suppressed due to a structural rearrangement, usually an inversion (Kirkpatrick and Barton 2006; Linksvayer et al. 2013; Faria et al. 2019; Gutiérrez-Valencia et al. 2021). Supergenes typically show a simple Mendelian inheritance and are often maintained by balancing selection. Examples of phenotypes associated with supergenes involving chromosomal inversions include Batesian mimicry in butterflies (Nishikawa et al. 2015), social organization in ants (Linksvayer et al. 2013; Brelsford et al. 2020), migration in trout (Pearse et al. 2019), mating systems in birds (Thomas et al. 2008), and ecological adaptation in Atlantic herring (Han et al. 2020), Atlantic cod (Matschiner et al. 2022), and sunflowers (Huang et al. 2022). Inversions may have a selective advantage because suppression of homologous recombination facilitates the maintenance of haplotypes composed of favorable combinations of linked gene variants. However, the presence of inversions may also lead to a negative impact on fitness because suppressed recombination reduces the efficacy of selection to purge deleterious mutations and is expected to result in the accumulation of mutational load (Faria et al. 2019; Charlesworth and Charlesworth 2020; Gutiérrez-Valencia et al. 2021). Inversions may also accumulate repetitive elements through “degenerative expansion” that results in a larger inversion haplotype than its ancestor (Stolle et al. 2019; Berdan et al. 2021; Gutiérrez-Valencia et al. 2021).
Inversion polymorphisms can have low fitness in homozygotes, and in the extreme, inversions may be recessive lethals when inversion breakpoints disrupt essential genes or deleterious mutations are captured by the inversion or accumulate in a nonrecombining region (Faria et al. 2019; Gutiérrez-Valencia et al. 2021). Only gene conversion and double crossovers can lead to genetic exchange between alternative chromosomal haplotypes (Navarro et al. 1997). Thus, inversion haplotypes with low fitness as homozygotes are expected to harbor a high mutation load due to the absence of purifying selection against recessive mutations in genes within the inversion. However, simulation studies suggest that mutational load can be mitigated by even moderate rates of gene conversion (Berdan et al. 2021) and deviations from mutation–selection equilibria can be influenced by the evolutionary history of the inversion. Despite predicted negative fitness effects, inversion supergenes are often implicated in local adaptation, leading to calls for a better understanding of mutational accumulation and the fate of supergenes.
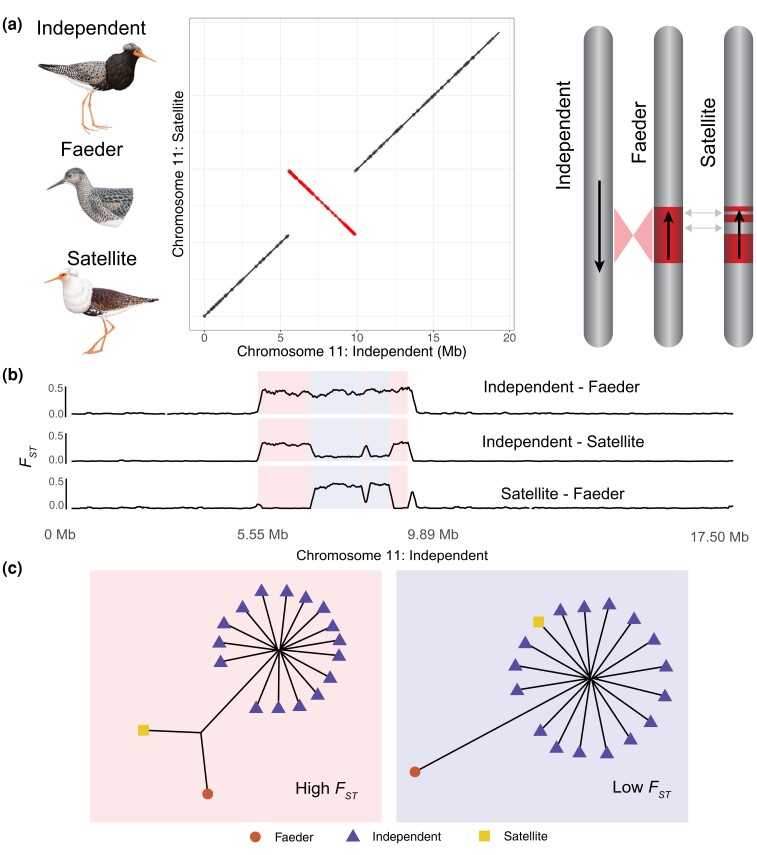
A supergene inversion polymorphism is responsible for variation in male mating and plumage phenotypes of the ruff (Calidris pugnax) (Küpper et al. 2016; Lamichhaney et al. 2016). In this lekking species, there are 3 types of male mating phenotypes (Fig. 1a): Independents have highly variable ornamental feathers and high testosterone and defend territories; satellites are smaller, have predominately light-colored ornamental feathers, and are nonterritorial but take part in the lek by associating with independents when females are present; and faeders resemble females in body size and plumage (Widemo 1998a; Jukema and Piersma 2006) and are also nonterritorial. The satellite (S) and faeder (F) haplotypes are dominant over the independent (I) haplotype (Lank et al. 1995, 2013), but their allele frequencies are about 5% and 1%, respectively (Höglund and Lundberg 1989; Widemo 1998a; Lamichhaney et al. 2016). I is ancestral, F is a full-length 4.5-Mb inversion with an estimated sequence divergence time from the I haplotype of about 4 million years, and S is the result of a single or a few recombination events between an inverted and a noninverted haplotype (Lamichhaney et al. 2016) (Fig. 1a). Both S and F are homozygous lethals, most likely because the inversion disrupts an essential centromere protein gene, centromere protein N (CENPN) (Küpper et al. 2016; Lamichhaney et al. 2016). Theoretical predictions and empirical data in other systems (Kirkpatrick and Barton 2006; Faria et al. 2019; Charlesworth and Charlesworth 2020; Berdan et al. 2021; Gutiérrez-Valencia et al. 2021; Jay et al. 2021) indicate that the inversion haplotypes should be burdened by a high mutational load and an expansion of transposable elements, but this was not possible to determine using the existing, highly fragmented, genome assemblies for ruff. The paradox of polymorphic inversions (Faria et al. 2019) is particularly apparent in the ruff because it is unclear how this inversion may have persisted over millions of years, despite homozygous lethality, low population frequency, and a predicted high genetic load.
Fig. 1.
Ruff male phenotypes and inversion haplotypes. a) Left: 3 male ruff phenotypes: independent, faeder, and satellite (illustrations reproduced with permission of Lynx Edicions). Chromosome 11 alignments between satellite and independent assemblies with the 4.3-Mb inversion highlighted in red. Right: a graphical representation of the ancestral independent chromosome 11, inverted faeder chromosome, and the recombined satellite chromosome. Satellites and faeders are heterozygous for inversion haplotypes and Independents are homozygous for the noninverted haplotype. b) Genetic divergence estimated by FST in 15-kb rolling windows between all 3 chromosomal arrangements across chromosome 11. The 4.3-Mb inverted region is highlighted by a red box marking the 5.55- and 9.89-Mb breakpoints. Blue boxes mark the recombinant regions in the satellite haplotype. c) Local phylogenetic trees for the nonrecombinant (high FST between independents and satellites) and recombinant regions (low FST between independents and satellites) based on 16 independents, a single satellite and a single faeder individual. The background colors used for the high and low FST trees are the same as used to highlight these regions in Fig. 1b.
In order to conduct detailed comparisons of molecular evolution between the inversion haplotypes, we combined a chromium 10× linked read assembly with PacBio long-read contigs to construct highly contiguous assemblies for both the independent and satellite haplotypes across the inverted region from 1 heterozygous individual. We use these assemblies and previously published whole-genome data to test the prediction that the satellite and faeder haplotypes show a high mutational load and expansion of repetitive elements. Specifically, we use estimates of synonymous and nonsynonymous substitution rates and tabulate repetitive elements to test the prediction of mutational load.
Results
Reassembly of the Region Harboring the Inversion Polymorphism
We generated a high-quality genome assembly from a satellite (S/I) individual collected in North Sweden. We first constructed a chromium 10× linked read library, which was sequenced to a depth of 90× for an estimated genome size of 1.23 Gb (Lamichhaney et al. 2016) and conducted phase-aware genome assembly using Supernova v2.0 (Supplementary Table S1). A 17-Mb scaffold was homologous to chicken chromosome 11 and harbored the inversion polymorphism (Fig. 1a). A comparison between the 2 versions of this scaffold representing the independent and satellite haplotypes identified a 4.3-Mb inversion in the satellite haplotype, similar in size to the 4.5-Mb inversion as previously described (Küpper et al. 2016; Lamichhaney et al. 2016).
We sequenced PacBio long-read library from the same individual to an estimated coverage of 82× and constructed a diploid assembly using an in-house pipeline (Supplementary Fig. S1). We polished the PacBio assembly using haplotype-aware chromium reads, which resulted in highly contiguous haplotype assemblies. We next replaced the 4.3-Mb region harboring the inversion region in the chromium assembly with the gapless genomic sequence of the matching haplotype from the PacBio assemblies (Supplementary Fig. S1). The satellite inversion haplotype was 14 kb shorter than the corresponding independent haplotype. We identified the inversion breakpoints of the hybrid genome assembly by mapping PacBio reads and chromium 10× reads to the final assembly (independent: 5,548,078 to 9,885,008; satellite: 5,546,737 to 9,869,715). We found that the independent haplotype assembly is approximately 200 kb shorter than the previous short-read only assembly (Lamichhaney et al. 2016). Evidence from genome and long-read alignments indicates that erroneously duplicated sequences account for ∼52 kb of unique sequence in the previous assembly and 115 kb represents gap sequences. No BUSCO genes are present within the inversion, but the BUSCO score (Manni et al. 2021) was high for the full assembly (94.8% completeness) demonstrating the quality of the base assembly. Together, these data suggest that the updated assembly reported here is both more contiguous and complete than the previous assembly.
An important reason for assembling the satellite haplotype was to examine if the gene content in the S and I haplotypes are identical despite the high sequence divergence (1.4%) (Lamichhaney et al. 2016). We found that gene content and gene order are identical and no gene has been recruited to the inversion haplotype or lost (Supplementary Table S2). We annotated the gene content of the inversion of both haplotypes using previously published transcriptome data from 5 tissues (n = 10 individuals) (Küpper et al. 2016) and 1 new sample from skin tissue of a male independent using the Maker (v3.01.2-beta) pipeline followed by manual refinement. We identified 107 full-length gene models, for which 99 have a known homology to annotated genes in other species, while 8 putative genes show no identifiable homology to known genes (Supplementary Table S2). These novel genes are supported by RNA-sequencing (RNA-seq) data, and the ratio of nonsynonymous to synonymous substitutions (dN/dS) between independent and satellite haplotypes for 4 of these genes was much lower than 1 (mean = 0.03) indicating that those genes are under strong purifying selection (Supplementary Table S3). Our annotation included 13 genes that were not previously reported in the ruff inversion, 1 that was identified as a different ortholog, and 2 that were annotated previously were not found (Supplementary Table S2).
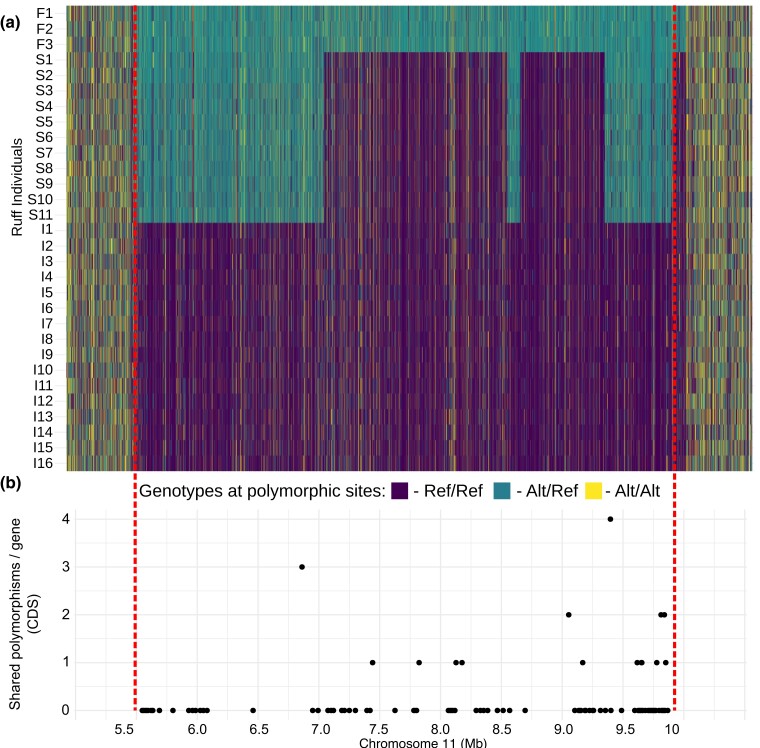
We next investigated genetic divergence and diversity using previously published (Küpper et al. 2016; Lamichhaney et al. 2016) resequencing data from 30 individuals: 16 independents, 11 satellites, and 3 faeders. Genetic divergence between the satellites, faeders, and independents on chromosome 11 revealed sharp boundaries separating the inversion region from the remainder of chromosome 11 in faeder individuals (Fig. 1b). The satellite haplotype is divided into high and low FST regions in comparison with the independent haplotype, which reflects the nonrecombinant regions and the regions that recombined with a noninverted haplotype, respectively, when the satellite haplotype was formed in the past. The satellite haplotype includes 3 distinct regions with strong genetic differentiation relative to the ancestral faeder haplotype but low or no genetic differentiation from the independent haplotype (Fig. 1b). One of these regions, the smallest, is present outside of the inversion breakpoint and begins at the 9.89-Mb breakpoint of the inversion and continues for 121 kb outside of the inversion. Despite being outside of the inversion, faeders were strongly differentiated from independents and satellites, indicating that suppressed recombination between the faeder and independent chromosome extends beyond the 9.89-Mb inversion breakpoint (Fig. 2a). This region contains 3 genes; NFAT5, NOB1, and WWP2. This region has apparently recombined with an I chromosome during the formation of the S haplotype. A notable exception to the genetic similarity between the I and S haplotypes in this region is a 2,082-bp long interspersed nuclear element (LINE) insertion at the 9.89-Mb inversion breakpoint in the S chromosome.
Fig. 2.
SNP genotypes in 3 male morphs in ruff across the inverted region. a) Genotypes for each SNP for the inversion region +500 kb outside each breakpoint (top panel): dark blue, homozygous for independent reference allele; light blue, heterozygous for a reference and alternate allele; and yellow, homozygous for an alternate allele. b) Number of shared SNPs between satellite and independent chromosomes for each gene within the inverted regions. These were identified as SNPs homozygous among satellites (S/I). Such shared SNPs are not expected to be present unless there is genetic exchange between the 2 haplotypes.
Estimating the Age of the Inversion and the Satellite Haplotype
We estimated the time of divergence between the independent and satellite haplotypes based on the net number of nucleotide substitutions (da) between them. da was calculated separately for the high and low FST regions under the assumption that the low FST regions are more recently diverged from each other due to recombination events occurring much later than the initial formation of the inversion. Within the high FST regions, we estimated sequence divergence (da) at 1.46% based on the average number of heterozygous sites in S/I heterozygotes and I/I homozygotes (see Materials and Methods), resulting in an estimated age of the inversion of 3.84 ± 0.08 million years (mean ± standard error of the mean), almost identical to the previous estimate of 3.87 million years (Lamichhaney et al. 2016). The net number of nucleotide substitutions for the low FST region was estimated at da = 0, because pairwise nucleotide diversity (dxy = 0.28%) between S and I haplotypes was identical to the average nucleotide diversity (dx) among I chromosomes (Supplementary Fig. S2). Thus, the recombination event(s) in satellite haplotypes must have occurred much more recently than the average divergence time among independent chromosomes, which was estimated at 740,000 yr before present using the estimated dx (0.28%). In other words, the recombined satellite haplotype in these regions is as divergent from the average independent haplotype as a randomly drawn independent haplotype is. We illustrate this using local phylogenetic trees for the high and low FST regions constructed with all independent samples and only single satellite and faeder males. The single satellite and faeder are on a long branch in a tree based on the high FST region but only the faeder is clearly separated from independents for the low FST regions within the inversion (Fig. 1c).
We therefore estimated the age of the satellite haplotype by calculating sequence divergence between the satellite and faeder haplotypes for the 2.15-Mb high FST region (i.e. the nonrecombinant regions). We used individual resequencing data and identified 411 nucleotide positions where all faeders (n = 3) were heterozygous for a single nucleotide polymorphism (SNP) not present in satellites (n = 11) or independents (n = 16) and the corresponding number for satellite-specific SNPs is 190. Based on the total number of SNP differences (601) between satellite and faeder haplotypes, we estimate sequence divergence (dxy) of 0.028% corresponding to an estimated divergence time of 73,000 ± 1,400 yr (mean ± standard error of the mean). This is the estimated divergence time for the satellite and faeder haplotypes, but it is possible that 1 or more recombination events (i.e. the 3 low FST regions) creating the current satellite haplotype occurred subsequent to this split.
Unexpected Low Mutation Load in the Satellite and Faeder Haplotypes
The lack of recombination and the reduced effective population size in inversion heterozygotes should lead to mutation load on inversion haplotypes, due to less efficient purifying selection. However, the same does not apply for the independent haplotype because I haplotypes are able to recombine freely in I/I homozygotes, which constitute ∼90% of the ruff population, meaning they have a large effective population size in this widespread Eurasian shorebird. This assumption is supported by similar patterns of linkage disequilibrium (LD) decay among independents inside and outside the inversion region (Supplementary Fig. S3). In contrast, an accumulation of genetic load is expected in the satellite and faeder haplotypes, because they are homozygous lethal and occur at low allele frequencies, about 5% and 1%, respectively. It has been proposed that the accumulation of genetic load in the faeder haplotype has contributed to intralocus sexual conflict and consequently low reproductive output in faeder females (Giraldo-Deck et al. 2022). We test 2 predictions of mutation load: first, the inverted haplotype should have expanded repeat content, and second, the inverted haplotype should have accumulated deleterious mutations as well as an excess of nonsynonymous substitutions.
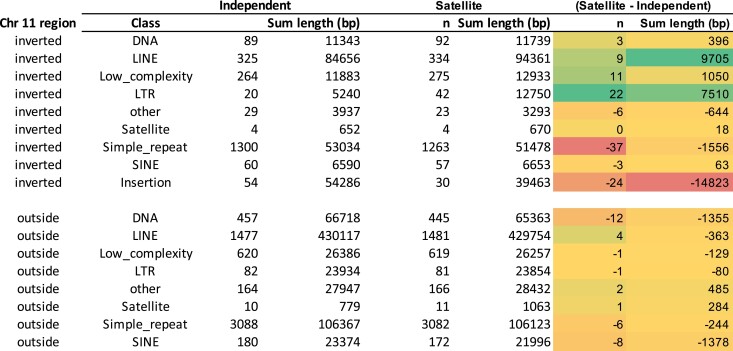
Reduced recombination due to the presence of inversions is expected to lead to the accumulation of transposable elements in chromosomes, and this has been noted in many taxa (Stolle et al. 2019; Gutiérrez-Valencia et al. 2021; Jay et al. 2021). However, consistent with the smaller size of the satellite haplotype relative to the independent haplotype, it included only about 187 kb of classified repetitive elements: less than 5% of the full inversion length (Table 1). Nevertheless, some repeat families have expanded in the satellite haplotype, including 9.7 kb of LINE elements and 7.5 kb of long terminal repeat (LTR) elements, but the smaller size of the satellite haplotype is explained by fewer simple repeats and the presence of intergenic deletions.
Table 1.
Number of repeats in the satellite and independent haplotypes
Summary of RepeatMasker annotated repeats within the inverted region and outside of the inverted region on chromosome 11. For the independent and satellite haplotype, the number of each repeat class and the total length of all elements are shown. The satellite–independent columns show the difference in number or length of elements between the 2 haplotypes. Insertions were tabulated only for the inverted region, because the remainder of chromosome 11 is not phased. The color codes for the columns representing the satellite–independent comparison highlight the most notable differences; green and red mean higher and lower numbers in the Satellite haplotype, respectively.
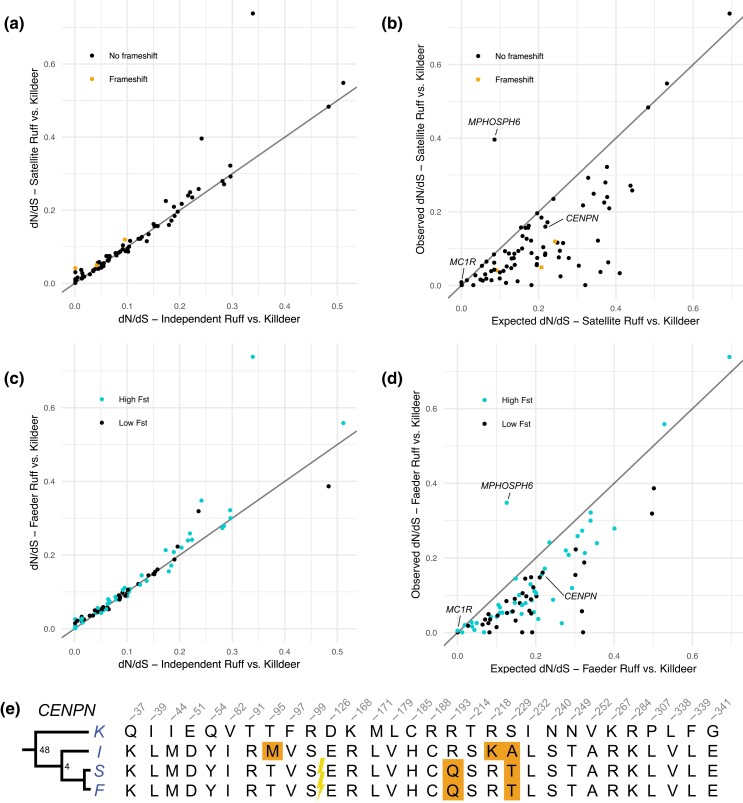
We did not identify a single premature stop codon on the satellite haplotype in any of the ∼100 genes within the inversion, but 4 genes (CMIP, TANGO6, COG8, and TMED6) carried frameshift mutations. To further quantify mutational load and patterns of molecular selection, we next calculated nonsynonymous (dN) and synonymous (dS) substitution rates for each of the genes within the inversion region comparing I versus S and I versus F haplotypes (Supplementary Fig. S4). We found that the great majority of genes had dN/dS ratios of <0.50. Six genes have dN/dS of >0.50 and <1.0, but each of these had just a few synonymous mutations, implying that this ratio is skewed by the lack of synonymous mutation (Supplementary Fig. S4). Two genes, melanocortin-1 receptor (MC1R) and M-phase phosphoprotein 6 (MPHOSPH6), had no synonymous mutations but multiple nonsynonymous changes.
The dN/dS ratio cannot determine if the observed missense mutations have occurred on the independent or the inversion haplotypes. We therefore calculated dN and dS rates for each of the genes with alignable orthologs between ruff haplotypes and killdeer (Charadrius vociferus), a related outgroup species with an annotated genome assembly (Supplementary Table S4). We first compared the I and S haplotypes to the corresponding sequences from killdeer. We restricted our analysis to the nonrecombinant high FST regions, because it is not meaningful to explore the presence of genetic load in the low FST regions with such high sequence identity between S and I haplotypes (Fig. 1c). We found very similar dN/dS in I and S orthologs compared with killdeer for the great majority of genes (Fig. 3a; Supplementary Table S4). Nevertheless, there was a significant excess of satellite orthologs with the highest dN/dS relative to killdeer; within the inversion, 45 genes had higher dN/dS in satellites compared with 25 genes showing the opposite trend (χ2 = 5.7, df = 1, P = 0.02), and dN/dS was identical for 12 genes. We next calculated the expected dN/dS relative to killdeer in the absence of purifying selection (i.e. dN = dS) for each satellite ortholog since the split from the independent allele (see Materials and Methods) and found that 68 of the 82 satellite orthologs had a lower dN/dS than expected (Fig. 3b), consistent with an effect of purifying selection on the evolution of protein-coding genes on the satellite haplotype. We performed the same analysis by comparing the faeder (using resequencing data) and independent haplotypes and found a very similar low mutation load on the faeder haplotype (Fig. 3c and d).
Fig. 3.
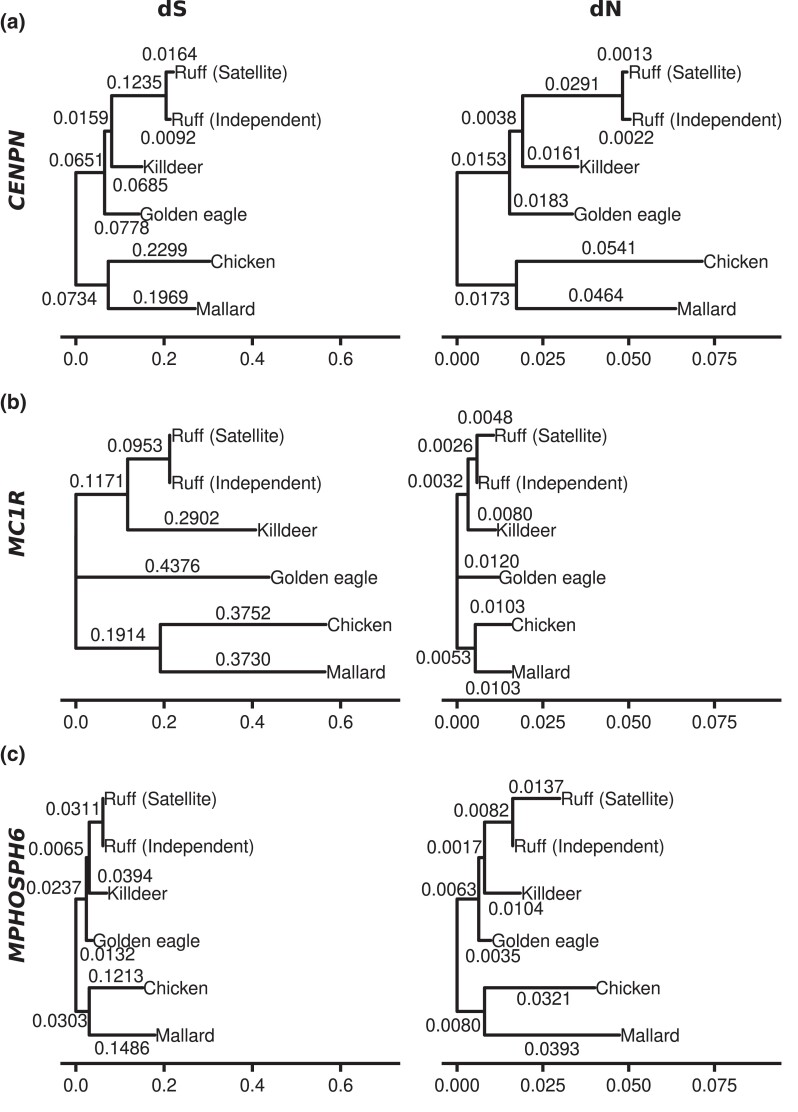
Evidence for purifying selection acting on satellite and faeder haplotypes. a) dN/dS ratios in pairwise comparisons of independent or satellite alleles versus killdeer (C. vociferus) for each gene ortholog located in regions with high FST in the comparison of satellites and independents. Genes with frameshift mutations in the satellite haplotype are marked in orange (CMIP, TMED6, and COG8), and all other genes are marked in black. b) The observed dN/dS ratios for satellite versus killdeer orthologs compared with expected under the assumption of no purifying selection acting on the satellite haplotype. c) dN/dS ratios in pairwise comparisons of independent and faeder alleles versus killdeer for each gene ortholog in the inversion. Each gene is colored by location in high FST (blue) or low FST (black) region. d) The observed dN/dS ratios for faeder versus killdeer orthologs compared with expected under the assumption of no purifying selection acting on the faeder haplotypes. e) Amino acid alignment of variable sites for the CENPN protein among killdeer and the 3 different ruff alleles. The N-terminal part of CENPN is encoded by exons located outside the inversion, and the inversion breakpoint is indicated with a flash. The derived T95M amino acid substitution was only found among independent chromosomes but is not a fixed difference.
The 5.55-Mb breakpoint of the inversion interrupts CENPN and is the predicted mechanism for the lethality of homozygous S/S individuals (Küpper et al. 2016; Lamichhaney et al. 2016) (Supplementary Fig. S5). We predicted that the 3′ fragment of the gene within the inversion would be a pseudogene as old as the inversion that has accumulated a roughly equal number of synonymous as nonsynonymous mutations. Unexpectedly, we found the same number of nonsynonymous substitutions (2) between killdeer and each of the 3 ruff haplotypes (Fig. 3e). We evaluated protein evolution under a phylogenetic context using 3 additional outgroups (Fig. 4a) using the Phylogenetic Analysis by Maximum Likelihood (PAML) package (Yang 2007). We found that the rate of nonsynonymous substitutions in the satellite branch did not differ significantly from the rate in the rest of the phylogeny (P(ω0 = ωS) = 0.32) (Fig. 4a). This is an unlikely finding if the gene was inactivated 4 million years ago or if it has evolved a novel function subsequent to the inversion event. Furthermore, a reverse transcription polymerase chain reaction (RT-PCR)-based study targeting the 3′ part within the inversion using kidney, muscle, and testis tissue from 2 adult satellite males revealed only expression of the independent allele (Supplementary Fig. S6). Similarly, Loveland et al. (2021) detected only full-length transcripts from the independent allele in satellite heterozygotes. This does not exclude the possibility that a truncated form of CENPN is expressed in a specific tissue or during a critical stage of development.
Fig. 4.
Comparison of the rate of nonsynonymous substitutions in the ruff satellite branch compared with other branches in the bird phylogeny. Shown here are phylogenetic trees for CENPN a), MC1R b), and MPHOSPH6 c) using a model where the substitution rate ω (dN/dS) was allowed to vary in the ruff Satellite branch. Branch lengths correspond to dS (left) and dN (right).
Identification of Candidate Genes under Selection
Two genes had a clear excess of derived amino acid substitutions on the inversion haplotypes, MC1R and MPHOSPH6 (Supplementary Figs. S4 and S7). Two missense mutations differ between killdeer and the ruff independent MC1R alleles that share a common ancestor about 50 million years before present (Prum et al. 2015), whereas independent and satellite alleles that separated not more than 4 million years ago differ by 4 (Supplementary Fig. S7a). Consistent with this observation, a PAML (Yang 2007) analysis supported an accelerated protein evolution in the satellite branch (P(ω0 = ωS) < 0.001) (Fig. 4b).
The faeder haplotype also has a high dN/dS ratio for MC1R (Fig. 3d); it shares 1 missense mutation with the satellite haplotype and has 2 unique missense mutations at residues 307 and 309 (Supplementary Fig. S7a). However, in this case, it may a reflect lack of purifying selection. Even if the F MC1R allele is nonfunctional, a single copy of an MC1R wild-type allele is expected to be sufficient for a wild-type phenotype. Faeders (I/F) show a plumage indistinguishable from the wild-type plumage in females as well to the wild-type male plumage outside the breeding season when independents and satellites do not have ornamental feathers.
MC1R has a fundamental function in pigmentation and determines pigment switching between black/brown eumelanin and red/yellow pheomelanin (Mundy 2005) and is therefore the leading candidate gene underlying light-colored ornamental feathers in satellites (Lamichhaney et al. 2016) (Fig. 1a). However, to explore the possibility that the satellite MC1R allele could have evolved a new function outside the pigment system, we screened the MC1R mRNA expression across 10 tissues including the brain in 2 independents and 2 satellites (Supplementary Fig. S8). MC1R expression was high in testis, brain, and wattle relative to other tissues, but there was no difference in MC1R expression between satellites and independents.
The satellite MPHOSPH6 allele also carries 4 derived missense mutations (Supplementary Fig. S7b) and not a single synonymous change (Fig. 3b; Supplementary Table S4) making this also a candidate gene under selection. In fact, PAML (Yang 2007) analysis supports accelerated evolution in the satellite branch also for MPHOSPH6 (P(ω0 = ωS) = 0.02) (Fig. 4c). The faeder allele shares 3 of the 4 nonsynonymous changes (Fig. 3d; Supplementary Fig. S7b). MPHOSPH6 is less well studied than MC1R but was identified as being phosphorylated during the M phase in the cell cycle (Matsumoto-Taniura et al. 1996) and is a component of the RNA exosome (Schilders et al. 2005). Genome-wide association studies in humans have found that MPHOSPH6 is associated with variation in leukocyte telomere length, a marker for genome aging (Li et al. 2020). Thus, the function of MPHOSPH6 may be relevant for chromosome function and the evolution of the ruff supergene because the inversion disrupts the CENPN gene.
Genetic Exchange between Haplotypes
Inversion haplotypes may exchange genetic material by double recombination and gene conversion (Navarro et al. 1997). Other than the 3 large recombinant regions described above, we did not find any evidence of frequent recombination events in the form of segments with high sequence identity between independent and satellite haplotypes in any pairwise comparison for the high FST region. In order to test the prediction of gene conversion, we counted shared polymorphisms in exons within the inversion by identifying SNPs for which both homozygous genotypes were present among satellites; in the absence of gene conversion, we expect none unless the same mutation occurred on both satellite and independent haplotypes. This analysis identified 14 genes that carried at least 1 shared polymorphism (Fig. 2b) with the most frequent occurring in Cadherin-14 (n = 4 shared polymorphisms). Notably, 7 of the 14 shared polymorphisms were found in the 500 kb immediately adjacent to the 9.89-Mb breakpoint. Genetic differentiation is high between satellites and independents in this region (Fig. 1b), suggesting that these shared polymorphisms were most likely produced through gene conversion rather than double crossovers.
Evaluation of Potential Donor Species for an Introgression Event
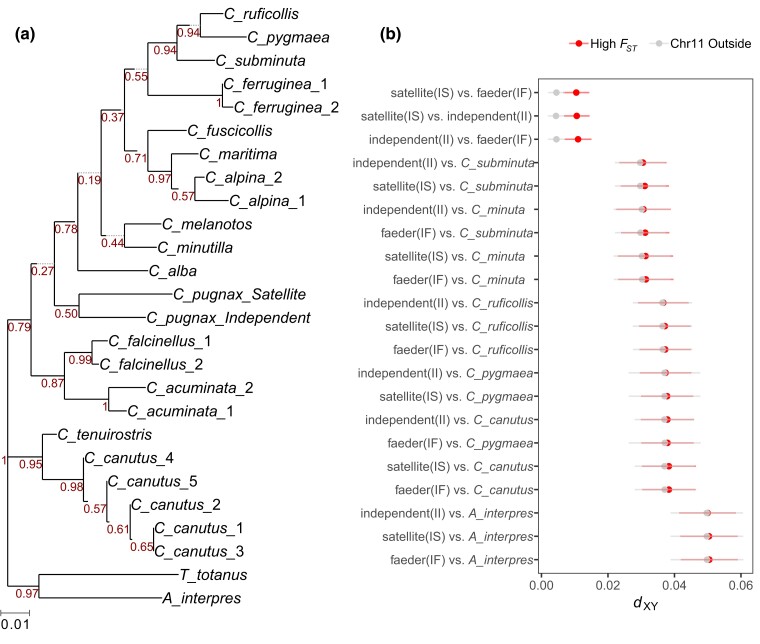
One possible explanation for the unexpected low mutational load in the ruff inversion polymorphism is that the inversion is much younger than indicated by the sequence divergence. This may occur if it was introgressed from another species, for example as has been suggested for an inversion polymorphism in the white-throated sparrow (Zonotrichia albicollis) (Schwander et al. 2014). We tested this hypothesis using 3 approaches: first, by sequencing a 965-bp amplicon from the high FST region (chr11: 5,695,640 to 5,696,604) from 15 other Calidris species and 2 outgroup species, Arenaria interpres and Tringa totanus (Supplementary Table S5); in this amplicon, the independent and satellite haplotypes differed by 43 single base substitutions. In a phylogenetic reconstruction, we found that the ruff independent and satellite sequences are monophyletic but on a long branch (Fig. 5a). Nevertheless, the genetic distance separating the independent and satellite sequences is larger than that between most sister species in this phylogeny and the independent sequence carries a derived 93-bp insertion. The branching order based on this amplicon sequence is not fully resolved, with low bootstrap values for all nodes. We next accessed publicly available whole-genome resequencing data for 4 Eurasian Calidris taxa and estimated sequence divergence among all pairwise comparisons across the inversion region. We found that dxy between any ruff haplotype and these other Calidris species were substantially larger (0.03 to 0.04) than between the independent and satellite/faeder haplotypes (0.01) and no species had reduced genetic divergence to the inversion haplotypes, as would be predicted by an introgression event (Fig. 5b). Finally, using an approach that does not require a donor species, we explored the possibility of an introgression event by searching the ruff genome for haplotypes that segregate at comparable sequence divergence as the ruff inversion and may harbor additional introgressed fragments. No other genomic regions show such an elevated sequence divergence among haplotypes matching the inversion (Supplementary Fig. S9). We conclude that although the mutational load data are consistent with an introgression event of an unsampled taxon, neither amplicon nor resequencing data provide evidence for introgression of the inversion from an extant species.
Fig. 5.
Evaluation of potential donor species for an introgression event. a) Maximum likelihood phylogenetic tree based on sequencing of a 1-kb amplicon from the high FST region (chr11: 5,695,640 to 5,696,604 bp in the independent assembly) from 15 Calidris species, 2 outgroup species in comparison with the corresponding independent and satellite sequences. Bootstrap values based on 500 repetitions are given at the nodes. b) Pairwise nucleotide divergence (dxy) among ruff mating phenotypes and 5 other Eurasian Calidris species and the outgroup A. interpres for which whole-genome sequencing data are available. Satellite and faeder are included as heterozygous genotypes, i.e. I/S and I/F. Mean dxy is shown for each pairwise comparison, and error bars represent the standard deviation among 15-kb windows either within high FST regions of the inversion or outside the inversion region on chromosome 11.
Discussion
Our assembly of the satellite inversion haplotype provides an opportunity to examine the evolutionary history of a chromosome segment that has been maintained in the heterozygous state for a long period of time, because the inversion is a recessive lethal and satellite homozygotes are not born (Küpper et al. 2016). The estimated sequence divergence of 1.46% between the nonrecombinant region of the satellite haplotype and the noninverted independent haplotype implies that the split from a common ancestral sequence occurred about 4 million years ago. The current understanding of the evolutionary trajectory of genomic regions that are homozygous lethal and nonrecombining is toward a fate resembling that of the Y chromosome in XY sex chromosome systems (Bachtrog 2008; Charlesworth and Charlesworth 2020). In these cases, the Y chromosome accumulates deleterious mutations that inactivate most of the genes found in the homologous X chromosome. Further, the accumulation of genetic load and transposable element expansion within supergenes associated with inversions is well documented empirically in several species of plants and insects (Nishikawa et al. 2015; Gutiérrez-Valencia et al. 2021; Jay et al. 2021).
The ruff inversion polymorphism is an unexpected exception to this model. First, we see no substantial accumulation of repetitive elements, a small expansion of LINE and LTR repeats amounts to less than 20 kb (<2% of the total inversion length; Table 1). This is in stark contrast to other examples of inversions in natural populations, ranging from the 340,000-yr-old inversion in Formica ant species composed of as much as 80% repeats (Brelsford et al. 2020) and an 1.8-million-yr-old inversion in Heliconius numata butterflies that is 10% larger than noninverted haplotypes mainly due to the recent insertions of transposable elements (Jay et al. 2021). The satellite inversion region is about 14 kb smaller than the ancestral haplotype, due to the presence of deletions that may be adaptive. Three of the deletions (total size = 26 kb) flank the HSD17B2 gene; HSD17B2 is an enzyme with a key role in testosterone metabolism (Lamichhaney et al. 2016); altered regulation of this gene may be related to the reported low levels of circulating testosterone in satellite and faeder males (Küpper et al. 2016). Second, we find low genetic load affecting coding sequences given the deep divergence time for the inversion polymorphism. A few genes carry frameshift mutations in the satellite haplotype, but the remaining 75 within the nonrecombinant region carry a low, if any, mutational burden in comparison to the independent chromosome. Double crossovers and gene conversion may cause some gene flow from independent to variant chromosomes but cannot explain the low dN/dS ratios in relation to the estimated time of divergence (Fig. 3). Furthermore, given the considerable sequence divergence between haplotypes, it was unexpected that the disrupted centromere protein gene CENPN is as well conserved on the satellite and faeder haplotypes as it is on the independent haplotype containing an intact CENPN copy (Fig. 3e). The 3′ part of the gene that is well separated from the promotor and located inside the inversion has a lower rate of nonsynonymous substitutions than expected if it was pseudogenized 4 million years ago or evolved an altered function subsequent to the inversion event (Figs. 3 and 4a).
The recombination event(s) that produced the satellite haplotype was previously estimated at 500,000 yr ago based on the average sequence divergence between the independent and satellite haplotypes (Lamichhaney et al. 2016). Here, we find a much more recent origin using a more accurate method taking into account sequence diversity among independent haplotypes (see Materials and Methods). This difference in the estimated time to divergence is a consequence of similar nucleotide divergence in the low FST region between the satellite and independent haplotypes to diversity among independent haplotypes. We refined our estimated age of the satellite haplotype to approximately 70,000 yr ago based on the sequence divergence between satellite and faeder haplotypes for the nonrecombinant region (high FST between satellites and independents and low FST between satellites and faeders) (Fig. 1c).
More than 100 missense mutations in genes within the inversion region on chromosome 11 distinguish the satellite and independent haplotypes, but only 3 of them occur in the recombinant regions (Lamichhaney et al. 2016). In contrast, the great majority of missense mutations that distinguish the satellite and faeder haplotypes are located in these recombinant regions where satellite and independent haplotypes are almost identical. Thus, the satellite haplotype constitutes a combination of 1 set of genetic variants shared with independents and another set shared with faeders, which is matched by the shared ornamental feathers between satellites and independents but for instance low levels of testosterone in blood in both satellites and faeders (Küpper et al. 2016).
The low mutation load in the satellite haplotype warrants speculation on the potential mechanistic drivers of the observed pattern. The accumulation of genetic load in a supergene that is recessive lethal can to some extent be counteracted by purifying selection acting in heterozygotes due to haploinsufficiency. However, this is an unlikely explanation in this case because this would imply that a large proportion of deleterious mutations are incompletely recessive since 68 out of 82 genes showed lower dN/dS ratios in satellite haplotypes than expected if mutations were fully recessive (Fig. 3b). Further, this interpretation does not apply for the truncated copy of CENPN located within the inversion, because selection to maintain its protein sequence could only occur if it has evolved an unknown functional role. If so, the expression of this 3′ fragment of the gene must be driven by a novel promoter, because the fragment is separated from its promoter sequence. However, we cannot support this model with our data, because we failed to detect any expression of the satellite allele in adult satellite heterozygotes (Supplementary Fig. S6). A more likely explanation is that the inversion is not as old as the sequence divergence data indicate. There are 2 main scenarios that would be consistent with existing data. First, that an old larger inversion has recombined much more recently and resulted in the disruption of CENPN and recessive lethality, similar to the recombination events creating the satellite haplotype (Fig. 1a) or the second inversion haplotype at the chicken Rose-comb locus (Imsland et al. 2012). If this hypothetical old version was fairly common and not a recessive lethal, efficient purifying selection against deleterious alleles could occur in inversion homozygotes, as is the case for inversions related to local adaptation in Atlantic herring (Han et al. 2020), Atlantic cod (Matschiner et al. 2022), and sunflowers (Huang et al. 2022). Second, the inversion may be the result of an introgression event from another species. Under this model, the inversion occurred before or after the introgression event and was favored by selection, because it kept together alleles at multiple loci contributing to the male mating phenotype. Such a scenario has been hypothesized for the mating system-linked supergene in white-throated sparrow (Z. albicollis) (Schwander et al. 2014). In the white-throated sparrow supergene, similar to the ruff supergene, dN/dS is the same on the wild-type and inversion chromosomes for all fixed differences between haplotypes. This was interpreted to reflect that the inversion is much younger than what the sequence divergence indicates (Schwander et al. 2014). Another example of an interspecies introgression of a supergene concerns the Y chromosome in the ninespine stickleback (Pungitius pungitius) that has been introgressed from the Amur stickleback (Pungitius sinensis) (Dixon et al. 2019).
We used 3 analyses to evaluate our hypothesis that the inversion originated from an introgression event from another species. However, amplicon data from 15 Calidris species, whole-genome resequencing data from 4 other Calidris species, and a search for other divergent haplotypes across the genome failed to identify candidate donor taxa or indications of introgression elsewhere in the genome. However, the question is complicated by the fact that gene conversion and double crossovers between wild-type and inversion haplotypes subsequent to the introgression could have blurred the phylogenetic signal making it challenging to identify a donor species among the large number of other extant Calidris species. Furthermore, the divergence between Ruff inversion haplotypes (∼4 million years ago) is younger than the estimated time to the most recent ancestor with other Calidris (∼15 million years ago) (Černý and Natale 2022), meaning any donor species may be extinct and would have been missed by our analyses. Nevertheless, we consider the introgression hypothesis as possible, because it is a simpler explanation for the low mutation load in the inversion than other potential models. We also note that the ancestral inversion phenotype exhibited by faeder male morphs is similar to other Calidris species, where all species lack prominent sexual dichromatism and the ornamental feathers of independent and satellite morphs.
Male mating success is highly skewed among territorial male ruff (Widemo and Owensi 1995), resulting in strong sexual selection for alternative male reproductive strategies. Our results extend the evolutionary model for the origins of the unique mating strategy in ruff. The evolution of a supergene may start because an inversion by itself results in a favorable phenotype, for instance by changing gene expression as documented for the Rose-comb inversion in chicken (Imsland et al. 2012) or because an inversion event captures multiple linked polymorphisms affecting fitness (Kirkpatrick and Barton 2006). Such a cluster of linked polymorphisms may have been introgressed into the population as discussed in this paper. The ancestral inversion faeder phenotype experienced a fitness gain and evolved an evolutionary stable strategy as a sneaker male competing for females attracted to leks by displaying independents (Jukema and Piersma 2006). Mutations enhancing fitness of the supergene have been under positive selection, and a prime example is the recombination events that led to the evolution of the satellite hybrid haplotype (Fig. 1a). The satellite male constitutes a second alternative strategy, relying on ritualized interactions with territory holding independents and scramble competition for females attracted to interacting independent–satellite pairs within leks. The independent, satellite, and faeder morphs are apparently under strong negative frequency-dependent selection as a mixed evolutionarily stable strategy (Widemo 1998a). The stability of the lekking system is maintained by females being attracted to independents displaying within established dominance hierarchies at traditional lek sites (Widemo 1997, 1998b). The success of nonterritorial faeders and satellites relies on there being sufficient displaying independents, and the frequency of the satellite and faeder haplotypes is low, usually about 5% and 1%, respectively (Widemo 1998a). Morphs carrying the inversion have a high fitness, compensating for homozygous lethality, and this fitness is high only when these nonterritorial morphs are rare. The low haplotype frequencies imply low effective population sizes compared with the independent haplotype, another argument why substantial genetic loads are expected for satellite and faeder.
Positive selection may lead to accelerated evolution as altered gene function evolves. A prime candidate is the evolution of the satellite MC1R allele (Fig. 4b) that is likely related to the light-colored ornamental feathers in satellite males (Fig. 1a). For a supergene that is recessive lethal like the faeder and satellite haplotypes, purifying selection will only affect dominant negative mutations as well as genes showing haploinsufficiency, i.e. when a deleterious mutation affects the fitness of heterozygotes.
Together, we show that the evolutionary history of the ruff inversion, which determines alternate male mating strategies, is more complex than previously thought. Molecular dating suggests that the inversion is very old (4 million years ago), but polymorphism data suggest that it neither carries the mutation load expected for a 4-million-yr-old recessive lethal nor an expansion of repeat sequences. A possible explanation for this apparent contradiction is that the inversion introgressed from another species much more recently or alternatively that the current organization with disruption of CENPN is relatively young. Furthermore, we show that the origin of the satellite haplotype is much more recent than previously thought and occurred about 70,000 yr before the present. Our comprehensive description of the evolution of the ruff mating polymorphism reveals that supergenes may have a complex history involving strong selection, recombination, and possibly introgression.
Materials and Methods
Genome Assembly
A male ruff individual with the satellite phenotype was collected in northern Sweden (coordinates WGS 84 [latitude, longitude] 68.1, 19.8) during the reproductive season of 2016 under permit Swedish Environmental Protection Agency NV 02900-16. Small muscle pieces were stored in RNAlater (Thermo Fisher) at 4 °C until DNA preparation using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacture's recommendation. A chromium 10× linked read library was produced according to the manufacturer's recommendation and sequenced on an Illumina HiSeqX to a target depth of 90×. Supernova (v2.0) (Weisenfeld et al. 2017) was used to build a diploid assembly with a phased contig N50 of 20.2 Mb. A PacBio long-read library was prepared from the same DNA sample and processed with Falcon (v0.5) (Chin et al. 2016) to produce a second diploid genome assembly. Assembly scaffolds were aligned to Gallus gallus and C. pugnax genomes using Satsuma2 (v2016-12-07, https://github.com/bioinfologics/satsuma2) chromosemble to identify the scaffold homologous to the part of chicken chromosome 11, which corresponds to the region of the ruff genome harboring the inversion. Chromium 10× linked reads were mapped to the genome assembly using longranger (v2.2.2) (Weisenfeld et al. 2017), and PacBio long reads were mapped with minimap2 (v2.14) (Li 2018). We compared the independent and satellite assemblies using Longranger to identify the inversion breakpoints as well as structural variants, which was further refined and confirmed with manual inspection of read alignments in IGV (v 2.5.3) (Robinson et al. 2011).
While the supernova assembly using linked reads provided a contiguous and well-phased genome assembly, the nature of the short-read genome assembly process left inevitable gaps in the sequence. To produce a complete assembly of both the independent and satellite haplotype of the inversion region the Falcon long-read assembly contigs were identified as independent or satellite using the phased SNPs from the supernova assembly. Falcon contigs were then aligned to each other using Satsuma2 and BLAST (v2.11.0) (Camacho et al. 2009) to resolve overlaps. Incorrect haplotype switching within Falcon contigs was identified using chromium 10× linked reads and manually corrected. Corrected Falcon contigs were merged into gapless haplotype assemblies and polished using Pilon (v1.22) (Walker et al. 2014), and the chromium 10× reads were split by haplotype. We assessed conserved gene completeness with BUSCO (v5.3.1) (Manni et al. 2021) using the aves_odb10 lineage data set, and results were consistent with a complete haploid assembly (C: 94.8% [S: 94.3%, D: 0.5%], F: 0.9%, M: 4.3%, n: 8,338).
Comparison to Previous Short-Read Assembly
The previously reported assembly of the ruff genome was based on short-read Illumina sequencing using fragment libraries with insert sizes ranging from 250 bp to 20 kb (Lamichhaney et al. 2016) and is thus more fragmented than the assembly reported in the present study; contig N50 length has increased from 106 to 294 kb, and scaffold N50 length has increased from 10.0 (Lamichhaney et al. 2016) to 27.7 Mb (Supplementary Table S1). Chromosome 11 of the PacBio-based assembly was aligned to Scaffold 28 of the previous short-read assembly using the MUMmer (v4.0.0rc1) (Marçais et al. 2018) dnadiff tool with default parameters. Gaps identified between the present assembly and the former short-read assembly in the inversion region were intersected with between-contig stretches of Ns in the short-read assembly using BEDTools (2.29.2) (Quinlan and Hall 2010). This exercise provided an estimate of the assembly size inflation due to the addition of Ns during the scaffolding of the short-read assembly. Long PacBio reads mapped to the current and previous assemblies as described above were used to determine support for apparent sequence duplication in the short read assembly as well as for visual inspection of uncategorized discrepancies greater than 1 kb.
Genome Alignments
Satellite and independent chromosome 11 were aligned to each other using MUMmer (nucmer) (Marçais et al. 2018). The corresponding alignments were uploaded to the Assemblytics (Nattestad and Schatz 2016) web portal and variants called with unique sequence length = 10,000, maximum variant size = 10,000, and minimum variant size = 50.
Genome Annotation
We annotated the independent and satellite haplotypes (of chromosome 11) using MAKER (3.01.2-beta) (Cantarel et al. 2008). Prior to annotation, we created a custom repeat library using Repeat Modeler (1.0.8, http://www.repeatmasker.org) and RepeatMasker (4.0.7, http://www.repeatmasker.org) and downloaded all protein sequences in the curated and reviewed UniProt database (Uniprot Consortium 2020) and for the previous assembly of ruff (GCF_001431845). We downloaded RNA-seq reads for 10 previously sequenced individuals (Küpper et al. 2016) across 5 tissues and additionally included a single skin tissue from the present study. Briefly, skin tissue was dissected in the field and immediately stored in RNAlater (Thermo Fisher) to stabilize the RNA. RNA was extracted and DNase was treated as described previously (Schwochow Thalmann et al. 2017). The RNA quality and concentration were measured by the RNA ScreenTape assay (TapeStation, Agilent Technologies). Strand-specific mRNA sequencing libraries were generated using the SENSE RNA-Seq Library Prep kit (Lexogen). Briefly, 1 µg of total RNA was poly-A selected using magnetic beads. Illumina-compatible linker sequences were introduced to the mRNA by random hybridization. The amplified libraries were size selected for an average insert size of ∼350 bp and sequenced using an Illumina HiSeq instrument at SciLifeLab, Uppsala, Sweden.
We mapped RNAseq reads to each genome assembly containing either the independent or satellite chromosome using HISAT2 v2.1.0 (Kim et al. 2019) and assembled transcripts using StringTie v1.3.3 (Pertea et al. 2016). We extracted splice junctions by mapping all reads with TopHat2 v2.1.1 (Kim et al. 2013) and converting to gff3 with MAKER's (Cantarel et al. 2008) tophat2gff3 script. To generate high-quality gene models on each chromosome version, we ran MAKER (v3.01.2-beta) using protein and RNAseq data as evidence, splice junction annotations, and lists of repeats to mask low-confidence regions. We ran MAKER with best practices recommendations and additionally included max_dna_len = 150,000 and split_hit = 50,000 to fine-tune annotation for an avian genome. All MAKER-predicted proteins were blasted (using BLAST 2.7.1+) (Camacho et al. 2009) against the given protein evidence to produce candidate ortholog annotations for each annotated gene and the resulting gff updated using maker_functional_gff.
Manual curation of the gene models was carried out using the web-based genomic annotation editing platform Web Apollo (Lee et al. 2013) for both the independent and satellite haplotype of each gene. An exon was included in a gene isoform if it was supported by at least 3 RNA-seq reads with identical splice boundaries in an individual. Exon boundaries were defined by the longest continuous block of RNA-seq reads. The longest isoform was chosen as the representative of a gene in further analysis.
LD Decay
We used the software PopLDdecay 3.4.2 (Zhang et al. 2018) with a max distance of 50 kb between SNPs to generate LD decay curves for independents in each of 3 genomic intervals: before inverted region (chr11: 1 to 5,548,078), within inversion interval (chr11: 5,548,079 to 9,885,008), and after inversion region (chr11: 9,885,009 to 19,330,666). This contrast, using independents only, was used to compare LD decay inside and outside the inversion on wild-type (independent) chromosomes that occur in the homozygous state with a frequency of about 90% in ruff populations.
Nucleotide Substitution Rates
We used the manually curated gene set to calculate nucleotide substitution rates between independent and satellite alleles of each gene and their orthologs in killdeer. We selected an outgroup species, the killdeer (C. vociferus, Genbank ID: 1184028) with a previously published genome annotation to polarize alleles. We identified 45 orthologs present within the high FST regions in the comparison of satellite and independent haplotypes using reciprocal best blast between protein sequences. All ortholog and allele pairs were aligned using ClustalO (v1.2.4) (Sievers et al. 2011) and alignments were given to PAML (v4.9e) (Yang 2007) CODEML to calculate dN and dS values for each pair. We calculated dN/dS only for genes within the inverted region, because we are interested in evaluating differences in genetic load between independent and satellite/faeder haplotypes within the inversion region. A comparison of genes outside the inversion is not meaningful, because there is no genetic differentiation among male morphs in the rest of the genome (Fig. 1c).
Relaxed purifying selection is expected for genes on the satellite haplotype because this haplotype always occurs in the presence of independent haplotypes in I/S heterozygotes. The deviation of the observed dN on the satellite haplotype from the expected dN in the absence of purifying selection was calculated by estimating the increase in expected nonsynonymous substitution on the satellite haplotype, ΔdNs, as follows.
Definitions
dNs = dN in the satellite branch since the formation of the inversion.
dNi = dN in the independent branch since the formation of the inversion.
dNis = dN between independent and satellite (dNis = dNs + dNi).
dSis = dS between independent and satellite.
The amount dN increases in satellites if there is no purifying selection can be estimated by
ΔdNs = (dSis/2) − dNi.
However, since we do not know the value of dNi directly, we can estimate dNi based on dNis and dSis:
dNis = dNi + (dSis/2).
Thus,
dNi = dNis − (dSis/2).
Thus,
ΔdNs = (dSis/2) − dNis + (dSis/2) = dSis − dNis.
Based on this, we calculated for each gene the expected dN (between satellite ruff vs. killdeer) as dN (between independent ruff vs. killdeer) + ΔdNs and compared this with observed dN (between satellite ruff vs. killdeer). Estimation of nucleotide substitution rates involving the faeder haplotype was calculated in a similar way, the only difference being that faeder gene models were constructed by manually modifying satellite gene models based on resequencing from faeder samples. Manually constructed faeder gene models were then aligned and analyzed in the same way as described for satellite.
The CODEML function of the PAML program (Yang 2007) was used to test the relative likelihood of models in which substitution rates varied on the branches leading to the ruff orthologs of CENPN, MC1R, and MPHOSPH6. For each gene, the orthologs for ruff, killdeer, golden eagle, chicken, and mallard were aligned as described above in the section on dN/dS. CODEML then calculated the log-likelihood for models in which the substitution rate ω (dN/dS) was held constant for the entire ortholog tree (model 0) or allowed to vary only on the branch leading to satellite ruff haplotype (model S). The significance of the difference between the log-likelihoods of the 2 models was calculated with the Pearson chi-squared test using a 1-tailed distribution.
Genetic Variation Analysis
Individual resequencing data from ruff were downloaded from 2 previous studies: 25 individuals from Lamichhaney et al. (2016) and 5 individuals from Küpper et al. (2016). The accession numbers and average sequence coverage per individual are summarized in Supplementary Table S6. For analysis of potential donor species for the introgression analysis, we additionally included all resequencing data from other Calidris species available from NCBI BioProject #PRJNA419629. This includes 13 Calidris pygmaea samples, 9 Calidris ruficollis, 1 Calidris minuta, and 1 Calidris subminuta. Finally, we included 1 outgroup species, A. interpres, available from NCBI BioProject #PRJNA545868 (SRA#: SRR9946666 and SRR9946658) that is part of the B10K project (Feng et al. 2020). Read sets were adapter trimmed with bbmap v38.61b (ktrim = r, qout = 33, k = 23, mink = 11, hdist = 1, qtrim = r, trimq = 10, maq = 10, tpe, ow = t, tbo) and mapped to the independent ruff genome assembly using BWA-MEM (v0.7.17) (Li 2013) and default settings. We called biallelic SNPs using bcftools mpileup and call (v1.17) (Danecek et al. 2021) filtering on mapping quality of ≥20 and genotype position quality of ≥20 (see Data availability). VCF files were analyzed in R (v4.0.3) (Team RC 2020) using the packages zoo (v1.8-8) (Zeileis and Grothendieck 2005), viridis (v0.6.1) (Garnier et al. 2021), tidyverse (v1.3.1) (Wickham et al. 2019), and vcfr (v1.12.0) (Knaus and Grünwald 2017) within custom scripts (see Data availability).
We estimated relative divergence (FST) using the software pixy v1.2.2.beta1 (Korunes and Samuk 2021) in 15-kb nonoverlapping sliding windows and per site by setting the window size in pixy to 1 bp. For pairwise comparisons involving the other 5 Calidris shorebird taxa (described above), we additionally calculated dxy using 15-kb windows using pixy. High-confidence invariant sites were included to prevent biases from omitting missing data in this calculation. Note that for this comparison, we calculated dxy between heterozygous satellite (I/S) and faeder (I/F) and the other taxa, meaning dxy is calculated including the independent allele in the calculation. If an introgression event has occurred in the past, the inclusion of the I allele in this estimate will increase dxy for comparisons involving satellite and faeder, but nevertheless we expect a donor species to show a pattern of reduced dxy compared with S and F within the inverted regions compared with outside the inversion region.
Estimating the Age of the Inversion and the Satellite Haplotype
Nucleotide diversity among satellite chromosomes cannot be calculated using standard software because these only occur in the heterozygous state. We therefore estimated dxy between satellite and independent chromosomes based on the average number of heterozygous sites among callable sites in satellite males (S/I) and nucleotide diversity (dx) among independents (I/I) in the same fashion. The classical way to calculate divergence time between DNA sequences from 2 populations is based on calculating the net number of nucleotide substitutions taking into account the intrapopulation nucleotide diversity at the splitting time T using the formula da = dxy − (dx + dy)/2 according to Nei (1987); dx and dy are the intrapopulation nucleotide differences within populations X and Y, and (dx + dy)/2 is the best estimate of nucleotide diversity in the ancestral population. For instance, at T = 0 for populations of infinite size dxy = dx = dy, then the standard formula gives da = 0. However, this is not applicable for an inversion polymorphism because it originates from population X and shows no intrapopulation nucleotide diversity (dy) at T = 0. Thus, in this case, dxy = dx while dy = 0. Applying the standard formula will give the erroneous estimate da = dxy − (dx + 0)/2 = 0.5dx. The correct estimate is obtained using da = dxy − dx, that is da = 0 since dxy = dx. This is illustrated in Fig. 1c (right) because in the satellite–independent comparison da << dx. A similar method has previously been used to estimate divergence times for inversion polymorphisms in Drosophila (Corbett-Detig and Hartl 2012).
Time since divergence between satellite and independent chromosomes was estimated using T = da/2λ and among independent chromosomes as T = dx/2 λ according to Nei (1987) where λ is the average nucleotide substitution rate per year, and here we used the average estimate for birds 1.9 × 10−9 (Zhang et al. 2014).
Time since divergence between the faeder and satellite haplotypes was calculated for the high FST region in a similar way; however, we were restricted to a lower bound divergence estimate of the age due to the unphased nature of resequencing data. A SNP was only considered present and unique in a particular haplotype if it was heterozygous in every individual carrying that haplotype (I/F or I/S) and homozygous for the reference allele in other individuals. We reasoned that applying these constraints made it improbable that polymorphisms present on the independent haplotype were mistakenly counted as a satellite or faeder substitution when calculating dxy. However, as shown in Supplementary Table S7, SNPs that were heterozygous in at least 1 satellite and homozygous in all independents were in fact heterozygous in most satellites. This shows that our approach has not seriously underestimated the sequence divergence between satellite and faeder haplotypes.
To estimate the standard error of the time since divergence, the inversion region was divided into 200-kb non-overlapping windows (9 in the high FST region and 10 in the low FST region). We then calculated dxy and da as well as the time since divergence as described above for each window. These were used to calculate the mean and standard error of the mean.
Introgression Analysis using HaploDistScan
Biallelic SNPs generated above were further filtered to exclude any sites with missing samples. This filtered SNP set was phased for the whole genome using BEAGLE (5.4) (Browning et al. 2021) with default parameters. Phased genotypes were then analyzed using HaploDistScan (Pettersson et al. 2017).
CENPN RT-PCR Analysis
Total RNA was isolated from testis, kidney, and muscle of 2 satellite and 2 independent individuals as described above for RNA-seq. First-strand cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Thermo Scientific). The parts of the CENPN transcript encoded by exons inside the inversion were amplified using the following primers (5′-3′): F: AGGATGTGGTTTATCTTTGTGAGGAAA; R: TCTCAAGCCTATATTGTGCAAATTC. The amplification program was as follows: 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min. PCR products were Sanger sequenced.
Screening of MC1R Expression across Tissues
RNA preparation was as described above for RNA-seq analysis. Given that MC1R includes a single exon, it is not possible to prevent contaminating genomic DNA amplification by primers spanning exon–intron junctions. Therefore, we included RT-qPCR reactions without reverse transcriptase in the cDNA synthesis process to assess the amount of DNA contamination present in RNA samples. Thus, cDNA was prepared in parallel; one reaction includes reverse transcriptase (RT+), while the other one would be enzyme free (RT−). Afterwards, RT-qPCR was performed with Applied Biosystems SYBR Green PCR Master Mix (Thermo Fisher Scientific) in 384-well plates using primers (5′-3′) for MC1R: F: TGTCCTCCCTCTCCTTCCTG; R: AGAGGAGGATGGCGTTGTTG and GAPDH: F: CGCTAAGCGTGTCATCATCT; R: CAAGAGGCATTGCTGACAATTT. The reaction mixture contained 1-µl cDNA, 5-µl SYBR Green PCR Master Mix (2×) and 0.3 µl of each primer (10 µM) in a total volume of 10 µl, and the following PCR cycle was used, denaturation for 10 min at 95 °C followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. After amplification, fluorescent data were converted to threshold cycle values (Ct) and all amplicons were visualized by gel electrophoresis. The Ct value of MC1R was normalized against that of reference gene GAPDH. To make the expression across tissues comparable, the average expression over all tissues were calculated and used for normalization to get the expression in specific tissue. The expression of MC1R was finally determined by gel electrophoresis. To be specific, if the amplicons were visualized by electrophoresis, the expression level would be calculated as (RT+) − (RT−), otherwise the level of RT+ would represent the expression.
Sequencing of Amplicons from Other Calidris Species
In order to identify potential donor species from a wide taxonomic breadth across Calidris, we amplified a 965-kb amplicon corresponding to the interval 5,695,640 to 5,696,604 bp on ruff chromosome 11 (coordinates according to the independent assembly) from 15 other Calidris species and 2 outgroup species (Supplementary Table S5). This interval was selected because it contains a large number of variable sites between the independent and satellite alleles. We designed the following primer sequences (F: GGGGATCTCGATACAGGTCAG; R: GTACGGCGAAGGTCCGATG) for PCR amplification and the following primers for Sanger sequencing: (Independentseq: AACCTCCTGTTACTTGTCTTCTTCC; Satelliteseq: ACTAATAACCTGTAGCTTGTCTTCT). PCR amplifications were carried out as for the CENPN RT-PCR using PCR Master Mix (2×, Thermo Fisher). We built a phylogenetic tree using the maximum likelihood method and with the Tamura–Nei substitution model (Tamura and Nei 1993). We present the tree with the highest log-likelihood (−2495.10). Initial trees for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura–Nei model and then selecting the topology with superior log-likelihood value. This analysis included sequences from satellite and independent alleles and 24 other sequences. All positions with less than 95% site coverage were eliminated; i.e. fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There were a total of 602 variable positions in the final data set. All phylogenetic analyses were conducted in MEGA11 (Tamura et al. 2021).
The tissue samples for the phylogenetic analysis of amplicon data were provided by the Swedish Museum of Natural History, Gothenburg Museum of Natural History, Sun Yat-sen University, Beijing Normal University, and Nanjing Normal University.
Supplementary material
Supplementary material is available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Russel Corbett-Detig, Peter Grant, and Rosemary Grant for comments on the manuscript, Mats Pettersson for technical advice, and Mateusz Konczal and Fyodor Kondrashov for access to Calidris data. The project was financially supported by Vetenskapsrådet (2017-02907 to L.A.), Knut and Alice Wallenberg Foundation (KAW 2016.0361 to L.A.), the National Science Foundation of China (81961128002 to Y.L.), and the Basal Research Grant of South China Institute of Environmental Sciences (No. 22060302001001124; to C.W.). The National Genomics Infrastructure (NGI)/Uppsala Genome Center provided service in massive parallel sequencing, and the computational infrastructure was provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX partially funded by the Swedish Research Council (2018-05973).
Contributor Information
Jason Hill, Department of Medical Biochemistry and Microbiology, Uppsala University, SE-75123 Uppsala, Sweden.
Erik D Enbody, Department of Medical Biochemistry and Microbiology, Uppsala University, SE-75123 Uppsala, Sweden; Department of Biomolecular Engineering, University of California Santa Cruz, Santa Cruz, CA 95060, USA.
Huijuan Bi, Department of Medical Biochemistry and Microbiology, Uppsala University, SE-75123 Uppsala, Sweden.
Sangeet Lamichhaney, Department of Medical Biochemistry and Microbiology, Uppsala University, SE-75123 Uppsala, Sweden; Department of Biological Sciences, Kent State University, Kent, OH 44241, USA.
Weipan Lei, Key Laboratory for Biodiversity Science and Ecological Engineering, National Demonstration Center for Experimental Life Sciences and Biotechnology Education, College of Life Sciences, Beijing Normal University, 100875 Beijing, China.
Juexin Chen, State Key Laboratory of Biocontrol, School of Ecology, Sun Yat-sen University, 510275 Guangzhou, China.
Chentao Wei, State Key Laboratory of Biocontrol, School of Ecology, Sun Yat-sen University, 510275 Guangzhou, China.
Yang Liu, State Key Laboratory of Biocontrol, School of Ecology, Sun Yat-sen University, 510275 Guangzhou, China.
Doreen Schwochow, Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences, SE-75007 Uppsala, Sweden.
Shady Younis, Department of Medical Biochemistry and Microbiology, Uppsala University, SE-75123 Uppsala, Sweden; Division of Immunology and Rheumatology, School of Medicine, Stanford University, Stanford, CA 94305, USA.
Fredrik Widemo, Department of Wildlife, Fish and Environmental Studies, Swedish University of Agricultural Sciences, SE-901 83 Umeå, Sweden.
Leif Andersson, Department of Medical Biochemistry and Microbiology, Uppsala University, SE-75123 Uppsala, Sweden; Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX 77843, USA.
Author contributions
L.A. conceived the study. J.H. and E.D.E. were responsible for the bioinformatic analyses and statistical testing. F.W. and S.L. performed the field work and collected the tissue samples. D.S. prepared the genomic DNA and mRNA. S.Y. prepared the RNAseq library. H.B. was responsible for experimental work. W.L., J.C., C.W., and Y.L. contributed with samples and amplicon sequencing of other Calidris species. J.H., E.D.E., and L.A. wrote the paper with input from other authors. All authors approved the manuscript before submission.
Data availability
The sequence data generated in this study and genome assemblies have been submitted to NCBI (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA816664). Genome annotation is available online (https://github.com/LeifAnderssonLab/Ruff_assembly_2022/Annotation_gff). We additionally used sequencing data from BioProjects PRJNA419629, PRJEB10677, SRP058220, and PRJNA545868. The analyses of data have been carried out with publicly available software and all are cited in the Materials and Methods section. Code associated with bioinformatic analyses are available online (https://github.com/LeifAnderssonLab/Ruff_assembly_2022). Correspondence and requests for materials should be addressed to L.A. (leif.andersson@imbim.uu.se).
References
- Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 2008:179(3):1513–1525. 10.1534/genetics.107.084012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdan EL, Blanckaert A, Butlin RK, Bank C. Deleterious mutation accumulation and the long-term fate of chromosomal inversions. PLoS Genet. 2021:17(3):e1009411. 10.1371/journal.pgen.1009411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford A, Purcell J, Avril A, Tran Van P, Zhang J, Brütsch T, Sundström L, Helanterä H, Chapuisat M. An ancient and eroded social supergene is widespread across Formica ants. Curr Biol. 2020:30(2):304–311.e304. 10.1016/j.cub.2019.11.032 [DOI] [PubMed] [Google Scholar]
- Browning BL, Tian X, Zhou Y, Browning SR. Fast two-stage phasing of large-scale sequence data. Am J Hum Genet. 2021:108(10):1880–1890. 10.1016/j.ajhg.2021.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics 2009:10(1):421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008:18(1):188–196. 10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černý D, Natale R. Comprehensive taxon sampling and vetted fossils help clarify the time tree of shorebirds (Aves, Charadriiformes). Mol Phylogenet Evol. 2022:177:107620. 10.1016/j.ympev.2022.107620 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Evolution: a new idea about the degeneration of Y and W chromosomes. Curr Biol. 2020:30(15):R871–R873. 10.1016/j.cub.2020.06.008 [DOI] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016:13(12):1050–1054. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 2012:8(12):e1003056. 10.1371/journal.pgen.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. Twelve years of SAMtools and BCFtools. GigaScience 2021:10(2):giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon G, Kitano J, Kirkpatrick M. The origin of a new sex chromosome by introgression between two stickleback fishes. Mol Biol Evol. 2019:36(1):28–38. 10.1093/molbev/msy181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria R, Johannesson K, Butlin RK, Westram AM. Evolving inversions. Trends Ecol Evol. 2019:34(3):239–248. 10.1016/j.tree.2018.12.005 [DOI] [PubMed] [Google Scholar]
- Feng S, Stiller J, Deng Y, Armstrong J, Fang Q, Reeve AH, Xie D, Chen G, Guo C, Faircloth BC, et al. Dense sampling of bird diversity increases power of comparative genomics. Nature 2020:587(7833):252–257. 10.1038/s41586-020-2873-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier S, Ross N, Rudis R, Camargo P, Sciaini M, Scherer C. 2021. viridis—Colorblind-Friendly Color Maps for R. R package version 0.6.2. https://sjmgarnier.github.io/viridis/.
- Giraldo-Deck LM, Loveland JL, Goymann W, Tschirren B, Burke T, Kempenaers B, Lank DB, Küpper C. Intralocus conflicts associated with a supergene. Nat Commun. 2022:13(1):1384. 10.1038/s41467-022-29033-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Valencia J, Hughes PW, Berdan EL, Slotte T. The genomic architecture and evolutionary fates of supergenes. Genome Biol Evol. 2021:13(5):evab057. 10.1093/gbe/evab057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Jamsandekar M, Pettersson ME, Su L, Fuentes-Pardo AP, Davis BW, Bekkevold D, Berg F, Casini M, Dahle G, et al. Ecological adaptation in Atlantic herring is associated with large shifts in allele frequencies at hundreds of loci. eLife 2020:9:e61076. 10.7554/eLife.61076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglund J, Lundberg A. Plumage color correlates with body size in the ruff (Philomachus pugnax). Auk. 1989:13:336–338. https://www.jstor.org/stable/4087731 [Google Scholar]
- Huang K, Ostevik KL, Elphinstone C, Todesco M, Bercovich N, Owens GL, Rieseberg LH. Mutation load in sunflower inversions is negatively correlated with inversion heterozygosity. Mol Biol Evol. 2022:39(5):msac101. 10.1093/molbev/msac101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsland F, Feng CG, Boije H, Bed'hom B, Fillon V, Dorshorst B, Rubin CJ, Liu RR, Gao Y, Gu XR, et al. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 2012:8(6):e1002775. 10.1371/journal.pgen.1002775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P, Chouteau M, Whibley A, Bastide H, Parrinello H, Llaurens V, Joron M. Mutation load at a mimicry supergene sheds new light on the evolution of inversion polymorphisms. Nat Genet. 2021:53(3):288–293. 10.1038/s41588-020-00771-1 [DOI] [PubMed] [Google Scholar]
- Jukema J, Piersma T. Permanent female mimics in a lekking shorebird. Biol Lett. 2006:2(2):161–164. 10.1098/rsbl.2005.0416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8):907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S. Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013:14(4):R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics 2006:173(1):419–434. 10.1534/genetics.105.047985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus BJ, Grünwald NJ. Vcfr: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour. 2017:17(1):44–53. 10.1111/1755-0998.12549 [DOI] [PubMed] [Google Scholar]
- Korunes KL, Samuk K. Pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol Ecol Resour. 2021:21(4):1359–1368. 10.1111/1755-0998.13326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper C, Stocks M, Risse JE, dos Remedios N, Farrell LL, McRae SB, Morgan TC, Karlionova N, Pinchuk P, Verkuil YI, et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 2016:48(1):79–83. 10.1038/ng.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Fan G, Widemo F, Gunnarsson U, Thalmann DS. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat Gen. 2016:48(1):84–88. 10.1038/ng.3430 [DOI] [PubMed] [Google Scholar]
- Lank DB, Farrell LL, Burke T, Piersma T, McRae SB. A dominant allele controls development into female mimic male and diminutive female ruffs. Biol Lett. 2013:9(6):20130653. 10.1098/rsbl.2013.0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lank DB, Smith CM, Hanotte O, Burke T, Cooke F. Genetic polymorphism for alternative mating behaviour in lekking male ruff Philomachus pugnax. Nature 1995:378(6552):59–62. 10.1038/378059a0 [DOI] [Google Scholar]
- Lee E, Helt GA, Reese JT, Munoz-Torres MC, Childers CP, Buels RM, Stein L, Holmes IH, Elsik CG, Lewis SE. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013:14(8):R93. 10.1186/gb-2013-14-8-r93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018:34(18):3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Stoma S, Lotta LA, Warner S, Albrecht E, Allione A, Arp PP, Broer L, Buxton JL, Da Silva Couto Alves A, et al. Genome-wide association analysis in humans links nucleotide metabolism to leukocyte telomere length. Am J Hum Genet. 2020:106(3):389–404. 10.1016/j.ajhg.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer TA, Busch JW, Smith CR. Social supergenes of superorganisms: do supergenes play important roles in social evolution? BioEssays 2013:35(8):683–689. 10.1002/bies.201300038 [DOI] [PubMed] [Google Scholar]
- Loveland JL, Lank DB, Küpper C. Gene expression modification by an autosomal inversion associated with three male mating morphs. Front Genet. 2021:12:641620. 10.3389/fgene.2021.641620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021:38(10):4647–4654. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018:14(1):e1005944. 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner M, Barth JMI, Tørresen OK, Star B, Baalsrud HT, Brieuc MSO, Pampoulie C, Bradbury I, Jakobsen KS, Jentoft S. Supergene origin and maintenance in Atlantic cod. Nat Ecol Evol. 2022:6(4):469–481. 10.1038/s41559-022-01661-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf JM. Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell. 1996:7(9):1455–1469. 10.1091/mbc.7.9.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy NI. 2005. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc Biol Sci. 272(1573):1633–1640. 10.1098/rspb.2005.3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattestad M, Schatz MC. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics 2016:32(19):3021–3023. 10.1093/bioinformatics/btw369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 1997:146(2):695–709. 10.1093/genetics/146.2.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, Sugano S, Fujiyama A, Kosugi S, Hirakawa H, et al. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet. 2015:47(4):405–409. 10.1038/ng.3241 [DOI] [PubMed] [Google Scholar]
- Pearse DE, Barson NJ, Nome T, Gao G, Campbell MA, Abadía-Cardoso A, Anderson EC, Rundio DE, Williams TH, Naish KA, et al. Sex-dependent dominance maintains migration supergene in rainbow trout. Nat Ecol Evol. 2019:3(12):1731–1742. 10.1038/s41559-019-1044-6 [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016:11(9):1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson ME, Kierczak M, Almén MS, Lamichhaney S, Andersson L. 2017. A model-free approach for detecting genomic regions of deep divergence using the distribution of haplotype distances. Biorxiv:144394.
- Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015:526(7574):569–573. 10.1038/nature15697 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010:26(6):841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotech. 2011:29(1):24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, Raijmakers R, Raats JM, Pruijn GJ. MPP6 Is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005:33(21):6795–6804. 10.1093/nar/gki982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T, Libbrecht R, Keller L. Supergenes and complex phenotypes. Curr Biol. 2014:24(7):R288–R294. 10.1016/j.cub.2014.01.056 [DOI] [PubMed] [Google Scholar]
- Schwochow Thalmann D, Ring H, Sundström E, Cao X, Larsson M, Kerje S, Höglund A, Fogelholm J, Wright D, Jemth P, et al. The evolution of Sex-linked barring alleles in chickens involves both regulatory and coding changes in CDKN2A. PLoS Genet. 2017:13(4):e1006665. 10.1371/journal.pgen.1006665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011:7(1):539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolle E, Pracana R, Howard P, Paris CI, Brown SJ, Castillo-Carrillo C, Rossiter SJ, Wurm Y. Degenerative expansion of a young supergene. Mol Biol Evol. 2019:36(3):553–561. 10.1093/molbev/msy236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993:10(3):512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021:38(7):3022–3027. 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics 2008:179(3):1455–1468. 10.1534/genetics.108.088229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniprot Consortium . Uniprot: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020:49:D480–D489. 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014:9(11):e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. Direct determination of diploid genome sequences. Genome Res. 2017:27(5):757–767. 10.1101/gr.214874.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, Mara Averick M, Bryan J, Winston Chang W, D’Agostino McGowan L, François R. 2019. Welcome to the Tidyverse Journal of Open Source Software 4:1686.
- Widemo F. The social implications of traditional use of lek sites in the ruff (Philomachus pugnax). Behav Ecol. 1997:13(2):211–217. 10.1093/beheco/8.2.211 [DOI] [Google Scholar]
- Widemo F. Alternative reproductive strategies in the ruff: a mixed ESS? Anim Behav. 1998a:13(2):329–336. 10.1006/anbe.1998.0792 [DOI] [PubMed] [Google Scholar]
- Widemo F. Competition for females on leks when male competitive abilities differ: empirical test of a model. Behav Ecol. 1998b:9(5):427–431. 10.1093/beheco/9.5.427 [DOI] [Google Scholar]
- Widemo F, Owensi IPF. Lek size, male mating skew and the evolution of lekking. Nature 1995:373(6510):148–151. 10.1038/373148a0 [DOI] [Google Scholar]
- Yang Z. PAML 4: a program package for Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007:24(8):1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zeileis A, Grothendieck G. . Zoo: s3 infrastructure for regular and irregular time series. J Stat Software. 2005:14(6):1–27. 10.18637/jss.v014.i06 [DOI] [Google Scholar]
- Zhang C, Dong S-S, Xu J-Y, He W-M, Yang T-L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2018:35(10):1786–1788. 10.1093/bioinformatics/bty875 [DOI] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin DM CL, S JF. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 2014:346(6215):1311–1320. 10.1126/science.1251385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data generated in this study and genome assemblies have been submitted to NCBI (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA816664). Genome annotation is available online (https://github.com/LeifAnderssonLab/Ruff_assembly_2022/Annotation_gff). We additionally used sequencing data from BioProjects PRJNA419629, PRJEB10677, SRP058220, and PRJNA545868. The analyses of data have been carried out with publicly available software and all are cited in the Materials and Methods section. Code associated with bioinformatic analyses are available online (https://github.com/LeifAnderssonLab/Ruff_assembly_2022). Correspondence and requests for materials should be addressed to L.A. (leif.andersson@imbim.uu.se).