Abstract
The narA locus required for nitrate reduction in Synechococcus sp. strain PCC 7942 is shown to consist of a cluster of genes, namely, moeA, moaC, moaD, moaE, and moaA, involved in molybdenum cofactor biosynthesis. The product of the moaC gene of strain PCC 7942 shows homology in its N-terminal half to MoaC from Escherichia coli and in its C-terminal half to MoaB or Mog. Overexpression of the Synechococcus moaC gene in E. coli resulted in the synthesis of a polypeptide of 36 kDa, a size that would conform to a protein resembling a fusion of the MoaC and MoaB or Mog polypeptides of E. coli. Insertional inactivation of the moeA, moaC, moaE, and moaA genes showed that the moeA-moa gene cluster is required for growth on nitrate and expression of nitrate reductase activity in strain PCC 7942. The moaCDEA genes constitute an operon which is transcribed divergently from the moeA gene. Expression of the moeA gene and the moa operon was little affected by the nitrogen source present in the culture medium.
Nitrate is probably the most abundant source of combined nitrogen for cyanobacterial nutrition, its assimilation being a process closely linked to photosynthesis (9). Nitrate is transported into the cyanobacterial cell by a multicomponent transport system of the ABC (ATP-binding cassette) type (34). Once inside the cell, nitrate is reduced to ammonium by two sequential reactions catalyzed by nitrate reductase and nitrite reductase, respectively. Ammonium is incorporated into carbon skeletons mainly via the glutamine synthetase/glutamate synthase cycle (9).
In Synechococcus sp. strain PCC 7942, the nir gene encoding nitrite reductase (22), the nrtABCD genes encoding the components of the nitrate transport system (34), and the narB gene encoding nitrate reductase (1, 45) are clustered together and constitute an operon (24, 51). Two other loci, narA and narC, involved in nitrate reduction in Synechococcus sp. strain PCC 7942 have been identified and cloned by means of complementation of nitrate reductase-deficient, Tn901-induced mutants with a gene library from strain PCC 7942 (20, 21). These loci are not clustered together in the Synechococcus genome. With regard to regulation, ammonium acts as a nutritional repressor of the nitrate assimilation system (9). The NtcA protein found in cyanobacteria (9, 10, 52) acts as a transcriptional activator that controls the expression of cyanobacterial genes subjected to repression by ammonium such as the nir operon (23).
Nitrate reductases from cyanobacteria are monomeric molybdoenzymes of about 75 kDa that use reduced ferredoxin as a physiological electron donor (9). Molybdoenzymes other than nitrogenase catalyze either oxidative hydroxylations or reductive dehydroxylations, and its molybdenum center is constituted by a molybdenum-pterin cofactor in the form of molybdopterin (MPT), molybdopterin guanine dinucleotide (MGD), molybdopterin cytosine dinucleotide, or others (40). In Escherichia coli, the pathway for molybdenum cofactor biosynthesis is the subject of intense research. The genes responsible for the transport of molybdate (modABC) (29), for MPT biosynthesis (moaABCDE and moeAB) (33, 36, 44), and for assembly of molybdenum into MPT (mog) (18) have been identified and sequenced, as is also the case for mobA, which is involved in the addition of GMP to MPT during the synthesis of the MGD form of the molybdenum cofactor (16, 17, 35).
In cyanobacteria, information about molybdenum cofactors is scarce. Some molybdenum cofactor, partially bound to a carrier protein that would stabilize the cofactor, has been reported to be present in the soluble fraction of Nostoc muscorum (3). In Anabaena variabilis, the existence of common and specific genes for the synthesis of the iron-molybdenum (of nitrogenase) and molybdenum cofactors has been inferred (28); in Anabaena sp. strain PCC 7120, inactivation of a moeA gene leads to loss of nitrate reductase activity (41), while the moeB-like hesA gene found downstream of the nifHDK operon seems to be necessary for attaining full nitrogenase activity (5). Recently, the entire genome of Synechocystis sp. strain PCC 6803 has been sequenced (19), and a cluster of open reading frames (ORFs) showing similarity to the moeA, moaA, moaC, and moaE genes of E. coli has been found close to the nir gene. Although not identified by the authors, ORF ssr1527, also found in this gene cluster, would encode a putative MoaD homolog (see below).
In this report, we describe a genetic analysis of the Synechococcus sp. strain PCC 7942 narA locus and show that it consists of five genes whose products are essential for nitrate reduction and would be involved in the biosynthesis of molybdopterin.
MATERIALS AND METHODS
Organisms, growth conditions, and plasmids.
Synechococcus sp. strain PCC 7942 was routinely grown photoautotrophically under white light with shaking (90 rpm) at 30°C in the BG11 medium (17.6 mM NaNO3 as the nitrogen source) described previously (43). When ammonium was used as the nitrogen source, nitrate was omitted and 4 mM NH4Cl and 8 mM N-tris(hydroxymethyl)methyl-2-aminoethane sulfonic acid (TES)-NaOH buffer (pH 7.5) were supplied, rendering medium BG110NH4+. For growth on plates, the medium was solidified with separately autoclaved 1% agar (Difco Laboratories). The plates were incubated at 30°C in the light. Synechococcus strains as well as plasmids used in this work are listed in Table 1. Mutant strain FM6 was grown in BG110NH4+ medium, and mutant strains CSLM26, CSLM27, CSLM35, and CSLM40 were grown in BG110NH4+ medium supplemented with 10 μg of kanamycin/ml. Mutant strains CSLM32 and CSLM34 were grown in BG110NH4+ medium supplemented with 2 μg of streptomycin and 2 μg of spectinomycin/ml. Mutant strain CSLM37 was grown in BG110NH4+ medium supplemented with 2 μg of streptomycin/ml, 2 μg of spectinomycin/ml and 10 μg of kanamycin/ml.
TABLE 1.
Cyanobacterial strains and plasmids used in this work
| Strain or plasmid | Origin and relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| PCC 7942 | Wild-type Synechococcus strain | 42 |
| CSLM26 | Kmr derivative of strain PCC 7942; moaA::lacZ-C.K3 gene fusion | This work |
| CSLM27 | Kmr derivative of strain PCC 7942; moeA::lacZ-C.K3 gene fusion | This work |
| CSLM32 | Smr Spr derivative of strain PCC 7942; gene cassette C.S3 inserted into the moeA gene | This work |
| CSLM34 | Smr Spr derivative of strain PCC 7942; gene cassette C.S3 inserted into the moaC gene | This work |
| CSLM35 | Kmr derivative of strain PCC 7942; moaC::lacZ-C.K3 gene fusion | This work |
| CSLM37 | Kmr Smr Spr derivative of strain CSLM26; double mutant (gene cassette C.S3 inserted into the moaC gene and moaA::lacZ-C.K3 gene fusion) | This work |
| CSLM40 | Kmr derivative of strain PCC 7942; nir::lacZ-C.K3 gene fusion | This work |
| FM6 | Derivative of strain PCC 7942; transposon Tn901 inserted into the narA locus; transformable to the wild-type phenotype by plasmid pNR12 | 26 |
| Plasmids | ||
| pBluescript SK(+) | Cloning vector | Stratagene |
| pCSLM6 | 4,758-bp XhoI DNA fragment from pNR1211 cloned into the XhoI site of pBluescript SK(+) | This work |
| pCSLM8 | Synechococcus sp. strain PCC 7942 nir gene cloned into the NcoI site of expression vector pTrc99A | 46 |
| pCSLM26 | Derivative of pCSLM6; 335-bp NheI DNA fragment internal to the moaA gene in pCSLM6 replaced by the lacZ-C.K3 transcriptional reporter cassette from pPE20 (see below); used to generate mutant strain CSLM26 | This work |
| pCSLM27 | Derivative of pCSLM6; lacZ-C.K3 transcriptional reporter cassette from pPE20 inserted into the StuI site of the moeA gene; used to generate mutant strain CSLM27 | This work |
| pCSLM32 | Derivative of pCSLM6; gene cassette C.S3 inserted into the StuI site of the moeA gene; used to generate mutant strain CSLM32 | This work |
| pCSLM34 | Derivative of pCSLM6; gene cassette C.S3 from pRL463 (see below) inserted into the HpaI site of the moaC gene; used to generate mutant strain CSLM34 | This work |
| pCSLM35 | Derivative of pCSLM6; transcriptional reporter cassette lacZ-C.K3 from pPE20 inserted into the HpaI site of the moaC gene; used to generate mutant strain CSLM35 | This work |
| pCSLM40 | Derivative of pCSLM8; a 99-bp NaeI DNA fragment internal to the nir gene in pCSLM8 substituted by the lacZ-C.K3 transcriptional reporter cassette from pPE20; used to generate mutant strain CSLM40 | This work |
| pCSLM43 | Synechococcus sp. strain PCC 7942 moaC gene cloned into the BamHI site of the expression vector pGEX-4T-2 (see below) | This work |
| pGEX-4T-2 | Expression vector for the production of GST-fused proteins | Pharmacia |
| pNR12 | Cosmid containing 19-kb SalI DNA fragment from the genome of strain PCC 7942 that includes the narA locus | 20 |
| pNR1211 | XhoI DNA fragment of 4,758 bp from pNR12 including the narA locus cloned into the XhoI site of pACYC177 | 20 |
| pPE20 | Promoterless lacZ gene followed by gene cassette C.K3 (transcriptional reporter cassette) cloned into the BamHI site of pUC18/19 | 48 |
| pRL463 | Plasmid pUC18/19 containing the C.S3 gene cassette cloned into the BamHI site of the L.EHE1 polylinker | 8 |
| pTrc99A | Expression vector | Pharmacia |
For β-galactosidase assays, Synechococcus strains were grown in 70-ml glass tubes containing 35 ml of the medium indicated in each experiment, bubbled with air-CO2 (98:2) at 30°C in the light. At the mid-exponential growth phase (cultures with about 3 to 5 μg of chlorophyll/ml), samples containing an amount of cells corresponding to about 2 μg of chlorophyll were withdrawn for determination of protein content and β-galactosidase activity.
For nitrate reductase assays and for growth rate determinations, Synechococcus strains were grown in 70-ml glass tubes containing 35 ml of BG110NH4+ medium without antibiotics; after extensive washing, the cells were transferred to and incubated in the medium in each experiment bubbled with air-CO2 (98:2) at 30°C in the light.
For isolation of DNA and RNA, Synechococcus strains were grown in 240-ml glass flasks containing 150 ml of BG11 medium bubbled with air at 30°C in the light. Cultures with a cell density corresponding to 3 to 5 μg chlorophyll/ml were used.
E. coli DH5α, GM48, BL21, and HB101 were grown in Luria-Bertani medium at 37°C with shaking (200 rpm). For growth of E. coli on plates, medium solidified with 1.5% agar was used. Antibiotics were used at standard concentrations (2).
Growth rates were estimated from the increase of protein concentration in the cultures. The growth rate constant corresponds to ln2/td, where td represents the doubling time.
Generation of mutant strains.
Insertions of either HincII-ended gene cassette C.S3 from plasmid pRL463 (8) or SmaI-ended gene cassette lacZ-C.K3 from plasmid pPE20 (48) at the StuI restriction site, which is internal to the Synechococcus moeA gene, of plasmid pCSLM6 were made to render plasmids pCSLM32 (moeA::C.S3) and pCSLM27 (moeA::lacZ-C.K3), respectively. Similar insertions were made into the moaC gene, disrupting it at the HpaI restriction site of pCSLM6 to render plasmids pCSLM34 (moaC::C.S3) and pCSLM35 (moaC::lacZ-C.K3). The moaA gene was also mutated by substitution of a 335-bp, NheI DNA fragment at the 3′ terminus of the gene, after digestion of pCSLM6 with NheI and treatment with the Klenow enzyme (2), by SmaI-ended gene cassette lacZ-C.K3, rendering plasmid pCSLM26. The nir gene was mutated by substitution of a 99-bp, NaeI DNA fragment by SmaI-ended gene cassette lacZ-C.K3, rendering plasmid pCSLM40 (Table 1). Restriction analysis of plasmids pCSLM26, pCSLM27, pCSLM35, and pCSLM40 confirmed that the antibiotic resistance genes present in the inserted gene cassettes were, in every case, in the same orientation as the Synechococcus genes disrupted by the gene cassette.
Plasmids pCSLM26, pCSLM27, pCSLM32, pCSLM34, pCSLM35, and pCSLM40 were transferred to Synechococcus sp. strain PCC 7942 by means of transformation (12) for generation of mutant strains CSLM26, CSLM27, CSLM32, CSLM34, CSLM35, and CSLM40, respectively. For generation of mutant strain CSLM37, pCSLM34 was transferred to strain CSLM26 (Table 1). After transformation, cells were spread onto nitrocellulose filters (Nuclepore; REC85) set successively atop BG110NH4+ solid medium (incubated for 48 h) and BG110NH4+ with the appropriate antibiotics (incubated for 3 weeks). Individual colonies were selected and, after recloning, maintained in BG110NH4+ solid medium with antibiotics.
DNA manipulations.
PCR using EcoTaq DNA polymerase (EcoGen S.R.L.), plasmid constructions, DNA electrophoresis, isolation of DNA fragments from agarose gels, ligation, and transformation of E. coli were carried out by standard methods (2). Restriction endonucleases were used according to the manufacturer’s recommendations or by standard methods (2). Sequencing was performed in double-stranded DNA by the chain termination method with a T7 Sequencing kit (Pharmacia LKB) and [35S]deoxyadenosine 5′-(α-thio)triphosphate (1,000 to 1,500 Ci/mmol). Both strands of the DNA were sequenced. Computer searching for homologies was made by using the FASTA and TFASTA algorithms included in the Genetics Computer Group package (7). Isolation of DNA from Synechococcus strains was performed essentially as described by Cai and Wolk (6). For Southern blots, restriction endonuclease-digested DNA or PCR products were subjected to electrophoresis in agarose gels and transferred to Genescreen Plus membranes (Dupont) as instructed by the manufacturer. Probes were labeled with [α-32P]dCTP (3,000 Ci/mmol). Prehybridization and hybridization were performed essentially as described by Frías et al. (10) in a solution containing 5× SSPE (0.8 M NaCl, 10 mM sodium phosphate, 1 mM EDTA [pH 7.4]), 5× Denhardt’s solution (47), 0.5% (wt/vol) sodium dodecyl sulfate (SDS), and 100 μg of nonhomologous DNA/ml, under high-stringency conditions at 65°C and under low-stringency conditions at 55°C.
RNA isolation and RT-PCR analysis.
Isolation of RNA from Synechococcus sp. strain PCC 7942 was performed as described by Mohamed and Jansson (32), with the modifications described in Luque et al. (23). Samples were treated with RNase-free DNase I (from bovine pancreas; Boehringer) for elimination of any remaining DNA. For retrotranscription-PCR (RT-PCR) experiments, 4 μg of strain PCC 7942 total RNA was mixed with 20 pmol of the oligonucleotide 5′-ATTGACCTTGAGGATCGGTAAGCG-3′ (complementary to nucleotides 479 to 456 with respect to the translation start of the moaA gene) in the presence of 50 mM Tris-HCl, 8 mM MgCl2, 30 mM KCl, and 1 mM dithiothreitol, pH 8.5 (AMV [avian myeloblastosis virus] buffer), heated for 2 min at 90°C, and immediately cooled down to 55°C. Then 1 mM each deoxynucleoside triphosphate, 20 U of RNA Guard (Pharmacia), and 50 U of AMV reverse transcriptase (Boehringer) were added, and the extension reaction was developed for 1 h at 54°C in a volume of 20 μl. To control for the presence of contaminating DNA, samples containing 4 μg of the RNA preparation, 20 pmol of the same oligonucleotide, and 1 μg of RNase A (DNase free; Boehringer) were incubated, in a 20-μl reaction volume, at 37°C for 1 h. PCR was carried out with 35 μl of retrotranscription mixture (diluted 10-fold with 10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) or RNase-treated sample (see above) as the template, and oligonucleotides 5′-ATTGACCTTGAGGATCGGTAAGCG-3′ (complementary to nucleotides 479 to 456 with respect to the moaA translation start; same as above) and 5′-GGTCTATCAGCGCGTTACTCAAGG-3′ (complementary to nucleotides 74 to 97 with respect to the moaC translation start) as primers. Control samples containing the same oligonucleotides and strain PCC 7942 genomic DNA as the template were run in parallel. PCR consisted of 35 cycles of template denaturation at 95°C for 1 min, annealing with the oligonucleotides for 1 min at 69°C, and DNA extension at 72°C for 2 min. One half of each sample was resolved by electrophoresis in 1% agarose gels and transferred to membranes for Southern blot analysis. A 0.53-kb, PvuII DNA fragment from plasmid pNR1211 (Table 1), internal to the moa operon (see Fig. 2), was used as the probe.
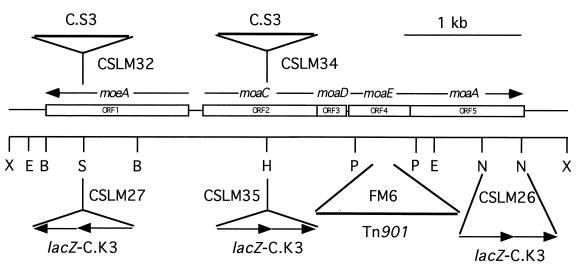
FIG. 2.
Structure of a genomic region of Synechococcus sp. strain PCC 7942 that contains moeA and several moa genes. The identities and orientations of gene cassettes inserted at some restriction sites for the generation of cyanobacterial mutants are indicated together with the CSLM denomination of the resulting mutant strain. The location of Tn901 in mutant strain FM6 is also indicated. B, BglII; E, EcoRV; H, HpaI; N, NheI; P, PvuII; S, StuI; X, XhoI.
Expression of the Synechococcus moaC gene in E. coli.
A 1,030-bp DNA fragment, containing the moaC gene and part of the moaD gene, from an unmethylated pCSLM6 plasmid restricted with ClaI and SacI and treated with Klenow enzyme was isolated and cloned into the BamHI site of plasmid pGEX-4T-2 (Pharmacia), made blunt ended with Klenow enzyme, to render plasmid pCSLM43. E. coli BL21 carrying plasmid pCSLM43 was used for overproduction of the glutathione S-transferase (GST)–MoaC fusion protein after induction with 1 mM isopropyl-β-d-thiogalactoside (IPTG). Preparation of cell extracts from BL21(pCSLM43), purification of GST-MoaC protein by using bulk glutathione-Sepharose 4B in batch, and thrombin cleavage of fusion proteins were carried out as recommended by the manufacturer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) as described by Schleif and Wensink (49), using a 12% acrylamide running gel with an upper 4% acrylamide stacking gel.
Enzyme assays and analytical procedures.
Nitrate reductase activity was determined by using dithionite-reduced methyl viologen as the reductant in alkyltrimethylammonium bromide-permeabilized Synechococcus cells (14). β-Galactosidase activity was determined as described by Schaefer and Golden (48) by colorimetric assay with o-nitrophenyl-β-d-galactopyranoside (ONPG) (31). One unit of enzymatic activity corresponds to the formation of 1 μmol of product (nitrite or o-nitrophenol) per min. Protein quantifications were made by a modified Lowry method (27), using bovine serum albumin as the standard. Chlorophyll a determinations were made in methanolic extracts as described by MacKinney (25).
Nucleotide sequence accession number.
The nucleotide sequence of the moeA and moa genes reported in this paper will appear in the EMBL/GeneBank/DDBJ nucleotide sequence data libraries under accession no. X99625.
RESULTS
Identification of the narA locus.
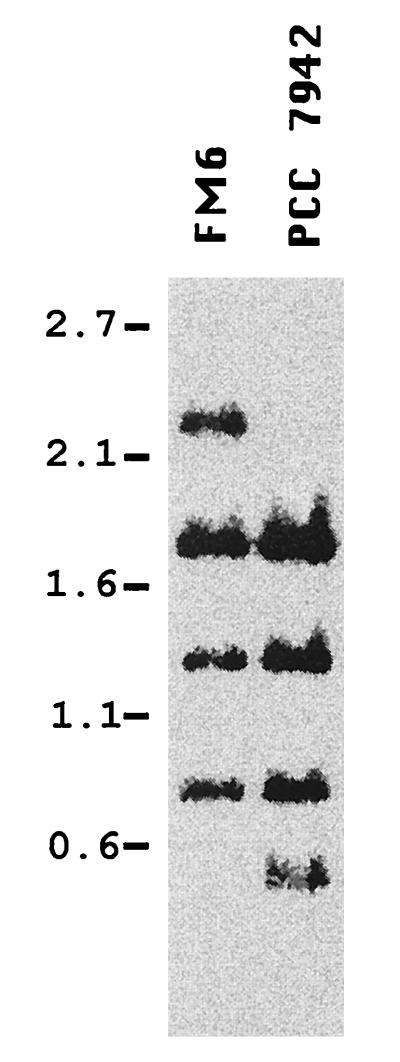
FM6 is a Tn901-induced mutant derived from Synechococcus sp. strain PCC 7942 that is impaired in nitrate reductase activity. This mutant is readily transformable to the wild-type phenotype by the 4.7-kb, XhoI DNA fragment from strain PCC 7942 that is cloned in plasmid pNR1211. This genetic locus has been named narA (20). For localization and identification of the nitrate reduction-related gene(s) corresponding to the narA locus, genomic DNA from mutant strain FM6 was restricted with the endonucleases XhoI, BglII, and PvuII and subjected to Southern blot analysis using as a probe the 4.7-kb, strain PCC 7942 DNA fragment of pNR1211. Compared to strain PCC 7942 DNA, mutant FM6 DNA showed a clear change in the hybridization pattern indicative of a Tn901 insertion into an 0.53-kb, PvuII DNA fragment (Fig. 1). Sequencing of this PvuII fragment revealed the existence of two ORFs. The putative product of one of them showed homology to the large subunit of the MPT-converting factor, the MoaE polypeptide of E. coli.
FIG. 1.
Localization of Tn901 in the narA locus of mutant strain FM6. Genomic DNA from the indicated strain was simultaneously digested with XhoI, BglII, and PvuII and subjected to Southern blot analysis using the 4.7-kb XhoI insert of plasmid pNR1211 as a probe. The positions and sizes (in kilobases) of some standards are indicated to the left.
Sequencing of the entire Synechococcus sp. strain PCC 7942 DNA fragment cloned in pNR1211 was carried out by using several synthetic oligonucleotides as primers and plasmid pNR1211 as the template. Sequence analysis revealed the existence of five ORFs in that fragment (Fig. 2). Putative ribosome binding sites could be found only in front of ORF1, ORF4, and ORF5. No other ORF was found after sequencing 300 bp of the Synechococcus DNA adjacent to the XhoI site closest to ORF5, using as template cosmid pNR12 (20), whose insert includes that of plasmid pNR1211.
As summarized in Table 2, ORF1 would encode a polypeptide of 403 amino acids that shows homology to MoeA polypeptides from E. coli (33) and Anabaena sp. strain PCC 7120 (41). ORF2 would encode a 319-amino-acid polypeptide whose N-terminal half shows homology to the MoaC polypeptide of E. coli (44) and whose C-terminal half shows homology to MoaB and Mog polypeptides of E. coli (it should be noted that MoaB and Mog are themselves homologous to each other) (44, 53). ORF3 would encode a polypeptide of 90 amino acids that in its C-terminal part (amino acids 60 through 90) shows homology to the MoaD polypeptide of E. coli (44). The putative product of ORF4 (165 amino acids) shows homology to the E. coli MoaE polypeptide (44). ORF5 would encode a polypeptide of 327 amino acids homologous to E. coli MoaA (44), to Bacillus subtilis NarA (11), and to Cnx2 from Arabidopsis thaliana (15). All Moa, Moe, and Mog polypeptides of E. coli have been shown to be involved in the biosynthesis of Mo-MPT (39). Because of the homologies described above, we propose to name ORF1 as moeA, ORF2 as moaC (whose product would bear a domain homologous to MoaC and another one homologous to MoaB and Mog from E. coli), ORF3 as moaD, ORF4 as moaE, and ORF5 as moaA.
TABLE 2.
ORFs in the narA locus of Synechococcus sp. strain PCC 7942
| ORF | Proposed gene name | Size (no. of amino acids) | Homologous protein (organism) | % Identitya | Overlapping fragmentb |
|---|---|---|---|---|---|
| ORF1 | moeA | 403 | MoeA (E. coli) | 36 | 1–403 |
| MoeA (Anabaena sp.) | 32 | 1–403 | |||
| ORF2 | moaC | 319 | MoaC (E. coli) | 47 | 1–145 |
| MoaB (E. coli) | 27 | 163–303 | |||
| Mog (E. coli) | 28 | 156–300 | |||
| ORF3 | moaD | 90 | MoaD (E. coli) | 34 | 60–90 |
| ORF4 | moaE | 165 | MoaE (E. coli) | 35 | 10–156 |
| ORF5 | moaA | 327 | MoaA (E. coli) | 46 | 1–327 |
| NarA (B. subtilis) | 31 | 1–327 | |||
| Cnx2 (A. thaliana) | 35 | 1–327 |
As deduced from the aligned fragment.
Position in the Synechococcus polypeptide of the amino acids that could be aligned with the indicated homologous protein (or with a fragment of that protein).
No evidence for the existence in strain PCC 7942 of other genes homologous to those present in the moeA-moa cluster here described could be obtained by means of Southern blot analysis under low-stringency conditions using a 3,458-bp, EcoRV DNA fragment containing most of the moa-moe gene cluster (Fig. 2) as a probe (not shown).
Overexpression of the Synechococcus moaC gene in E. coli.
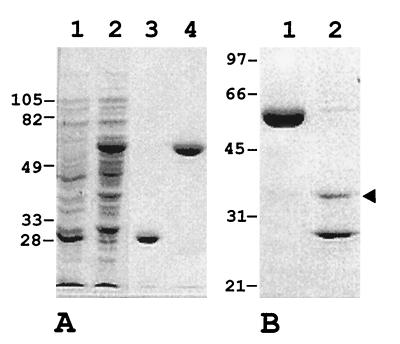
The MoaC polypeptide from strain PCC 7942 was produced in E. coli BL21(pCSLM43) cells as a GST-MoaC fusion protein of about 60 kDa. After purification of the GST-MoaC protein and cleavage with thrombin, a 36-kDa MoaC protein was released (Fig. 3). This protein would differ from the native MoaC only in the first two amino acids, which were changed from Met-Ile in the native protein to Gly-Ser in the recombinant protein. The size of MoaC polypeptide derived from the nucleotide sequence of the Synechococcus moaC gene would be 33 kDa.
FIG. 3.
Expression in E. coli and purification of a GST-MoaC fusion protein. (A) SDS-PAGE of a cell extract from strain BL21(pGEX-4T-2) containing GST (lane 1), cell extract from BL21(pCSLM43) containing GST-MoaC (lane 2), purified GST (lane 3), and purified GST-MoaC protein (lane 4). (B) SDS-PAGE of purified GST-MoaC protein (lane 1) and thrombin-treated GST-MoaC protein (lane 2). The arrowhead points to the ca. 36-kDa Synechococcus MoaC protein. Positions and sizes (in kilodaltons) of some molecular weight markers are indicated to the left of each panel.
Mutational analysis of the moeA and moa genes.
Three of the genes found in the insert of pNR1211 were mutated by in vitro gene cassette insertion (Fig. 2 and Table 1) to test their involvement in expression of nitrate reductase activity. Synechococcus strains bearing those mutations in the moeA or moa genes were obtained by genetic transformation of strain PCC 7942 with plasmids bearing the inactivated genes (see Fig. 2 and Materials and Methods for details). Strain CSLM26 bears gene cassette lacZ-C.K3, which does not carry transcriptional terminators (30), substituting for the 335-bp NheI fragment internal to the moaA gene; strain CSLM27 bears gene cassette lacZ-C.K3 inserted into the StuI site of the moeA gene; strain CSLM32 bears gene cassette C.S3, which carries transcriptional terminators (38), inserted into the StuI site within the moeA gene; strain CSLM34 bears gene cassette C.S3 inserted into the HpaI site within the moaC gene; strain CSLM35 bears gene cassette lacZ-C.K3 inserted at the same HpaI site within the moaC gene. We also constructed a double mutant, strain CSLM37, that bears the mutations present in strains CSLM34 and CSLM26. The genetic structure of each of the mutant strains in the moeA-moa region, as well as the absence of wild-type chromosomes in them, was confirmed by PCR analysis using primers flanking each mutation (not shown). In addition, strain FM6, which bears Tn901 inserted into the 0.53-kb PvuII fragment, was analyzed by PCR using several primers internal to the moaE and moaA genes. Results obtained (not shown) indicated that Tn901 in strain FM6 is inserted into the moaE gene (Fig. 2).
Strains CSLM26, CSLM27, CSLM32, CSLM34, CSLM35, and CSLM37 were unable to grow with nitrate as the sole nitrogen source (Table 3). Moreover, in contrast to strain PCC 7942, none of the mutants exhibited nitrate reductase activity upon incubation in medium containing nitrate as the sole nitrogen source (Table 3).
TABLE 3.
Growth rates and nitrate reductase activities of mutant strains derived from Synechococcus sp. strain PCC 7942
| Strain | Relevant genotype | Growth rate (days−1)a
|
Nitrate reductase (mU/mg of protein)b | |
|---|---|---|---|---|
| Ammonium | Nitrate | |||
| PCC 7942 | Wild type | 2.76 | 3.07 | 36.5 |
| CSLM26 | moaA::lacZ-C.K3 | 2.10 | <0.01 | <0.1 |
| CSLM27 | moeA::lacZ-C.K3 | 1.61 | <0.01 | <0.1 |
| CSLM32 | moeA::C.S3 | 2.90 | <0.01 | <0.1 |
| CSLM34 | moaC::C.S3 | 2.06 | <0.01 | <0.1 |
| CSLM35 | moaC::lacZ-C.K3 | 3.26 | <0.01 | <0.1 |
| CSLM37 | moaC::C.S3, moaA::lacZ-C.K3 | 1.80 | <0.01 | <0.1 |
| FM6c | moaE::Tn901 | 1.80 | <0.01 | <0.1 |
Ammonium-grown cells of the wild-type and mutant strains were washed twice with medium lacking combined nitrogen and used to inoculate, at a final concentration of 0.2 μg of chlorophyll/ml, cultures with the indicated nitrogen source. The cultures were bubbled with air-CO2 (98:2) at 30°C in the light for 24 h and then diluted eightfold with fresh culture medium with the same nitrogen source. To estimate the growth rate, protein content was measured in aliquots sampled periodically from the last set of cultures.
For nitrate reductase activity determinations, ammonium-grown cells of the wild-type and mutant strains were washed twice with medium lacking combined nitrogen and incubated for 6 h in medium containing nitrate as the sole nitrogen source, bubbled with air-CO2 (98:2) at 30°C in the light. Nitrate reductase activity was then determined in aliquots of each culture.
Although the phenotype of this strain has been previously reported (26), it is included in this experiment for the sake of comparison.
Expression of the moeA and moa genes.
Because we were unable to detect any moa transcript in Synechococcus sp. strain PCC 7942 by means of Northern analysis, we studied the expression of moa genes by subjecting mRNA to RT-PCR (see Materials and Methods for details). For retrotranscription, an oligonucleotide complementary to sequences internal to the moaA gene was used as the primer. The resulting cDNA was then amplified by PCR using the same primer used for retrotranscription and another one that should anneal at the beginning of the moaC gene. After electrophoresis of the RT-PCR products on an agarose gel, a band of the expected size, 2.1 kb, was observed. This band was verified by Southern blot analysis to hybridize to a probe of the moaE gene (Fig. 4, lane 3). Since this RT-PCR product was strictly dependent on the presence of RNA (Fig. 4, lane 2), it cannot be due to amplification of contaminating genomic DNA. These results suggest that the moaC, moaD, moaE, and moaA genes of strain PCC 7942 (Fig. 2) are cotranscribed into a single mRNA species. It is worth noting that overlapping of termination and start codons is found between the moaC and moaD genes, as well as between the moaE and moaA genes, of strain PCC 7942.
FIG. 4.
Southern analysis of RT-PCR products of the moa gene cluster of Synechococcus sp. strain PCC 7942, using a moaE gene probe. The primers used for the RT-PCR corresponded to DNA sequences of the moaA and moaC genes (see Materials and Methods for primers used). Lane 1, PCR-amplified strain PCC 7942 genomic DNA; lane 2, PCR-amplified, RNase-treated strain PCC 7942 total RNA; lane 3, RT-PCR-amplified RNA. Samples of lanes 2 and 3 were incubated with DNase before the RNase treatment or the retrotranscription reaction. The size of the amplified DNA fragment is indicated to the left.
The effect of the presence of ammonium or nitrate in the extracellular medium on the expression of the moeA and moa genes of strain PCC 7942 was studied by measuring β-galactosidase activity in mutant strains bearing gene fusions to the lacZ-C.K3 gene cassette. Ammonium-grown cells of mutant strains CSLM26 (moaA::lacZ-C.K3), CSLM27 (moeA::lacZ-C.K3), CSLM35 (moaC::lacZ-C.K3) (Fig. 2), and, as a control, CSLM40 (nir::lacZ-C.K3) (Table 1) were incubated for 14 h in media containing either ammonium or nitrate as the sole nitrogen source, and protein and β-galactosidase activities were determined (Table 4). While the activity level of β-galactosidase in CSLM40 (carrying the nir::lacZ-C.K3 fusion) was about 4.2-fold higher in nitrate- than in ammonium-incubated cells, only 1.7- to 1.9-fold-higher levels were found in nitrate- than in ammonium-incubated cells of the strains carrying lacZ fused to the moeA or moa genes. Data in Table 4 should be interpreted with caution, however, since β-galactosidase basal levels can be relatively high in Synechococcus cells carrying lacZ. As an example, β-galactosidase activity in ammonium-grown cells of strain CSLM37, in which lacZ is inserted within the moa operon about 1.8 kb downstream from the transcriptional terminator present in the C.S3 gene cassette, was about 60 mU/mg of protein. No β-galactosidase activity was detected in wild-type strain PCC 7942.
TABLE 4.
β-Galactosidase activities of Synechococcus sp. mutant strains CSLM40, CSLM26, CSLM27, and CSLM35 incubated with ammonium or nitratea
| Strain | Relevant genotype | β-Galactosidase (mU/mg of protein)
|
|
|---|---|---|---|
| Ammonium | Nitrate | ||
| CSLM40 | nir::lacZ-C.K3 | 113 (18) | 475 (90) |
| CSLM26 | moaA::lacZ-C.K3 | 110 (11) | 190 (40) |
| CSLM27 | moeA::lacZ-C.K3 | 106 (10) | 200 (31) |
| CSLM35 | moaC::lacZ-C.K3 | 116 (22) | 216 (19) |
Ammonium-grown cells were collected and transferred to media containing the indicated nitrogen source. After 14 h of incubation under culture conditions, total protein and β-galactosidase activity were measured. Data are the medians of four independent experiments (standard deviations are shown into parentheses).
DISCUSSION
Some years ago, three genetic loci, narA, narB, and narC, whose mutation leads to impairment of nitrate reductase activity in the unicellular cyanobacterium Synechococcus sp. strain PCC 7942 were identified and cloned (20, 21). The narB locus was later identified as the structural gene for nitrate reductase (1, 45) that is part of an operon of nitrate assimilation genes which includes, in addition to narB, the nir and nrtABCD genes (22, 24, 34). Up to now, nothing was known about the actual function of the genes in the narA or narC locus.
We have mapped the site of insertion of Tn901 in a previously reported narA mutant of Synechococcus sp. strain PCC 7942, strain FM6 (26), and have determined that the inactivated genomic region carries a cluster of genes whose putative polypeptide products show similarity to genes involved in the biosynthesis of the molybdenum cofactor of nitrate reductase and other molybdoenzymes. The genes identified include homologs of the moaA, moaB and mog, moaC, moaD, moaE, and moeA genes from E. coli and some other biological sources.
The synthesis of all molybdenum cofactors of molybdoenzymes, except the iron-molybdenum cofactor of nitrogenase, comprises the synthesis of Mo-MPT, which presumably is common to all molybdoenzyme-containing organisms, and, in some cases, the posterior formation of different dinucleotide variants. Synthesis of MPT takes place through the formation of a sulfur-free pterin precursor, termed precursor Z, that is then sulfurylated, leading to MPT and, after incorporation of molybdenum, to the Mo-MPT complex (39). The moaABC genes of E. coli, which are part of the moaABCDE operon, are involved in the biosynthesis of precursor Z. The moa operon of Synechococcus sp. strain PCC 7942 described in this work contains genes homologous to moaABC. While Synechococcus MoaA would be similar to other MoaA (or Cnx2) proteins, Synechococcus MoaC is unique in that it resembles a fusion protein of MoaC (N-terminal half) and MoaB or Mog (C-terminal half). Sequence similarities do not allow us to conclude whether the C-terminal half of Synechococcus MoaC represents a MoaB or a Mog domain. In Synechocystis sp. strain PCC 6803, ORFs showing similarity to moaA (slr0901) and moaC (slr0902) are also clustered together (19). In this case, moaC would be fused to mobA (a gene required in a late step of MGD biosynthesis), but the actual assignments of these genes to the corresponding ORFs awaits experimental confirmation. The moaDE genes of E. coli encode the two subunits of the so-called converting factor or MPT synthase that adds dithiolene sulfurs to precursor Z, thus generating MPT (37). The Synechococcus moa operon also contains moaDE homologs (Table 2 and Fig. 2), as is also the case for the Synechocystis genome (19). The putative cyanobacterial MoaD polypeptides are peculiar in that they show appreciable identity to E. coli MoaD only in the 30 C-terminal amino acids; notably, however, these include the C-terminal Gly-Gly sequence that is thought to be essential for MoaD function (39). On the other hand, the moeB gene, which in E. coli encodes MPT synthase sulfurylase that catalyzes the transfer of sulfur to the MoaD subunit of MPT synthase, is not present in the Synechococcus moeA-moa gene cluster. Finally, the MoeA protein from strain PCC 7942 would be similar to MoeA from E. coli that has recently been suggested to be involved in activation of molybdate (13). The fact that Synechococcus strains bearing mutations in the moeA-moa gene cluster are devoid of nitrate reductase activity indicates that this gene cluster is involved in the synthesis of the Mo cofactor of nitrate reductase. In particular, four of the genes in the cluster (moeA, moaC, moaE, and moaA) have been inactivated, the phenotype of the corresponding mutants showing the involvement of these genes in production of an active nitrate reductase. It should be noted that both the lacZ-C.K3 gene cassette and transposon Tn901 allow transcription of genes located downstream from them in a transcriptional unit (30, 46, 50).
Results of RT-PCR presented in this work show that Synechococcus sp. strain PCC 7942 synthesizes mRNA molecules containing a message for both the moaC and moaA genes. This finding indicates that the moa genes in the identified gene cluster can be expressed as a single mRNA molecule from a promoter located upstream from moaC, thus constituting an operon. On the other hand, moeA, which is located in the complementary DNA strand, would be expressed independently.
In Synechococcus sp. strain PCC 7942, structural genes for nitrate assimilation proteins including nitrite reductase, the components of the nitrate/nitrite transport system, and nitrate reductase, which constitute the nir operon, are subjected to repression by ammonium. Results presented here on the expression of the moeA gene and the moa operon, compared to that of the nir operon, using β-galactosidase as a transcriptional reporter indicate that the expression of these Mo cofactor biosynthesis genes is not regulated by the nitrogen source to the same extent as the nir operon is. This resembles the situation with the moa genes in E. coli, whose expression is not affected by the regulatory element NarL, a nitrate-responsive activator of the synthesis of nitrate reductase, or by high levels of nitrate in the growth medium (4). Lack of regulation by the nitrogen source of the moeA and moa genes in Synechococcus sp. strain PCC 7942 would be consistent with a role of these genes in this cyanobacterium, as is also the case for E. coli, in the synthesis of the Mo cofactor not only of nitrate reductase but also of some other molybdoenzymes involved in processes other than nitrogen assimilation.
ACKNOWLEDGMENTS
This work was supported by grant PB95-1267 from the Dirección General de Enseñanza Superior (Spain).
We thank T. Thiel for providing us with a lacZ-C.K3 gene cassette.
REFERENCES
- 1.Andriesse X, Bakker H, Weisbeek P. Analysis of nitrate reduction genes in cyanobacteria. In: Ullrich W R, Rigano C, Fuggi A, Aparicio P J, editors. Inorganic nitrogen in plants and microorganisms: uptake and metabolism. Berlin, Germany: Springer-Verlag; 1990. pp. 303–307. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1997. [Google Scholar]
- 3.Bagchi S N, Sherman T D, Funkhouser E A. Biochemical characterization of molybdenum-cofactor in a cyanobacterium, Nostoc muscorum. Plant Cell Physiol. 1987;28:1411–1419. [Google Scholar]
- 4.Baker K P, Boxer D H. Regulation of the chlA locus of Escherichia coli K12: involvement of molybdenum cofactor. Mol Microbiol. 1991;5:901–907. doi: 10.1111/j.1365-2958.1991.tb00764.x. [DOI] [PubMed] [Google Scholar]
- 5.Borthakur D, Basche M, Buikema W J, Borthakur P B, Haselkorn R. Expression, nucleotide sequence and mutational analysis of two open reading frames in the nif gene region of Anabaena sp. strain PCC 7120. Mol Gen Genet. 1990;221:227–234. doi: 10.1007/BF00261725. [DOI] [PubMed] [Google Scholar]
- 6.Cai Y, Wolk C P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequence. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 9.Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 487–517. [Google Scholar]
- 10.Frías J E, Mérida A, Herrero A, Martín-Nieto J, Flores E. General distribution of the nitrogen control gene ntcA in cyanobacteria. J Bacteriol. 1993;175:5710–5713. doi: 10.1128/jb.175.17.5710-5713.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser P, Danchin A, Kunst F, Zuber P, Nakano M M. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J Bacteriol. 1995;177:1112–1115. doi: 10.1128/jb.177.4.1112-1115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golden S S, Sherman L A. Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J Bacteriol. 1984;158:36–42. doi: 10.1128/jb.158.1.36-42.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasona, A., R. M. Ray, and K. T. Shanmugam. Physiological and genetic analysis leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 14.Herrero A, Flores E, Guerrero M G. Regulation of nitrate reductase cellular levels in the cyanobacteria Anabaena variabilis and Synechocystis sp. FEMS Microbiol Lett. 1985;26:21–25. [Google Scholar]
- 15.Hoff T, Schnorr K M, Meyer C, Caboche M. Isolation of two Arabidopsis cDNAs involved in early steps of molybdenum cofactor biosynthesis by functional complementation of Escherichia coli mutants. J Biol Chem. 1995;270:6100–6107. doi: 10.1074/jbc.270.11.6100. [DOI] [PubMed] [Google Scholar]
- 16.James R, Dean D, Debbage J. Five open reading frames upstream of the dnaK gene of E. coli. J DNA Sequencing Mapping. 1993;3:327–332. doi: 10.3109/10425179309020832. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J L, Indermaur L W, Rajagopalan K V. Molybdenum cofactor biosynthesis in Escherichia coli. Requirement of the chlB gene product for the formation of molybdopterin guanine dinucleotide. J Biol Chem. 1991;266:12140–12145. [PubMed] [Google Scholar]
- 18.Joshi M S, Johnson J L, Rajagopalan K V. Molybdenum cofactor biosynthesis in Escherichia coli mod and mog mutants. J Bacteriol. 1996;178:4310–4312. doi: 10.1128/jb.178.14.4310-4312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 20.Kuhlemeier C J, Logtenberg T, Stoorvogel W, van Heugten H A A, Borrias W E, van Arkel G A. Cloning of nitrate reductase genes from the cyanobacterium Anacystis nidulans. J Bacteriol. 1984;159:36–41. doi: 10.1128/jb.159.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlemeier C J, Teeuwsen V J P, Janssen M J T, van Arkel G A. Cloning of a third nitrate reductase gene from the cyanobacterium Anacystis nidulans R2 using a shuttle cosmid library. Gene. 1984;31:109–116. doi: 10.1016/0378-1119(84)90200-2. [DOI] [PubMed] [Google Scholar]
- 22.Luque I, Flores E, Herrero A. Nitrite reductase gene from Synechococcus sp. PCC 7942: homology between cyanobacterial and higher-plant nitrite reductases. Plant Mol Biol. 1993;21:1201–1205. doi: 10.1007/BF00023618. [DOI] [PubMed] [Google Scholar]
- 23.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luque I, Herrero A, Flores E, Madueño F. Clustering of genes involved in nitrate assimilation in the cyanobacterium Synechococcus. Mol Gen Genet. 1992;232:7–11. doi: 10.1007/BF00299130. [DOI] [PubMed] [Google Scholar]
- 25.MacKinney G. Absorption of light by chlorophyll solutions. J Biol Chem. 1941;140:315–322. [Google Scholar]
- 26.Madueño F, Borrias W E, van Arkel G A, Guerrero M G. Isolation and characterization of Anacystis nidulans R2 mutants affected in nitrate assimilation: establishment of two new mutant types. Mol Gen Genet. 1988;213:223–228. [Google Scholar]
- 27.Markwell M A K, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 28.Martín-Nieto J, Flores E, Herrero A. Mutants of Anabaena variabilis requiring high levels of molybdate for nitrate reductase and nitrogenase activities. FEMS Microbiol Lett. 1990;67:1–4. [Google Scholar]
- 29.Maupin-Furlow J A, Rosentel J K, Lee J H, Deppenmeier U, Gunsalus R P, Shanmugam K T. Genetic analysis of the modABCD (molybdate transport) operon of Escherichia coli. J Bacteriol. 1995;177:4851–4856. doi: 10.1128/jb.177.17.4851-4856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazodier P, Cossart P, Giraud E, Gasser F. Completion of the nucleotide sequence of the central region of Tn5 confirms the presence of three resistance genes. Nucleic Acids Res. 1985;13:195–205. doi: 10.1093/nar/13.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- 33.Nohno T, Kasai Y, Saito T. Cloning and sequencing of the Escherichia coli chlEN operon involved in molybdopterin biosynthesis. J Bacteriol. 1988;170:4097–4102. doi: 10.1128/jb.170.9.4097-4102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omata T, Andriesse X, Hirano A. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC 7942. Mol Gen Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 35.Palmer T, Santini C L, Iobbi-Nivol C, Eaves D J, Boxer D H, Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol Microbiol. 1996;20:875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 36.Pitterle D M, Rajagopalan K V. The biosynthesis of molybdopterin in Escherichia coli. Purification and characterization of the converting factor. J Biol Chem. 1993;268:13499–13505. [PubMed] [Google Scholar]
- 37.Pitterle D M, Johnson J L, Rajagopalan K V. In vitro synthesis of molybdopterin from precursor Z using purified converting factor: role of protein-bound sulfur in formation of the dithiolene. J Biol Chem. 1993;268:13506–13509. [PubMed] [Google Scholar]
- 38.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 39.Rajagopalan K V. Biosynthesis of the molybdenum cofactor. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 674–679. [Google Scholar]
- 40.Rajagopalan K V, Johnson J L. The pterin molybdenum cofactors. J Biol Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- 41.Ramaswamy K S, Endley S, Golden J W. Nitrate reductase activity and heterocyst suppression on nitrate in Anabaena sp. strain PCC 7120 require moeA. J Bacteriol. 1996;178:3893–3898. doi: 10.1128/jb.178.13.3893-3898.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rippka R, Herdman M. /1993. 1992. Pasteur Culture Collection of Cyanobacterial Strains. Catalogue and taxonomic handbook, vol. I. Institute Pasteur, Paris, France. [Google Scholar]
- 43.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strains histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 44.Rivers S L, McNairn E, Blasco F, Giordano G, Boxer D H. Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol Microbiol. 1993;8:1071–1081. doi: 10.1111/j.1365-2958.1993.tb01652.x. [DOI] [PubMed] [Google Scholar]
- 45.Rubio L M, Herrero A, Flores E. A cyanobacterial narB gene encodes a ferredoxin-dependent nitrate reductase. Plant Mol Biol. 1996;30:845–850. doi: 10.1007/BF00019017. [DOI] [PubMed] [Google Scholar]
- 46.Rubio, L. M., A. Herrero, and E. Flores. Unpublished results.
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Schaefer M R, Golden S S. Differential expression of members of a cyanobacterial psbA gene family in response to light. J Bacteriol. 1989;171:3973–3981. doi: 10.1128/jb.171.7.3973-3981.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schleif R F, Wensink P C. Practical methods in molecular biology. New York, N.Y: Springer-Verlag; 1981. [Google Scholar]
- 50.Sherratt D. Tn3 and related transposable elements: site-specific recombination and transposition. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: ASM Press; 1989. pp. 163–184. [Google Scholar]
- 51.Suzuki I, Sugiyama T, Omata T. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 52.Vega-Palas M A, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 53.Yura H Mori, Nagai H, Nagata T, Ishihama A, Fujita N, Isono K, Mizobuchi K, Nakata A. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Nucleic Acids Res. 1992;20:3305–3308. doi: 10.1093/nar/20.13.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]