Abstract
Termination of DNA replication, complete topological unlinking of the parental template DNA strands, partition of the daughter chromosomes, and cell division follow in an ordered and interdependent sequence during normal bacterial growth. In Escherichia coli, topoisomerase IV (Topo IV), encoded by parE and parC, is responsible for decatenation of the two newly formed chromosomes. In an effort to uncover the pathway of information flow between the macromolecular processes that describe these events, we identified dnaX, encoding the τ and γ subunits of the DNA polymerase III holoenzyme, as a high-copy suppressor of the temperature-sensitive phenotype of the parE10 allele. We show that suppression derives from overexpression of the γ, but not the τ, subunit of the holoenzyme and that the partition defect of parE10 cells is nearly completely reverted at the nonpermissive temperature as well. These observations suggest a possible association between Topo IV and the replication machinery.
In all cells, DNA replication results in daughter chromosomes that are topologically interlinked, or catenated. The activity of a type II topoisomerase is essential for the resolution of these interlinked daughter molecules (8, 16, 45, 49, 55, 56). In Escherichia coli, strong genetic and biochemical evidence indicates that topoisomerase IV (Topo IV) is the enzyme that performs this task (1, 18, 19, 42, 64).
Topo IV is a heterotetramer composed of the ParE and ParC proteins. The genes encoding ParE and ParC were first identified in a cytological screen for temperature-sensitive mutations conferring a partition phenotype (18, 19, 31, 44). This phenotype is characterized by chromosomes that can be replicated but not partitioned, resulting in the accumulation of large nucleoids in the middle of a filamentous cell. In parE and parC mutant cells, this phenotype results from failure of the newly replicated chromosomes to be decatenated, establishing the role of Topo IV as the primary decatenase in the cell (18, 19).
Topo IV has the ability to relax positive and negative supercoils, as well as catenate and decatenate double-stranded circular DNAs (19, 20, 42, 43). Topo IV is also capable of relaxing the positive superhelicity generated during DNA replication of a closed circular template, as demonstrated by its ability to support nascent chain elongation in vitro (12). However, the extent to which Topo IV participates during elongation in vivo is uncertain. Topo IV has a high degree of sequence homology with the other type II topoisomerase in E. coli, DNA gyrase (19). Biochemical and genetic studies suggest that there is a division of labor between these two homologous enzymes in the cell, with gyrase providing the essential topoisomerase activity during the elongation stage of DNA replication and Topo IV providing the essential decatenase activity at the termination of DNA replication (1, 10, 13, 14, 18, 19, 28, 41, 42, 57, 64).
Although much has been learned so far, many questions surround the role played by Topo IV during cell growth. For example, what determines the proposed division of labor between DNA gyrase and Topo IV? Is it based purely on their biochemical differences? Or is there some sort of physical barrier generating the division? Perhaps Topo IV is part of a multiprotein termination complex, sequestered from the replicating chromosome. Furthermore, in E. coli, chromosome decatenation and partitioning are tightly coupled with cell division (9), raising the intriguing possibility that Topo IV functions in some way as a link between these cell cycle events.
In order to begin to answer these questions and to further define the role played by Topo IV, we sought to identify proteins that may physically or functionally interact with this enzyme. A genetic screening for high-copy suppressors of a temperature-sensitive parE10 allele (19) was performed. Four independent suppressor clones containing dnaX were isolated. dnaX encodes both the τ and γ subunits of the DNA polymerase III holoenzyme, the replicative polymerase (26, 38). τ is the full-length protein product of dnaX, responsible for dimerizing the core polymerase (37, 47), coupling the leading- and lagging-strand polymerases (22), and interacting with the replication fork helicase, DnaB (23, 63). This interaction is required for rapid replication fork movement (23) and determines which of the two catalytic cores of the holoenzyme becomes the leading-strand polymerase (24, 63). γ is a truncated protein product of dnaX, resulting from a ribosomal frameshifting event (4, 11, 52), and is part of the γ complex, a five-subunit protein complex that acts to load β, the processivity factor, onto a primer template (33, 34, 60). Expression of γ alone, but not τ alone, could rescue the temperature-sensitive phenotype of parE10 cells. In addition, expression of γ alone resulted in near-complete reversion of the partition phenotype at the nonpermissive temperature.
These observations suggest that Topo IV may not act independently in the cell and potentially position it in association with proteins of the replication apparatus.
MATERIALS AND METHODS
Bacterial strains and microbiological techniques.
W3110parE10 and its wild-type isogenic parent, W3110, were described previously by Kato et al. (19) and were obtained from the laboratory of N. Cozzarelli (University of California, Berkeley). DH5-α, which was used to prepare all plasmid DNAs, was from Gibco BRL. Cultures were grown in Luria (L) broth or on Luria-Bertani (LB) agar plates (35). Antibiotics, when added, were at the following concentrations: ampicillin, 100 μg/ml, and kanamycin, 50 μg/ml. Competent cells were prepared by CaCl2 treatment (6) and were used for transformation as described in product information for Library Efficiency DH5-α competent cells from Gibco BRL.
To determine the efficiency of plating, cultures were grown at 32°C to an optical density at 600 nm (OD600) of 0.5 to 1.0, diluted by 10−5 in L broth containing drugs as required, plated (0.1 ml), and grown overnight on LB agar plates containing drugs at both 32 and 42°C.
Comparative growth rates of cultures on plates were determined as follows. The cultures were grown at 32°C to an OD600 of approximately 1.0. The cultures were then adjusted to equal concentrations and diluted serially by a factor of 10 in L broth containing drugs as required in sterile 96-well plates. Aliquots (5 μl) were then plated with a multi-pipette on LB agar plates containing drugs and incubated overnight at both 32 and 42°C.
Enzymes, reagents, proteins, and antibodies.
Restriction enzymes and bacteriophage T4 DNA ligase were from New England Biolabs. Pfu polymerase was from Stratagene. DNA polymerase I and the Klenow fragment were from Boehringer Mannheim. SeaKem ME agarose was from FMC. Acrylamide was from Bio-Rad. [α-32P]ATP, Hybond enhanced chemiluminescence (ECL) nitrocellulose membrane, and ECL-Western blotting detection reagents were from Amersham. γ and τ were the gifts of Charles McHenry (University of Colorado) and were prepared as described previously (7). Monoclonal antisera against τ and γ were also from Charles McHenry. ParE and ParC were as described by Peng and Marians (43). Polyclonal antisera against ParC and ParE were raised in rabbits. Goat anti-rabbit and goat anti-mouse immunoglobulin G antibodies conjugated to horseradish peroxidase were from Bio-Rad.
DNA manipulations, Southern hybridization, and DNA sequencing.
Large-scale plasmid DNA purification was by alkaline lysis and cesium chloride-ethidium bromide density gradient centrifugation as described previously (35). Small-scale plasmid purification was performed with the Quantum Prep plasmid miniprep kit (Bio-Rad). DNA fragments for cloning purposes were isolated with the Gene-Clean II kit (Bio 101).
Conversion of fragments with 5′ overhanging ends to blunt ends was done as follows. Reaction mixtures (100 μl) containing 50 mM Tris-HCl (pH 7.5 at 23°C), 10 mM MgSO4, 1 mM dithiothreitol, 50 μg of bovine serum albumin/ml, 80 μM deoxynucleoside triphosphates, 100 nM [α-32P]dATP, 50 μg of DNA/ml, and 40 U of Klenow fragment/ml were incubated at room temperature for 30 min. The DNA fragment was then gel purified with the Gene-Clean II kit.
Nick translation of plasmids was done as follows. Reaction mixtures (50 μl) containing 50 mM Tris-HCl (pH 7.5 at 23°C), 10 mM MgCl2, 1 mM dithiothreitol, 80 μM deoxynucleoside triphosphates, 330 nM [α-32P]dATP, 5.3 mU of DNase I, 2.5 U of DNA polymerase I, and 1 μg of plasmid DNA were incubated at 37°C for 2 h. Radiolabeled plasmids were purified through CentriSep DNA spin columns (Princeton Separations).
Southern hybridization was carried out essentially as described previously (39). The Kohara DNA membrane was from TaKaRa Biomedical.
DNA sequencing reactions were performed with an Amplitaq DNA polymerase FS DNA sequencing kit from Perkin-Elmer. Reactions were analyzed on an ABI 373A Stretch DNA sequencer, and data analysis was done with ABI PRISM version 2.1.1 software.
pBR-parE was constructed as follows. A 3.5-kbp PvuII fragment containing parE was excised from a pBS+/− plasmid carrying a 5.2-kbp EcoRI-BglII fragment from Kohara λ phage 506 (which spans the parCEF region) (27) and ligated into EcoRV-digested pBR322-kan-inc#3 vector DNA.
Generation of an E. coli genomic DNA library. (i) pBR322-kan-inc#3 vector.
A PstI fragment containing a kanamycin resistance cassette was inserted into PstI-digested pBR322 DNA. The inc#3 mutation, located within the ColE1 origin region (50), which increases the plasmid copy number from approximately 30 to 50 per chromosome and renders the plasmid compatible with other ColE1 origin plasmids, was introduced by site-directed mutagenesis with the PCR technique of splicing by overlap extension (17).
(ii) Genomic DNA fragments.
Genomic DNA was prepared from E. coli C600 and partially digested with Sau3AI. Digested DNA was fractionated by sedimentation through 10 to 40% neutral sucrose gradients, and fractions containing DNA fragments ranging from 3 to 6 kbp were pooled.
(iii) Library construction.
Large-scale ligations were performed with BamHI-digested pBR322-kan-inc#3 DNA and Sau3AI-digested, size-selected genomic DNA fragments. Ligation mixtures were transformed into DH5-α competent cells. Approximately 400,000 colonies were combined (by scraping from LB agar plates), and plasmid library DNA was isolated as described above. The final DNA concentration was 480 μg/ml.
ECL-Western analysis.
Cell cultures were grown at the indicated temperatures to an OD600 of 0.5 to 1.0 and chilled, and an aliquot (1 ml) was pelleted in a microcentrifuge. The cell pellet was resuspended at 250 OD units/μl in Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (29). Aliquots (5 μl for τ and γ expression and 20 μl for ParC and ParE expression) were subjected to SDS-PAGE through 10% gels (29). The gels were equilibrated in transfer buffer (47.8 mM Tris base, 386 mM glycine, and 0.02% SDS) for 20 min and then transferred to a Hybond-ECL membrane with a Bio-Rad Trans-Blot electrophoretic transfer cell. The membranes were blocked overnight in a solution containing 1× phosphate-buffered saline (PBS), 0.1% Tween 20, and 5% nonfat milk, incubated with the appropriate primary antibodies, washed in a solution containing 1× PBS and 0.1% Tween 20, incubated with the appropriate secondary antibody conjugated to horseradish peroxidase (in blocking solution), washed again, developed with ECL-Western blotting detection reagents as described by the manufacturer, and immediately exposed to X-ray film.
DAPI staining and fluorescence microscopy.
Cell cultures were grown overnight at 32°C, diluted to an OD600 of approximately 0.01, and then grown at 42°C for approximately 3 h. Aliquots (5 ml) were pelleted in a microcentrifuge and resuspended in either 0.84% NaCl or 1× PBS at a 1.2-fold concentration. Aliquots (30 μl) of cell suspension were spread on Superfrost/Plus microscope slides (Fisher Scientific) and allowed to air dry. Samples were fixed by soaking in methanol (at −20°C) for 10 min and were then air dried. Slides were rinsed by dunking in tap water 10 times, air dried, and stored at −80°C. Before examination, the samples were fixed again, rinsed in tap water, and then stained by being incubated in 5 μg of DAPI (4′,6-diamidino-2-phenylindole)/ml in either 0.84% NaCl or 1× PBS for 15 min in the dark. Slides were then rinsed three times for 5 min each in either 0.84% NaCl or 1× PBS, and a drop of glycerol-based fluorescent mounting medium containing antiquenching agents was placed on top of the sample before the coverslip was positioned. Photomicroscopy was done with an Axiophot microscope (Zeiss). Images were recorded on slide film, scanned with an AGFA Duoscan slide scanner, and processed with Adobe Photoshop version 3.0 software.
RESULTS
Identification of dnaX as a high-copy suppressor of parE10.
The E. coli strain W3110parE10 (19) was used in a genetic screening for high-copy suppressors of the temperature-sensitive phenotype of the parE10 allele. A plasmid library was generated by inserting size-selected E. coli genomic DNA into a pBR322 vector (pBR322-kan-inc#3) that carried a mutation giving it a slightly higher copy number than normal (50). All E. coli genes present in this library were expressed from their natural promoters. Library DNA was transformed into W3110parE10, and about 20,000 transformants were plated at 42°C. Of the approximately 300 colonies that grew, 50 were retested for growth at both 32 and 42°C. Five colonies regrew at both temperatures. Plasmid DNA was isolated from these five clones and retransformed into W3110parE10. Four of the five, 31, 37, 39, and 40, conferred the ability to grow at 42°C. These were therefore designated as suppressor clones.
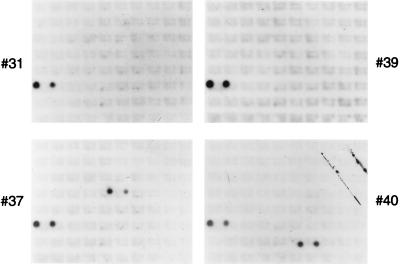
In order to determine what region of the E. coli chromosome was contained within the suppressor clones, the chromosomal DNA carried on the plasmids was mapped to the E. coli genome by hybridization to a membrane carrying the λ phage genomic library developed by Kohara et al. (27). Suppressor clone DNAs were radiolabeled by nick translation and hybridized to the Kohara membrane. All four suppressor clones carried genomic DNA that mapped to the same two λ clones, 151 and 152, which contained overlapping regions of the E. coli chromosome spanning min 10.5 to 10.8 (Fig. 1). Two of the four suppressor clones, 37 and 40, also carried DNA that hybridized to unique second sets of contiguous λ clones that were noncontiguous with λ clones 151 and 152. Sequence data showed that this resulted from two noncontiguous pieces of E. coli genomic DNA becoming joined during creation of the DNA library.
FIG. 1.
Mapping of suppressor clones to the E. coli chromosome. Suppressor plasmid clones (31, 37, 39, and 40), as well as a small amount of λ DNA, were radiolabeled by nick translation. Southern hybridization of a Kohara DNA membrane was then carried out as described in Materials and Methods. All four suppressor clones hybridized to the same two λ clones, 151 and 152 (spots on lower left-hand side of each blot), containing overlapping regions of the E. coli chromosome. Suppressor clones 37 and 40 also hybridized to unique second sets of contiguous λ clones (spots on right-hand side of each blot).
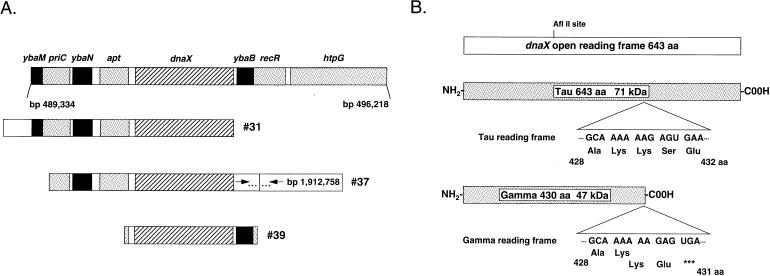
In order to identify which gene within the 10.5- to 10.8-min region, spanning roughly nucleotide coordinates 487,000 to 501,000 of the E. coli chromosome (2), was responsible for suppression, maps of the genomic inserts within the suppressor clones were constructed. The ends of the genomic DNA inserts were sequenced with primers flanking the BamHI genomic DNA insertion site within the pBR322-kan-inc#3 vector. This sequence data was then used to position the genomic inserts on the sequence of the E. coli chromosome. The only intact gene within the overlapping genomic regions of the suppressor clones was dnaX (Fig. 2A).
FIG. 2.
dnaX, encoding both the τ and γ subunits of the DNA polymerase III holoenzyme, is the only intact gene within the overlapping genomic regions of the suppressor clones. (A) Shown in the top bar are all known genes (shaded boxes) and open reading frames (black and hatched boxes) that lie within the region potentially contained within the suppressor clones (nucleotides 4.87 × 105 to 5.01 × 105 of the E. coli chromosome). The genomic regions contained on suppressor clones 31, 37, and 39 are shown below. (B) dnaX open reading frame with an exploded view of the region where the ribosomal frameshifting event that generates the γ subunit (codons 428 to 430) occurs. Also indicated is the AflII restriction endonuclease site used to generate the dnaX open reading frame disruption. aa, amino acids.
dnaX encodes both the τ and γ subunits of the DNA polymerase III holoenzyme (26, 38). τ is the full-length, 71-kDa (643-amino-acid) protein product, whereas γ is a 47-kDa (431-amino-acid) truncated protein product generated as a result of a −1 ribosomal frameshift that occurs at a heptanucleotide repeat (AAAAAAG) at codons 428 to 430 of the dnaX open reading frame (4, 11, 52, 55). γ is thus the amino-terminal 430 amino acids of τ with the addition of a unique C-terminal glutamate at amino acid residue 431 (Fig. 2B).
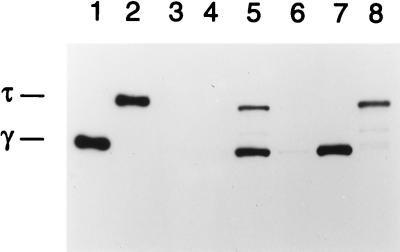
To confirm that dnaX expression was responsible for the suppressor phenotype, the dnaX open reading frame within one of the suppressor clones, 39 (subsequently referred to as pBR-dnaX), was disrupted. pBR-dnaX was digested with the AflII restriction enzyme that cuts the plasmid once, 651 bp downstream from the start of dnaX. The 4-bp overhang was repaired with the Klenow enzyme, and the DNA was religated. This reading frame disruption affected both the γ and τ gene products by generating an opal stop codon at amino acid position 230. The disrupted plasmid, designated pBR-dnaXG230Op, was transformed into W3110parE10 to test for suppressor activity. The lack of either γ or τ expression from this plasmid, compared to the expression of both proteins from the pBR-dnaX parent plasmid, was confirmed by ECL-Western analysis with a monoclonal antibody that recognized both τ and γ (Fig. 3, compare lanes 5 and 6).
FIG. 3.
ECL-Western analysis of τ and γ expression. W3110parE10 cells carrying various plasmids were grown in culture at 32°C, and ECL-Western analysis was performed as described in Materials and Methods. Lane 1, 15 ng of γ; lane 2, 15 ng of τ; lanes 3 to 8, W3110parE10 cells containing either pBR322 (lane 3), pBR-parE (lane 4), pBR-dnaX (lane 5), pBR-dnaXG230Op (lane 6), pBR-dnaX-γ (lane 7), or pBR-dnaX-τ (lane 8).
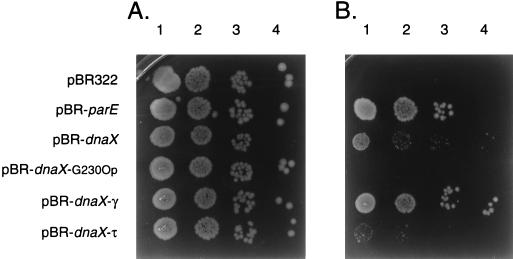
W3110parE10 carrying pBR-dnaX grew fairly well at 42°C, although colony size was smaller than when the vector plasmid carried wild-type parE (pBR-parE) (Fig. 4). The plating efficiency of W3110parE10(pBR-dnaX) at the nonpermissive temperature was 0.80 compared to that at the permissive temperature (Table 1). pBR-dnaXG230Op failed to rescue the growth of W3110parE10 at the nonpermissive temperature (Fig. 4 and Table 1), indicating that expression of the dnaX gene products was required to observe the suppressor phenotype.
FIG. 4.
Comparative growth of W3110parE10 cells containing various plasmids at 32 (A) and 42°C (B). Cultures were grown, serially diluted, and plated as described in Materials and Methods. Lanes 1 to 4, 10−3, 10−4, 10−5, and 10−6 dilutions, respectively, of the indicated cultures.
TABLE 1.
Efficiency of plating of W3110parE10 containing various plasmids
| Plasmid | Efficiency of platinga |
|---|---|
| pBR322 | <0.003 |
| pBR-parE | 1.09 |
| pBR-dnaX | 0.8 |
| pBR-dnaXG230Op | <0.002 |
| pBR-dnaX-γ | 0.96 |
| pBR-dnaX-τ | 0.08 |
Averages of at least five independent experiments. The numbers represent the efficiency of plating at 42°C compared to that at 32°C.
Expression of γ alone, but not of τ alone, is sufficient for elaboration of the suppression phenotype.
Although both τ and γ are subunits of the DNA polymerase III holoenzyme and are encoded by the same gene, biochemical analyses argue that they have different functions during DNA replication. τ plays a central role in cementing the replisome together via protein-protein interactions between polymerase components and with the replication fork helicase (22–24, 37, 47, 63). γ participates in the loading of the processivity subunit, β, onto the primer template (33, 34, 48, 60). It was therefore of interest to determine whether γ alone or τ alone was sufficient for rescue of the growth of W3110parE10 at 42°C or whether both proteins were required.
Plasmid DNAs expressing either only γ (pRT610B) or only τ (pRT610A) were obtained from the laboratory of C. McHenry (7). These plasmids contain point mutations in and around the ribosomal frameshifting site within the dnaX open reading frame. In the case of the γ-only overexpression plasmid, point mutations were introduced that resulted in an altered codon and an obligatory stop codon, which are also present in the frameshifted open reading frame; in the case of the τ-only overexpression plasmid, point mutations abolishing the heptanucleotide repeat essential for ribosomal slippage were introduced. AflII-SplI fragments from these plasmid DNAs encompassing the region of the point mutations were cloned into pBR-dnaX, creating pBR-dnaX-γ and pBR-dnaX-τ. Sequence analysis confirmed the presence of the correct sequences, and ECL-Western analysis confirmed the expression of either γ or τ alone from the respective plasmids (Fig. 3, lanes 7 and 8).
pBR-dnaX-γ and pBR-dnaX-τ were transformed into W3110parE10 to test for suppressor activity. W3110parE10(pBR-dnaX-γ) grew essentially like W3110parE10(pBR-parE), with a plating efficiency of 0.96 and extensive growth at 42°C on LB agar. In contrast, W3110parE10(pBR-dnaX-τ) hardly grew at all at 42°C, with a plating efficiency of 0.08 and minimal growth on LB agar (Fig. 4 and Table 1).
All known biochemical activities of γ can be duplicated by τ (7, 40); in addition, it is τ, not γ, that has a unique polypeptide domain. Thus, these results were unexpected. Expression of the two proteins at different levels from their respective plasmid constructs was considered as a possible explanation. However, although the Western blot pictured in Fig. 3 does appear to show higher expression of γ than of τ, multiple repetitions of this experiment failed to demonstrate such differences consistently. To investigate the issue of rescue by γ and not τ more closely, growth of W3110parE10 and W3110, the wild-type isogenic parent strain, carrying the various plasmid constructs was examined in liquid media at the permissive and nonpermissive temperatures.
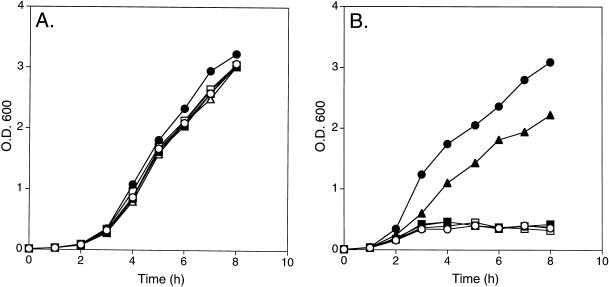
W3110parE10 carrying either the modified pBR322 vector alone (pBR322), pBR-parE, pBR-dnaX, pBR-dnaXG230Op, pBR-dnaX-γ, or pBR-dnaX-τ all grew well, with similar growth rates, at 32°C (Fig. 5A). At 42°C, analogous with previous results, cells expressing either wild-type ParE or γ alone grew very well, with similar growth rates, whereas cells that did not express either τ or γ (either vector alone or pBR-dnaXG230Op) and those expressing τ alone failed to grow to any significant extent (Fig. 5B). Interestingly, cells carrying the original suppressor plasmid, pBR-dnaX, also failed to grow (although some growth was occasionally observed above background levels). This result correlates with the observation that the colony size for W3110parE10 rescued by pBR-dnaX was smaller than that for cells rescued by pBR-dnaX-γ and suggested that either the level of γ expression correlates with the degree of rescue or the expression of τ might be toxic to the cell. The growth of W3110 carrying either pBR322 (vector alone), pBR-dnaX, pBR-dnaX-γ, or pBR-dnaX-τ was therefore examined at 32 and 42°C.
FIG. 5.
Growth curves of W3110parE10 cells containing various plasmids at 32 and 42°C. Cultures were grown overnight at 32°C, diluted to an OD600 of approximately 0.01, and grown at 32 (A) and 42°C (B). ○, pBR322; •, pBR-parE; ▪, pBR-dnaX; □, pBR-dnaXG230Op; ▴, pBR-dnaX-γ; ▵, pBR-dnaX-τ.
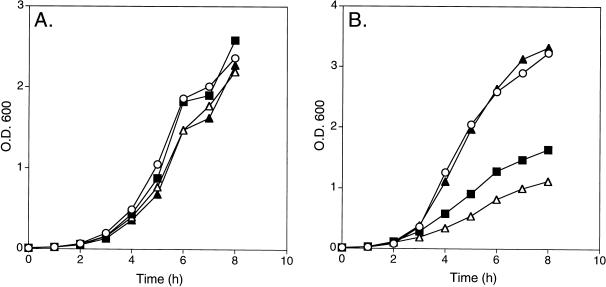
All the cells grew well at 32°C, with similar growth rates (Fig. 6A). However, at 42°C, cells that did not express either τ or γ and those expressing γ alone grew identically, whereas cells expressing either both γ and τ or those expressing τ alone had a significantly decreased growth rate, with cells expressing τ alone being the most inhibited (Fig. 6B). Why this differential growth rate was observed only at 42°C is unclear, because τ was expressed to similar extents at both 32 and 42°C (data not shown); however, it is consistent with the expression of τ being toxic. In any event, these observations cloud our ability to reach a definitive conclusion as to the ability of τ to rescue the parE10 mutation.
FIG. 6.
Growth curves of W3110 cells containing various plasmids at 32 and 42°C. Cultures were grown overnight at 32°C, diluted to an OD600 of approximately 0.01, and grown at 32 (A) and 42°C (B). ○, pBR322; ▪, pBR-dnaX; ▴, pBR-dnaX-γ; ▵, pBR-dnaX-τ.
Overexpression of γ at the nonpermissive temperature results in near-complete reversion of the partition defect of the parE10 allele.
In order to determine to what extent the partition phenotype was suppressed at the cellular level, W3110parE10 cells carrying various plasmid constructs were visualized by fluorescence microscopy. Cells were grown overnight at 32°C, diluted to an OD600 of approximately 0.01, grown at 42°C for approximately 3 h, fixed on slides, stained with DAPI, and visualized by fluorescence microscopy.
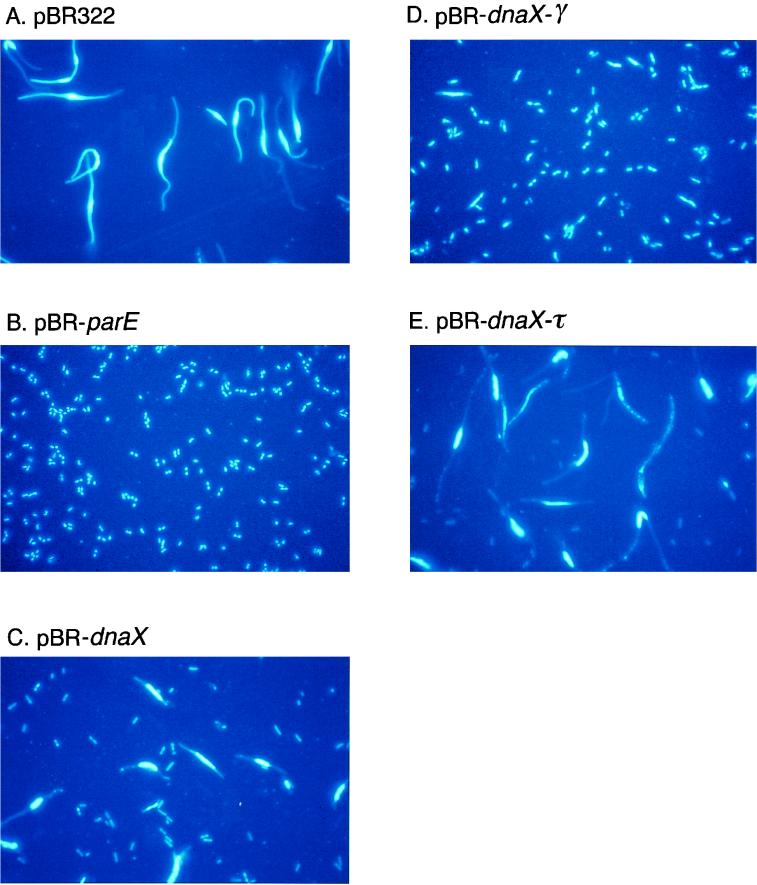
W3110parE10 cells carrying the vector alone (pBR322) exhibited a striking partition phenotype, containing large nucleoid masses in the centers of extremely large, elongated cells (Fig. 7A). In contrast, W3110parE10(pBR-parE) cells appeared to be essentially wild type. Cells were small, of wild-type size, and contained either one or two discrete nucleoids (Fig. 7B).
FIG. 7.
Fluorescence microscopy of DAPI-stained W3110parE10 cells containing various plasmids grown at 42°C. Cultures were grown, fixed, DAPI stained, and visualized as described in Materials and Methods.
W3110parE10(pBR-dnaX) cells displayed a somewhat intermediate phenotype, with a few cells containing discrete nucleoids and many cells exhibiting a partition phenotype (Fig. 7C). Additionally, there appeared to be many chromosomeless cells present. This phenotype was consistent with the inability of these cells to grow in liquid medium at 42°C (Fig. 5B).
W3110parE10 cells carrying either pBR-dnaX-γ or pBR-dnaX-τ are shown in Fig. 7D and E, respectively. As expected, cells expressing γ alone exhibited a near-wild-type appearance, with the majority of cells containing one or two discrete nucleoids, characteristic of successful chromosome partition. These cells did, however, appear to have on average slightly larger than wild-type cell size as well as nucleoid mass, suggesting a possible delay in chromosome partition and therefore in cell division. In contrast, cells expressing τ alone exhibited a typical partition phenotype, with enlarged nucleoid masses in the center of an elongated cell. Additionally, there also appeared to be many chromosomeless cells, as in the cultures expressing τ and γ together. Furthermore, cells expressing τ alone appeared to show some signs of membrane disruption and/or nucleoid fragmentation, supporting the suggestion that τ expression may be toxic for cell growth.
DISCUSSION
Topoisomerases play essential roles in all aspects of DNA metabolism. In all organisms, type II topoisomerases are required for the proper topological separation of newly replicated chromosomal DNA. In E. coli, Topo IV is the enzyme responsible for accomplishing this task (1, 18, 19, 42, 64). In its absence, cells arrest with a partition phenotype, characterized by a large nucleoid mass of intertwined chromosomes in the center of a filamentous cell (19, 44).
In an attempt to further delineate the flow of information during the terminal stages of DNA replication and cell division, we carried out a genetic screening to detect high-copy suppressors of the temperature-sensitive phenotype of the parE10 allele. This screening yielded dnaX, which encodes the τ and γ subunits of the DNA polymerase III holoenzyme, the replication fork polymerase. Additional characterization of the suppression by dnaX revealed that expression of γ alone, but not expression of τ alone, could almost completely rescue both the temperature-sensitive and partition phenotypes of parE10 E. coli.
However, at this time, we cannot conclude that we have uncovered a distinct role for γ that cannot be accomplished by τ. The expression of τ appears to be toxic to some extent, which could limit the possibility of observing rescue by expression of τ alone. Additionally, rescue of W3110parE10 cells by γ alone was more complete than when τ and γ were expressed together. This increased ability of γ alone to rescue could be the result of either a higher level of expression of γ from the pBR-dnaX-γ construct than from the pBR-dnaX construct, the absence of the toxic effect of τ, or a combination of both factors.
The question of whether γ and τ have distinct roles is an intriguing one that has yet to be answered satisfactorily. Biochemically, τ can substitute for γ, forming a β-loading τ complex (7, 40), and it has been suggested that only τ, not γ, is required for viability of E. coli (3). This is consistent with the observation that γ is not required for replication fork action in vitro during rolling circle DNA replication reconstituted with purified proteins (25). On the other hand, γ is clearly associated with holoenzyme purified from bulk E. coli and only γ complex is found free in the cell; τ complex has never been detected (21, 33, 34, 36).
How might overproduction of γ result in the observed suppression? There are two obvious possibilities that we have been able to eliminate. (i) Overexpression of γ could cause, either directly or indirectly, overexpression of the ParE10 protein. This might stabilize the polypeptide against denaturation at the nonpermissive temperature. This has been observed, for example, for the ssb-1 temperature-sensitive allele (5, 61). However, ECL-Western blot analysis has shown that the levels of both ParE and ParC remain constant in W3110parE10 in either the presence or absence of the pBR-dnaX-γ plasmid at both 32 and 42°C (data not shown). (ii) γ, which is an ATPase that is involved in opening the ring of the β dimer to allow it to encircle the DNA template (33, 34, 48, 52, 60), might form a novel topoisomerase with either ParC or GyrA that is capable of decatenating the daughter chromosomes. We have assessed this directly by testing whether the purified proteins exhibit such an activity in vitro and have not detected it, even at protein concentrations far in excess of what would be required to observe activity with wild-type Topo IV or DNA gyrase (data not shown).
A third interesting possibility is that overexpression of γ interferes with DNA replication in some way. If so, replication may proceed at a lower rate, thereby reducing the need for rapid unlinking of the replicated duplex DNA and allowing other topoisomerases, such as gyrase, to assist or replace the weakened Topo IV. However, a number of our observations suggest that this is not a likely possibility. If this explanation were true, and DNA replication were slowed to a point where topoisomerases that can decatenate less efficiently than Topo IV could take over, we would expect an obligatory delay in cell division with a concomitant increase in cell size (assuming the rate of DNA replication is the limiting factor for cell division during rapid growth in rich medium). Upon microscopic examination, however, parE10 cells expressing γ alone that were grown at the permissive temperature were indistinguishable from cells expressing wild-type ParE. Additionally, the growth rates at the permissive temperature, based on OD, as well as viable CFU (data not shown) of parE10 cells expressing γ alone or wild-type ParE were similar. These data suggest that overexpression of γ does not significantly delay the average time between cell divisions and therefore probably does not slow DNA replication significantly.
Another interesting possibility is that γ, which has some characteristics of a chaperone, might be healing the damaged ParE protein at the nonpermissive temperature. This has been more difficult to test, because it requires the purification of the ParE10 protein, which, unlike the wild type, is insoluble when overexpressed (data not shown).
High-copy suppression of a temperature-sensitive allele is generally taken to indicate the existence of a complex between the two gene products, where the overexpressed protein stabilizes the temperature-sensitive protein at the nonpermissive temperature. However, we have been unable to detect an interaction between γ and ParE by gel filtration chromatography when the two proteins were mixed together at micromolar concentrations (data not shown). This does not eliminate the possibility of an interaction. The interaction between DnaG and DnaB at the replication fork, which can be detected functionally (51, 62) and by affinity chromatography (30), cannot be detected by gel filtration.
The possibility of a physical interaction between γ and Topo IV could be eliminated if overexpression of dnaX was found to suppress a parE null allele, but to our knowledge no such allele exists, and our attempts to create one have been unsuccessful. However, neither pBR-dnaX nor pBR-dnaX-γ can rescue the parC1215 (18) temperature-sensitive allele (data not shown). This suggests that γ expression is not compensating for a complete lack of Topo IV activity. It would be informative if dnaX suppression of parE was shown to be allele specific, but additional alleles are not known.
Given all this, our current working hypothesis is that γ and ParE interact. If this interaction occurs, it could take one of two forms. Topo IV could associate with the γ complex at the replication fork, or with free γ complex. γ complex can be isolated from the holoenzyme and may therefore exist in free form as well as forming part of the holoenzyme (33, 36). This excess γ complex may be essential for recycling β that is left behind on the nascent duplex DNA after the lagging-strand polymerase moves to a new primer during lagging-strand synthesis (58). If there were an interaction between Topo IV and free γ complex, it would suggest a novel function for the latter.
In support of a Topo IV-γ interaction at the replication fork, interallelic suppressors of a temperature-sensitive parE mutation in Salmonella typhimurium have been mapped to dnaE (46), encoding the α subunit of the holoenzyme (59). If there is a Topo IV-replication fork interaction, why might this interaction take place?
Within the cell, Topo IV may not have free access to the replicating DNA. Double-stranded DNA binding proteins may limit the access of Topo IV to the DNA, and/or Topo IV may be sequestered in some manner from the replicating chromosome. Topo IV may be membrane associated, as suggested by the observation that under certain conditions of isolation ParC has been shown to be associated with the inner cell membrane (18, 20). As previously suggested, the membrane association of Topo IV may be via ParF, an inner membrane protein first identified in a screening for partition mutations (along with parE and parC) in Salmonella (31, 44). Additional support for the idea that Topo IV does not have free access to the replicating DNA is the observation that only gyrA and gyrB mutations show an immediate-stop DNA replication phenotype, as expected for the enzyme that supports this reaction in vivo (10, 28), even though Topo IV is as capable as DNA gyrase of supporting nascent chain elongation during theta-type DNA replication in vitro (12) and there are roughly equivalent amounts of Topo IV and gyrase in the cell (about 400 tetramers [12, 15]).
Association with the replication fork could serve as an entry point for Topo IV to the DNA. After association, Topo IV could ride along with the advancing replication fork or be dropped off behind the fork to relax the positive windings that arise between the newly replicated daughter duplexes. Alternatively, the interaction could take place between Topo IV and the replication fork as it is nearing completion of replication, forming a termination complex. A membrane-associated termination complex would be strategically positioned for signaling to partition or septation proteins that DNA replication and decatenation were complete.
ACKNOWLEDGMENT
These studies were supported by NIH grant GM34558.
REFERENCES
- 1.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1476. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Blinkowa A, Hervas C, Stukenberg P T, Onrust R, O’Donnell M E, Walker J R. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, τ and γ, but only τ is essential. J Bacteriol. 1993;175:6018–6027. doi: 10.1128/jb.175.18.6018-6027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blinkowa A L, Walker J R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chase J W, Murphy J B, Whittier R F, Lorensen E, Sninsky J J. Amplification of ssb-1 mutant single-stranded DNA-binding protein in Escherichia coli. J Mol Biol. 1983;164:193–211. doi: 10.1016/0022-2836(83)90075-x. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S N, Chang A C Y, Hus L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci USA. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallmann H G, Thimmig R L, McHenry C S. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Central role of τ in initiation complex assembly and in determining the functional asymmetry of holoenzyme. J Biol Chem. 1995;270:29555–29562. [PubMed] [Google Scholar]
- 8.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donochie W D, Begg K J, Vincente M. Cell length, cell growth, and cell division. Nature. 1976;264:328–333. doi: 10.1038/264328a0. [DOI] [PubMed] [Google Scholar]
- 10.Filutowicz M, Jonczyk P. The gyrB gene product functions in both initiation and chain polymerization of Escherichia coli chromosome replication: suppression of the initiation deficiency in gyrB-ts mutants by a class of rpoB mutations. Mol Gen Genet. 1983;191:282–287. doi: 10.1007/BF00334827. [DOI] [PubMed] [Google Scholar]
- 11.Flower A M, McHenry C S. The γ subunit of DNA polymerase II holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci USA. 1990;87:3713–3716. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiasa H, Marians K J. Topoisomerase IV can support oriC DNA replication in vitro. J Biol Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 13.Hiasa H, Marians K J. Two distinct modes of unlinking during theta-type DNA replication. J Biol Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 14.Hiasa H, DiGate R J, Marians K J. Decatenating activity of Escherichia coli DNA gyrase and topoisomerase I and III during oriC and pBR322 DNA replication in vitro. J Biol Chem. 1994;269:2093–2099. [PubMed] [Google Scholar]
- 15.Higgins N P, Peebles C L, Sugino A, Cozzarelli N R. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci USA. 1978;75:1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm C, Goto T, Wang J C, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 17.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 18.Kato J-I, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato J-I, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 20.Kato J-I, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- 21.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replication machine. Annu Rev Biochem. 1995;64:1171–1200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Dallmann H G, McHenry C S, Marians K J. τ couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J Biol Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Dallmann H G, McHenry C S, Marians K J. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Dallmann H G, McHenry C S, Marians K J. τ protects β in the leading-strand polymerase complex at the replication fork. J Biol Chem. 1996;271:4315–4318. doi: 10.1074/jbc.271.8.4315. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S., C. S. McHenry, and K. J. Marians. Unpublished data.
- 26.Kodaira M, Biswas S B, Kornberg A. The dnaX gene encodes the DNA polymerase III holoenzyme τ subunit, precursor of the γ subunit, the dnaZ gene product. Mol Gen Genet. 1983;192:80–96. doi: 10.1007/BF00327650. [DOI] [PubMed] [Google Scholar]
- 27.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 28.Kreuzer K N, Cozzarelli N R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979;140:424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y B, Ratnakar P V, Mohanty B K, Bastia D. Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc Natl Acad Sci USA. 1996;93:12902–12907. doi: 10.1073/pnas.93.23.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luttinger A L, Springer A L, Schmid M B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991;3:687–697. [PubMed] [Google Scholar]
- 32.Maki H, Maki S, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J Biol Chem. 1988;263:6570–6578. [PubMed] [Google Scholar]
- 33.Maki S, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the γ subunit essential for processive synthesis. J Biol Chem. 1988;263:6555–6560. [PubMed] [Google Scholar]
- 34.Maki S, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerase reconstituted from purified subunits. J Biol Chem. 1988;263:6561–6569. [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 36.McHenry C, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J Biol Chem. 1977;252:6478–6484. [PubMed] [Google Scholar]
- 37.McHenry C S. Purification and characterization of DNA polymerase III′. Identification of τ as a subunit of the DNA polymerase III holoenzyme. J Biol Chem. 1982;257:2657–2663. [PubMed] [Google Scholar]
- 38.Mullin D A, Woldringh C L, Henson J M, Walker J R. Cloning of the Escherichia coli dnaZX region and identification of its products. Mol Gen Genet. 1993;192:73–79. doi: 10.1007/BF00327649. [DOI] [PubMed] [Google Scholar]
- 39.Nurse P, DiGate R J, Zavitz K H, Marians K J. Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc Natl Acad Sci USA. 1990;87:4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O’Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. J Biol Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 41.Orr E, Staudenbauer W L. An Escherichia coli mutant thermosensitive in the B subunit of DNA gyrase: effect on the structure and replication of the colicin E1 plasmid in vitro. Mol Gen Genet. 1981;181:52–56. doi: 10.1007/BF00339004. [DOI] [PubMed] [Google Scholar]
- 42.Peng H, Marians K J. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc Natl Acad Sci USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng H, Marians K J. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 44.Schmid M B. A locus affecting nucleoid segregation in Salmonella typhimurium. J Bacteriol. 1990;172:5416–5424. doi: 10.1128/jb.172.9.5416-5424.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shamu C E, Murray A W. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Springer A L. Ph.D. thesis. Princeton, N.J: Princeton University; 1992. [Google Scholar]
- 47.Studwell-Vaughan P S, O’Donnell M. Constitution of the twin polymerase of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:19833–19841. [PubMed] [Google Scholar]
- 48.Stukenberg P T, Studwell-Vaughan P S, O’Donnell M. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:17790–17795. [PubMed] [Google Scholar]
- 49.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 50.Tomizawa J-I, Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci USA. 1981;78:6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tougu K, Peng H, Marians K J. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 52.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuchihashi Z, Kornberg A. ATP interactions of the tau and gamma subunits of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1989;264:17790–17795. [PubMed] [Google Scholar]
- 54.Tsuchihashi Z, Brown P O. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNALYS and an AAG lysine codon. Genes Dev. 1992;65:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- 55.Uemura T, Yanagida M. Isolation of type I and type II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotype in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uemura T, Yanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 1986;5:1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ullsperger C, Cozzarelli N R. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J Biol Chem. 1966;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 58.Vytautas N, Turner J, O’Donnell M. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- 59.Welch M M, McHenry C S. Cloning and identification of the product of the dnaE gene of Escherichia coli. J Bacteriol. 1982;152:351–356. doi: 10.1128/jb.152.1.351-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickner S. Mechanism of DNA elongation catalyzed by Escherichia coli DNA polymerase III, dnaZ protein, and DNA elongation factors I and III. Proc Natl Acad Sci USA. 1976;73:3511–3515. doi: 10.1073/pnas.73.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams K R, Murphy J N, Chase J W. Characterization of the structural and functional defect in the Escherichia coli single-stranded DNA binding protein encoded by the ssb-1 mutant gene. J Biol Chem. 1984;25:11804–11811. [PubMed] [Google Scholar]
- 62.Wu C A, Zechner E L, Marians K J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem. 1992;267:4030–4044. [PubMed] [Google Scholar]
- 63.Yuzhakov A, Turner J, O’Donnell M. Replisome assembly reveals the basis for asymmetric function in leading- and lagging-strand replication. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 64.Zechiedrich E L, Cozzarelli N R. Roles of topoisomerase IV and DNA gyrase in DNA replication unlinking in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]