Abstract
Stigmatella aurantiaca is a gram-negative bacterium which forms, under conditions of starvation in a multicellular process, characteristic three-dimensional structures: the fruiting bodies. For studying this complex process, mutants impaired in fruiting body formation have been induced by transposon insertion with a Tn5-derived transposon. The gene affected (fbfB) in one of the mutants (AP182) was studied further. Inactivation of fbfB results in mutants which form only clumps during starvation instead of wild-type fruiting bodies. This mutant phenotype can be partially rescued, if cells of mutants impaired in fbfB function are mixed with those of some independent mutants defective in fruiting before starvation. The fbfB gene is expressed about 14 h after induction of fruiting body formation as determined by measuring β-galactosidase activity in a merodiploid strain harboring the wild-type gene and an fbfB-Δtrp-lacZ fusion gene or by Northern (RNA) analysis with the Rhodobacter capsulatus pufBA fragment fused to fbfB as an indicator. The predicted polypeptide FbfB has a molecular mass of 57.8 kDa and shows a significant homology to the galactose oxidase (GaoA) of the fungus Dactylium dendroides. Galactose oxidase catalyzes the oxidation of galactose and primary alcohols to the corresponding aldehydes.
Stigmatella aurantiaca is a member of the order Myxobacterales. Myxobacteria are gram-negative, rod-shaped soil bacteria which are distinguished from most other bacteria mainly by two properties. First, they are able to move by gliding, a property which they share with a few other prokaryotes. Second, under conditions of starvation they form fruiting bodies, a property which is unique to the myxobacteria. The life cycle of the myxobacteria is bipartite. It is composed of the vegetative growth cycle, during which cells divide by transverse fission, and the developmental cycle, into which cells enter under conditions of starvation. At the end of this cycle, cells form the multicellular fruiting body which encloses 104 to 105 dormant cells, the myxospores (8, 32). The myxobacterial fruiting body is species specific. Whereas Myxococcus xanthus forms simple mounds during fruiting, the fruiting body of S. aurantiaca is composed of a stem bearing several sporangioles on delicate pedicels at its top.
Qualls et al. (31) monitored the synchronous aggregation and fruiting body formation of S. aurantiaca cells by electron microscopy when starved on an agar surface. They defined different stages of S. aurantiaca development: the cells form early aggregates, early stalks, late stalks, and mature fruits about 9, 12, 15, and 24 h, respectively, after the beginning of starvation (cf. Fig. 2A to D in reference 31). The early and the late stalks look like a morel and a champignon, respectively. The myxospores appear between 17 and 24 h after the beginning of starvation (31). Sporulation of vegetative cells may be induced independently of fruiting body formation by indole or some of its derivatives, which leads to independent single spores (9).
FIG. 2.
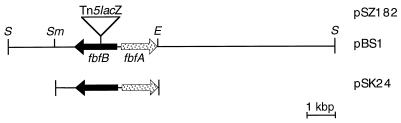
Schematic depiction of the fbfA-fbfB locus and various plasmid constructs. The genes fbfA and fbfB are arranged in a divergent orientation, and the distance between their start codons is 153 bp. The Tn5lacZ insertion in the fbfB gene of the mutant AP182 is 550 bp downstream of the start codon. The insert of pSZ182 is identical to the insert of pBS1 containing Tn5lacZ. E, EcoRI; S, SalI; Sm, SmaI.
The development of the fruiting body is strictly coupled to a time- and compartment-specific synthesis of regulatory factors, which stimulate the expression of several genes or gene families (18, 23, 24). Inactivation of the genes involved in the synthesis of these regulatory factors would lead to a defect in fruiting. The myxobacteria’s capacity to glide permits a tight cell-cell contact and an efficient intercellular communication via diffusible signal molecules. These features allow the transmission of positional information about the single cell which is needed for the coordination of the metabolism and of the movement of the cells during fruiting.
To detect genes involved in fruiting body formation of S. aurantiaca, Tn5 insertional mutagenesis was performed with the transposon Tn5lacZ (29). The defect in one of the mutants obtained, AP182, in which the fbfB (fruiting body formation) gene was inactivated, can be rescued partially by mixing the cells of this mutant strain with cells of the nonaggregating mutant AP191. Recently, we described the inactivation of an independent gene involved in fruiting body formation, fbfA, which is located near fbfB and encodes a putative chitin synthase (37). The mutant phenotype is partially rescued by mixing the cells of this mutant with those of AP191 before inducing fruiting body formation. In this communication, the characterization of the fbfB gene encoding a polypeptide with sequence homologies to the galactose oxidase (GaoA) of Dactylium dendroides is reported.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are described in Table 1. S. aurantiaca DW4/3-1 and its derivatives (30) were grown at 32°C in incandescent light. For cultivation, Casitone medium (1% Casitone [Difco], 0.15% MgSO4 · 7H2O, pH 7.0) or tryptone medium (1% tryptone [Difco], 0.2% MgSO4 · 7H2O, pH 7.2) was used and supplemented with streptomycin sulfate (120 μg/ml) and, when necessary, with kanamycin sulfate (50 μg/ml). To obtain colonies from single cells of S. aurantiaca after conjugation, Trypticase peptone agar was used (0.025% Trypticase peptone [Becton Dickinson], 0.05% MgSO4 · 7H2O, 0.05% CaCl2 · 2H2O, pH 7.2). For fruiting body formation assay, starvation agar was used (containing only 0.1% CaCl2 · 2H2O) (29). Escherichia coli strains were grown in Luria broth at 37°C, supplemented when necessary with chloramphenicol (34 μg/ml), kanamycin sulfate (50 μg/ml), and tetracycline base (10 μg/ml). If necessary, media were solidified with 1.5% agar or, in the case of soft agar, with 0.75% agar (Difco).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli S17-1 | E. coli K-12 thi pro hsdR mutant hsdM+ recA (RP4 Smr Tpr) | 39 |
| S. aurantiaca strains | ||
| AP182 | DW4/3-1, fbfB::Tn5lacZ (Kmr Smr) | 29 |
| AP191 | DW4/3-1, fbf::Tn5lacZ (Kmr Smr) | 29 |
| BS14 | DW4/3-1, fbfA::neo (Kmr Smr) | 37 |
| BS34 | DW4/3-1, fbfB::(Δ5′Δ3′ fbfB-pufBA-neo) (merodiploid for fbfB) (Kmr Smr) | This work |
| BS35 | DW4/3-1, fbfB::(Δ3′ fbfB-Δtrp-lacZ-neo) (merodiploid for fbfB) (Kmr Smr) | This work |
| DW4/3-1 | Wild type (Smr) | 30 |
| Plasmids | ||
| pBS7 | fbfA::(pufBA-neo) in pBS SK(−) (Ampr Kmr) | 37 |
| pBS21 | pSUP102 harboring an XbaI-SalI fragment containing Δ3′Δ5′ fbfB-pufBA-neo (Cmr Kmr) | This work |
| pBS22 | pSUP102 harboring a HindIII-SalI fragment containing Δ3′ fbfB-Δtrp-lacZ-neo (Cmr Kmr) | This work |
| pSK24 | 3.6-kbp EcoRI-SmaI fragment containing fbfA and fbfB in pBS SK(−) (Ampr) | 37 |
| pBS SK(−) | Ampr | Stratagene |
| pSUP102 | Cmr Tetr | 38 |
| pSZ182 | pSUP102 containing a 20-kbp SalI fragment from AP182 harboring Tn5lacZ (Cmr Kmr) | 37 |
| pUC4KIXX | Ampr Kmr | Pharmacia |
| Mini-Tn5lacZ1 | Ampr Kmr | 5 |
Transfer of conjugable plasmids from E. coli to S. aurantiaca (10).
A total of 5 × 108 exponentially growing S. aurantiaca cells were mixed with a total of 5 × 108 exponentially growing cells of E. coli and filtered onto a membrane filter (0.45-μm pore size; 25-mm diameter; Schleicher & Schuell, Dassel, Germany). The cells were washed twice with 5 ml of Casitone medium. The filter was placed onto a Casitone plate and incubated overnight at 32°C. Cells were scraped off the filter and suspended in 5 ml of Casitone medium. Portions were plated in soft agar onto Trypticase peptone plates containing antibiotics for selection and incubated for 7 days at 32°C. Resistant clones were transferred into 3 ml of Casitone medium and further incubated to obtain the required cell density.
Fruiting body formation assay (29) and germination assay (32).
Exponentially growing S. aurantiaca cells were sedimented, washed in HEPES buffer (100 mM HEPES, 10 mM CaCl2, pH 7.2), sedimented again, and resuspended in HEPES buffer to a concentration of 4 × 1010 cells per ml. Aliquots of 5 μl were spotted onto starvation agar and incubated at 32°C for 24 h in incandescent light. For the phenotypic complementation assay, an equal number of cells of two different mutants were mixed for fruiting body formation.
For the germination assay, the 5-μl cell suspensions were spotted onto filter paper (Schleicher & Schuell) about 1 cm2 in size. The filter papers were shifted onto starvation agar plates and incubated at 32°C for 10 days in incandescent light. Then the filter pads were placed into screw-cap tubes and dried in an evacuated desiccator over silica gel for 7 days at room temperature. For germination, filter papers were placed upside down on CY agar plates (0.3% Casitone [Difco], 0.3% yeast extract [Difco], 0.3% CaCl2 · 2H2O, pH 7.2) at 32°C. After 2 days, the filter papers were shifted to another place on the plate for a further incubation for 2 days.
Induction of spore formation by indole.
Spore formation was induced by addition of indole (Sigma) to a final concentration of 0.5 mM to late-log-phase culture (2 × 108 to 3 × 108 cells per ml in shake flasks) in tryptone medium at 32°C. About 2 h after the addition of indole, the cells formed shortened rods, and they started to become refractile after about 4 h. A total of 60 to 80% of the initial cells converted into sonication-resistant cells (9, 12).
β-Galactosidase assay (37).
Fruiting bodies which had been scraped off starvation agar plates or vegetative cells were suspended in a buffer containing 50 mM 3-N-morpholinopropanesulfonic acid (MOPS) at pH 7.5, 10 mM MgCl2, 10 mM dithiothreitol, and 1 mM phenylmethanesulfonyl fluoride and sonicated (Branson sonifier; cell disrupter B15) with glass beads (diameter, 0.1 mm) at 4°C for 1 min in an Eppendorf tube with a cup horn (Branson EDP 101-151-003). To remove cell debris, the samples were centrifuged at 15,000 × g at 4°C for 10 min. The supernatant was assayed for β-galactosidase activity with the substrate 4-methylumbelliferyl-β-d-galactopyranoside (4-MUG) (Sigma). A total of 0.1 ml of the supernatant containing 10 μg of protein was mixed with 0.3 ml of 10 mM sodium phosphate buffer (pH 7.0) containing 0.1 M NaCl, 1 mM MgCl2, 0.1% bovine serum albumin, and 10 μg of 4-MUG and incubated for 30 min at 37°C. The reaction was stopped with 3 ml of 0.1 M glycine buffer (pH 10.3). The fluorescence intensity was measured with a Shimadzu RF-5000 fluorescence spectrophotometer with wavelengths of 360 nm for excitation and 450 nm for emission.
DNA manipulations, sequencing, and PCR.
Restriction analysis and plasmid subcloning were performed according to standard protocols (35). Chromosomal DNA from S. aurantiaca was prepared as described previously (27). The sequence of pSK24, containing the fbfB gene, was determined with exonuclease III-generated directed deletions (13) and synthetic oligonucleotides. PCR was carried out with Vent DNA polymerase (New England Biolabs). Amplification was performed at final concentrations of 0.1 nM template, 1 μM each primer, 300 μM each deoxynucleoside triphosphate, and 2 U of Vent DNA polymerase in a total volume of 100 μl. The reaction mixture was overlaid with 100 μl of mineral oil. The conditions for the amplification with Trio-Thermoblock (Biometra) were as follows: the initial denaturation step was at 94°C for 3 min, annealing was at 65°C for 1 min, polymerization was at 72°C for 2 min, subsequent denaturation was at 94°C for 1 min, and there were 30 cycles. The PCR products were purified with the Qiaquick PCR purification kit (Qiagen).
Southern hybridization.
Southern blot analysis was performed according to standard protocols (35). Prehybridization was carried out for 2 h at 60°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s solution–0.5% sodium dodecyl sulfate (SDS)–50 μg of denatured herring sperm DNA per ml. Hybridization was performed overnight at 60°C after addition of the DNA probes, which were 32P labelled with a nick translation kit (Boehringer Mannheim, Mannheim, Germany). The filters were washed in 0.1× SSC–0.1% SDS twice for 30 min at 60°C.
RNA isolation and Northern hybridization.
S. aurantiaca RNA from vegetative cells and developing cells was isolated as described previously (4). RNA electrophoresis was performed as follows. A suspension of 1 g of agarose in 72 ml of diethylpyrocarbonate-treated H2O was melted. After the suspension was cooled to 65°C, 10 ml of 10× MOPS running buffer (200 mM MOPS, 10 mM EDTA, 50 mM NaAc, 100 mM NaOH) and 18 ml of formaldehyde (37%) were added. The RNA to be electrophoresed was dried and suspended in 9 μl of sample buffer (10% 10× MOPS running buffer, 50% formamide, 18% formaldehyde). After the mixture was heated to 65°C for 10 min, 1 μl of stop buffer (50% glycerol, 6 mM EDTA, 0.05% bromophenol blue) was added. Electrophoresis was performed at 70 V for 4 h. The RNA was transferred to a nylon membrane (Biodyne B; Pall) with a vacuum blotter (Appligene). After UV cross-linking with the Stratalinker (Stratagene), the prehybridization of the filters was at 42°C for 5 h in 50% formamide–4× Denhardt’s solution–0.1% glycine–5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% SDS–0.25 mg of denatured herring sperm DNA per ml. Hybridization was carried out overnight at 42°C in 55% formamide–1.1% Denhardt’s solution–5.5× SSPE–0.1% SDS–10% dextran sulfate–4 mM sodium diphosphate–0.1 mg of denatured herring sperm DNA per ml, containing the 32P-labelled DNA probe. The membrane was washed at room temperature twice for 15 min in 2× SSPE–0.1% SDS and at 37°C twice for 15 min in 0.1× SSPE–0.1% SDS.
Construction of S. aurantiaca insertional mutants. (i) Mutant BS34: construction of an fbfB deletion mutant.
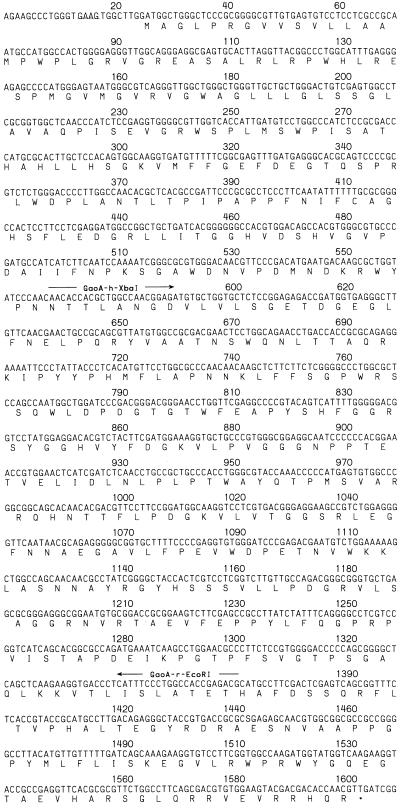
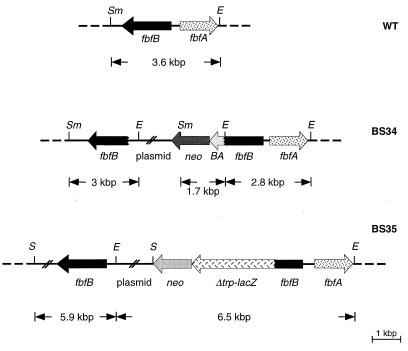
An internal sequence (806 bp) of the fbfB gene was amplified by PCR with the oligonucleotides GaoA-r-EcoRI (CCGGAATTCCGTCTCGGTGGCCAGGGAAATG) and GaoA-h-XbaI (CTAGTCTAGAAACACCACGCTGGCCAACGGAG) (see Fig. 3). After restriction with EcoRI and XbaI, the PCR fragment was directly ligated to the EcoRI site of a SalI-EcoRI fragment, composed of the pufBA fragment from the puf operon from Rhodobacter capsulatus (20, 21) and the kanamycin resistance gene (neo) of Tn5, which was isolated from plasmid pBS7 (37). This fragment was inserted in the XbaI-SalI sites of the plasmid pSUP102, generating pBS21. For the conjugation of pBS21 to S. aurantiaca, E. coli S17-1 was transformed. Kanamycin-resistant transconjugants of S. aurantiaca were obtained with a frequency of 1.2 × 10−7. One of the transconjugants, mutant BS34, contains two truncated fbfB genes (see Fig. 5 and 6).
FIG. 3.
Nucleotide sequence of the fbfB gene. The putative ribosome binding site is underlined. The letters below the nucleotide sequence indicate the deduced amino acid sequence of the putative FbfB, in single-letter code. ∗, stop codon. For construction of the fbfB deletion strain BS34, the internal part of fbfB was amplified by PCR with the oligonucleotides GaoA-r-EcoRI and GaoA-h-XbaI. The nucleotide sequence data are available in the GenBank database under accession no. Z11601.
FIG. 5.
Schematic depiction of the fbfA-fbfB region of the S. aurantiaca wild-type (WT) strain DW4/3-1 and of the strains BS34 and BS35. BS34 has two truncated fbfB genes: one is shortened at its 5′ end and the other is fused at its 3′ end with the Rhodobacter pufBA fragment and the neo gene. BS35 has a wild-type fbfB gene and a Δtrp-lacZ-fbfB fusion gene, which is under the control of the fbfB promoter. E, EcoRI; S, SalI; Sm, SmaI.
FIG. 6.
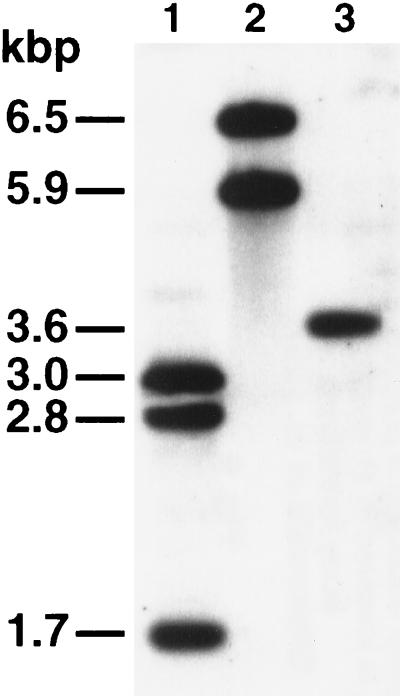
Southern analysis of restricted chromosomal DNA of the S. aurantiaca wild-type strain DW4/3-1 and of the strains BS34 and BS35 with the fbfB gene and the neo gene as probes (Fig. 5). Lane 1, chromosomal DNA of BS34 restricted by EcoRI and SmaI. Three fragments (3.0, 2.8, and 1.7 kbp) were detected. Lane 2, chromosomal DNA of BS35 restricted by EcoRI and SalI. Two fragments (6.5 and 5.9 kbp) were detected. Lane 3, chromosomal DNA of the wild-type strain DW4/3-1 restricted by EcoRI and SmaI. Only a 3.6-kbp fragment was detected. Five micrograms of DNA per lane was analyzed. The sizes of the fragments were estimated by using HindIII-restricted λ DNA as a reference.
(ii) Mutant BS35: construction of the Δtrp-lacZ-fbfB fusion gene.
A 762-bp fragment, composed of 140 bp upstream and 619 bp downstream of the ATG start codon of the fbfB gene, was amplified by PCR with the oligonucleotides BSPCR15 (CCGGGGATCCCGGCAGTTCGTTGAACAAGC) and BSPCR16 (CCGGAAGCTTAGGGAGGGGAGCAGCTGTCC). After restriction with BamHI-HindIII, the fragment was cloned into the corresponding sites of the plasmid pSUP102. The Δtrp-lacZ indicator gene construct (isolated from the mini-Tn5lacZ1 [5]), which has no functional promoter, was fused to the neo gene from pUC4KIXX and inserted into the BamHI-SalI sites of the former plasmid, generating plasmid pBS22. For the conjugation of pBS22 to S. aurantiaca, E. coli S17-1 was transformed with pBS22. Kanamycin-resistant S. aurantiaca transconjugants were obtained with a frequency of 6 × 10−8. One of the transconjugants, BS35, contains a wild-type fbfB gene and a Δtrp-lacZ-fbfB fusion gene (see Fig. 5 and 6).
Nucleotide sequence accession number.
The nucleotide sequence of fbfB is available in the GenBank database under the accession no. Z11601.
RESULTS AND DISCUSSION
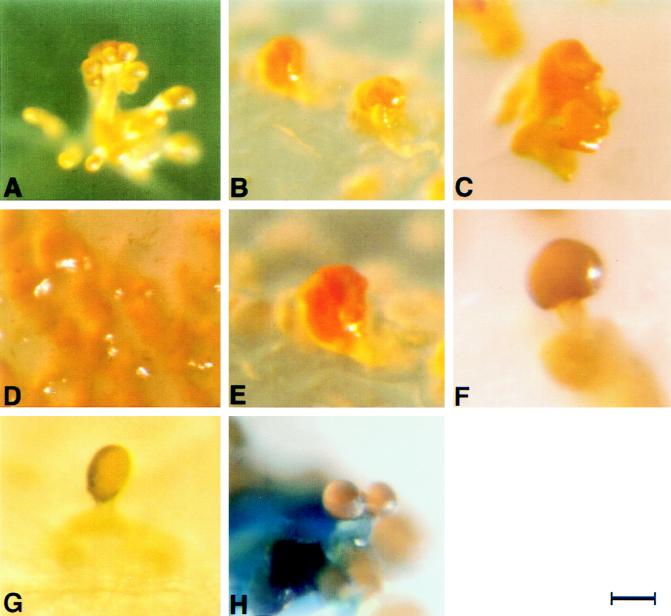
For the identification of genes involved in S. aurantiaca fruiting body formation, transposon mutagenesis with Tn5lacZ was performed. Three mutant types affected in fruiting body formation have been obtained. These include mutants which form neither fruiting bodies nor aggregates, mutants which are able only to aggregate into nonstructured clumps, and mutants which undergo only a part of the differentiation process (29). One of the mutants (AP182) which forms only clumps during starvation (Fig. 1B) was selected for further analysis. Mixing the AP182 cells with those of strain AP191 prior to starvation leads to a partial phenotypic complementation. Mutant AP191 forms neither fruiting bodies nor aggregates (Fig. 1D) but shows a normal gliding behavior. Instead of the clumps, a mushroom-like structure, similar to a champignon (cf. Fig. 1F), has been obtained.
FIG. 1.
Fruiting body morphologies of various S. aurantiaca strains. (A) Fruiting body of the wild-type strain DW4/3-1 is differentiated into a stalk, branches, and sporangioles. (B) The Tn5lacZ insertional mutant AP182 can form only unstructured clumps. The transposon is inserted into the fbfB gene. (C) The fbfB mutant BS34 is able only to aggregate into clumps, like AP182. (D) The Tn5lacZ insertional mutant AP191 shows no cell aggregation. (E) The fbfA mutant BS14 aggregates only into clumps. (F) Mixing of cells of BS34 and AP191 following starvation leads to a mushroom-like structure (champignon). (G) Mixing of cells of BS34 and fbfA mutant BS14, which is able only to aggregate into clumps, following starvation leads to a mushroom-like structure (morel). (H) Fruiting body of the merodiploid strain BS35, harboring a functional fbfB gene and a Δtrp-lacZ-fbfB fusion gene, on starvation plates containing 20 μg of X-Gal per ml shows blue staining of the stem. Bars, about 40 μm for panels A, D, and H and about 30 μm for panels B, C, E, F, and G.
In a λ EMBL3 library of SalI-restricted chromosomal DNA of the mutant strain AP182 (29, 37), a 20-kbp SalI restriction fragment harboring the Tn5lacZ insertion was detected with the neo gene derived from Tn5 (3) as a probe. After subcloning of this fragment into the SalI site of pSUP102 (resulting in plasmid pSZ182), the position and orientation of the transposon were determined by restriction analyses and sequencing starting with the insertional sequence elements (Fig. 2). With this 20-kbp SalI fragment as probe, the corresponding 12-kbp SalI fragment was detected in a λ EMBL3 library of SalI-restricted chromosomal DNA of the wild-type strain DW4/3-1. This 12-kbp fragment was cloned into the SalI site of the conjugable plasmid pSUP102, resulting in plasmid pBS1 (Fig. 2).
A 3.6-kbp SmaI-EcoRI fragment containing the site of transposon insertion of strain AP182 was cloned into pBS SK(−), resulting in pSK24 (Fig. 2). After sequencing, two putative open reading frames (ORFs) named fbfA (37) and fbfB, which are arranged in a divergent orientation on the fragment, have been detected (Fig. 2). Both genes have been localized on an 862.5-kbp SpeI and a 676.8-kbp AseI restriction fragment of the S. aurantiaca genome (28). The distance between the start codons of fbfA and fbfB is 153 bp. The size of the fbfB gene is 1,581 bp (Fig. 3). The site of the transposon insertion in mutant AP182 is 550 bp downstream of the ATG start codon of the fbfB gene. A putative Shine-Dalgarno sequence (GAAG) is detected 8 bp upstream of the start codon (Fig. 3). fbfB, which we suggest to be involved in fruiting body formation, encodes a putative polypeptide (FbfB) composed of 526 amino acid residues with a molecular mass of 57.8 kDa. Protein database searches for the deduced polypeptide with BLASTP 2.0.3 (1) revealed a significant homology between FbfB and the secreted copper enzyme galactose oxidase (GaoA) from the deuteromycete fungus D. dendroides (Fig. 4) (25). In addition, some similarity to the copper enzyme glyoxal oxidase (Glx) from the lignin-degrading Basidiomycete Phanerochaete chrysosporium was found (19, 44). GaoA catalyzes the oxidation of primary alcohols and of the C-6 hydroxyl group of galactose to aldehydes. During this reaction, molecular oxygen is reduced to hydrogen peroxide by a radical mechanism (42). The four amino acid residues Tyr-313, Tyr-536, His-537, and His-622 of GaoA, which form the copper binding site, and Cys-269, which forms the thioether cysteinyltyrosine with Tyr-313 (17), are conserved in FbfB (Fig. 4). As FbfB may act on the outside of the bacterial cell during development, the N-terminal domain of the putative polypeptide was screened for a signal sequence. The N-terminal sequence (40 amino acids) of FbfB (MAGLPRGVVSVLL̂AMPWPLGRVGREAŜLRLRPWHLRES) was analyzed with the program SignalSeq of the Heidelberg Unix Sequence Analysis Resources (43). Two hypothetical cleavage sites, indicated by ^ in the above sequence, have been proposed, of which, if any, the site for the longer signal sequence seems more probable.
FIG. 4.
Alignment of the amino acid residues of the putative polypeptide FbfB from S. aurantiaca with the galactose oxidase, GaoA, from D. dendroides. The amino acid residues are numbered as indicated. The sequences start in the case of FbfB at position 67 and in the case of GaoA at position 194. Identical amino acid residues are boxed. The amino acid residues Tyr-313, Tyr-536, His-537, and His-622, representing the four copper binding sites, are shaded. They participate in binding one copper ion in GaoA. The alignment was performed with BLASTP 2.0.3 (1). The score (bits) is 143, and the E value is 3e-33.
To rule out the possibility that the phenotype of mutant AP182 is due to a second-site mutation, fbfB had to be inactivated. For this purpose, the conjugable plasmid pBS21 was constructed. It harbors an insertion of a 5′- and 3′-truncated fbfB gene fused to the Rhodobacter pufBA fragment and the Tn5 neo gene (see Materials and Methods). pBS21 was transferred into E. coli S17-1 and subsequently conjugated into wild-type S. aurantiaca to obtain strains in which the wild-type gene is replaced by two genes truncated at the 5′ or 3′ end, respectively. One of the kanamycin-resistant transconjugants, BS34, was used for further analyses. As expected, Southern blot analysis showed that BS34 is a merodiploid strain containing two truncated fbfB genes (Fig. 5 and 6). Wild-type cells form well-defined fruiting bodies during starvation (Fig. 1A), whereas mutant BS34 cells generate only nonstructured aggregates (Fig. 1C). Fruiting body formation of mutant BS34 is partially restored by mixing the mutant cells with cells of mutant AP191, which though competent for gliding are not able to form aggregates during starvation (Fig. 1D). Mixing of the mutant cells before starvation leads to the formation of a structure composed of a stem and a cap which looks like a champignon (Fig. 1F). A fruiting body structure resembling that of a morel (Fig. 1G) is obtained if cells of BS34 are mixed with those of the fbfA mutant BS14, defective in fruiting (Fig. 1E). Mutants BS14 and BS34 generate spores during the development of their fruiting bodies. These spores have the capability of germinating in our germination assay (see Materials and Methods).
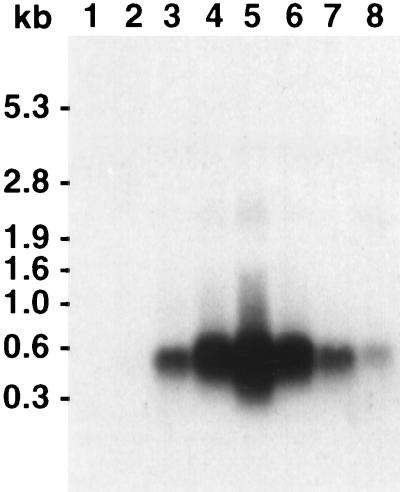
We have not been able to detect the transcript of the fbfB gene generated during fruiting body formation by using an fbfB-derived probe for Northern analysis. To prove fbfB transcription, strain BS34 was constructed. BS34 contains a 3′-truncated fbfB gene fused to the pufBA fragment from R. capsulatus and a 5′-truncated fbfB fragment (Fig. 5). pufBA encodes an mRNA that has a half-life of about 30 min (20, 21). It has been shown recently that the time at which the transcription of a pufBA fusion gene starts can be easily determined by Northern analysis with the pufBA gene as a probe (37). The pufBA transcript was detected in mutant BS34 about 14 h after the beginning of starvation (Fig. 7) but not in vegetative cells. No significant amount of fbfB gene transcript or part of it was detected. This suggests that the expression of fbfB is low and/or that its transcript is very unstable.
FIG. 7.
Northern analysis of RNA isolated from strain BS34 at different times after beginning of starvation with the 567-bp radiolabelled R. capsulatus pufBA fragment as a probe. A signal with the size of the pufBA fragment was detected after 14 h of starvation. Lane 1, RNA isolated from vegetative cells of BS34. Lanes 2 to 8, RNA isolated from cells 8, 10, 14, 16, 17, 18, and 24 h, respectively, after the beginning of starvation. A total of 10 μg of RNA from each sample was used. The sizes of the fragments were estimated with the RNA molecular weight marker I (Boehringer Mannheim).
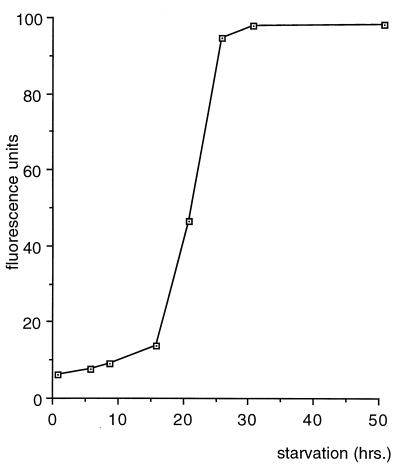
For analyzing the progression of fbfB expression during fruiting body formation or indole-induced sporulation (9), the merodiploid strain BS35 was constructed. It contains the wild-type fbfB gene and a 3′-truncated fbfB gene to which a promoterless Δtrp-lacZ gene fusion and the neo cassette (2) for transconjugant selection were fused (Fig. 5 and 6). Starvation of BS35 cells on water agar containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) resulted in fruiting bodies which had the same form as those of the wild type but whose stems were stained blue in the course of 2 days after the beginning of development (Fig. 1H). For the determination of fbfB fusion gene expression during fruiting body formation, cells were scraped off the agar dish and broken by sonication. β-Galactosidase activity was determined in the cell extract with the fluorescent substrate 4-MUG (34), as β-galactosidase activity of BS35 cells was low. β-Galactosidase activity starts to increase about 14 h after the beginning of starvation and reaches its maximum level after about 30 h (Fig. 8). No β-galactosidase activity was detected during indole-induced sporulation and in vegetative cells.
FIG. 8.
Determination of the β-galactosidase activity of strain BS35 during fruiting body formation. After 16 h of development, β-galactosidase activity is detectable; after 30 h of starvation, it reaches its maximum.
Fruiting body formation of the myxobacteria is a multicellular process. Multicellular development requires intercellular signalling for the coordination of the physiology of the single cell as a function of both the location in the swarm and the progress of fruiting. Signalling substances may be diffusible compounds or may be attached to or associated with the cell surface.
McVittie et al. isolated mutants of M. xanthus impaired in fruiting body formation and observed that it was possible to rescue development by mixing certain mutants with others before starvation (26). They demonstrated that the complementation was not genetic and suggested this extracellular complementation to be based on a synergistic interaction, i.e., an exchange of substances involved in developmental interaction. A large number of M. xanthus mutants defective in development and showing synergistic interaction were isolated (11). They fell into four groups. Mixing a member of one group with one belonging to another group or with the wild type resulted in extracellular complementation of fruiting body formation. The authors concluded that fruiting of M. xanthus was governed by at least four developmental signals and that each mutant group lost the ability to produce one of these signals (11). Meanwhile, a fifth factor involved in M. xanthus developmental cell-cell signaling has been detected (6, 7).
The fruiting body of S. aurantiaca is much more complex than that of M. xanthus. Studies of S. aurantiaca development will eventually provide the opportunity to learn more about the formation of complex multicellular structures and about the signals which coordinate the physiology of the single cell as a function of the morphogenetic process. During the first hours of development, S. aurantiaca forms a diffusible signal, the pheromone, which induces aggregation of the starving cells (40). The structure of the compound has been elucidated recently (16).
Mutants impaired in S. aurantiaca fruiting body formation were induced by insertional mutagenesis (29). Two of these mutant strains, AP182 and AP191 (Fig. 1B and D), were selected for further analysis because they showed a synergistic interaction. Mixing of the mutant cells before the beginning of starvation resulted in a partial phenotypic complementation of fruiting (Fig. 1F). The gene impaired in AP182, fbfB, was modified in vitro and crossed back into the wild-type strain to obtain a merodiploid derivative, BS34, harboring two truncated copies of fbfB.
Strain BS34 showed the same phenotype as did mutant AP182. A partial rescue of fruiting body formation was observed after mixing the BS34 cells with those of AP191 before starvation (Fig. 1C and F). The fruiting body obtained after mixing the cells of the fbfB mutants BS34 and AP191 has a champignon-like shape. This form is also found 15 h after the beginning of starvation of wild-type cells (cf. Fig. 2C in reference 31). Fruiting body formation with the mixture of the mutant cells is obviously blocked at this 15-h stage. With the merodiploid strain BS35, which harbors an indicator gene fused to fbfB, and by Northern analysis with strain BS34, it was shown that fbfB expression starts about 14 h after the beginning of starvation. Interestingly, fruiting body formation in the mutant BS14, in which another gene involved in fruiting, fbfA, is inactivated, can be partially rescued by mixing the cells with those of mutant AP191 (37). The fruiting body looks like a morel. fbfA is expressed after 8 h of development, and the shape of the fruiting body obtained in the mixing experiment corresponds to that 12 h after the induction of fruiting body formation of the wild type (cf. Fig. 2B in reference 31). Mixing of cells of an fbfA and an fbfB mutant before starvation resulted only in fruiting bodies with a morel-like shape (Fig. 1G).
The partial phenotypic complementation suggests that factors involved in fruiting and which are lacking in one mutant may be obtained from the other. The shapes of the fruiting bodies obtained in the mixing experiments correspond to those observed during development of the wild type. Expression of fbfA or fbfB is a prerequisite for early or late stalk formation, respectively. A reason for the incomplete phenotypic complementation may be that not all substances involved in fruiting (e.g., intracellular macromolecules) which are lacking in one of the strains can be supplemented by the other mutant and vice versa.
Both fbfA and fbfB are not expressed during vegetative growth or indole-induced sporulation. Obviously, fbfA and fbfB are development-specific genes that are involved in the morphogenetic process of fruiting body, and not of spore, formation. It is tempting to speculate that sporulation and the formation of the structural parts of the fruiting body are partially independent processes which coincide in the late stage. It was shown for M. xanthus that vegetative cells starved in liquid culture may efficiently convert to spores which seem to be identical with those formed in fruiting bodies (33). This suggests that the formation of the fruiting body structure is not tightly coupled to the formation of starvation-induced spores.
During sequence analyses downstream of the fbfB region, two ORFs, hesA and pksA, were detected (36). The disruption of these ORFs by the insertion of the neo gene has no effect on fruiting. No coding sequence was detected between fbfB and hesA, which is located about 800 bp downstream of the stop codon of fbfB. These results prove that the inactivation of fbfB and not a polar effect of the mutation on downstream sequences leads to the defect in fruiting.
The putative polypeptide encoded by hesA shows homology with some polypeptides probably involved in the export of antibiotics (14, 15, 22). Most interestingly, the second ORF, pksA (about 2 kbp downstream of the stop codon of fbfB), encodes a putative polyketide synthase which probably is involved in the synthesis of myxothiazol (31a). This compound is an inhibitor of the b-c1 complex of the respiratory chain (41) which possibly protects the fruiting body of S. aurantiaca against fungal attack or vegetative cells against fungal competition for food.
ACKNOWLEDGMENTS
This work was supported by grants Scha 150/8-1 and Scha 150/8-2 of the Deutsche Forschungsgemeinschaft and by the Fonds der Chemischen Industrie.
We thank Yves Cully for image processing and Berta Reiner for database searches.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barany F. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene. 1985;37:111–123. doi: 10.1016/0378-1119(85)90263-x. [DOI] [PubMed] [Google Scholar]
- 3.Barany F. Two-codon insertion mutagenesis of plasmid genes by using single-stranded hexameric oligonucleotides. Proc Natl Acad Sci USA. 1985;82:4202–4206. doi: 10.1073/pnas.82.12.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Downard J, Ramaswamy S V, Kil K S. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Mol Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- 8.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerth K, Metzger R, Reichenbach H. Induction of myxospores in Stigmatella aurantiaca (myxobacteria)—inducers and inhibitors of myxospore formation, and mutants with a changed sporulation behaviour. J Gen Microbiol. 1993;139:865–871. [Google Scholar]
- 10.Glomp I, Saulnier P, Guespin-Michel J, Schairer H U. Transfer of IncP plasmids into Stigmatella aurantiaca leading to insertional mutants affected in spore development. Mol Gen Genet. 1988;214:213–217. doi: 10.1007/BF00337713. [DOI] [PubMed] [Google Scholar]
- 11.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 12.Heidelbach M, Skladny H, Schairer H U. Purification and characterization of SP21, a development-specific protein of the myxobacterium Stigmatella aurantiaca. J Bacteriol. 1993;175:905–908. doi: 10.1128/jb.175.3.905-908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–166. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- 14.Hiraoka H, Ano T, Shoda M. Molecular cloning of a gene responsible for the biosynthesis of the lipopeptide antibiotics iturin and surfactin. J Ferment Bioeng. 1992;74:323–326. [Google Scholar]
- 15.Huang C-C, Ano T, Shoda M. Nucleotide sequence and characteristics of the gene, lpa-14, responsible for biosynthesis of the lipopeptide antibiotics iturin A and surfactin from Bacillus subtilis RB14. J Ferment Bioeng. 1993;76:445–450. [Google Scholar]
- 16.Hull W, Berkessel A, Stamm I, Plaga W. Abstracts of the 24th Annual Meeting on the Biology of the Myxobacteria. 1997. Intercellular signalling in Stigmatella aurantiaca: proof, purification and structure of a myxobacterial pheromone; p. 25. [Google Scholar]
- 17.Ito N, Knowles P F, Phillips S E. X-ray crystallographic studies of cofactors in galactose oxidase. Methods Enzymol. 1995;258:235–262. doi: 10.1016/0076-6879(95)58050-6. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser D, Losick R. How and why bacteria talk to each other. Cell. 1993;73:873–885. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- 19.Kersten P J, Witek C, van den Wymelenberg A, Cullen D. Phanerochaete chrysosporium glyoxal oxidase is encoded by two allelic variants: structure, genomic organization, and heterologous expression of glx1 and glx2. J Bacteriol. 1995;177:6106–6110. doi: 10.1128/jb.177.21.6106-6110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klug G. The role of mRNA degradation in the regulated expression of bacterial photosynthesis genes. Mol Microbiol. 1993;9:1–7. doi: 10.1111/j.1365-2958.1993.tb01663.x. [DOI] [PubMed] [Google Scholar]
- 21.Klug G, Jock S, Rothfuchs R. The rate of decay of Rhodobacter capsulatus-specific puf-mRNA segments is differentially affected by RNaseE activity in Escherichia coli. Gene. 1992;121:95–102. doi: 10.1016/0378-1119(92)90166-m. [DOI] [PubMed] [Google Scholar]
- 22.Krätzschmar J, Krause M, Marahiel M A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J Bacteriol. 1989;171:5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus xanthus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 24.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 25.McPherson M J, Ogel Z B, Stevens C, Yadav K D, Keen J N, Knowles P F. Galactose oxidase of Dactylium dendroides. Gene cloning and sequence analysis. J Biol Chem. 1992;267:8146–8152. [PubMed] [Google Scholar]
- 26.McVittie A, Messik F, Zahler S A. Developmental biology of Myxococcus. J Bacteriol. 1962;84:546–551. doi: 10.1128/jb.84.3.546-551.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann B, Pospiech A, Schairer H U. Rapid isolation of genomic DNA from gram negative bacteria. Trends Genet. 1992;8:332–333. doi: 10.1016/0168-9525(92)90269-a. [DOI] [PubMed] [Google Scholar]
- 28.Neumann B, Pospiech A, Schairer H U. A physical and genetic map of the Stigmatella aurantiaca DW4/3.1 chromosome. Mol Microbiol. 1993;10:1087–1099. doi: 10.1111/j.1365-2958.1993.tb00979.x. [DOI] [PubMed] [Google Scholar]
- 29.Pospiech A, Neumann B, Silakowski B, Schairer H U. Detection of developmentally regulated genes of the myxobacterium Stigmatella aurantiaca with the transposon Tn5lacZ. Arch Microbiol. 1993;159:201–206. [Google Scholar]
- 30.Qualls G T, Stephens K, White D. Light-stimulated morphogenesis in the fruiting myxobacterium Stigmatella aurantiaca. Science. 1978;201:444–445. doi: 10.1126/science.96528. [DOI] [PubMed] [Google Scholar]
- 31.Qualls G T, Stephens K, White D. Morphogenetic movements and multicellular development in the fruiting myxobacterium, Stigmatella aurantiaca. Dev Biol. 1978;66:270–274. doi: 10.1016/0012-1606(78)90291-9. [DOI] [PubMed] [Google Scholar]
- 31a.Reichenbach, H. Personal communication.
- 32.Reichenbach H, Dworkin M. The myxobacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 3416–3487. [Google Scholar]
- 33.Rosenbluh A, Rosenberg E. Sporulation of Myxococcus xanthus in liquid shake flask cultures. J Bacteriol. 1989;171:4521–4524. doi: 10.1128/jb.171.8.4521-4524.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruan K-H, Kulmasz R J, Wilson A, Wu K K. Highly sensitive fluorimetric enzyme immunoassay for prostaglandin H synthase solubilized from cultured cells. J Immunol Methods. 1993;162:23–30. doi: 10.1016/0022-1759(93)90403-t. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Silakowski B, Ehret H, Schairer H U. Abstracts of the 24th Annual Meeting on the Biology of the Myxobacteria. 1997. Genes involved in fruiting body formation of S. aurantiaca; p. 24. [Google Scholar]
- 37.Silakowski B, Pospiech A, Neumann B, Schairer H U. Stigmatella aurantiaca fruiting body formation is dependent on the fbfA gene encoding a polypeptide homologous to chitin synthases. J Bacteriol. 1996;178:6706–6713. doi: 10.1128/jb.178.23.6706-6713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon R, O’Connell M, Labes M, Pühler A. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other gram negative bacteria. Methods Enzymol. 1986;118:643–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 40.Stephens K, Hegeman G D, White D. Pheromone produced by the myxobacterium Stigmatella aurantiaca. J Bacteriol. 1982;149:739–747. doi: 10.1128/jb.149.2.739-747.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thierbach G, Reichenbach H. Myxothiazol, a new inhibitor of the cytochrome b-c1 segment of the respiratory chain. Biochim Biophys Acta. 1981;638:282–289. doi: 10.1016/0005-2728(81)90238-3. [DOI] [PubMed] [Google Scholar]
- 42.Tressel P S, Daniel J K. Galactose oxidase from Dactylium dendroides. Methods Enzymol. 1982;89:163–171. doi: 10.1016/s0076-6879(82)89029-0. [DOI] [PubMed] [Google Scholar]
- 43.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whittaker M M, Kersten P J, Nakamura N, Sanders Loehr J, Schweizer E S, Whittaker J W. Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J Biol Chem. 1996;271:681–687. doi: 10.1074/jbc.271.2.681. [DOI] [PubMed] [Google Scholar]