Summary
Cellular Src tyrosine kinase (c-Src) exists in the secretomes of several human cancers (extracellular, e-Src). Phosphoproteomics has demonstrated the existence of 114 potential extracellular e-Src substrates in addition to Tissue Inhibitor of Metalloproteinases 2. Here, we present a protocol to characterize secreted tyrosine-phosphorylated substrates as a result of c-Src expression and secretion. We describe steps for collecting cell secretomes and extracts, performing antibody treatment and Ni-NTA pull-down, and detecting protein-protein interaction and substrate Y-phosphorylation. This protocol is adaptable for studies examining the function of other extracellular kinases.
For complete details on the use and execution of this protocol, please refer to Backe et al. (2023)1 and Sánchez-Pozo et al. (2018).2
Subject areas: Cell Biology, Cancer, Molecular Biology
Graphical abstract

Highlights
-
•
Isolation and characterization of tyrosine-phosphorylated proteins from cell secretomes
-
•
Protocol that is adaptable for studies characterizing the function of secreted kinases
-
•
Kinase-specific antibodies are utilized to demonstrate extracellular c-Src signaling
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
c-Src tyrosine kinase exists in the secretomes of several human cancers (e-Src). Phosphoproteomics has demonstrated the existence of 114 potential extracellular e-Src substrates in addition to tissue inhibitor metalloproteinase 2. Here, we present a protocol to characterize secreted tyrosine-phosphorylated substrates as a result of c-Src expression and secretion. We describe steps for collecting cell secretomes and extracts, performing antibody treatment and Ni-NTA pull-down, and detecting protein-protein interaction and substrate Y-phosphorylation. This protocol is adaptable for studies examining the function of other extracellular kinases.
Before you begin
Institutional permissions
Research performed using biological specimens or cell lines that originate from any species or organisms requires the approval of a research protocol by the Institutional Biosafety Committee. Manipulation of human tissues must follow local, state and federal ethical guidelines. Handling of biological material should agree with the biosafety level 2 (BSL2) guidelines. Culture, propagation and maintenance of mammalian cell cultures should take place in sterile conditions under a class II biosafety cabinet (hood).
Key points
-
1.
In here, we describe processes and specific steps using murine SYF and SYF+c-Src cell lines. However, we have also used this protocol using HEK293 cells, human cancer cell lines including fibrosarcoma HT1080, prostate LNCaP, DU145 and PC3, breast MCF7 and MDA-MB231 and lung A549 cell lines.1,2
-
2.
We have previously characterized these cell lines for presence (in SYF+c-Src) and absence (in SYF) of c-Src kinase protein1,2 (Figure 1).
-
3.
Similarly, we recommend to have all cell lines (wild type and knock-out) tested for the expression and secretion of the kinase protein(s) of interest, before starting any experiments. Methods to use for protein expression include western blotting (WB, qualitative assay) and enzyme-linked immunosorbent assay (ELISA, quantitative assay).
-
4.
Cell lines resuscitated from frozen stocks require approximately one week to adapt to culture conditions. We advise using these cells for up to 4–6 weeks, during which time experiments will be performed. Following that, new cultures will have to be established from frozen stocks.
-
5.
In order to collect cell secretomes, cells need to be cultured in serum-free media (i.e., in media without fetal bovine serum or growth factors). It is essential to pre-determine the length of time for cells to be kept in serum-free conditions in order to maintain maximum cell viability. This characteristic is cell-dependent and differs from cell type to cell type. We have previously used a method to determine cell viability following serum starvation for HEK293 cells.3 We provide some details below (see cell cytotoxicity assay).
-
6.
Follow company guidelines for storage of antibodies (e.g., storing temperature, aliquoting, etc.). Check the antibody buffer formulation: presence and high concentration of sodium azide may affect cell viability if used to treat cells for long period of time (>12 h).
-
7.
Prepare and store accordingly all reagents and media needed for cell culture e.g., DMEM without serum, DMEM with 10% FBS, PBS and trypsin.
Figure 1.

Western Blotting of total protein extracts from SYF and SYF+c-Src cells
Probing for c-Src and total input GAPDH.
Establish SYF and SYF+c-Src cultures from frozen stocks
Timing: 1 week
Note: Cells should be in culture and passaged at least once before setting up an experiment. This may take at least 1 week from thawing cells.
-
1.Thaw cells at 37°C using a water bath.
-
a.Swirl vial with frozen cells gently and observe if ice crystals are still present every 10 s. When very few crystals are present, remove from water bath and transfer cells into a 15 mL sterile conical tube.
-
a.
Note: Spray vial with 70% Ethanol to sterilize before opening in the hood.
-
2.
Dropwise, add cell-line specific cell culture medium (containing growth factors and/or serum) to cells (prewarmed to 23°C) filling up to 10 mL volume.
-
3.
Centrifuge tubes at 290 g for 5 min at 23°C.
-
4.
Aspirate medium making sure not to disturb the cell pellet.
-
5.
Resuspend the pellet in 10 mL fresh cell culture medium, e.g., DMEM containing 10% Fetal Bovine Serum (FBS) and transfer to a sterile 10 cm cell culture treated plate.
-
6.
Repeat until all required cell lines are thawed.
Note: Different cell lines may grow in different cell culture media.
-
7.
Allow cells to recover and adhere for 16 h in a humidified 37°C incubator containing 5% CO2.
-
8.
Next day, replace medium with fresh growth factor supplemented medium prewarmed to 23°C.
-
9.Propagation.
-
a.Observe cells under the microscope and when they reach 70%–90% confluency, split as appropriate to maintain or to start the experiment as planned.
-
a.
CRITICAL: Do not allow cells to overgrow in culture. This will result in extensive cell apoptosis and cell death. Cells should look healthy prior to start the experimental procedure. Always observe cells under the microscope BEFORE starting any cell processing step (e.g. splitting, treatment, protein isolation etc.).
Cell cytotoxicity assay
Timing: 1.5–3 days
Note: Prior to antibody treatment, perform a cell cytotoxicity assay on tested cell lines (e.g. SYF and SYF+c-Src) to determine whether culture conditions (e.g. serum starvation) have an impact on cell viability for the duration of a tested treatment.
Note: Perform the Cell Cytotoxicity Assay or LDH (Lactate Dehydrogenase) assay in all mammalian cell lines studied. This assay allows for colorimetric measurements of lactate dehydrogenase (LDH) which is a compound released from cells as the plasma membrane is damaged or destroyed.
Note: Our lab used the Pierce Scientific LHD assay kit (Thermo Fisher Scientific 88953). https://www.fishersci.com/shop/products/pierce-ldh-cytotoxicity-assay-kit/PI88953.
This kit is currently discontinued, but there are numerous such kits available from different suppliers. Follow manufacturer’s protocol for “Determination of Optimum Cell Number for LDH Cytotoxicity Assay”. For example, use the CyQUANT LDH Cytotoxicity Assay (Thermo Fisher Scientific C20300).
Key steps of the assay (for complete details of this protocol, refer to Figure S1D-E in Baker-Williams et al.3):
-
10.Determine optimal number of cells for each cell line to test for spontaneous LDH release.
-
a.Count cells using a dead-cell discrimination dye.
-
b.Plate different number of cells at least in triplicate in a 96-well cell culture plate.
-
c.Incubate for the appropriate time, according to experimental conditions (e.g., 24 h of serum-starvation).
-
d.Add manufacturer’s reagents to cells (control treatment, spontaneous release and maximum LDH release control cells).
-
e.Measure absorbance using a plate reader.
-
a.
-
11.
Determine the number of cells that shows no change in LDH release.
-
12.
Proceed with the actual Cell Cytotoxicity Assay using own reagents (e.g., serum starvation effect, drug treatment, antibody treatment).
Note: Include a control of the diluent of the compounds being tested (e.g. DMSO/PBS/BSA etc.).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Recombinant rabbit IgG, monoclonal (EPR25A) isotype control Dilution: 1 μg/mL |
Cell Signaling Technology | Cat #ab172730; RRID: AB_2687931 |
| Mouse IgG2A isotype control, clone # 20102 Dilution: 1 μg/mL |
R&D Systems | Cat #MAB003; RRID: AB_357345 |
| Rabbit anti-Src mAb (32G6) Dilution: 1:5,000 (WB); 1 μg/mL for treatment |
Cell Signaling Technology | Cat #2123; RRID: AB_2106047 |
| Mouse anti-Src mAb (L4A1) Dilution: 1:10,000 (WB); 1 μg/mL for treatment |
Cell Signaling Technology | Cat #2110; RRID: AB_10691385 |
| Rabbit anti-phospho-Src family (Tyr416) Dilution: 1:2,000 (WB) |
Cell Signaling Technology | Cat #2101; RRID: AB_331697 |
| Mouse anti-6x-His epitope tag (HIS.H8) Dilution: 1:10,000 (WB) |
Thermo Fisher Scientific | Cat #MA1-21315; RRID: AB_557403 |
| Mouse anti-GAPDH (1D4) Dilution: 1:10,000 (WB) |
Enzo Life Sciences | Cat #ADI-CSA-335; RRID: AB_10617247 |
| Goat anti-rabbit IgG-HRP Dilution: 1:2,000 (WB) |
Santa Cruz Biotechnology, Inc. | Cat #sc-2004; RRID: AB_631746 |
| Goat anti-mouse IgG-HRP Dilution: 1:2,000 (WB) |
Santa Cruz Biotechnology, Inc. | Cat #sc-2005; RRID: AB_631736 |
| Chemicals, peptides, and recombinant proteins | ||
| TIMP2-His6 Working concentration 1 μg/mL |
NCI, Dr W. Stetler-Stevenson | Sánchez-Pozo et al.,2 Baker-Williams et al.,3 Chowdhury et al.4 |
| HisPur Ni-NTA resin | Thermo Fisher Scientific | Cat #88221 |
| Pierce ECL2 western blotting substrate | Thermo Fisher Scientific | Cat #PI80196 |
| Pierce Femto western blotting substrate | Thermo Fisher Scientific | Cat #34094 |
| Trans-Blot Turbo RTA Nitrocellulose Transfer Kit | Bio-Rad | Cat #1704271 |
| Dried low-fat milk | Carnation | Nestle 12428935 |
| Bovine serum albumin (optional) | Sigma | Cat #A3294 |
| Dulbecco’s modified Eagle’s medium (DMEM) for SYF and SYF+c-Src cells (Gibco) | Thermo Fisher Scientific | Cat #11965092 |
| Fetal bovine serum (FBS) (Corning) | Thermo Fisher Scientific | Cat #MT35016CV |
| Criterion Tris-HCl precast gel 4%–20% gradient 12 + 2 wells | Bio-Rad | Cat #3450032 |
| Centrifugal filter units | Sigma | Cat #UFC8010 (4 mL unit) |
| Coomassie Brilliant Blue R-250 | Pierce | Cat #20278 |
| Trizma base | Sigma | Cat #T1503 |
| Sodium chloride (NaCl) | Sigma | Cat #S7653 |
| Hydrochloric acid (HCl) | Thermo Fisher Scientific | Cat #SA56-1 |
| Glycine | Thermo Fisher Scientific | Cat #BP381-5 |
| Sodium dodecyl sulfate (SDS) | Sigma | Cat #L3771 |
| Ethanol (EtOH) | Thermo Fisher Scientific | Cat #BP2818500 |
| NP-40 (Igepal CA-630) | Sigma | Cat #I8896 |
| Magnesium chloride (MgCl2) | Sigma | Cat #M8266 |
| 1 M Tris-HCl solution pH 7.4 | Thermo Fisher Scientific | Cat #J60202-K2 |
| Sodium molybdate dihydrate (Na2MoO4 · 2H2O) | Sigma | Cat #331058 |
| cOmplete ULTRA, mini, EDTA-free EASYpack protease inhibitor cocktail tablets | Sigma | Cat #05892791001 |
| PhosStop EASYpack phosphatase inhibitor cocktail tablets | Sigma | Cat #PHOSS-RO |
| Glycerol (C3H8O3) | Sigma | Cat #G5516 |
| β-mercaptoethanol (BME) | Sigma | Cat #M6250 |
| Glacial acetic acid (CH3COOH) | Thermo Fisher Scientific | Cat #18-602-911 |
| Methanol (MeOH) | Sigma | Cat #179337 |
| Imidazole (C3H4N2) | Sigma | Cat #I2399 |
| Ponceau S staining solution | Thermo Fisher Scientific | Cat #A40000279 |
| Phosphate-buffered saline (PBS) | Sigma | Cat #MFCD00131855 |
| Critical commercial assays | ||
| Bradford assay | Bio-Rad | Cat #5000205 |
| Experimental models: Cell lines | ||
| SYF (mouse embryonic fibroblast cells without Src, Fin, or Yes kinases) | ATCC (American Type Culture Collection) | Cat #CRL-2459; (RRID: CVCL_6461) |
| SYF + c-Src (mouse embryonic fibroblast cells without Fin, or Yes kinases) | ATCC (American Type Culture Collection) | Cat #CRL-2498; (RRID: CVCL_8976) |
| Software and algorithms | ||
| BioRender | https://biorender.com/ | |
| Other | ||
| Trans-Blot Turbo transfer system | Bio-Rad | Cat #1704150EDU |
| Criterion vertical electrophoresis cell | Bio-Rad | Cat #1656001 |
| Criterion cell and PowerPac basic power supply | Bio-Rad | Cat #1656019 |
Materials and equipment
10X TBS
| Reagent | Final concentration | Add to 2 L |
|---|---|---|
| Trizma | 50 mM | 48.4 g |
| NaCl | 150 mM | 160 g |
| HCl | 10% | 20 mL |
| ddH2O | - | 1974 mL |
Store at 19°C–23°C for up to 24 months.
For use, dilute the 10X TBS with ddH2O to 1X and add 0.1% Tween (TBST). Store at 4°C for up to 1 month.
10x protein running buffer
| Reagent | Final concentration | Add to 2 L |
|---|---|---|
| Glycine | 2 M | 288.4 g |
| Trizma | 250 mM | 60.55 g |
| ddH2O | - | Up to 2 L |
| SDS | 10% | 20 g |
| Dilute with ddH2O and use at 1X | N/A | N/A |
Store at 19°C–23°C for up to 6 months.
Protein Transfer Buffer
| Reagent | Final concentration | Add to 2 L |
|---|---|---|
| 5x Transfer Buffer (Bio-Rad, see key resources table) | 20% | 400 mL |
| Ethanol | 20% | 400 mL |
| ddH2O | 60% | 1200 mL |
Store at 4°C for up to 1 week.
0.1% NP-40 Extraction Buffer
| Reagent | Final concentration | Add to 50 mL |
|---|---|---|
| NP-40 (Ipegal) | 0.1% | 50 μL |
| MgCl2 (1 M) | 1 mM | 50 μL |
| NaCl (5 M) | 100 mM | 1 mL |
| Tris pH 7.4 (1 M) | 20 mM | 1 mL |
| Sodium Molybdate (500 mM) | 0.2 mM | 2 mL |
| PhosStop | - | 1 tablet |
| EDTA-Free Protease Inhibitor | - | 1 tablet |
| ddH2O | - | Up to 50 mL |
Store at 4°C for up to 1 month. PhosStop phosphatase inhibitor and EDTA-Free Protease inhibitor tablets are added to the Buffer. Verify that the tablets are completely dissolved.
5x Protein Loading Buffer
| Reagent | Final concentration | Add to 20 mL |
|---|---|---|
| Tris pH 6.8 (1 M) | 12.5 mM (12.5%) | 2.5 mL |
| 20% SDS | 12.3% | 2 mL |
| Glycerol | 10% | 2 mL |
| β-mercaptoethanol | 5% | 1 mL |
| Bromophenol Blue (1% Solution in water) | 0.22% | 430 μL |
| ddH2O | 60% | 12 mL |
Store at 4°C for up to 2 weeks. The reducing agent β-mercaptoethanol is unstable and, therefore, we recommend to replenish it weekly.
Coomassie Blue Staining Solution/Destain
| Reagent | Final concentration | Add to 1 L |
|---|---|---|
| Coomassie Brilliant Blue | 0.1% | 1 g |
| Glacial Acetic Acid | 10% | 100 mL |
| Methanol | 50% | 500 mL |
| ddH2O | 40% | 400 mL |
Store at 19°C–23°C for up to 24 months. Filter Coomassie Blue Staining Solution to reuse one more time.
To make the Destain, repeat this recipe omitting the Coomassie Blue dye.
His6 -tagged protein purification buffer
| Reagent | Final concentration | Add to 50 mL |
|---|---|---|
| NP-40 (Igepal) | 0.1% | 50 μL |
| MgCl2 (1 M) | 1 mM | 50 μL |
| NaCl (5 M) | 500 mM | 5 mL |
| Tris pH 7.4 (1 M) | 20 mM | 1 mL |
| Sodium Molybdate (500 mM) | 0.2 mM | 2 mL |
| Imidazole (1 M) | 50 mM | 2.5 mL |
| PhosStopa | - | 1 tablet |
| EDTA-Free Protease Inhibitora | - | 1 tablet |
| ddH2O | - | Up to 50 mL |
a Store at 4°C for up to 1 month. Add PhosStop phosphatase inhibitor and EDTA-Free Protease inhibitor tablets to the Buffer and verify that the tablets are completely dissolved.
CRITICAL: The following table details the reagents that require special handling
| Reagent | Hazard |
|---|---|
| HCl (Hydrochloric Acid) | Corrosive/Toxic (use PPE, avoid inhalation) |
| SDS (Dodecyl Sodium Sulfate) | Toxic (avoid inhalation) |
| Ethanol | Flammable |
| NP-40 (Igepal) | Toxic (do not ingest) |
| Glacial Acetic Acid | Corrosive/Toxic (use PPE, avoid inhalation) |
| Methanol | Flammable/Toxic (do not ingest, avoid inhalation) |
| Imidazole | Toxic (use PPE, avoid contact with skin, do not ingest) |
| β-mercaptoethanol | Toxic/Flammable (use PPE, use in fume hood, avoid contact with skin, do not ingest) |
Step-by-step method details
Cell culture and processing of cell extracts and conditioned media (secretome)
Timing: ∼3 days
Timing: 16 h (for step 1)
Timing: 24–36 h (for step 2)
Timing: 1–2 h (for step 3)
Timing: ∼1 h (for step 4)
Timing: ∼2–4 h (for steps 5 and 6)
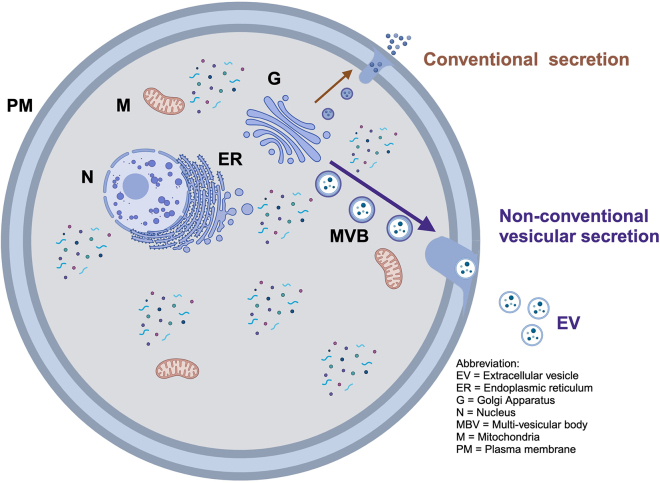
Note: This section describes step by step procedures for culturing, collecting and processing of cell extracts and conditioned media (CM) from cells expressing c-Src kinase (SYF+c-Src) and genetically c-Src-knockout cells (SYF). This is the first major and critical process for the collection of cell-secreted proteins or cell secretome (Figure 2).
Note: This multi-step process is easily adaptable to collect secretomes from any mammalian cells which can then be used for various downstream applications and proteomic analyses (Figure 3).
-
1.Cell seeding.
-
a.DAY 0 – Grow cells in 10 cm tissue culture dishes until they reach 70%–90% confluency.
-
b.For each cell line to be studied, split cells from one dish in two 10 cm dishes and culture for ∼16 h.
-
a.
Note: Resuspend cells homogenously to ensure even seeding. Alternatively, count and seed a defined number of cells.
CRITICAL: Seeding fewer cells may result in low abundance of secreted proteins in the CM, whereas, too many cells will result in cell apoptosis and death.
CRITICAL: Do not allow cells to reach 100% confluency prior to serum starvation. Addition of serum-free media on 100% confluent cultures will result in apoptotic and dead cells, as well as release of cytosolic proteins into the CM.
CRITICAL: While cell confluency of 70%–90% is the optimum density for SYF and SYF+c-Src cells, different cell lines may proliferate at different rates. Therefore, it is best to optimize cell density and duration of serum-starvation for each cell line. See above (cell cytotoxicity assay) the method we use to evaluate cell death in different conditions.
-
2.Serum starvation.
-
a.DAY 1 – Next morning and while cells are at ∼80% confluency, gently wash attached cells twice with sterile PBS suitable for cell culture or serum-free medium pre-warmed to 23°C.Note: Some cell lines attach weakly to plastic and can easily detach following washes with PBS.
-
b.Aspirate out the second wash of PBS/medium and add 4 mL of fresh serum-free medium by gently tilting the plate and adding medium at the edge so to avoid the mechanical detachment of cells.
-
c.Incubate cells in a humidified 37°C cell culture incubator containing 5% CO2 for 24–36 h.
-
a.
-
3.DAY 2 – Collect cells and conditioned media (CM) in the afternoon.
-
a.Transfer CM from each replicate dish in a sterile 15 mL conical tube. Combine CM from both dishes. Total volume of CM should be 8 mL. Keep CM on ice.
-
b.Centrifuge CM at 290 g to pellet any floating detached cells at 23°C.
-
c.Transfer CM to a new 15 mL tube and repeat centrifugation of at 3,220 g for 5 min at 4°C to remove dead cells.
 Pause point: Store media at ‒80°C until ready to proceed to step 4.
Pause point: Store media at ‒80°C until ready to proceed to step 4. -
d.For cell extracts, rinse gently cells once with 7 mL of ice-cold PBS. Remove and repeat with another 7 mL of ice-cold PBS and place dishes on ice for protein extraction.
-
e.Leave PBS on cells until ready for step 5.Note: Be cautious because some cells may easily detach following washes with PBS.
-
a.
-
4.Concentration of conditioned media (CM).Note: We concentrate media at least 10x using Amicon Ultra Centrifugal Filter Units (Millipore Sigma). We use 3 kDa or 10 kDa cut-off units which eliminate small molecular weight proteins from the collected media.Note: Ensure unit cut-off is appropriate for the size of proteins tested.Note: To concentrate the CM, follow manufacturer’s protocol (https://www.emdmillipore.com/US/en/product/Amicon-Ultra-Centrifugal-Filters,MM_NF-C134281?ReferrerURL = https%3A%2F%2Fwww.google.com%2F).Note: Perform centrifugations at 4°C and chill all materials on wet ice.When using the Amicon Ultra-4 Centrifugal Filter Unit.
-
a.Add up to 4 mL of CM (3.5 mL if using 23° fixed-angle rotor) to the filter device, cap the tubes and place them in a centrifuge rotor.
-
b.Make sure to counterbalance the centrifuge.
-
c.If you use a swinging-bucket rotor, spin the device at 4,000 g maximum for approximately 10–15 min (to achieve 10x concentration of CM).
-
d.For 10x concentration of 8 mL CM you will end up with 800 μL CM.Note: If total CM volume is 8 mL one may use the Amicon Ultra-15 Centrifugal Filter Unit instead.
-
e.Spin CM in 5-min intervals at 3,220 g and stop when the volume of media in the concentrator reaches 800 μL.Note: Observe the volume that remains in the concentrator after each centrifugation and repeat centrifugation step if needed. Record the final volume of the concentrated media (Figure 4).
-
f.Remove concentrated CM immediately after final centrifugation for optimal recovery.
-
g.Aliquot unused CM media and store at ‒80°C.
 Pause point: Store concentrated CM in 1.5 mL microcentrifuge tubes at ‒80°C until ready for downstream analyses.Note: Units can be reused up to 3 times. Follow manufacturer’s instructions.
Pause point: Store concentrated CM in 1.5 mL microcentrifuge tubes at ‒80°C until ready for downstream analyses.Note: Units can be reused up to 3 times. Follow manufacturer’s instructions.
-
a.
-
5.Protein extraction from serum-starved cells.Note: Protein quantification is routinely used as a method to determine the concentration of protein collected from cell extracts and to equalize protein levels for western blot analyses. We do not use the same method to equalize total protein from conditioned media (CM).Note: Although cells are plated in serum-free conditions, traces of albumin may remain in CM and will account for the majority of protein concentration. As an alternative, serum-free cultured cells are collected, lysed, and protein is extracted. The resulting concentration from these extracts is determined and subsequently used to normalize and equalize CM input for western blot analyses.Note: CM protein levels can also be normalized based on the cell number; specifically, following collection of CM, serum-starved cells may be trypsinized and counted.
 CRITICAL: Cool down a refrigerated bench-top microcentrifuge to 4°C to spin down protein lysates. Protein degradation occurs if samples are allowed to warm up.Note: Include protease and phosphatase inhibitors in the 0.1% NP-40 Protein Extraction Buffer (see key resources table).
CRITICAL: Cool down a refrigerated bench-top microcentrifuge to 4°C to spin down protein lysates. Protein degradation occurs if samples are allowed to warm up.Note: Include protease and phosphatase inhibitors in the 0.1% NP-40 Protein Extraction Buffer (see key resources table).-
a.Gently aspirate the ice-cold PBS from the dishes placed on ice in step 3d.
-
b.Discard remaining PBS by tilting the dishes and collecting the PBS at the bottom side of the plate.
-
c.Wait 30 sec-1 min and aspirate out the collected liquid.
-
d.Add up to 200 μL of 0.1% NP-40 Protein Extraction Buffer to each plate and allow to incubate on ice for 30 sec-1 min.Note: Add 200 μL of 0.1% NP-40 Protein Extraction Buffer in >80% confluent cells. If cell confluency is <80%, add less.
-
e.Use a cell scraper to collect all cell lysate to one side of the dish.
-
f.Pipet the cell lysate into a labeled polypropylene microcentrifuge tube.
-
g.Collect any remaining cell lysate and add this to the tube on ice.
-
h.After collecting all samples, sonicate protein lysates for 3 s per each sample (∼65 kHz). Samples remain on ice.
 CRITICAL: Wear hearing protection during sonication.
CRITICAL: Wear hearing protection during sonication. -
i.Place the tubes in a microcentrifuge and spin at 21,300 g at 4°C for 7 min.
-
j.Transfer supernatant into a clean microcentrifuge tube and place on ice.
-
a.
-
6.
Quantify protein concentration for each cell lysate sample per manufacturer’s instructions.
Note: Colorimetric protein quantification assays include the Bradford or the Bicinchoninic Acid (BCA) Assays, both available for purchase from many commercial sources. You will need a spectrophotometer that reads in the recommended wavelength.
Note: Some detergents included in Protein Extraction Buffers may not be compatible with these kits. It is advisable to consult the protein compatibility guide, if detergents other that NP-40 are used, see: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2F23246_23246S_deter_compat_bradford_UG.pdf
-
7.
Identify the protein sample that has the lowest concentration.
-
8.
Calculate the ratios between the least concentrated sample and each of the remaining samples.
-
9.
Use the same ratios to determine the volumes of the concentrated CM for loading in an SDS-PAGE.
-
10.
To load equal sample volumes in an SDS-PAGE, add 0.1% NP-40 Protein Extraction Buffer in those samples. In our case, this volume is 40 μL per sample.
-
11.
Add 10 μL of 5X Protein Loading Buffer for a final volume of 50 μL.
-
12.
Boil the samples on a heat block at 95°C for 5 min.
CRITICAL: Use caution when removing samples from the heat block as built-up pressure may cause lids to suddenly pop.
-
13.
Spin samples in a microcentrifuge for 1 min at full speed.
Pause point: Chill samples on wet ice or store at ‒20°C until use.
Figure 2.
Schematic of protein secretion pathways
Conventional (ER/Golgi) and non-conventional (multivesicular bodies/MVB) pathways.
Figure 3.
Secretomes and downstream applications
Figure 4.
Concentrating cell-conditioned media (CM) using a 4 mL - 10K cutoff unit
(A) One Amicon Ultra Centrifugal Filter 10K-cutoff unit.
(B) 4 mL CM were added into the concentrator.
(C) CM is centrifuged at 3,220 g and at 4°C.
(D) After concentration, top is the concentrated CM and bottom is the flowthrough.
Western blotting of CM from SYF and SYF+c-Src cells
Timing: 12–24 h
Note: Western blotting is the qualitative method we use to detect proteins of interest expressed inside (cell extracts) or outside (secreted, CM) cells. Detection of substrate phosphorylation and its interaction with the secreted kinase will then follow (with and without anti-kinase antibody treatment).
Note: Secretory proteins trafficked through the ER/Golgi pathway are overall abundant and easily detectable in the CM (e.g. TIMPs, MMPs) (Figure 2). Non-conventionally secreted proteins (e.g. c-Src, Hsp90) that predominantly function inside cells generally exist at much lower levels in the extracellular space.
Note: Non-conventionally secreted proteins are less abundant and hard to detect using standard immunoblotting. One way to increase protein detectability in CM is to collect and analyze the maximum volume of concentrated CM possible in subsequent experiments.
-
14.For SDS-PAGE run an appropriate volume of concentrated CM prepared as described above in steps 7–8.
-
a.Include protein ladder and load samples on an SDS-PAGE gradient 4%–20% gel.
-
b.Volume of sample load is determined by the capacity of the gel.Note: Run the gel long enough to resolve the smallest size protein for which you are probing.
-
c.Use the semi-dry transfer system to transfer the proteins to a nitrocellulose membrane (see conditions below in Table 1: SDS Page and Protein Transfer).
-
a.
-
15.
At this point, continue with probing for proteins of interest e.g., c-Src kinase or TIMP2 substrate or other interacting protein candidates (e.g., Hsp90) identified in Backe et al.1
Note: Probing for a ‘housekeeping’ protein, e.g. GAPDH, known to be a cytosolic protein is also advisable. We provide examples of western blots in Figure 1 in Backe et al.1
Table 1.
SDS-PAGE and protein transfer
| SDS-PAGE amperage | 200 mA |
|---|---|
| SDS-PAGE Time | 45 min- 1 h (resolve smallest band of interest) |
| Trans-Blot Turbo Amperage | 2.5 A Constant |
| Trans-Blot Turbo Time (High Molecular Weight) | 10 min |
| Trans-Blot Turbo Time (Mixed Molecular Weight) | 7 min |
| Trans-Blot Turbo Time (Low Molecular Weight) | 5 min |
Cell treatment with anti-c-Src antibodies
Timing: 2–3 days
Timing: 1 ½ days (for step 16)
Timing: ∼3–6 h (for steps 17–25)
Note: We use anti-kinase (c-Src) antibodies to block formation of, and/or cause disruption of kinase-substrate complexes. We pre-treat SYF+c-Src cell cultures with anti-c-Src antibodies or the relevant IgG isotype controls for 1 h. This step is followed by addition of an extracellular kinase substrate of interest. In here, we test the human recombinant protein TIMP2-his6.
Note: c-Src antibodies are commercially available, and details on formulation, specificity and sensitivity are found in the company’s antibody datasheet. Two such antibodies are listed in the key resources table. For the whole list of c-Src antibodies see key resources table in Backe et al.1
Note: Because antibodies are added to cells for a short period of time (1–2 h), the presence of sodium azide in the antibody buffer will not affect cell viability. Antibody stocks will also be further diluted to working concentrations (at least 100x).
CRITICAL: Nevertheless, if presence of sodium azide is unfavorable, perform buffer exchange to remove any salts or unwanted molecules from the antibody or simply ensure that azide- and carrier-free forms of antibodies are purchased.
Note: If stock antibody concentration is not indicated in the company’s datasheet, it is necessary to call the technical support ahead of time and request that information.
-
16.For cell seeding and serum starvation follow steps 1 and 2 described earlier.
-
a.DAY 0 - Prepare four 10 cm dishes with cells growing at 70%–90% confluency and culture for ∼16 h.
-
b.DAY 1 - Remove media, wash cells with PBS and add 4 mL of fresh serum-free medium to each dish by gently tilting the plate and adding medium at the edge so to avoid the mechanical detachment of cells.
-
c.Incubate in a humidified 37°C incubator containing 5% CO2 for 24 h.
-
a.
-
17.Cell treatments – stock and working antibodies.Note: Treat cells with the anti-c-Src antibody or relevant IgG control at the final concentration of 1 μg/mL.
-
a.DAY 2 – Observe cell cultures under the microscope. Note cell viability and confluency and return cultures back into the incubator.
-
b.Determine the amounts of antibodies to use for each treatment and prepare antibody stocks.
 CRITICAL: Keep antibodies on wet ice.
CRITICAL: Keep antibodies on wet ice.
-
a.
-
18.Label four cell culture dishes as follows:
-
a.Plate 1 (negative control): add 1x PBS equal to the experimental antibody volume.
-
b.Plate 2: add 1x PBS equal to the experimental antibody volume.
-
c.Plate 3: add IgG control so that the final concentration is 1 μg/mL.
-
d.Plate 4: add anti c-Src antibody so that the final concentration is 1 μg/mL.
-
a.
-
19.
Gently swirl plates to mix.
-
20.
Return cultures back to the 37°C incubator for 1 h.
-
21.
Prepare stock solutions of substrate of interest in sterile microcentrifuge tubes.
Note: We study human recombinant TIMP2-his6 at working concentration of 50 ng/mL, while the more concentrated stock is prepared in sterile PBS containing 0.1% fetal bovine serum (FBS).
-
22.Remove the four dishes from the incubator and add the following:
-
a.Plate 1 (negative control): add PBS containing 0.1% FBS at the volume equal to the TIMP2-his6 volume.
-
b.Plates 2–4: Add recombinant TIMP2-his6 at final concentration of 50 ng/mL.
-
a.
-
23.
Swirl dishes gently and return to the incubator for 1–2 h.
-
24.
Remove dishes from the incubator; collect conditioned media (CM).
-
25.
Concentrate media 10X as described in steps 4 and 5 (collect cells and determine protein concentration to equalize CM in western blot).
Pause point: Store concentrated media at ‒80°C until ready for downstream analyses.
Note: Coomassie blue staining of SDS-PAGE gels or Ponceau staining of the nitrocellulose membrane is required to visualize equal protein loading and transfer (respectively) (Figure 5).
Figure 5.

Coomassie blue dye staining
SDS-PAGE gel showing total protein loading from concentrated 10X CM with indicated treatments.
Detection of kinase-substrate interaction
Timing: 1–2 days
Note: Verify the detection of extracellular kinase-substrate interaction in the absence of c-Src antibodies. However, pre-treatment of cells with anti-c-Src antibodies will convincingly disrupt protein-protein interactions, as we have previously shown to occur between c-Src and TIMP2 in Backe et al.1).
Note: Treatment of cells with anti-c-Src antibodies also provides valuable information on the impact of extracellular phosphorylation on cellular function (e.g. cell growth, survival, migration, invasion, etc.) (see Backe et al.1).
CRITICAL: Keep everything on ice unless otherwise specified.
Note: We use HisPur Ni-NTA Resin to pull down TIMP2-His6 from concentrated CM prepared in step 25.
Note: Trim pipette tips to facilitate accurate pipetting of resin slurry.
CRITICAL: Be extra careful not to disturb the resin during washes.
-
26.
Aliquot 40 μL of Ni-NTA resin per sample into one microcentrifuge tube per experimental condition. For example, for four pull-downs, combine 160 μL resin in one microcentrifuge tube.
-
27.
Wash resin with 500 μL of fresh 0.1% NP-40 Protein Extraction Buffer.
-
28.
Centrifuge the tubes at 21,300 g for 30 s and repeat steps 27–28 four times.
Note: Vortex resin in 0.1% NP-40 Protein Extraction Buffer before centrifuging the tubes.
CRITICAL: The 0.1% NP-40 Protein Extraction Buffer should be freshly made and contain protease and phosphatase inhibitors.
-
29.
After the final wash, remove the supernatant and add 100 μL of 0.1% NP-40 Protein Extraction Buffer per tested sample. Here, we add 400 μL 0.1% NP-40 Protein Extraction Buffer.
-
30.
Aliquot 100 μL resin mixture in four microcentrifuge tubes (one tube per sample).
-
31.
Centrifuge tubes at 21,300 g for 30 s then visually check for equal distribution of the resin among the four tubes.
-
32.
Gently aspirate out the excess of 0.1% NP-40 Protein Extraction Buffer.
-
33.
Continue with the pull-down process (step 34).
CRITICAL: Be extra careful not to disturb the resin during washes.
-
34.
Thaw all tubes with concentrated CM on wet- ice.
-
35.
Add 100 μL–200 μL concentrated CM to the HisPur Ni-NTA Resin (from step 32) and place on rotator for 1 h–16 h at 4°C.
-
36.
Wash four times with fresh 0.1% NP-40 Protein Extraction Buffer containing 50 mM imidazole to reduce non-specific binding of proteins to resin.
-
37.
After removing final wash, add 5x Protein Loading Buffer to the resin and boil samples in heating block for 5 min.
-
38.
Centrifuge at 21,300 g for 30 s and transfer the supernatant containing the proteins to a clean microcentrifuge tube.
-
39.Run a test-pull-down on an SDS-PAGE.
-
a.Include protein ladder and load 5 μL of volume per pull-down sample.
-
b.Probe with anti-his antibody to detect his-tagged protein (TIMP2) in all samples where TIMP2 was added in.
-
c.Verify all bands have equal intensity for TIMP2-his6. If they do not appear equal adjust volumes and run the samples again.
-
a.
-
40.Co-pull-down for interacting proteins.
-
a.Load up to 40 μL of pull-down sample prepared in step 38.
-
b.When loading 40 μL of samples for the co-pull-down, normalize as in step 39c.
-
a.
-
41.
Proceed with western blotting and probe for c-Src or any other proteins of interest.
Note: If co-pull-down is not detected due to weak protein-protein interactions, consider increasing the volume of concentrated CM for pull-down.
Detection of extracellular substrate TIMP2 tyrosine phosphorylation
Timing: 1–2 days
Note: Below we describe a method that detects tyrosine phosphorylation of extracellular substrate TIMP2-his6 by c-Src-mediated tyrosine kinase in concentrated SYF+c-Src cell CM.
CRITICAL: Keep everything cold or on ice unless specified.
-
42.
Refer to steps 25 to 35 to prepare the Ni-NTA resin-CM mixture.
-
43.
Transfer the tubes to a microcentrifuge and centrifuge at 94 g for 30 s.
-
44.
Wash the resin-CM mixture with 500 μL of high salt His6 -tagged protein purification buffer (contains 500 mM NaCl and 50 mM imidazole) to strip the his6-interacting proteins.
-
45.
Repeat step 44.
-
46.
Wash the resin-CM mixture once with 500 μL 0.1% NP40 protein extraction buffer (for this step, add 50 mM imidazole).
-
47.
Spin down at 94 g for 30 s.
-
48.
Repeat step 46.
-
49.
Centrifuge at max speed (21,300 g) for 30 s.
-
50.
Remove supernatant then re-suspend the resin in 50–100 μL of 5x protein loading buffer (for this step, add 300 mM imidazole in the 5x protein loading buffer).
-
51.
Boil the samples for 5 min.
-
52.
Spin the tubes down for 30 s at 21,300 g then transfer the supernatant to new microcentrifuge tubes.
CRITICAL: Be extra careful not to transfer resin with the supernatant.
-
53.
Run 5 μL on SDS-PAGE to test the success of the pull-down.
-
54.
Probe for his6 (to detect TIMP2) to confirm equal loading.
-
55.
Repeat SDS-PAGE using 30 μL of sample in another SDS-PAGE and probe for total tyrosine phosphorylation of TIMP2-his6 using a pan-pY antibody (see key resources table).
Expected outcomes
In this protocol, we describe methods to isolate and characterize the function of cell-secreted proteins, some being naturally secretory while others are released upon cell stress. However, dying cells can also release their intracellular protein contents to the extracellular space when their plasma membrane has been compromised and damaged. It is, therefore, essential that the cell cytotoxicity assay on tested cells is performed before collecting the cell secretome for downstream analyses. The cell cytotoxicity assay should show no increase of baseline cell death when cells are cultured in serum-free conditions as was shown in Baker-Williams e al.3 Furthermore, when testing the wild type and kinase knockout cells for protein expression, do so in both cells extracts and concentrated CM (see1,2,5). It is expected that when probing for endogenous c-Src kinase, the protein band is absent from the knockout cell line SYF (Figure 1). In our previous studies, we tested a number of commercially available anti-c-Src antibodies as an approach to characterize secreted kinase e-Src interaction with its substrate TIMP2.1,2 These antibodies are monoclonal, targeting different epitopes on c-Src protein domains. All tested antibodies blocked c-Src-TIMP2 interaction and disrupted TIMP2 tyrosine phosphorylation except one (see Figure 5 in Backe et al.1). Furthermore, these findings were specific to the TIMP2 substrate. Therefore, the outcomes may be different for other e-Src substrates.
Limitations
When testing kinase knockout cell lines, it is important to know whether other members of the same family of proteins are present and expressed in the cells. For instance, Yes and Fyn tyrosine kinases are structurally similar to c-Src and, fortunately, they are also absent from SYF cells. This is beneficial for our studies because many commercially available anti-c-Src antibodies cross-react with Yes and Fyn. It is therefore critical to select appropriate knockout cell lines and specific antibodies to c-Src.
Troubleshooting
Problem 1
Serum starvation results in increased cell death or damage.
Potential solution
-
•
Design a time-course experiment where cells are cultured in serum-free conditions. Collect cells and CM at different time points and perform cell cytotoxicity assay3 to determine the optimum cell culture conditions.
Problem 2
Proteins are not detected in immunoblots performed using CM.
Potential solutions
-
•
Concentrate CM more than 10x (20x would be our next choice).
-
•
If possible, increase the volume of the concentrated CM run in the western blot.
-
•
Probe with more concentrated primary antibody.
-
•
Use an ultra-sensitive enhanced chemiluminescence substrate (ECL) for detection (e.g., Femto).
Problem 3
Absence of protein-protein interaction or phosphorylation of a potential substrate.
Potential solutions
-
•
Test kinase interaction with a known substrate (e.g., TIMP2) as a positive control.
-
•
Try different salt concentration of buffers.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dimitra Bourboulia (bourmpod@upstate.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
This protocol does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award no. R01GM139932 (D.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional funding sources include the SUNY Upstate Medical University, SUNY Upstate Cancer Center, the Upstate Foundation. Some figure panels were prepared using BioRender software (https://biorender.com/).
Author contributions
Conceptualization and experimental design, D.B..; experimental investigation, M.P.O., D.A., S.D.V., and D.B.; writing – original draft, D.B.; contributions to manuscript writing – review and editing, M.P.O., D.A., S.D.V., and D.B.; supervision, D.B. All authors read the manuscript and provided their final approval for the content.
Declaration of interests
The authors declare no competing interests.
References
- 1.Backe S.J., Votra S.D., Stokes M.P., Sebestyén E., Castelli M., Torielli L., Colombo G., Woodford M.R., Mollapour M., Bourboulia D. PhosY-secretome profiling combined with kinase-substrate interaction screening defines active c-Src-driven extracellular signaling. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sánchez-Pozo J., Baker-Williams A.J., Woodford M.R., Bullard R., Wei B., Mollapour M., Stetler-Stevenson W.G., Bratslavsky G., Bourboulia D. Extracellular Phosphorylation of TIMP-2 by Secreted c-Src Tyrosine Kinase Controls MMP-2 Activity. iScience. 2018;1:87–96. doi: 10.1016/j.isci.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker-Williams A.J., Hashmi F., Budzynski M.A., Woodford M.R., Gleicher S., Himanen S.V., Makedon A.M., Friedman D., Cortes S., Namek S., et al. Co-chaperones TIMP2 and AHA1 Competitively Regulate Extracellular HSP90:Client MMP2 Activity and Matrix Proteolysis. Cell Rep. 2019;28:1894–1906.e6. doi: 10.1016/j.celrep.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury A., Brinson R., Wei B., Stetler-Stevenson W.G. Tissue Inhibitor of Metalloprotease-2 (TIMP-2): Bioprocess Development, Physicochemical, Biochemical, and Biological Characterization of Highly Expressed Recombinant Protein. Biochemistry. 2017;56:6423–6433. doi: 10.1021/acs.biochem.7b00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Votra S.D., Alsalih D., Bourboulia D. Methods to Assess the Impact of Hsp90 Chaperone Function on Extracellular Client MMP2 Activity. Methods Mol. Biol. 2023;2693:221–232. doi: 10.1007/978-1-0716-3342-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.