Abstract
The entomopathogenic bacterium Photorhabdus luminescens exhibits phase variation when cultured in vitro. The variant forms of P. luminescens are pleiotropic and are designated phase I and phase II variants. One of the characteristic phenotypes of phase I cells is the production of two types of intracellular protein inclusions. The genes encoding the protein monomers that form these inclusions, designated cipA and cipB, were cloned and characterized. cipA and cipB encode hydrophobic proteins of 11,648 and 11,308 Da, respectively. The deduced amino acid sequences of CipA and CipB have no significant amino acid sequence similarity to any other known protein but have 25% identity and 49% similarity to each other. Insertional inactivation of cipA or cipB in phase I cells of P. luminescens produced mutants that differ from phase I cells in bioluminescence, the pattern and activities of extracellular products, biochemical traits, adsorption of dyes, and ability to support nematode growth and reproduction. In general, the cip mutants were phenotypically more similar to each other than to either phase I or phase II variants.
Bacteria of the genera Photorhabdus and Xenorhabdus are mutualistically associated with entomopathogenic rhabditid nematodes of the families Heterorhabditidae and Steinernematidae, respectively (for a review, see reference 18). Photorhabdus and Xenorhabdus spp. exist in two forms, designated phase I and phase II variants, which differ in many phenotypic traits. Phase I variants, which are isolated from infective-stage nematodes, produce an extracellular protease (3, 33), antibiotic substances (2, 28, 31), extracellular lipase (3, 20, 39), and intracellular protein crystals (12, 14, 15) and are bioluminescent (in Photorhabdus sp.). The phase II variant, which appears following prolonged growth in vitro, lacks detectable protease, lipase, and antibiotic activity (2, 9, 10, 22). The phase I and phase II variants also exhibit differences in colony morphology, pigmentation, bioluminescence, dye adsorption, metabolism, and the ability to support the growth and reproduction of a mutualistic nematode species (1, 10).
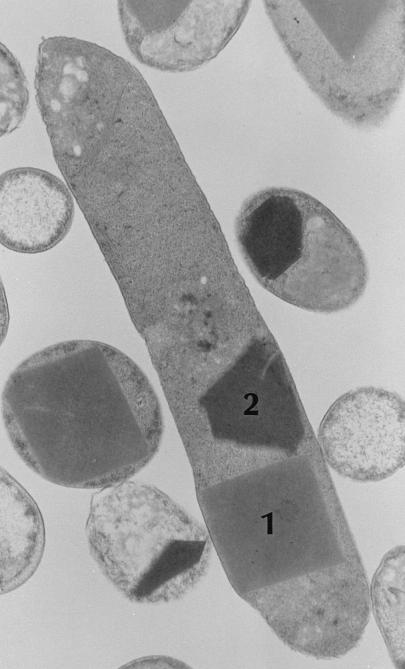
One of the characteristic phenotypes of phase I but not phase II variant cells is the presence of two types of intracellular protein inclusions (Fig. 1) that can account for 40% of the total protein content of stationary-phase cells (12). These inclusion proteins were partially characterized in Xenorhabdus nematophilus (14, 15) and Photorhabdus luminescens (12), but their biological significance has not been determined. Since this phenomenon is observed in two distinct genera of bacteria that inhabit similar ecological niches, the implication is that the crystalline inclusion proteins have a biological role. The primary objective of this study was to characterize the genes encoding the crystalline inclusion proteins. It was hoped that this information would prove useful in addressing the biological role(s) of the crystalline inclusion proteins in the mutualistic/pathogenic life cycle of P. luminescens. To accomplish these objectives, we first cloned and characterized the genes encoding these proteins, which have been designated cipA (for crystalline inclusion protein) and cipB. Then, cipA and cipB mutants were constructed via allelic exchange. Finally, these cip mutants were characterized with regard to bacterial phenotypes, insect pathogenesis, and growth and reproduction of a mutualistic nematode. In this study, we present the molecular analysis of cipA and cipB as well as the phenotypic characterization of cip mutants of P. luminescens NC1.
FIG. 1.
Micrograph of sectioned P. luminescens cells. The two different inclusion types (12), designated type 1 and type 2, are composed of CipB and CipA, respectively. Stationary-phase cells of P. luminescens NC1/1 were prepared according to standard methods and examined by transmission electron microscopy at the University of Wisconsin—Madison Electron Microscope Facility. Magnification, ×36,000.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth or on LB agar (1.5% agar). For E. coli, ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), streptomycin (50 μg/ml), spectinomycin (50 μg/ml), kanamycin (50 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml), and sucrose (5%) were added to the media as required. Micrococcus luteus was grown in nutrient broth or on nutrient agar at 30°C. P. luminescens strains were grown in the dark at 30°C in 2% Proteose Peptone no. 3 (PP3) broth or on PP3 agar. For P. luminescens, chloramphenicol (20 μg/ml), streptomycin (20 μg/ml), spectinomycin (20 μg/ml), kanamycin (25 μg/ml), and sucrose (5%) were added to the media as required. Phase variants of Photorhabdus species were distinguished as previously described (1).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH5α | Cloning strain | Gibco BRL |
| XL1-Blue MRF | Cloning strain | Stratagene |
| EC109 | DH5α with pSB109; AprcipA | This study |
| EC211 | DH5α with pSB211; AprcipB | This study |
| P. luminescens | ||
| Hm/1 | Strain Hm, phase I variant | G. M. Thomas |
| NC1/1 | Strain NC1, phase I variant | This study |
| NC1/2 | Strain NC1, phase II variant | This study |
| NP5-1 | NC1/1 with pSB5-1 | This study |
| NP7-3 | NC1/1 with pSB7-3 | This study |
| NP151 | NC1/1; cipB1::Ω-Km Kanr | This study |
| NP173 | NC1/1; cipA1::Ω Strr Specr | This study |
| Plasmid vectors | ||
| pGEM3Z(+) | Cloning vector; Apr, ColE1 | Promega |
| pBCSK(+) | Cloning vector; Cmr, ColE1 | Stratagene |
| pUM24 | Source of 2.6-kb PstI fragment containing sacB | 30 |
| pCA9 | pGEM3Z(+) with 10-kb genomic insert containing cipA | This study |
| pSB109 | pGEM3Z(+) with 1.4-kb EcoRI-XbaI fragment containing cipA | This study |
| pSB211 | pGEM3Z(+) with 1.1-kb DraI fragment containing cipB | This study |
| pCB11 | pGEM3Z(+) with 3.4-kb genomic insert containing cipB | This study |
| pBS101 | pBC SK(+) with 2.6-kb PstI fragment containing sacB; Cmr Sucs | This study |
| pHP45Ω | Source of interposon conferring Strr; Strr Specr | 17 |
| pHP45Ω-Km | Source of interposon conferring Kanr; Kanr | 17 |
| pSB5-1 | pBS101 with cipB1::Ω-Km; Sucs Cmr Kanr | This study |
| pSB7-3 | pBS101 with cipA1::Ω; Sucs Cmr Strr Specr | This study |
| Nematode strain H. bacteriophora NC1 | H. Kaya |
Dyes, antibiotics, and Tween detergents used in this study were purchased from Sigma Chemical Co. (St. Louis, Mo.). All culture media used in this study were purchased from Difco (Detroit, Mich.). Media used for phenotypic characterization of the cip mutants were nutrient agar supplemented with bromthymol blue and 2,3,5-triphenyltetrazolium at 25 and 40 mg/liter, respectively, blood agar (5% [vol/vol] sheep erythrocytes in Trypticase soy agar), Congo red agar (nutrient agar plus 0.01% [wt/vol] Congo red), egg yolk agar (5% [vol/vol] egg yolk in nutrient agar), EB agar (eosin Y and methylene blue at 400 and 65 mg/liter, respectively, in 2% PP3 agar), MacConkey agar, and Tween agars (0.5% [vol/vol] Tween 20, 40, 60, or 80 in nutrient agar) (35). Antibiotic medium no. 3 was used for antibiotic assays. For chrome azurol S (CAS) medium, CAS dye solution was prepared exactly as described (34) and added to 2% PP3 agar.
Transformation of E. coli and P. luminescens.
E. coli was transformed as previously described (32). P. luminescens was transformed by a modified CaCl2-RbCl2 procedure. A 50-ml sample of LB in a 250-ml Erlenmyer flask was inoculated with 1.25 ml of a 16-h culture of P. luminescens NC1/1 and grown until cells were in mid-log growth (approximately 5 h). Cells were pelleted by centrifugation (5 min, 4,000 × g, 4°C), resuspended in 50 ml of buffer A (20 mM morpholinepropanesulfonic acid [MOPS; pH 7.0], 50 mM RbCl2, 25 mM CaCl2), incubated on ice for 15 min, and pelleted by centrifugation (5 min, 4,000 × g, 4°C). Cells were then resuspended in buffer B (100 mM MOPS [pH 6.5], 25 mM RbCl2, 50 mM CaCl2). DNA, in a volume of 1 μl, was added to 100 μl of competent cells. This mixture was incubated on ice for 30 min, heat shocked at 42°C for 1 min, and placed on ice for 5 min. A 900-μl volume of LB was added, and the cells were incubated at 30°C on a rotary shaker for 1 h before plating on selective media. Transformation efficiency of P. luminescens NC1/1 competent cells obtained with this procedure was routinely 1 to 10 transformants per μg of plasmid DNA with pBCSK(+).
Genomic library construction and screening.
For cloning of cipA and cipB, a genomic library was constructed by ligation of gel-purified Sau3A partial digests of genomic DNA isolated from P. luminescens Hm/1, in the size range of 5 to 8 kb, into BamHI-cut pGEM3Z(+) and transformed into E. coli XL1-Blue MRF. Screening was performed by plating the library on LB agar with ampicillin (100 μg/ml), blotting the colonies to 0.45-μm-pore-size nitrocellulose filters as previously described (21), and performing immunodetection with polyclonal CipA or CipB antiserum (8).
Protein analyses.
Protein inclusions were purified by density centrifugation on Percoll (Sigma, St. Louis, Mo.) gradients as described previously (12). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was carried out essentially as described by Laemmli (25) under reducing conditions with either 12 or 18% polyacrylamide gels. Protein concentration was determined with a bicinchoninic acid assay kit (Pierce, Rockford, Ill.). Western blotting was performed by electrophoretically transferring proteins from polyacrylamide gels to 0.45-μm-pore-size nitrocellulose membranes with a Genie electroblotter (Idea Scientific Co., Corvallis, Oreg.). Detection of antigen was performed with the ProtoBlot II AP system (Promega, Madison, Wis.), using alkaline phosphatase-conjugated secondary antibodies.
DNA manipulations.
Isolation of chromosomal DNA was performed as previously described (6). Restriction enzymes, T4 DNA ligase, and calf intestinal alkaline phosphatase were used as directed by the supplier (Stratagene, La Jolla, Calif.). DNA fragments used for subcloning and probes were isolated from agarose gels with a Qiaex kit (Qiagen Inc., Chatworth, Calif.). DNA sequencing was carried out by using an fmol DNA sequencing kit (Promega). Southern blotting was performed as previously described (6), using an Illuminator detection system (Stratagene) under high-stringency conditions. DNA fragments used as probes in this study were the 0.6-kb SspI fragment containing cipA, the 1.1-kb DraI fragment containing cipB, the 1.8-kb BamHI fragment containing the interposon from pHP45Ω (interposon used for construction of mutant cipA allele), the 2.0-kb BamHI fragment containing the interposon from pHP45Ω-KmΩ (interposon used for construction of mutant cipB allele), and the 5.8-kb SacI fragment of pSB101 (delivery vector used for allelic exchange mutagenesis).
Nucleotide and protein sequence analysis.
Nucleotide and protein sequence analyses were performed by using the sequence analysis software package (version 8.0) of the Genetics Computer Group (GCG) (16). BLAST (Basic Local Alignment Search Tool) (5) analysis of the nonredundant protein database at the National Center for Biotechnology Information at the National Library of Medicine was used for database searches.
Construction of cipA and cipB mutants.
To construct a delivery vector for allelic exchange mutagenesis, a 2.6-kb PstI fragment containing sacB was isolated from pUM24 (30) and ligated into PstI-digested pBCSK(+). The resulting plasmid is designated pSB101. To construct a mutant cipA allele, pCA9 was digested with BclI and ligated to a BamHI-cut interposon encoding streptomycin resistance (Strr) (17). Approximately 700 bp of P. luminescens DNA flanks each end of the interposon. The resultant plasmid was cut with XbaI and SalI and ligated to similarly cut pGEM3Z+. The resultant plasmid was digested with EcoRI, and the insertionally inactivated cipA allele was removed as an EcoRI fragment which was subsequently ligated to an EcoRI partial digest of pSB101. This plasmid, designated pSB7-3, was used to construct a P. luminescens cipA null mutant via even-numbered homologous recombination. pSB7-3 was transformed into P. luminescens NC1/1, and transformants were tested for chloramphenicol resistance (Cmr), Strr, and sucrose sensitivity (Sucs). A single Cmr Strr Sucs transformant was picked and grown overnight in LB broth without any antibiotic selection to allow loss of the plasmid. The culture was serially diluted and plated onto LB agar containing streptomycin, spectinomycin, and sucrose. Strr Sucr colonies were patched onto LB agar containing chloramphenicol. Strr Sucr Cms colonies, which should have arisen via a reciprocal exchange of the mutated cipA allele for the wild-type cipA allele with subsequent loss of the vector, were identified as putative cipA null mutants. These were further verified with Western and Southern blotting.
To construct a mutant cipB allele, pCB11 was partially digested with BglII and ligated to a BamHI-cut interposon encoding kanamycin resistance (Kmr) (17). Approximately 1.4 kb of P. luminescens DNA flanks each end of the interposon. The resultant plasmid was digested with SacI, and the insertionally inactivated cipB allele was removed and subsequently ligated to SacI-digested pSB101. This plasmid, designated pSB5-1, was transformed into NC1/1, and cipB null mutants were identified as described for cipA null mutants above except that Kmr was scored instead of Strr.
Phenotypic characterization of cip mutants.
Assays used for the phenotypic characterization of the cip mutants were interpreted as follows. Bioluminescence was visually determined by examining the bacterial colonies in the dark. Extracellular lipase activity was indicated by a halo of precipitated material surrounding the colony cultured on Tween agar. Hemolytic activity was determined by a clearing surrounding the bacterial colonies cultured on blood agar. Production of compounds with siderophore-like activity was determined by the formation of an orange halo surrounding the bacterial colonies cultured on CAS agar. Production of antimicrobial compounds was assayed by overlaying chloroform-killed colonies of the test strains with M. luteus as previously described (1). Zones of growth inhibition of M. luteus were interpreted as the result of production of antimicrobial compounds by the test strain. For all assays, both phase I and phase II variants of P. luminescens were characterized on the same plates to provide positive and negative controls. Three independent clones of each test strain were analyzed on two plates for each medium assayed. All plates were cultured for 3 days at 30°C before assays were interpreted.
Biochemical traits of the test strains were determined by using API 20E strips (bioMerieux Vitek, Inc., Hazelwood, Mo.). Single bacterial colonies, resuspended in 0.85% saline, were added to the test strips and incubated for 2 days at 30°C before tests were interpreted as instructed by the supplier. All testing was performed in triplicate, using three isolated colonies of each test strain.
Insecticidal assays.
Manduca sexta eggs were purchased from Carolina Biological Supply Co. (Burlington, N.C.). Eggs were hatched, and larva were reared under a 16-h light/8-h dark photoperiod at 25°C, using a Gypsy moth wheat germ diet (ICN Biomedicals, Inc., Aurora, Ohio). For assays involving injection of whole cells, bacterial cultures grown overnight in 2% PP3 were used. One to 100 CFU, in a volume of 10 μl, was injected through the first proleg of fourth- or fifth-instar M. sexta larvae, using a 25-μl Gastight syringe (Hamilton Co., Reno, Nev.). For assays of extracellular insecticidal activity, bacterial cultures were grown in 2% PP3 broth for 48 h and pelleted by centrifugation. Supernatant fluid was filter sterilized by using a 0.2-μm-pore-size membrane filter (Schleicher & Schuell, Inc., Keene, N.H.) and concentrated 30-fold by using a 30,000-molecular-weight-cutoff filtration device (Alltech, Deerfield, Ill.). A 10-μl sample of the concentrated filtrate, containing 100 μg of protein, was injected though the first proleg of fourth- or fifth-instar M. sexta larvae, using a 25-μl Gastight syringe (Hamilton). The weights and survival of the larva were recorded at 24-h intervals for 10 days.
Nematode growth and reproduction assays.
Infective juvenile (IJ)-stage Heterorhabditis bacteriophora NC1 nematodes were maintained by passing IJs through M. sexta larva. IJs were surface sterilized as previously described (27), resuspended in 0.85% NaCl, and applied to filter paper on which second- or third-instar M. sexta larva were placed. Nematode-infected larva died within 2 to 3 days and became orange in color and bioluminescent. IJs emerged 7 to 9 days after the death of the insect.
For determination of nematode growth and reproduction on test strains, 100-μl samples of an overnight culture of the test strain was spread onto nematode growth medium (13) agar and incubated at 30°C overnight. Approximately 25 surface-sterilized IJ-stage H. bacteriophora NC1 nematodes were added to the lawns of test strains, and the plates were incubated at 25°C. Nematode cultures were observed daily for development, production of eggs, and hatching of eggs into second generation IJs, using a inverted dissecting microscope. All assays were performed in triplicate, using three independent cultures of the test strains.
Nucleotide sequence accession numbers.
The nucleotide sequence of the 1,106-bp fragment containing cipA and the 1,160-bp fragment containing cipB presented in this report have been deposited with GenBank and have the accession numbers M97630 and U89925, respectively.
RESULTS
Cloning and nucleotide sequence determination of cipA and cipB.
Polyclonal antisera, which was previously generated for CipA and CipB (8), was used to screen a genomic library to identify recombinant E. coli expressing either CipA or CipB. Of approximately 8,000 recombinants screened, 11 that expressed detectable levels of either CipA (6 recombinants) or CipB (5 recombinants) were isolated. No recombinants that contained detectable levels of both CipA and CipB were isolated.
One recombinant plasmid, designated pCA9, was isolated from an E. coli transformant that expressed CipA and contained an insert of approximately 10 kb. Subcloning of this insert DNA indicated that a 1.4-kb XbaI-EcoRI fragment of pCA9 was sufficient for expression of antigen recognized by CipA antiserum (Fig. 2). The nucleotide sequence of the 1,404-bp XbaI-EcoRI fragment of pCA9 revealed only one significant (>20 amino acid residues) open reading frame (ORF) of 312 nucleotides (nt) which was preceded by a putative ribosome binding site (-GGAG-) (36) (Fig. 3A). The deduced amino acid sequence of this ORF contained an N-terminal sequence of 20 amino acids which was identical to that of CipA (12). Further evidence that this ORF encoded CipA was the high amount of methionine (13.3%), leucine (10.5%), and lysine (10.5%) as predicted from amino acid compositional analysis of CipA. The ORF, which we have designated cipA, encodes a hypothetical protein of 104 amino acids with a molecular size (11,648 Da) which is in good agreement with that predicted for CipA by amino acid compositional and SDS-PAGE analysis.
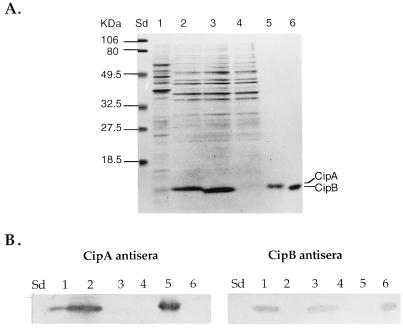
FIG. 2.
Protein and immunoblot analyses of E. coli recombinants expressing CipA and CipB. (A) SDS-PAGE analysis of whole-cell lysates and purified CipA and CipB on a 12% acrylamide gel. Lanes: Sd, molecular weight standards; 1, P. luminescens Hm/1; 2, E. coli DH5α expressing CipA (EC109); 3, E. coli DH5α expressing CipB (EC211); 4, E. coli DH5α control (EC30); 5, purified CipA; 6, purified CipB. The positions of CipA and CipB are indicated. Lanes containing cell lysates and purified inclusion proteins contained 10 and 1 μg of protein, respectively. (B) Corresponding immunoblot analysis of the same samples, using either CipA or CipB antiserum. Lanes are the same as in panel A.
FIG. 3.
(A) Partial nucleotide sequence of the 1,404-bp EcoRI-XbaI fragment of pCA9 which encodes CipA. Amino acids deduced from the nucleotide sequence are specified by standard one-letter abbreviations. The N-terminal amino acid sequence previously determined for CipA is underlined. The positions of putative promoters (−35 and −10 regions) (38), putative ribosome-binding site (36), and the BclI restriction site used for construction of the cipA mutant allele are indicated. Underlined nucleotide sequence downstream of cipA corresponds to the ERIC sequence (23). The positions of a putative stem-loop structure is marked by dashed arrows. (B) Operator-like region identified upstream of cipA. The positions of a putative promoter and ribosome-binding site are indicated. The regions of twofold symmetry are boxed.
A putative promoter with the sequence -TTCAGA--17 bp--TATTAA- was identified 36 bp upstream from the initiation codon of cipA. Overlapping the −10 region of this putative promoter is a 36-bp operator-like region consisting of an imperfect inverted repeat with a twofold axis of symmetry (Fig. 3B). Further nucleotide sequence analysis identified a region (nt 870 to 975) downstream from the termination codon of cipA that had high nucleotide sequence identity (76% over an 84-bp stretch) with an enterobacterial repetitive intergenic consensus (ERIC) sequence (23) and has the potential to form a stem-loop structure with a ΔG of −25.8 kcal/mol.
One recombinant plasmid, designated pCB11, was isolated from a E. coli transformant that expressed CipB and contained an insert of 3.4 kb. Subcloning of the insert DNA from pCB11 revealed that a 1.1-kb DraI fragment was sufficient for expression of antigen recognized by CipB antiserum (Fig. 2). Nucleotide sequence analysis of this fragment revealed only one significant complete ORF preceded by a potential ribosome-binding site (-GGAG-) (36) (Fig. 4). Residues 2 to 21 of the deduced amino acid sequence of this ORF were identical to the N-terminal sequence obtained for the CipB N-terminal peptide (8). The ORF, which we have designated cipB, encodes a hypothetical protein of 101 amino acids with a molecular size (11,308 Da) which is in good agreement with that predicted for CipB by amino acid compositional and SDS-PAGE analysis (12).
FIG. 4.
Partial nucleotide sequence of the 1,624-bp DraI-Sau3AI fragment that contains cipB. Amino acids deduced from the nucleotide sequence are indicated by standard one-letter abbreviations. The N-terminal amino acid sequence previously determined for CipB is underlined. The positions of a putative promoter (−35 and −10 regions) (38), putative ribosome-binding site (36), and the BglII site used for construction of a mutant cipB allele are indicated. The positions of potential stem-loop structures are marked by dashed arrows.
Three potential stem-loop structures were identified in the DNA flanking cipB (Fig. 4). The first is 32 bp upstream of the initiation codon of cipB (nt 185 to 232) and has a ΔG of −11.9 kcal/mol. The second potential stem-loop structure is 192 bp downstream of the termination codon of cipB (nt 760 to 795) as has a ΔG of −15.2 kcal/mol. This structure is fairly G+C rich and is characteristic of a factor-independent transcriptional terminator. The third potential stem-loop structure (nt 997 to 1130), which is analogous to the putative stem-loop structure identified downstream of cipA, is 389 bp downstream of the termination codon of cipB and has a ΔG of −27.4 kcal/mol.
Analysis of the deduced amino acid sequences of CipA and CipB.
The deduced amino acid sequences of CipA and CipB were analyzed by BLAST analysis to determine if they have similarity to any known proteins. No significant amino acid sequence similarity (>25% identity over a 30-amino-acid residue stretch) between CipA or CipB and any other protein in these databases was detected. However, between each other, the deduced amino acid sequences of CipA and CipB had 25% identity and 49% similarity over the entire lengths of the proteins (Fig. 5).
FIG. 5.
Comparison of the deduced amino acid sequences of CipA and CipB. Alignments were constructed using the program BESTFIT from the Genetics Computer Group software package. Identical residues are indicated with vertical lines, and similar residues are indicated with colons. Gaps are represented by dashes.
Selection of P. luminescens NC1 for construction of cip mutants.
The cipA and cipB genes characterized in this study were isolated from P. luminescens Hm/1. However, the unidentified Heterorhabditis sp. nematode from which P. luminescens Hm/1 was isolated has unfortunately been lost (37). We and others (4) have been unable to isolate a Heterorhabditis sp. that will form a mutualistic association with P. luminescens Hm. For this reason, it was decided to construct cip mutants in a P. luminescens strain other than Hm. P. luminescens NC1 was chosen for several reasons. First, Southern analysis using cipA- and cipB-specific probes performed under high-stringency conditions indicated a high degree of nucleotide sequence identity between the cip genes of Hm and NC1 (data not shown). Second, the mutualistic nematode strain H. bacteriophora NC1 was available and could be maintained in vitro. Third, these particular nematode and bacterial strains have been previously described in several studies (10, 11).
Isolation of cipA and cipB mutants.
Allelic exchange was used to construct cipA and cipB mutants. Five independent putative cipA or cipB mutants were characterized by Western blot and Southern blot analyses to ensure correct resolution of the allelic exchange. Western blotting of cell lysates with a polyclonal antiserum to CipA or CipB revealed detectable quantities of only one inclusion protein in the putative cipA and cipB mutants (Fig. 6). Southern blotting of genomic DNA from the these isolates with cipA-, cipB-, interposon-, and delivery vector-specific probes ensured that the mutant cip allele containing the interposon was exchanged with the wild-type cip allele, without integration of the delivery vector, and resulted in the predicted hybridization pattern (data not shown). These data indicate that all of the isolates examined resulted from gene replacement at either cipA or cipB. The cipA and cipB mutants of P. luminescens NC1 were designated NP173 and NP151, respectively.
FIG. 6.
Protein and immunoblot analyses of cipA and cipB mutants of P. luminescens NC1. (A) SDS-PAGE analysis of cell lysates on a 12% acrylamide gel. Lanes: Sd, molecular weight standards; 1, P. luminescens phase I variant (NC1/1); 2, P. luminescens NC1 phase II variant (NC1/2); 3, cipA mutant (NP173); 4, cipB mutant (NP151). Twenty micrograms of each cell lysate was loaded per lane. The positions of CipA and CipB are indicated. (B) Corresponding immunoblot analysis of the same samples, using either CipA or CipB antiserum. Lanes are the same as in panel A.
Construction of a cipAcipB double-mutant strain was attempted by the method used to construct the single cip mutants. NP151 and NP173 were transformed with pSB7-3 and pSB5-1, respectively. Isolation of mutant strains was performed exactly as described, but no double-mutant strains were isolated in three independent experiments. If the frequency of the double-mutant strains was similar to that of the single cip mutant strains, sufficient cells were plated to obtain >104 double mutants.
Phenotypic characterization of cip mutants.
The cip mutants were found to differ from phase I cells in physiological and biochemical traits that have been used to distinguish between phase I or phase II cells of P. luminescens (1, 9, 10, 22). The phenotypes of P. luminescens phase I (NC1/1), P. luminescens phase II (NC1/2), cipA mutant (NP173), and cipB mutant (NP151) in these assays are summarized in Table 2.
TABLE 2.
Phenotypes of phase I variant, phase II variant, and cip mutants of P. luminescens NC1
| Phenotype assayed | Phenotypea of strain:
|
|||
|---|---|---|---|---|
| NC1/1 (phase I variant) | NC1/2 (phase II variant) | NP151 (cipB1::Ω-Km) | NP173 (cipA1::Ω) | |
| Dye adsorption | ||||
| Neutral red | + | w+ | w+ | w+ |
| Eosin Y-methylene blue | + | − | w+ | w+ |
| Bromthymol blue | + | − | − | − |
| Congo red | + | − | − | − |
| Bioluminescence | + | − | − | − |
| Extracellular products | ||||
| Lipase activity | + | − | − | − |
| Hemolytic activity | + | − | w+ | w+ |
| Siderophore activity | + | − | w+ | w+ |
| Protease activity | + | − | − | − |
| Antibiotic production | + | − | + | w+ |
| Biochemical traits | ||||
| Indole production | + | − | − | − |
| Citrate utilization | + | − | + | + |
| Glucose utilization | − | + | + | + |
| Colony morphology on: | ||||
| 2% PP3 agar | Convex, mucoid | Flat, nonmucoid | Convex, mucoid | Convex, mucoid |
| Antibiotic medium no. 3 | Flat, rough | Flat, rough | Flat, rough | Flat, spreading |
| Pigmentation on: | ||||
| 2% PP3 agar | Yellow | Light yellow | Yellow | Yellow |
| 0.5% Tween 20 agar | Yellow | Nonpigmented | Nonpigmented | Nonpigmented |
| 0.5% Tween 60 agar | Yellow | Light yellow | Light yellow | Light yellow |
| Egg yolk agar | Yellow | Yellow | Orange | Orange |
| CAS agar | Yellow | White | White | White |
| Support of nematode growth and reproduction | + | − | − | − |
Assayed as described in Materials and Methods. +, positive; −, negative; w+, weakly positive.
The cip mutants were first assayed on agar-based media for dye adsorption, bioluminescence, and extracellular products. In many of these assays, which include adsorption of several dyes (neutral red, bromthymol blue, and Congo red), bioluminescence, and extracellular lipase activity, the cip mutants phenotypically resembled NC1/2. In the remaining assays (adsorption of eosinY-methylene blue, extracellular hemolytic activity, extracellular siderophore activity, and production of antimicrobial compounds), the cip mutants had phenotypes different from those of both NC1/1 and NC1/2.
The cip mutants were also assayed in 23 standard biochemical tests using API 20E biochemical identification strips. In 19 of these biochemical tests, no differences were observed among NC1/1, NC1/2, NP151, and NP173. Phenotypic differences among these strains were observed in four tests; citrate utilization, glucose utilization, production of indole from tryptophan, and gelatin hydrolysis. In these tests, the cip mutants phenotypically resembled either NC1/1 (positive for citrate utilization) or NC1/2 (positive for glucose utilization; negative for gelatin hydrolysis and indole production).
The colony morphology and pigmentation of NP151 and NP173 were comparable to those observed with NC1/1 on most of the culture media used. However, on some media, NP151 and/or NP173 differed in colony morphology or pigmentation from either NC1/1, NC1/2, or each other.
Effect of cip mutants on growth and reproduction of a mutualistic nematode.
Since NP151 and NP173 exhibited several phenotypic traits that differed from those of NC1/1, we wanted to determine if the cip mutants were able to provide the nutritional requirements necessary for nematode growth and reproduction. Nematode development and reproduction were detected only on IJs cultured with NC1/1 as a nutrient source. After 6 days, gravid females were observed in these cultures; second-generation IJs were observed 2 to 3 days later. Nematodes cultured on NC1/2, NP151, or NP173 did not develop into adults, and no viable nematodes were visually observed 4 days after inoculation with the IJs. In this assay, NP151 and NP173 phenotypically resembled NC1/2 and did not support nematode growth and reproduction.
Insecticidal activity of cip mutants.
Intact cells of P. luminescens are highly insecticidal when injected into the hemolymph of an insect; the 50% lethal dose for Galleria mellonella larva is 1 to 10 cells (7, 29). To determine if whole cells of the mutant strains differed in insect pathogenesis, 1 to 100 CFU of either NC1/1, NC1/2, NP151, or NP173 was injected into the hemolymph of M. sexta larvae. At this cell dose, all strains were lethal to all of the injected larva within 48 h (data not shown). Two days after larval death, the cadavers infected with NC1/1 were orange in color and bioluminescent, which are characteristics of larva infected with Photorhabdus sp. The cadavers infected with NC1/2, NP151, or NP173 were brown in color and were not noticeably bioluminescent. From this experiment, it was concluded that whole cells of the cip mutants are not grossly affected, if affected at all, in insect pathogenesis compared to NC1/1 and NC1/2.
P. luminescens was also shown to produce one or more extracellular factors with potent insecticidal activity (7, 12). To assay this, concentrated culture filtrates of NC1/1, NC1/2, NP151, and NP173 were injected into M. sexta larva. Starting at 24 h after injection, all of the larva injected with each of these filtrates began to exhibit effects associated with insecticidal activity such as discoloration, cessation of feeding, and death. Even though injected filtrates from all four strains exhibited insecticidal activity, the filtrates from NP151 and NP173 were considerably more lethal to the larva than filtrates of NC1/1 or NC1/2 (Table 3). When these culture filtrates were examined by SDS-PAGE analysis, different protein patterns were observed. The filtrates of NP151 and NP173 had protein patterns that were essentially indistinguishable from each other but were different from those of either NC1/1 or NC1/2 culture filtrates (Fig. 7).
TABLE 3.
Insecticidal activities of culture filtrates of cip mutants of P. luminescens to M. sexta larvae
| Culture filtrate of: | Cumulative % of larvae killed/daya
|
|||
|---|---|---|---|---|
| Day 1 | Day 3 | Day 5 | Day 7 | |
| NC1/1 (phase I variant) | 5 | 15 | 20 | 30 |
| NC1/2 (phase II variant) | 0 | 0 | 10 | 20 |
| NP151 (cipB1::Ω-Km) | 55 | 100 | 100 | 100 |
| NP173 (cipA1::Ω) | 35 | 75 | 95 | 100 |
| Control | 0 | 0 | 0 | 0 |
Culture filtrates of test strains were prepared as described in Materials and Methods; 10 μl of each of the culture filtrates (containing 100 μg of total protein) was injected into the hemolymph of 10 M. sexta larvae. For the control, 10 μl of 2% PP3 was used. Data are averages of two experiments.
FIG. 7.
SDS-PAGE analysis of culture filtrates on a 12% acrylamide gel. Lanes: Sd, molecular weight standards; 1, P. luminescens NC1 phase I variant (NC1/1); 2, P. luminescens NC1 phase II variant (NC1/2); 3, cipA mutant (NP173); 4, cipB mutant (NP151). Thirty micrograms of each culture filtrate was loaded per lane.
DISCUSSION
Characterization of the deduced amino acid sequences of CipA and CipB has provided three main observations. First, CipA and CipB do not have significant amino acid sequence identity with any other known protein but do have low amino acid sequence identity to each other. Second, both CipA and CipB are composed of a high percentage of hydrophobic residues such as methionine, leucine, isoleucine, and valine. Compositions of hydrophobic residues in CipA and CipB are 42.3 and 47.5%, respectively. It is possible that the high percentage of hydrophobic residues is important for properties of CipA and CipB, such as crystalline inclusion formation or interaction with other cellular components. Third, it is interesting that CipA has a higher methionine content (13.3%/mol) than most proteins (1.7%/mol) (24). However, it is possible that the high methionine content of CipA has biological relevance other than hydrophobicity. For example, the high methionine content of CipA is comparable to that of larval storage proteins (10.8%/mol), which are involved in the storage of methionine in insect larva (26). Also, one of the inclusion proteins of X. nematophilus, designated IP-1, is also relatively methionine rich (8.5%/mol) (14). These observations suggest the interesting idea that the high methionine content of CipA inclusion protein may have biological relevance, such as sulfur storage or as an indication of nutrient availability via the methionine pool.
The construction of cipA and cipB mutants via allelic exchange is, to our knowledge, the first study in which gene replacement has been described for Photorhabdus sp. Characterization of the cip mutants revealed that insertional inactivation of either cipA or cipB resulted in mutants that were altered in many different phenotypic traits such as dye adsorption, bioluminescence, production of extracellular products, biochemical traits, colony morphology and pigmentation, and interactions with a mutualistic nematode relative to the phase I cells. In general, the phenotypes of the cip mutants were more similar to each other than to those of either phase I or phase II cells.
Since the cip mutants exhibited pleiotropic phenotypes, we suggest that the cipA and cipB loci may be involved in the expression of phenotypic traits. Also, the similarity of phenotypes of the cipA and cipB mutants implies that the absence of either CipA or CipB causes similar physiological responses. The mechanism by which the loci that encode these proteins are involved in the expression of phenotypic traits is not yet clear. One hypothesis is that CipA or CipB is directly involved in the expression of these phenotypic traits, perhaps by interacting with other cellular factors, such as positive/negative effectors of gene regulation. Another hypothesis is that the involvement of the inclusion proteins is indirect. It is possible, for example, that the production of these proteins to such high concentrations induces physiological changes in the bacterial cell, such as changes in nutrient or amino acid availability, which in turn influence the regulation of these phenotypic traits. The possible involvement of CipA in the storage or sensing of methionine is consistent with this hypothesis. Another hypothesis, in which the Cip proteins are indirectly involved in the expression of phenotypic traits, is that the introduction of a polar mutation (such as an interposon) into either cip gene affects the expression of a downstream gene which results in the phenotypes observed for the cip mutants. Experiments designed to determine if the cip genes are transcribed on a monocistronic transcript will test this hypothesis.
The mechanism of phase variation in Photorhabdus spp. is unknown (for a review, see reference 18). It has been hypothesized that a form of global regulatory control is responsible for the regulation of phenotypic traits associated with phase variation. Interestingly, both posttranscriptional and posttranslational regulation are involved in the expression of genes necessary for bioluminescence and secreted lipase in the phase variation exhibited by P. luminescens. Since the cip mutants characterized in this study are altered in many phenotypic traits, most of which are associated with the phase variation of P. luminescens, it is tempting to speculate that the CipA and CipB proteins may be involved in such a form of global regulatory control. The primary objective of this study was to characterize the cip genes and to address the biological role(s) of the crystalline inclusion proteins in the mutualistic/pathogenic life cycle of P. luminescens. Our data suggest that CipA and CipB are involved in an unknown manner with the expression of phenotypic traits of P. luminescens. Further studies of the transcriptional regulation of the cip genes and elucidation of the specific role(s) of CipA and CipB in cellular regulation and physiology are continuing.
ACKNOWLEDGMENTS
We thank David Bowen, Susan Frackman, and Kenneth Nealson for assistance in cloning cipA. We also thank Timothy J. Donohue and Gary P. Roberts for helpful discussions and for critically reviewing the manuscript.
This work was supported by a U.S. Department of Agriculture Hatch grant through the College of Agriculture and Life Sciences, University of Wisconsin—Madison.
REFERENCES
- 1.Akhurst R J. Morphological and functional phase variation in Xenorhabdus spp.: bacteria symbiotically associated with the insect pathogenic nematodes Neoplectana and Heterorhabditis. J Gen Microbiol. 1980;121:303–309. [Google Scholar]
- 2.Akhurst R J. Antibiotic activity of Xenorhabdus spp.: bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J Gen Microbiol. 1982;128:3061–3065. doi: 10.1099/00221287-128-12-3061. [DOI] [PubMed] [Google Scholar]
- 3.Akhurst R J, Boemare N E. A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J Gen Microbiol. 1988;134:1835–1845. doi: 10.1099/00221287-134-7-1835. [DOI] [PubMed] [Google Scholar]
- 4.Akhurst, R. J. Personal communication.
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 7.Balcerzak M. Comparative studies on parasitism caused by entomogenous nematodes, Steinernema feltiae and Heterorhabditis bacteriophora. I. The roles of the nematode-bacterial complex, and of the associated bacteria alone, in pathogenesis. Acta Parasitol Pol. 1991;36:175–181. [Google Scholar]
- 8.Bintrim S. A study of the crystalline inclusion proteins of Photorhabdus luminescens. Ph.D. thesis. Madison, Wis: University of Wisconsin—Madison; 1995. [Google Scholar]
- 9.Bleakly B, Nealson K H. Characterization of primary and secondary phase cells of Xenorhabdus luminescens strain Hm. FEMS Microbiol Ecol. 1988;53:241–250. [Google Scholar]
- 10.Boemare N E, Akhurst R J. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. J Gen Microbiol. 1988;134:751–756. [Google Scholar]
- 11.Boemare N E, Akhurst R J, Mourant R G. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int J Syst Bacteriol. 1993;43:249–255. [Google Scholar]
- 12.Bowen D. Characterization of a high molecular weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Ph.D. thesis. Madison, Wis: University of Wisconsin—Madison; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couche G, Gregson R. Protein inclusions produced by the entomopathogenic bacterium Xenorhabdus nematophilus subsp. nematophilus. J Bacteriol. 1987;169:5279–5288. doi: 10.1128/jb.169.11.5279-5288.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couche G A, Lehbach P R, Forage R G, Cooney G C, Smith D R, Gregson R P. Occurrence of intracellular inclusions and plasmids in Xenorhabdus spp. J Gen Microbiol. 1987;133:967–973. [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 18.Forst S, Nealson K. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol Rev. 1996;60:21–43. doi: 10.1128/mr.60.1.21-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimont P A D, Steigerwalt A G, Boemare N, Hickman-Brenner F W, Deval C, Grimont F, Brenner D J. Deoxyribonucleotide acid relatedness and phenotypic study of the genus Xenorhabdus. Int J Syst Bacteriol. 1984;34:378–388. [Google Scholar]
- 21.Helfman D M, Hughes S H. Use of antibodies to screen cDNA expression libraries prepared in plasmid vectors. Methods Enzymol. 1987;152:451–457. doi: 10.1016/0076-6879(87)52053-5. [DOI] [PubMed] [Google Scholar]
- 22.Hurlbert R E, Xu J, Small C L. Colonial and cellular polymorphism in Xenorhabdus luminescens. Appl Environ Microbiol. 1989;55:1136–1143. doi: 10.1128/aem.55.5.1136-1143.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulton C, Higgins C F, Sharp P M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991;5:825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 24.Klapper M H. Frequency of occurrence of each amino acid residue in the primary structures of 207 unrelated proteins of known sequence. Biochem Biophys Res Commun. 1977;78:1018–1024. doi: 10.1016/0006-291x(77)90523-x. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Levenbook L. Insect storage proteins. In: Kerkut G A, Gilbert L I, editors. Comprehensive insect physiology, biochemistry and pharmacology. Vol. 10. Oxford, England: Pergamon Press; 1985. pp. 307–346. [Google Scholar]
- 27.Lunau S, Stoessal S, Schmidt-Peisker A J, Ehlers R-U. Establishment of monoxenic inocula for scaling up in vitro cultures of the entomopathogenic nematodes Steinernema spp. and Heterorhabditis spp. Nematologica. 1993;39:385–399. [Google Scholar]
- 28.Paul V J, Frautschy S, Fenical W, Nealson K H. Antibiotics in microbial ecology: isolation and structure assignment of several new antibacterial compounds from the insect-symbiotic bacteria Xenorhabdus spp. J Chem Ecol. 1981;7:588–597. doi: 10.1007/BF00987707. [DOI] [PubMed] [Google Scholar]
- 29.Poinar G O, Jr, Thomas G M. The nature of Achromobacter nematophilus as an insect pathogen. J Invertebr Pathol. 1967;9:510–514. [Google Scholar]
- 30.Reid J, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 31.Richardson W H, Schmidt T M, Nealson K H. Identification of an anthraquinone pigment and hydroxystilbene antibiotic from Xenorhabdus luminescens. Appl Environ Microbiol. 1988;54:1602–1605. doi: 10.1128/aem.54.6.1602-1605.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schmidt T M, Bleakley B, Nealson K H. Characterization of an extracellular protease from the insect pathogen Xenorhabdus luminescens. Appl Environ Microbiol. 1988;54:2793–2797. doi: 10.1128/aem.54.11.2793-2797.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 35.Sierra G. A simple method for the detection of lipolytic activity of microorganisms and some observations on the influence of the contact between cells and fatty acid substrates. J Microbiol Serol. 1957;23:15–22. doi: 10.1007/BF02545855. [DOI] [PubMed] [Google Scholar]
- 36.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas G M, Poinar G O. Xenorhabdus gen. nov., a genus of entomopathogenic, nematophilic bacteria of the family Enterobacteriaceae. Int J Syst Bacteriol. 1979;29:352–360. [Google Scholar]
- 38.von Hippel P, Bear D, Morgan W, McSwiggen J. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Dowds B C. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J Bacteriol. 1993;175:1665–1673. doi: 10.1128/jb.175.6.1665-1673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]