Abstract
Escherichia coli genes regulated by environmental inorganic phosphate (Pi) levels form the phosphate (Pho) regulon. This regulation requires seven proteins, whose synthesis is under autogenous control, including response regulator PhoB, its partner, histidine sensor kinase PhoR, all four components of the Pi-specific transport (Pst) system (PstA, PstB, PstC, and PstS), and a protein of unknown function called PhoU. Here we examined the effects of uncoupling PhoB synthesis and PhoR synthesis from their normal controls by placing each under the tight control of the arabinose-regulated ParaB promoter or the rhamnose-regulated PrhaB promoter. To do this, we made allele replacement plasmids that may be generally useful for construction of ParaB or PrhaB fusions and for recombination of them onto the E. coli chromosome at the araCBAD or rhaRSBAD locus, respectively. Using strains carrying such single-copy fusions, we showed that a PrhaB fusion is more tightly regulated than a ParaB fusion in that a PrhaB-phoR+ fusion but not a ParaB-phoR+ fusion shows a null phenotype in the absence of its specific inducer. Yet in the absence of induction, both ParaB-phoB+ and PrhaB-phoB+ fusions exhibit a null phenotype. These data indicate that less PhoR than PhoB is required for transcriptional activation of the Pho regulon, which is consistent with their respective modes of action. We also used these fusions to study PhoU. Previously, we had constructed strains with precise ΔphoU mutations. However, we unexpectedly found that such ΔphoU mutants have a severe growth defect (P. M. Steed and B. L. Wanner, J. Bacteriol. 175:6797–6809, 1993). They also readily give rise to compensatory mutants with lesions in phoB, phoR, or a pst gene, making their study particularly difficult. Here we found that, by using ParaB-phoB+, PrhaB-phoB+, or PrhaB-phoR+ fusions, we were able to overcome the extremely deleterious growth defect of a Pst+ ΔphoU mutant. The growth defect is apparently a consequence of high-level Pst synthesis resulting from autogenous control of PhoB and PhoR synthesis in the absence of PhoU.
The control of the Escherichia coli phosphate (Pho) regulon by environmental inorganic phosphate (Pi) levels is a paradigm of a bacterial signal transduction pathway in which occupancy of a cell surface receptor (the Pi-specific binding protein PstS) regulates gene expression in the cytoplasm (reference 29 and references therein). This signal transduction pathway requires seven proteins, all of which probably interact in a membrane-associated signaling complex. The Pi signaling proteins include (i) two members of the large family of two-component regulatory systems, namely, response regulator PhoB (a transcriptional activator) and its partner, histidine sensor kinase PhoR (itself an integral-membrane protein); (ii) four components of the ATP-binding cassette family Pi-specific transport (Pst) machinery (PstA, PstB, PstC, and PstS); and (iii) a negative regulator of unknown function called PhoU.
We proposed elsewhere that the Pi signaling response involves three processes: activation, deactivation, and inhibition (30). Accordingly, activation occurs under conditions of Pi limitation and requires both PhoB and PhoR; activation involves autophosphorylation of PhoR by ATP, phosphotransfer to PhoB, and transcriptional activation of Pho regulon promoters by phospho-PhoB (P-PhoB). Deactivation is a distinct intermediate step-down process that occurs upon a growth shift from Pi-limiting to Pi excess conditions. It is required to reestablish inhibition and leads to the dephosphorylation of P-PhoB in a process requiring PhoR and an excess of either PhoU, a Pst component(s), or both. Inhibition prevents phosphorylation of PhoB when Pi is in excess; it requires all seven Pi signaling proteins (PhoB, PhoR, PhoU, PstA, PstB, PstC, and PstS) in an “inhibition complex” that, by insulating PhoB, interferes with its phosphorylation.
In order to study how PhoU and the Pst system interact with PhoB and PhoR, we had previously constructed mutants with defined deletions of the pstSCAB-phoU operon. This led to our discovery that a ΔphoU (unlike a phoU missense) mutation causes a severe growth defect, resulting from an apparent Pi sensitivity phenotype (26). Growth inhibition by Pi has also been seen in mutants of the Pst transporter in which the Pi transport channel is permanently “switched on” (open), although in that case the growth defect is much less severe (34). The deleterious growth phenotype resulting from a ΔphoU mutation also leads to the accumulation of compensatory mutants with lesions in phoB, phoR, or a pst gene under normal growth conditions (26). Indeed, the poor growth of a ΔphoU mutant was apparently responsible for another laboratory concluding incorrectly that a ΔphoU mutation abolishes Pi uptake (19), as their “ΔphoU mutant” carries also a linked pst mutation (29). On the contrary, we proved that a ΔphoU mutation has no effect on Pi uptake (26).
We have now found ways to overcome the difficulties of studying phoU and (presumably) open-channel pst mutations as well. The growth defect due to a ΔphoU mutation is apparent only when the Pst system is synthesized at a high level, which in turn requires increased amounts of PhoB and PhoR, whose synthesis is autogenously controlled (10). We uncoupled phoB expression and phoR expression from their normal controls by placing them behind the foreign, arabinose-regulated ParaB or rhamnose-regulated PrhaB promoter. Upon induction of the corresponding ΔphoB or ΔphoR mutant with arabinose or rhamnose, such strains show nearly normal Pi control of the Pho regulon. However, the fold induction is lowered. Importantly, a ΔphoU mutation has no deleterious effect on growth of these strains, even in the presence of the respective inducer. Our methods for constructing and characterizing ParaB and PrhaB fusions may also be generally useful.
MATERIALS AND METHODS
Media and culture conditions.
Luria-Bertani broth, tryptone-yeast extract, and M63 were routinely used as complex and minimal media. These were prepared as described elsewhere (28). To maintain plasmids, antibiotics (Sigma, St. Louis, Mo.) were added as follows: ampicillin at 100 μg/ml, gentamicin at 15 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 12.5 μg/ml. Recombinants with a single-copy plasmid were selected with gentamicin at 4 μg/ml or kanamycin at 10 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Bachem, Torrance, Calif.) was used at 40 μg/ml to detect β-galactosidase. Tetracycline-sensitive (Tets) cells were selected on tetracycline-sensitive-selective (TSS) agar as described elsewhere (17). MacConkey agar (Difco, Detroit, Mich.) containing 1% l-arabinose, lactose, or l-rhamnose (Sigma) was used to test for the use of these carbon sources. Ara− and Rha− strains were verified by their inability to grow on M63 media containing 0.2% l-arabinose or l-rhamnose, respectively. Ara− and Rha− recombinants are distinguishable from arabinose- or rhamnose-sensitive ones by the inability of the latter to grow on MacConkey or glycerol agar in the presence of arabinose or rhamnose, respectively. In contrast, Ara− and Rha− recombinants are insensitive to these carbohydrates.
MOPS (morpholinepropanesulfonic acid) media containing different carbon sources were used to study gene regulation in ParaB, PrhaB, and PrhaS fusion strains. Cells were grown on MOPS agar containing the same carbon source at 0.2% for d-glucose, d-fructose, and d-mannitol or 0.4% for glycerol, but without an inducer. Fresh isolated colonies (after less than 20 h of growth) were used to inoculate 0.06% glucose–, fructose–, or mannitol–MOPS medium or 0.1% glycerol–MOPS medium without or with an inducer. Cultures were incubated at 37°C for 16 to 24 h prior to the assay. Such carbon-limited cultures yield highly reproducible values that are qualitatively similar to ones obtained for logarithmic-growth phase cultures (27). l-Arabinose, l-rhamnose, and isopropyl-β-d-galactopyranoside (IPTG; Sigma) were used at 1.3, 1.1, and 0.2 mM, respectively, for induction.
Bacteria.
All strains assayed are described in Table 1. Others included BT333 (endA::tetAR; from W. Wackernagel [4]), BW5045 (srlC300::Tn10; [18]), BW8078 (λrecA+ recA1; [28]), BW21391 (leu-63::Tn10; [12]), BW22773 (lacIq rrnBT14 ΔlacZWJ16 proC::Tn5-132; [14]), CA10 (galU95; from M. Berlyn), JW383 (metF159 zii-510::Tn10 thi-1; from M. Berlyn), W3110trpB114 (trpB114::Tn10; from C. Yanofsky), and ZK1001 (cysC95::Tn10 rpoS::kan; from R. Kolter). Only relevant markers are given in parentheses.
TABLE 1.
Bacterial strains
| Straina | Genotype | Pedigreeb | Source or reference(s) |
|---|---|---|---|
| BW17142 | DE3(lac)X74 ΔphoU559::kan rpoS(Am) | BD792 via BW13711 | 26 |
| BW17335 | DE3(lac)X74 Δ(pstSCAB-phoU)560::kan rpoS(Am) | BD792 via BW13711 | 26 |
| BW18897 | DE3(lac)X74 ΔphoU559 rpoS(Am) phn(EcoB) | BD792 via BW18834 | Ilv+ with P1 on BW18833 (26) |
| BW21578 | rrnBT14 ΔlacZWJ16 | BD792 via BW21342 | 15 |
| BW22831 | lacIqrrnBT14ParaB-lacZAH31 ΔaraBADAH33rpoS(Am) | BD792 via BW22766 | Leu+ Ara− with P1 on BW22826c (12) |
| BW22861 | lacIqrrnBT14PphnC-lacZWJ19 ΔphoR574 ΔcreBCD153 Δ(pta ackA hisQ hisP)TA3516phn(EcoB) ΔaraBADAH33::ParaB-phoR[M1-D431]AH35rpoS(Am) | BD792 via BW22829 | Leu+ Ara− with P1 on BW22835 |
| BW22886 | rrnBT14PrhaB-lacZLD68 ΔrhaBADLD78 | BD792 via BW22779 | Met+ Rha− with P1 on BW22875d |
| BW22887 | rrnBT14PrhaB-lacZLD68 ΔrhaBADLD78rpoS(Am) | BD792 via BW22780 | Met+ Rha− with P1 on BW22875 |
| BW22888 | rrnBT14PrhaS-lacZLD69 ΔrhaBADLD78 | BD792 via BW22781 | Met+ Rha− with P1 on BW22875 |
| BW23473 | Δlac-169 rpoS(Am) robA1 creC510 hsdR514 uidA(ΔMluI)::pir+endABT333recA1 | BD792 via BW23438 | 12, 13 |
| BW24249 | lacIqrrnBT14 ΔlacZWJ16 ΔphoBR580 ΔcreABCD154 hsdR514 Δ(pta ackA hisQ hisP)TA3516phn(EcoB) ΔaraBADAH33 ΔrhaBADLD78uidA(ΔMluI)::pir+rposS(Am) endABT333galU95 recA1 | BD792 via BW24217 | Srl+recA with P1 on BW8078 (28) |
| BW24486 | lacIqrrnBT14PphnC-lacZWJ19 ΔphoBR580 ΔcreBCD153 Δ(pta ackA hisQ hisP)TA3516phn(EcoB) ΔaraBADAH33::ParaB-phoR[M1-D431]AH35rpoS(Am) Δ(pstSCAB-phoU)560::Ω | BD792 via BW23424 | Spcr and Strr with P1 on BW17636 (26) |
| BW24508 | Like BW24486 except attλ::pSK58(PphoB-phoB+) | BD792 via BW24486 | pSK58 integrant |
| BW24962 | lacIqrrnBT14 ΔlacZWJ16 ΔphoB578 ΔcreABDC154 rpoS(Am) ΔaraBADAH33 ΔrhaBADLD78 Δ(ackA pta)160AH25 | BD792 via BW24651 | Pro+ with P1 on BW23562 (13) |
| BW24739 | lacIqrrnBT14 ΔlacZWJ16 ΔcreABDC154 rpoS(Am) ΔaraBADAH33 ΔrhaBADLD78 Δ(ackA pta)160AH25 Δ(pstSCAB-phoR)560::Ω | BD792 via BW24713 | Ilv+ with P1 on BW17646 (26) |
| BW24740 | lacIqrrnBT14 ΔlacZWJ16 ΔphoB574 ΔcreABDC154 rpoS(Am) ΔaraBADAH33 ΔrhaBADLD78 Δ(ackA pta)160AH25 Δ(pstSCAB-phoU)560::Ω | BD792 via BW24715 | Ilv+ with P1 on BW17646 (26) |
| BW24741 | lacIqrrnBT14 ΔlacZWJ16 ΔphoB578 ΔcreABDC154 rpoS(Am) ΔaraBADAH33 ΔrhaBADLD78 Δ(ackA pta)160AH25 Δ(pstSCAB-phoU)560::Ω | BD792 via BW24717 | Ilv+ with P1 on BW17646 (26) |
| BW24768 | Like BW24740 except attHK022::pAH151(PrhaB-phoR+) | BD792 via BW24740 | pAH151 integrant |
| BW24770 | Like BW24741 except attλ::pAH85(PphoB-phoB+) | BD792 via BW24741 | pAH85 integrant |
| BW24773 | Like BW24741 except attλ::pAH136(PrhaB-phoB+) | BD792 via BW24741 | pAH136 integrant |
| BW24774 | Like BW24741 except attλ::pAH150(ParaB-phoB+) | BD792 via BW24741 | pAH150 integrant |
| BW24775 | Like BW24741 except attλ::pSK49(Ptac-phoB+) | BD792 via BW24741 | pSK49 integrant |
| BW24777 | lacIqrrnBT14 ΔlacZWJ16 ΔphoB578 ΔcreABCD154 rpoS(Am) ΔaraBADAH33 Δ(ackA pta)160AH25 Δ(pstSCAB-phoU)560::Ω ΔrhaBADLD78::PrhaB-phoBLD79 | BD792 via BW24754 | Met+ Rha− with P1 on BW22876 |
| BW24803 | Like BW24692 except attλ::pAH150(ParaB-phoB+) | BD792 via BW24692 | pAH150 integrant |
| BW24858 | Like BW24740 except attHK022::pAH152(PrhaS-phoR+) | BD792 via BW24740 | pAH152 integrant |
| BW24936 | lacIqrrnBT14 ΔlacZWJ16 ΔphoR574 ΔcreABCD154 rpoS(Am) ΔaraBADAH33 ΔrhaBADLD78 Δ(ackA pta)160AH25 ΔphoU559 | BD792 via BW24714 | Ilv+ Spcs and Strs with P1 on BW24627 |
| BW24937 | lacIqrrnBT14 ΔlacZWJ16 ΔphoB578 ΔcreABCD154 rpoS(Am) ΔaraBADAH33 ΔrhaBADLD78 Δ(ackA pta)160AH25 ΔphoU559 | BD792 via BW24716 | Ilv+ Spcs and Strs with P1 on BW24627 |
| BW24948 | Like BW24936 except attHK022::pAH151(PrhaB-phoR+) | BD792 via BW24936 | pAH151 integrant |
| BW24949 | Like BW24937 except attλ::pAH136(PrhaB-phoB+) | BD792 via BW24937 | pAH136 integrant |
| BW24950 | Like BW24937 except attλ::pAH150(ParaB-phoB+) | BD792 via BW24937 | pAH150 integrant |
All are derivatives of E. coli K-12, except that the phn(EcoB) locus is from E. coli B (31).
Pedigree gives the parental strain from another laboratory and its most immediate ancestor in this laboratory. Many of our strains are descendents of E. coli K-12 strain BD792 (CGSC6159), which, like MG1655 (CGSC6300), is an unmutagenized and direct descendent of E. coli W1485 (CGSC5024) (1, 2).
The ara and leu-63::Tn10 markers are about 55% linked in P1kc crosses (data not shown).
The relevant gene order is zii-510::Tn10 metF159 ΔrhaBADLD78 in which Tn10 and metF are ca. 70% linked and metF and rha are ca. 10% linked (data not shown).
Plasmids and phage.
pAH85, pSK49, and pSK58 were constructed in an earlier study; each has the same backbone as pSK50ΔuidA2 (13). pAH85 and pSK58 are similar, except that the former has a silent SpeI site at codon 113 of phoB. Each expresses phoB from its native promoter. pSK49 expresses phoB from Ptac. pAH136 and pAH150 are derivatives of pAH85 and pSK49 carrying PrhaB and ParaB in place of PphoB and the lacIq-Ptac region, respectively; pAH151 and pAH152 are similar plasmids carrying PrhaB-phoR+ and PrhaS-phoR+ except that they have the attachment site of HK022 and encode gentamicin resistance (14). pBAD32 and pBAD33 (11) were from L.-M. Guzman. They are similar except that pBAD32 has an undefined deletion of ca. 0.6 kbp between the polylinker and the cat gene of pBAD33 (data not shown). pBC35 is a derivative of pBGS19+ (25) carrying the phoBR operon within a 4.7-kbp PstI-to-EcoRI fragment (32) (Fig. 1). pSK47 (16) has the same phoBR fragment cloned into the backbone of pSK50ΔuidA2 (13). pSLF13 and pSLF22 were constructed in an earlier study (8). pSLF13 is a derivative of pSPORT1 (Gibco-BRL, Bethesda, Md.) that encodes a segment of VanS within a 0.4-kbp EcoRI-to-BamHI insert that has a start codon overlapping an NdeI site immediately downstream of a synthetic ribosome-binding site. pSLF22 is a derivative of pBAD32 containing a similar insert. pWJ17 and pWJ18 are derivatives of the lacZ transcriptional fusion vector pWJ13 containing a kanamycin resistance cassette as a PstI fragment, whose loss facilitates recognizing promoter-lacZ fusion plasmids as kanamycin-sensitive (Kans) derivatives (15). Fusions made with these plasmids were recombined onto the chromosome by allele replacement as described below.
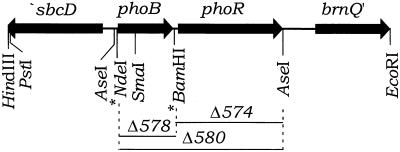
FIG. 1.
Structure of the phoBR chromosomal region. The 4.7-kbp PstI-to-EcoRI fragment in pBC35 and pSK47 is shown. Asterisks mark the NdeI and BamHI sites that were previously introduced by site-directed mutagenesis upstream and downstream, respectively, of the phoB coding region. The ΔphoB578 mutation was made in pLD82 (Table 2) and recombined onto the chromosome by allele replacement as described in the text. The construction of ΔphoR574 and ΔphoBR580 is described elsewhere (12). Arrows show gene orientations.
All plasmids constructed in this study are described in Table 2. Many have the γ replication origin of R6K, denoted oriRR6Kγ, which requires the Π protein (encoded by pir) for replication as a plasmid. Many also contain tetAR, which can be deleterious when present at high copy number. Therefore, these plasmids were routinely maintained in BW23473 or BW24249 or in similar pir+ hosts (17). λRZ5lacP-phoU+ (PS15) has been described previously (26). Generalized transduction was carried out with P1kc from a laboratory stock.
TABLE 2.
Plasmid constructions
| Plasmid | Descriptiona | Constructionb |
|---|---|---|
| pAH16 | Plac-phoR+bla oriRMB1lacI+ | Replaced 0.4-kbp EcoRI-to-BamHI vanS fragment of pSLF13 with similar 1.3-kbp phoR+ fragment generated by using P109 and P110 with pBC35 as the template |
| pAH21c | ParaB-phoR+cat oriR15AaraC+ | Replaced 0.4-kbp NdeI-to-SacI vanS fragment of pSLF22 with similar 1.3-kbp phoR+ fragment from pAH16 |
| pAH31 | ParaB-lacZ tetAR bla oriRR6Kγ | Replaced 0.5-kbp XhoI-to-PstI kan fragment of pWJ18 with 0.5-kbp ParaB PCR fragment generated by using P125 and P126 with pBAD33 as the template |
| pAH33d | ParaB-′araD′ tetAR bla oriRR6Kγ | Replaced 3.0-kbp SalI-to-BsiWI lacZ fragment of pAH31 with 0.6-kbp ′araD′ PCR fragment generated by using P127 and P128 with BW13711 chromosomal DNA as the template |
| pAH35 | ParaB-phoR+-′araD′ tetAR bla oriRR6Kγ | Subcloned 1.4-kbp BamHI phoR+ fragment from pAH21 into pAH33 |
| pAH54 | Like pAH33 except lacking NdeI2 | Digested pAH33 with BsiWI and partially with NdeI followed by filling-in with T4 DNA polymerase and religation |
| pLD68 | PrhaB-lacZ tetAR bla oriRR6Kγ | Replaced 1.3-kbp XhoI-to-SalI kan fragment of pWJ18 with similar 0.3-kbp PrhaSB fragment generated by using P119 and P120 with pLD1 (3) as the template |
| pLD69 | PrhaS-lacZ tetAR bla oriRR6Kγ | Same as pLD68, except fragment is in opposite orientation |
| pLD70 | ′rhaD tetAR bla oriRR6Kγ | Replaced 3.0-kbp BamHI-to-BsiWI lacZ fragment of pWJ17 with 0.6-kbp ′rhaD fragment generated by using P121 and P122 with BW13711 chromosomal DNA as the template |
| pLD71 | PrhaB-′rhaD tetAR bla oriRR6Kγ | Subcloned 0.7-kbp XhoI-to-BamHI PrhaB fragment from pLD68 into pLD70 |
| pLD77 | Like pWJ17 except lacking NdeI2 | Digested pWJ17 partially with NdeI followed by filling-in with T4 DNA polymerase and religation |
| pLD78 | Like PLD71 except lacking NdeI2 | Replaced 4.3-kbp XhoI-to-BsiWI lacZ fragment of pLD77 with similar 0.8-kbp PrhaB-′rhaD fragment from pLD71 |
| pLD79 | PrhaB-phoB+-′rhaD tetAR bla oriRR6Kγ | Subcloned 0.7-kbp NdeI-to-BamHI phoB+ fragment from pSK2 (9) into pLD78 digested with BamHI and partially with NdeI |
| pLD82 | ΔphoB578 kan oriRR6Kγ | Digested pSK47 with NdeI and BamHI followed by filling-in with T4 DNA polymerase and religation |
| pLD83 | ΔphoB578 tetAR bla oriRR6Kγ | Subcloned 3.4-kbp NcoI fragment from pLD82 that was made blunt with T4 DNA polymerase into SmaI-cut pLD55 (17) |
The replication origins of pMB1 and p15A are denoted oriRMB1 and oriR15A, respectively.
Additional information is given in the text. Oligonucleotide primers are shown in Table 3.
pAH21 has a unique NdeI site at the phoR start codon and unique downstream SacI, SphI, PstI, and HindIII sites.
Restriction analysis showed that the SphI and BglII sites in P127 were absent from the cloned fragment.
Molecular biology methods.
PCR amplifications of ara, rha, and phoR sequences were carried out with Vent DNA polymerase (New England Biolabs, Beverly, Mass.) and oligonucleotide primers (Table 3; IDT Inc., Coralville, Iowa). Other enzymes were from New England Biolabs or Promega (Madison, Wis.). QIAGEN (Hilden, Germany) products were used for isolation of plasmid DNA, extraction of DNA fragments from agarose gels, and purification of PCR fragments. The phoR, ParaB, and PrhaSB fragments were sequenced on both strands at the Dana Farber Cancer Institute Molecular Biology Core Facility (Harvard Medical School, Boston, Mass.). The ‵araD′ and ‵rhaD fragments (the prime indicates that a gene portion is missing on the side of the prime) were verified only by restriction enzyme analysis.
TABLE 3.
Oligonucleotide primers
| Name | Description | Sequencea |
|---|---|---|
| P109 | phoR N terminus | 5′-GGAATTCGTCATATGCTGGAACGGC-3′ |
| P110 | phoR C terminus | 5′-CGGGATCCGCTGAGCTCAGTCGCTGTTTTTGGC-3′ |
| P119 | PrhaSB upstream | 5′-CCGCTCGAGGCCATGGTGGCCTCCTGATGTCG-3′ |
| P120 | PrhaSB downstream | 5′-CCGGTCGACATATGGTGATCCTGCTGAATTTC-3′ |
| P121 | rhaD′ upstream | 5′-CGGGATCCGCATGCGTCGACAGCGACGG-3′ |
| P122 | rhaD′ downstream | 5′-CGTACGTACGCAGCGCCAGCGCAC-3′ |
| P125 | ParaB downstream | 5′-GCGCGCTGCAGGTATGGAGAAACAGTAGAG-3′ |
| P126 | ParaB upstream | 5′-GGTCCTCGAGCGAATGGTGAGATTG-3′ |
| P127 | ′araD′ upstream | 5′-GACACGTCGACGCATGCAGATCTCAAACGCCAG-3′ |
| P128 | ′araD′ downstream | 5′-CGTACGTACGACCTCTTCCAGCAC-3′ |
Underlined bases are complementary to the template(s). Those in boldface indicate restriction enzyme sites.
Molecular genetics.
Many new strains were constructed by generalized P1 transduction as described elsewhere (28). BW23321 (Δlac-169 hsdR514 uidA(ΔMluI)::pir+ endABT333) was made by transduction of BW21116 (17) to tetracycline resistance (Tetr) with P1 grown on BT333 followed by transformation of resultant transductant BW23298 with FLP expression plasmid pCP20 and excision of the tetAR genes as described elsewhere (4). The notation endABT333 signifies the resultant allele. In order to construct strains carrying a minimum number of antibiotic resistance markers, new mutations were usually introduced in two steps. In the first step, a linked auxotrophic marker was introduced by selecting Tetr transductants. These were then made prototrophic with P1 grown on an appropriate donor; the resultant transductants were scored for loss of antibiotic resistance and other relevant phenotypes. Several new mutations and fusions were constructed on plasmids as described above and then recombined onto the chromosome by allele replacement as described elsewhere (17). The resultant chromosomal allele is often given a designation corresponding to the plasmid used in its construction. As necessary, transductants carrying a ΔphoU mutation were verified by complementation with λRZ5lacP-phoU+ upon introduction of phoB+ or phoR+ or by backcross of the ΔphoU mutation into an appropriate recipient.
The ΔphoB578 mutation (Fig. 1) was recombined onto the chromosome of BW21016 [DE3(lac)X74] by using pLD83 to make BW22901. The PrhaB-lacZLD68 and PrhaS-lacZLD69 fusions were recombined onto the chromosome of BW21578 (rrnBT14 ΔlacZWJ16) by using pLD68 and pLD69 to make BW22716 and BW22721, respectively. The ParaB-lacZAH31 fusion was recombined onto the chromosome of BW21480 (lacIq rrnBT14 ΔlacZWJ16) by using pAH31 to make BW22746. The desired lacZ fusion recombinants were recognized as ones showing a rhamnose- or arabinose-dependent Lac+ phenotype on X-Gal agar. These fusions have the following on the chromosome at the lac locus in a counterclockwise orientation: lacI, four tandem copies of the rrnB transcriptional terminator (denoted rrnBT14), and the foreign promoter preceding the lacZYA operon. The construction of these and similar strains with lacIq rrnBT14 PphnC-lacZWJ19, lacIq rrnBT14 ΔlacZWJ16, rrnBT14 ΔlacZWJ16, and other promoter-lacZ fusions is unpublished (15). Site-specific recombination of oriRR6Kγ attP plasmids onto the chromosome has been described previously (13). All integrants were verified by PCR as described elsewhere (12).
The ParaB and PrhaB fusions were recombined onto the chromosome by using derivatives of new allele replacement plasmids pAH33, pAH54, and pLD78 (Fig. 2). This was done with an Ara+ or Rha+ parental strain, so that the resulting segregants with an allele replacement were recognizable as Ara− or Rha− recombinants, respectively. Because these plasmids contain a segment of araD or rhaD, most integrants formed by the initial recombination event are AraD− or RhaD−. Such recombinants are arabinose or rhamnose sensitive, respectively. All selections are therefore carried out in the absence of arabinose or rhamnose.
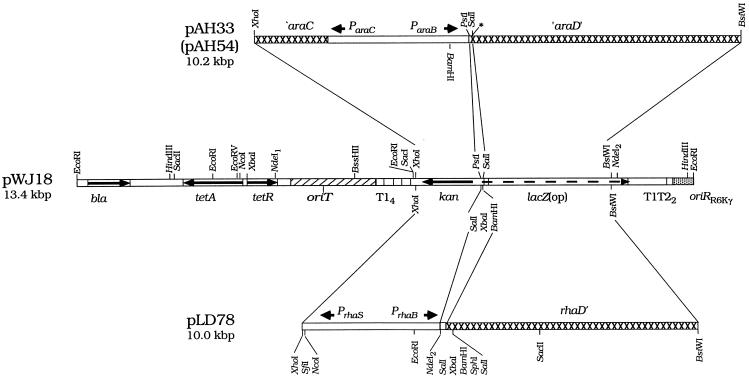
FIG. 2.
Structures of the ParaB and PrhaB allele replacement plasmids. Arrows show gene and promoter orientations. Open boxes in pAH33, pAH54, and pLD78 are promoter regions; hatched boxes are coding regions. All sites are shown for the enzymes indicated. NdeI sites are subscripted to correspond to those indicated in Table 2. pAH33 and pAH54 contain unique PstI and SalI sites downstream of ParaB for construction of a ParaB fusion. Genes cloned into these sites require also a ribosome binding site. pLD78 has SalI, XbaI, BamHI, and SphI sites downstream of PrhaB for construction of a PrhaB fusion. pLD78 has the native rhaB ribosome-binding site upstream of the NdeI2 site, which corresponds to the normal met start codon of rhaB. Therefore, genes cloned into this site do not require a ribosome-binding site, although partial digestions are required to use this site. An asterisk marks SphI and BglII sites that were lost during cloning of the ‵araD′ fragment. Arrows show gene and promoter orientations. bla, β-lactamase gene; tetA and tetR, tetracycline resistance and repressor genes, respectively; oriT, origin of transfer from RP4; T14 and T1T22, transcription terminators; kan, kanamycin resistance gene; lacZ (op), promoterless lacZ gene for construction of transcriptional, i.e., operon (op), fusions (17). See text.
The ΔaraBADAH33 mutation was recombined onto the chromosome of BW13711 [DE3(lac)X74] by using pAH33. Integrants carrying pAH33 were selected as Tetr transformants; they were purified once nonselectively, after which Tets segregants were selected on TSS agar. One showing the expected Ara− phenotype was verified and named BW22826. Similarly, the ParaB-phoR[M1-D431]AH35 fusion was recombined onto the chromosome of BW21555 (ΔphoR574) by using pAH35 to make BW22835. Recombination of the ParaB-phoR[M1-D431]AH35 fusion onto the chromosome results also in removal of the same sequences eliminated by the ΔaraBADAH33 mutation. Hence, the resultant allele is denoted ΔaraBADAH33::ParaB-phoR[M1-D431]AH35. The ΔrhaBADLD78 mutation and ΔrhaBADLD78::PrhaB-phoBLD79 fusion were recombined onto the chromosome of BW22860 [Δ(phoBR brnQ)525 Δcya-161] by using pLD78 and pLD79 to make BW22875 and BW22876, respectively. The desired recombinants were recognized as Rha− ones.
Enzyme assays.
β-Galactosidase and bacterial alkaline phosphatase (BAP) assays were carried out as described elsewhere (28).
RESULTS
Construction of allele replacement vectors for tightly regulated protein synthesis from arabinose- and rhamnose-regulated promoters.
Since expression of the phoBR operon is subject to positive autogenous control (10), we considered that it would be advantageous to uncouple PhoB synthesis and PhoR synthesis from their normal controls for new studies on the Pho regulon. We therefore constructed a derivative of ParaB plasmid pBAD32 (11) carrying a ParaB-phoR+ fusion (pAH21; Table 2). However, in preliminary studies, we found that substantial amounts of PhoR were apparently made in the absence of inducer. A ΔphoR mutant carrying pAH21 synthesized BAP upon Pi limitation in the absence of arabinose even under conditions of catabolite repression due to glucose (data not shown). No doubt the “leakiness” of ParaB under these conditions reflects the small amount of PhoR required for activation (phosphorylation) of PhoB. To further reduce PhoR synthesis, we assembled an allele replacement, “suicide” vector system to recombine the ParaB-phoR+ fusion onto the chromosome in single copy at the ara locus. We also constructed an analogous allele replacement vector carrying the rhamnose-regulated PrhaB promoter (6) for construction and recombination of a PrhaB-phoB+ fusion onto the chromosome in single copy at the rha locus.
Our vectors for constructing chromosomal ParaB and PrhaB fusions are illustrated in Fig. 2. pAH33 and pAH54 are useful for making a ParaB fusion; pLD78 is useful for making a PrhaB fusion. These plasmids contain sequences of the araCBAD or rhaRSBAD locus flanking cloning sites for construction of the respective promoter fusion. pAH33 and pAH54 have a 0.5-kbp fragment containing ParaB and a 0.6-kbp fragment of araD upstream and downstream of the cloning region, respectively. They differ in that pAH33 has two NdeI sites (one in tetR and the other in a segment of lacZ) and pAH54 has only one NdeI site (in tetR; Table 2). Derivatives of the latter have facilitated the use of NdeI in particular plasmid constructions (data not shown). pLD78 has a 0.3-kbp fragment containing PrhaB and a 0.6-kbp fragment of rhaD upstream and downstream of its cloning region, respectively. The flanking upstream and downstream sequences provide homologous regions for recombining the fusions onto the chromosome by allele replacement.
pAH33, pAH54, and pLD78 are derivatives of pWJ18 (or pWJ17; Fig. 2), which is in turn a derivative of pir-dependent, counterselectable (tetAR) allele replacement vector pLD53 (17). Upon introduction of these plasmids into a normal (non-pir) Ara+ host (by transformation, electroporation, or conjugation), they cannot replicate and therefore integrate into the chromosome via homologous recombination. Figure 3 shows the integration of ParaB-phoR+ plasmid pAH35 (Table 2) at the araCBAD locus. Integrants are selectable as Tetr colonies. A subsequent recombination event occurring in the absence of selection leads to the loss of plasmid sequences; this event results in restoration of the parental (wild-type) chromosomal structure or an allele replacement. Recombinants that have lost the plasmid backbone are selectable as Tets ones. Those carrying the desired chromosomal ParaB or PrhaB fusion have the fused gene in single copy at the araCBAD or rhaRSCAB locus in place of araBAD or rhaBAD sequences (Fig. 4); they are therefore recognizable as Ara− or Rha− ones, respectively. Ones that are also antibiotic sensitive are then verified by genetic linkage or PCR tests. Because the appropriate segregants are Ara− or Rha−, the recombinants provide the additional advantage of not being able to catabolize the respective inducer.
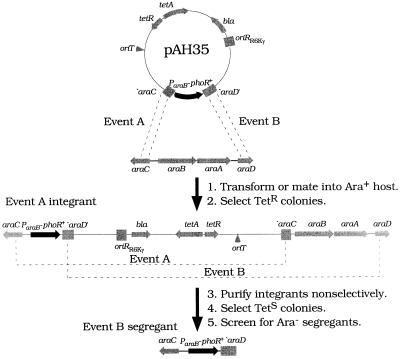
FIG. 3.
Recombination of a ParaB-phoR+ fusion onto the chromosome at the araCBAD locus. The construction of pAH35 is described in Table 2. pAH35 can integrate into the chromosome via either of two homologous recombination events (event A or event B). Only an event A integrant is shown for simplicity. Subsequent recombination events result in loss of the plasmid, regenerating the parental strain (event A) or an allele replacement (event B segregant). These are selectable on TSS agar; ones with an allele replacement are recognizable as Ara− colonies (see Materials and Methods). A ΔphoR mutant was used to recombine the ParaB-phoR+ fusion onto the chromosome to avoid recombination at the phoBR locus; a wild-type strain can also be used. A black arrow shows the orientation of phoR. Grey arrows show the orientations of other genes. Grey boxes show araC or araD gene fragments and the pir-dependent vector origin (oriRR6Kγ). A prime indicates that a gene portion is missing on the side of the prime. An arrowhead marks the nick site of oriT, the RP4 conjugative transfer origin.
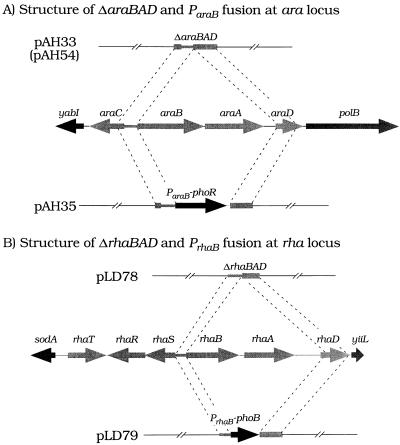
FIG. 4.
Allele replacements at the chromosomal araCBAD and rhaRSBAD regions. (A) Structure of the ΔaraBAD mutant and a ParaB-phoR+ fusion at the araCBAD locus. (B) Structure of the ΔrhaBAD mutant and a PrhaB-phoB+ fusion at the rhaRSBAD locus. Black arrows represent ParaB-phoR and PrhaB-phoB fusions. Light grey arrows signify genes belonging to the ara and rha gene clusters. Dark grey arrows show the yabI and polB genes flanking the ara locus and the sodA and yiiL genes flanking the rha locus. Dotted lines indicate regions where homologous recombination between the plasmid and the chromosome can occur. See text.
Measurements of arabinose and rhamnose regulation of single-copy promoter-lacZ fusions.
To judge the regulatory capabilities of the arabinose- and rhamnose-regulated promoters in this system, we constructed ParaB-, PrhaB-, and PrhaS-lacZ fusions and recombined them onto the chromosome in single copy at the lac locus (Materials and Methods). We then examined lacZ expression by measuring β-galactosidase levels when strains were grown in the presence and absence of the respective inducer on different carbon sources. Different carbon sources were used because these promoters are subject to catabolite repression. The results are shown in Table 4. In brief, arabinose or rhamnose led to induction of >100-fold to several thousandfold, depending upon the promoter, inducer, and carbon source. As expected, the lowest expression levels were obtained during growth on glucose, for which catabolite repression is the strongest; intermediate expression levels were obtained during growth on fructose, for which catabolite repression is less severe; the highest expression levels were obtained during growth on glycerol, for which catabolite repression is the weakest (7). Differences in both the basal (uninduced) and induced levels exist. In the absence of inducer, the basal level was about 10-fold higher for ParaB than for PrhaB or PrhaS. The induced levels were similar for ParaB and PrhaB (compare BW22831 with BW22887), while the induced levels for PrhaS were ca. 25% of those for PrhaB (compare BW22886 with BW22888).
TABLE 4.
Regulation of ParaB-, PrhaB-, and PrhaS-lacZ fusions
| Strain | Relevant genotypea | Carbon source | β-Galactosidase sp act onb:
|
Fold induction | |
|---|---|---|---|---|---|
| −Ind | +Ind | ||||
| BW22831 | ParaB-lacZ rpoS(Am) | d-Glucose | 6.6 ± 0.1 | 1,270 ± 114 | 190 |
| d-Fructose | 6.7 ± 0.6 | 3,861 ± 313 | 580 | ||
| Glycerol | 6.5 ± 0.4 | 3,853 ± 119 | 590 | ||
| BW22887 | PrhaB-lacZ rpoS(Am) | d-Glucose | 0.5 ± 0.0 | 743 ± 44 | 1,600 |
| d-Fructose | 0.5 ± 0.0 | 3,300 ± 67 | 6,900 | ||
| Glycerol | 0.5 ± 0.0 | 3,689 ± 72 | 7,800 | ||
| BW22886 | PrhaB-lacZ rpoS+ | d-Glucose | 0.4 ± 0.0 | 185 ± 10 | 430 |
| d-Fructose | 0.4 ± 0.0 | 1,158 ± 33 | 2,700 | ||
| Glycerol | 0.4 ± 0.0 | 2,209 ± 50 | 5,800 | ||
| BW22888 | PrhaS-lacZ rpoS+ | d-Glucose | 0.4 ± 0.0 | 56 ± 2 | 130 |
| d-Fructose | 0.5 ± 0.1 | 253 ± 5 | 490 | ||
| Glycerol | 0.8 ± 0.1 | 619 ± 21 | 810 | ||
All strains are also ΔaraBAD or ΔrhaBAD, as appropriate; complete genotypes are given in Table 1.
Cells were assayed after 18 h of growth in 0.06% glucose–, 0.06% fructose–, or 0.1% glycerol–MOPS–2 mM Pi without (−Ind) or with (+Ind) the inducer arabinose or rhamnose. Values are nanomoles of o-nitrophenol made per unit of cell culture optical density at 420 nm (means ± standard deviations). Strains were grown and assayed in triplicate.
In the course of this study, we found that several of our strains carry an rpoS(Am) mutation (Table 1), which is also present in many lines of E. coli K-12, including progenitor K-12 strains EMG2 and W1485 (1, 22). As shown in Table 4, the induced level of PrhaB expression is significantly lower in an rpoS+ strain. Apparently, this rpoS effect is related to catabolite repression, as the magnitude varies with the carbon source. The reason for this is unknown. The induction levels in this study are therefore directly comparable only among strains having the same rpoS allele.
Strategy for using ParaB and PrhaB fusions to study regulation of the Pho regulon.
We constructed a number of ΔphoB and ΔphoR mutants in which PhoB or PhoR is synthesized under the control of a foreign inducible promoter(s). We did this for two reasons. First, we considered that such strains may be useful in our studies on protein-protein interactions between their gene products, as reported elsewhere (13). Second, we suspected that the deleterious effects of a ΔphoU mutation were in part due to high-level expression of the Pho regulon. Because expression of the phoBR operon is under positive autogenous control, high-level expression of the Pho regulon probably requires also high-level synthesis of PhoB and PhoR. We therefore expected to overcome these deleterious effects by down regulating PhoB or PhoR synthesis by using these foreign-regulated promoters.
Arabinose-independent expression of ParaB fusions.
The above results show the ParaB promoter to be tightly regulated for expression of the lacZ structural gene. To assess whether ParaB is useful for tight control of a regulatory gene under similar conditions, we constructed strains carrying chromosomal ParaB-phoB+ and ParaB-phoR+ fusions. Each strain has also a precise deletion of the respective regulatory gene. They have in addition a deletion of creC (ΔcreBCD or ΔcreABCD) as well as genes for acetyl phosphate synthesis [Δ(ackA pta)], thus eliminating activation of PhoB by the kinase CreC or acetyl phosphate (33). We then examined PhoB- and PhoR-dependent control by measuring BAP levels under conditions of Pi limitation in the absence of arabinose during growth on different carbon sources. As shown in Table 5, ParaB-phoB+ ΔphoB strain BW24803 exhibits a null phenotype in the absence of arabinose on all carbon sources. In contrast, ParaB-phoR+ ΔphoR strain BW22861 exhibits a PhoR+ phenotype in the absence of arabinose, even during growth on glucose. These results are consistent with less PhoR than PhoB being required for transcriptional activation of phoA. Therefore, ParaB appears to be sufficiently tightly controlled for the expression of phoB, but not for the expression of phoR. Yet, phoR expression is clearly limiting under these conditions, as much higher BAP levels are seen in the presence of arabinose (data not shown).
TABLE 5.
Inducer-independent expression of ParaB-phoB+ and ParaB-phoR+ fusions
| Strain | Relevant genotypea | BAP sp act onb:
|
||
|---|---|---|---|---|
| d-Glucose | d-Fructose | Glycerol | ||
| BW24803 | ParaB-phoB+ ΔphoB phoR+ | 0.3 ± 0.0 | 0.4 ± 0.0 | 0.4 ± 0.0 |
| BW22861 | ParaB-phoR+phoB+ ΔphoR | 26.4 ± 4.9 | 47.7 ± 5.1 | 58.8 ± 4.4 |
Strains are ΔcreABCD Δ(ackA pta) ΔaraBAD; complete genotypes are given in Table 1. The ParaB-phoB+ fusion is in single copy at attλ; the ParaB-phoR+ fusion is in single copy at the araCBAD locus.
Cells were assayed after 18 h of growth in 0.06% glucose–, 0.06% fructose–, or 0.1% glycerol–MOPS medium containing 0.1 mM Pi without arabinose. Values are nanomoles of p-nitrophenol made per unit of cell culture optical density at 420 nm (means ± standard deviations). Strains were grown and assayed in triplicate.
An eventual goal was to express both phoB and phoR independently and simultaneously from a foreign promoter(s). In order to find appropriate conditions to do this, we used strains with a deletion of the pstSCAB-phoU operon that leads to constitutive expression of the Pho regulon. Individual ones are also ΔphoB or ΔphoR as well as ΔcreC and Δ(ackA pta), as described above. As shown in Table 6, phoA expression is abolished in the absence of PhoB or PhoR (compare BW24741 and BW24740 with BW24739). Introduction of a PphoB-phoB+ fusion in single copy elsewhere on the chromosome restores normal phoA expression (compare BW24770 with BW24739). Introduction of a Ptac-phoB+ fusion in single copy restores phoA expression partially in the absence of IPTG and fully in the presence of IPTG (compare BW24775 without and with IPTG). This apparent leakiness of Ptac was expected, as proper regulation by LacI requires additional upstream and downstream operator sequences (20) that are absent in the Ptac-phoB+ fusion. Yet partial inducer dependence is observed even for expression of a regulatory gene from Ptac.
TABLE 6.
Inducer-dependent control by using a Ptac-phoB+ fusion
| Strain | Relevant genotypea | BAP sp act onb:
|
|
|---|---|---|---|
| −IPTG | +IPTG | ||
| BW24739 | phoBR+ Δ(pstSCAB-phoU) | 376 ± 19 | N.D. |
| BW24741 | ΔphoB phoR+ Δ(pstSCAB-phoU) | 0.2 ± 0.0 | N.D. |
| BW24740 | phoB+ ΔphoR Δ(pstSCAB-phoU) | 0.2 ± 0.0 | N.D. |
| BW24770 | PphoB-phoB+ ΔphoB phoR+ Δ(pstSCAB-phoU) | 376 ± 17 | N.D. |
| BW24775 | Ptac-phoB+ ΔphoB phoR+ Δ(pstSCAB-phoU) | 21.6 ± 0.9 | 355 ± 12 |
All strains are lacIq ΔcreABCD Δ(ackA pta); complete genotypes are given in Table 1. The PphoB-phoB+ and Ptac-phoB+ fusions are in single copy at attλ.
Cells were assayed after 18 h of growth in 0.06% glucose–MOPS–2 mM Pi without (−) or with (+) IPTG. BAP specific activity values are as defined for Table 5. N.D., not determined.
Arabinose- and rhamnose-dependent expression of ParaB, PrhaB, and PrhaS fusions.
We compared strains carrying chromosomal ParaB-phoB+, PrhaB-phoB+, ParaB-phoR+, PrhaB-phoR+, and PrhaS-phoR+ fusions to determine which promoter fusion(s) and growth conditions were appropriate for studying gene regulation in the Pho regulon. This was done with strains that are otherwise similar to ones described above. We examined PhoB- and PhoR-dependent control of phoA expression by measuring BAP levels when strains were grown on one of four carbon sources (glucose, mannitol, fructose, or glycerol), which were expected to result in different levels of catabolite repression (7). As shown in Table 7, strains carrying a ParaB-phoB+ or PrhaB-phoB+ fusion express a null phenotype in the absence of inducer. The same ones synthesized ca. 12-fold to 2,400-fold more BAP in the presence of inducer, depending upon the carbon source and promoter. The relative expression levels correlate well with expectation in regards to catabolite repression. Catabolite repression is more severe with glucose than mannitol, more severe with mannitol than fructose, and more severe with fructose than glycerol. The BAP levels in these ParaB-phoB+ and PrhaB-phoB+ fusion strains (ca. 280 to 490 U; BW24774, BW24777, and BW24773) (Table 7) during growth on glycerol are similar to that in an otherwise isogenic wild-type strain (ca. 375 U; BW24739) (Table 6). Strains with a PrhaB-phoR+ fusion at two chromosomal sites were examined; no difference between them was seen.
TABLE 7.
Inducer-dependent control by using ParaB-phoB+, PrhaB-phoB+, ParaB-phoR+, PrhaB-phoR+, and PrhaS-phoR+ fusions
| Strain | Relevant genotypea | Inducerb | BAP sp act onc:
|
|||
|---|---|---|---|---|---|---|
| d-Glucose | d-Mannitol | d-Fructose | Glycerol | |||
| BW24774 | ParaB-phoB+ ΔphoB Δ(pstSCAB-phoU) | None | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 |
| Ara | 4.0 ± 0.4 | 103 ± 0 | 174 ± 6 | 281 ± 5 | ||
| BW24777 | PrhaB-phoB+ ΔphoB Δ(pstSCAB-phoU) | None | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 |
| Rha | 2.6 ± 0.3 | 120 ± 23 | 350 ± 36 | 442 ± 5 | ||
| BW24773 | PrhaB-phoB+ ΔphoB Δ(pstSCAB-phoU) | None | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 |
| Rha | 2.4 ± 0.3 | 104 ± 9 | 359 ± 11 | 486 ± 2 | ||
| BW24508 | ParaB-phoR+ ΔphoR Δ(pstSCAB-phoU) | None | 24.8 ± 0.5 | 36.3 ± 1.6 | 75.5 ± 7.4 | 118 ± 3 |
| Ara | 149 ± 6 | 483 ± 9 | 336 ± 29 | 400 ± 27 | ||
| BW24768 | PrhaB-phoB+ ΔphoR Δ(pstSCAB-phoU) | None | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.0 |
| Rha | 3.4 ± 0.8 | 108 ± 13 | 357 ± 4 | 464 ± 2 | ||
| BW24858 | PrhaS-phoR+ ΔphoR Δ(pstSCAB-phoU) | None | 78.5 ± 5.4 | 85.7 ± 8.1 | 161 ± 11 | 257 ± 15 |
| Rha | 92.4 ± 4.5 | 175 ± 13 | 398 ± 14 | 673 ± 5 | ||
All strains are ΔcreABCD Δ(ackA pta) and ΔaraBAD or ΔrhaBAD, as appropriate; complete genotypes are given in Table 1. The ParaB-phoB+ and PrhaB-phoB+ fusions are in single copy at attλ; the PrhaB-phoB+ and ParaB-phoR+ fusions are in single copy at the rha and ara loci, respectively; the PrhaB-phoR+ and PrhaS-phoR+ fusions are in single copy at attHK022.
Cells were assayed after 18 h of growth in 0.06% glucose–, 0.06% mannitol–, 0.06% fructose–, or 0.1% glycerol–MOPS medium containing 2 mM Pi without (none) or with arabinose (Ara) or rhamnose (Rha) for induction.
BAP specific activity values are as defined for Table 5.
Of the three phoR fusion strains examined, only one (PrhaB-phoR+ fusion strain BW24768) showed a null phenotype in the absence of inducer. Substantial activation was apparent in both the ParaB-phoR+ and PrhaS-phoR+ fusion strains (BW24508 and BW24858) in the absence of inducer even during growth on glucose, indicating that both ParaB and PrhaS are somewhat leaky. Their basal levels of expression are also subject to catabolite repression, as they vary with the carbon source. Upon induction with rhamnose, the PrhaB-phoR+ fusion strain synthesized ca. 17-fold to 2,300-fold more BAP; these BAP levels also correlate well with the carbon source. The addition of the respective inducer resulted in increased BAP synthesis in the ParaB-phoR+ and PrhaS-phoR+ fusion strains as well.
Use of foreign promoters to bypass growth defect due to a ΔphoU mutation.
We showed above that we can modulate expression of the Pho regulon in our ParaB-phoB+, PrhaB-phoB+, and PrhaB-phoR+ fusion strains by growing them on different carbon sources in the presence of the respective inducer. We therefore tested whether these conditions can be used to overcome the deleterious effect of a ΔphoU mutation. Whereas a ΔphoU mutant such as BW17142 grows extremely poorly under all conditions tested, an otherwise isogenic Δ(pstSCAB-phoU) mutant such as BW17335 grows reasonably well (Table 8) (26). These strains have a kanamycin resistance cassette in place of phoU and pstSCAB-phoU sequences, respectively. A similar strain with an unmarked ΔphoU mutation (BW18897; Table 8) also shows a severe growth defect. Yet all these strains synthesize similar amounts of BAP. To determine whether modulating expression of phoB or phoR can be used to overcome the growth inhibition due to a ΔphoU mutation, we constructed otherwise isogenic ΔphoU and Δ(pstSCAB-phoU) strains with a ΔphoB and ΔphoR mutation carrying the respective fusions. These strains have an unmarked ΔphoU mutation, as each has a kanamycin resistance marker elsewhere (Table 1).
TABLE 8.
Pho regulon control in ΔphoU mutants by using ParaB-phoB+, PrhaB-phoB+, and PrhaB-phoR+ fusions
| Strain | Relevant genotypea | Growthb | BAP sp act onc:
|
|
|---|---|---|---|---|
| −Ind | +Ind | |||
| BW17142 | ΔphoU | +/− | 419d | N.D. |
| BW17335 | Δ(pstSCAB-phoU) | +++ | 428d | N.D. |
| BW18897 | ΔphoU | + | 444 ± 33 | N.D. |
| BW24739 | Δ(pstSCAB-phoU) Δ(ackA pta) | +++ | 443 ± 20 | N.D. |
| BW24774 | ParaB-phoB+ ΔphoB Δ(pstSCAB-phoU) Δ(ackA pta) | ++++ | 0.2 ± 0.0 | 94.8 ± 3.5 |
| BW24950 | ParaB-phoB+ ΔphoB ΔphoU Δ(ackA pta) | ++++ | 0.2 ± 0.0 | 113 ± 3 |
| BW24773 | PrhaB-phoB+ ΔphoB Δ(pstSCAB-phoU) Δ(ackA pta) | ++++ | 0.2 ± 0.0 | 109 ± 27 |
| BW24949 | PrhaB-phoB+ ΔphoB ΔphoU Δ(ackA pta) | ++++ | 0.2 ± 0.0 | 89.7 ± 6.4 |
| BW24768 | PrhaB-phoR+ ΔphoR Δ(pstSCAB-phoU) Δ(ackA pta) | ++++ | 0.2 ± 0.0 | 104 ± 2 |
| BW24948 | PrhaB-phoR+ ΔphoR ΔphoU Δ(ackA pta) | ++++ | 0.2 ± 0.0 | 83.4 ± 8.9 |
Complete genotypes are given in Table 1. The ParaB-phoB+ and PrhaB-phoB+ fusions are in single copy at attλ; the PrhaB-phoR+ fusion is in single copy at attHK022.
Colony size after 24 h of incubation +/−, barely detectable; +, tiny; +++, ca. 1 mm in diameter; ++++, ca. 1.5 mm in diameter.
Cells were assayed after 18 h of growth in 0.06% mannitol–MOPS–2 mM Pi without (−Ind) or with (+Ind) arabinose or rhamnose for induction. BAP specific activity values are as defined for Table 5. N.D., not determined.
BAP specific activity value is an average of four independent determinations (unpublished data cited in reference 26).
As shown in Table 8, our ΔphoU ΔphoB, Δ(pstSCAB-phoU) ΔphoB, ΔphoU ΔphoR, and Δ(pstSCAB-phoU) ΔphoR strains carrying a ParaB-phoB+, PrhaB-phoB+, or PrhaB-phoR+ fusion grow equally well in the absence or presence of the respective inducer. The ParaB-phoB+ and PrhaB-phoB+ fusion strains (BW24774, BW24950, BW24773, and BW24949) show a null phenotype in the absence of inducer. Each synthesizes ca. 500-fold more BAP in the presence of the respective inducer. Likewise, the PrhaB-phoR+ fusion strains (BW24768 and BW24948) show a null phenotype in the absence of rhamnose; each synthesizes ca. 500-fold more BAP in its presence. Upon induction these strains synthesize ca. 25% of the normal amount of BAP (see BW24739 for comparison), suggesting that this level of Pho regulon gene expression is apparently insufficient to result in a severe growth defect due to a ΔphoU mutation.
DISCUSSION
Our primary goal was to develop a method(s) to study PhoU function. The severe growth defect of a ΔphoU mutant is especially problematic due to the rapid accumulation of compensatory mutants with lesions in phoB, phoR, or a pst gene. A Pst+ ΔphoU mutant is also exquisitely sensitive to extracellular Pi, suggesting that PhoU has, in addition to its function in Pi signaling, a role as an enzyme in intracellular Pi metabolism (26). As shown here, we were able to overcome the deleterious effect of a ΔphoU mutation by uncoupling PhoB or PhoR synthesis from its normal autogenous control and expressing phoB or phoR from a foreign promoter(s). To do this, we constructed strains carrying a ParaB-phoB+, PrhaB-phoB+, ParaB-phoR+, or PrhaB-phoR+ fusion on the chromosome. We used strains with single-copy chromosomal fusions to avoid problems resulting from the use of plasmids, such as an antibiotic requirement, variable plasmid copy number, and even plasmid loss. We also examined ways for modulating expression levels from the ParaB and PrhaB promoters. We already reported work with similar strains and growth conditions to study the regulatory genes of the Enterococcus faecium vancomycin resistance gene cluster in an E. coli model system (12).
We constructed the corresponding ParaB and PrhaB fusion strains with conditionally replicative (pir-dependent) allele replacement plasmids pAH33, pAH54, and pLD78 (Fig. 2). Strains carrying these fusions are made via a simple two-step procedure as illustrated in Fig. 3. The resulting ParaB and PrhaB fusions reside on the chromosome at the araCBAD and rhaRSBAD loci, respectively, as shown in Fig. 4. We made similar strains with the respective ΔaraBADAH33 and ΔrhaBADLD78 chromosomal mutations as controls. In these ways, we constructed a number of strains that express various normal or mutant pho, cre, or van genes under the control of ParaB or PrhaB in single copy on the chromosome at the respective loci (12–14, 16).
The ParaB promoter (also called PBAD) has been shown elsewhere to be tightly regulated (11, 21). In those studies, genes that are normally expressed at moderate levels were examined. In preliminary studies, we found that a pBAD plasmid (11) carrying phoR did not provide sufficiently tight control for our purposes, suggesting that these plasmids are somewhat leaky (14). This is consistent with phoR normally being expressed at a very low level. In order to reduce this basal level of expression, we recombined several ParaB-phoR fusions onto the chromosome by using derivatives of ParaB fusion allele replacement plasmid pAH33. When single-copy fusion strains were used, particular ParaB-phoR fusions, e.g., one expressing only the C-terminal kinase domain of PhoR (13), no longer appeared to be leaky. Yet a single-copy ParaB-phoR+ fusion strain synthesizes sufficient PhoR for (partial) activation in the absence of arabinose even in the presence of glucose (Table 5). These results suggest that full-length PhoR is a more active kinase than its C-terminal domain. However, in the absence of arabinose, the amount of PhoR synthesis from a ParaB-phoR+ fusion is also clearly limiting because upon Pi limitation only ca. 10% as much BAP is made in this strain as in a wild-type strain. In contrast, a single-copy ParaB-phoB+ fusion strain shows no leakiness as it exhibits a PhoB− phenotype in the absence of inducer on all carbon sources tested (Table 7). Therefore, more PhoB than PhoR is apparently required for expression of the Pho regulon. These data are consistent with PhoR acting catalytically as an autokinase and phosphotransferase in the activation (phosphorylation) of PhoB, which in turn acts as a transcription factor.
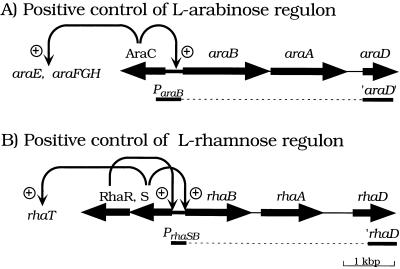
In order to control PhoB and PhoR synthesis independently, we also constructed strains with a PrhaB-phoB+ or PrhaB-phoR+ fusion. Although both l-arabinose and l-rhamnose act directly as inducers for expression of regulons for their catabolism, important differences exist in regard to the regulatory mechanisms (Fig. 5). l-Arabinose acts as inducer with the activator AraC in the positive control of the arabinose regulon (23). However, the l-rhamnose regulon is subject to a regulatory cascade; it is therefore subject to an even tighter control. l-Rhamnose acts as an inducer with the activator RhaR for synthesis of RhaS, which in turn acts as an activator in the positive control of the rhamnose regulon (6). As shown in Table 7, both the PrhaB-phoB+ and PrhaB-phoR+ fusion strains show a PhoB− and PhoR− phenotype, respectively, in the absence of rhamnose. We also examined a PrhaS-phoR+ fusion. Like the ParaB-phoR+ fusion strain, the PrhaS-phoR+ fusion strain synthesized a substantial, though limited, amount of BAP in the absence of induction.
FIG. 5.
Transcriptional activation of the arabinose and rhamnose regulons. (A) Positive control of the l-arabinose regulon. The araCBAD region near 1.5 min on the E. coli chromosome is shown. The unlinked araE and araFGH genes encode arabinose transport systems (23). (B) Positive control of the l-rhamnose regulon. The rhaRSBAD region near 88.2 min is shown. The nearby rhaT gene is transcribed towards rhaR (Fig. 4) and encodes a rhamnose permease (3). Thick arrows show gene orientations. Curved ones with circled pluses indicate sites for transcriptional activation by AraC (A) or RhaR and RhaS (B). The lower bars show the ara and rha sequences in the respective ParaB and PrhaB fusion allele replacement vectors illustrated in Fig. 2. The dashed lines show the chromosomal regions deleted in the resultant recombinants. See text.
The l-arabinose and l-rhamnose regulons are also regulated by catabolite repression. We therefore modulated expression of ParaB, PrhaB, and PrhaS fusions by using various carbon sources (glucose, mannitol, fructose, and glycerol) that lead to different levels of catabolite repression (7). When strains are grown on these carbon sources with the inducer in excess, the expression of the Pho regulon can be modulated 70- to 200-fold in our ParaB-phoB+, PrhaB-phoB+, and PrhaB-phoR+ fusion strains (Table 7). As expected, the differences are much smaller in the ParaB-phoR+ and PrhaS-phoR+ fusion strains as the expression of these fusions is also leaky under these conditions. In preliminary experiments, we had also attempted to modulate expression levels by using different inducer concentrations. However, we were unable to maintain a constant, steady state level of induction (data not shown) (5). An inability to maintain steady-state induction at intermediate levels by the limiting inducer concentration is expected as the presence of arabinose and rhamnose results also in increased synthesis of their respective transport systems (Fig. 5). This has now been substantiated by monitoring ParaB expression levels in cells grown in the presence of subsaturating inducer concentrations (24). Importantly, as shown here, expression of these promoters can be modulated over a wide range by using different carbon sources in the presence of saturating inducer concentrations.
Both PhoU and the Pst transporter have been implicated in the negative control of the Pho regulon, as mutations of either result in high constitutive Pho regulon gene expression. Presumably, they somehow act together. To study how they interact with the PhoB-PhoR system, we made strains that synthesize PhoB or PhoR under control of the tightly regulated ParaB and PrhaB promoters. By using these strains, we were able to express phoB or phoR at a reduced level and thereby overcome the harmful effect of a ΔphoU mutation. In these ways, it should now be possible to study further the function(s) of PhoU.
ACKNOWLEDGMENTS
We thank individuals cited in the text for providing strains and plasmids.
This research is supported by NIH grant GM57695. The Dana Farber Cancer Institute Molecular Biology Core Facility is supported by NIH grants CA06516 and AI28691.
REFERENCES
- 1.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 2.Bachmann, B. J. Personal communication.
- 3.Baldoma L, Badia J, Sweet G, Aguilar J. Cloning, mapping and gene product identification of rhaT from Escherichia coli K12. FEMS Microbiol Lett. 1990;72:103–108. doi: 10.1016/0378-1097(90)90353-r. [DOI] [PubMed] [Google Scholar]
- 4.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, L. L., and B. L. Wanner. Unpublished data.
- 6.Egan S M, Schleif R F. A regulatory cascade in the induction of rhaBAD. J Mol Biol. 1993;234:87–98. doi: 10.1006/jmbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- 7.Epstein W, Rothman-Denes L B, Hesse J. Adenosine 3′:5′-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:2300–2304. doi: 10.1073/pnas.72.6.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher S L, Jiang W, Wanner B L, Walsh C T. Cross-talk between the histidine protein kinase VanS and the response regulator PhoB: characterization and identification of a VanS domain that inhibits activation of PhoB. J Biol Chem. 1995;270:23143–23149. doi: 10.1074/jbc.270.39.23143. [DOI] [PubMed] [Google Scholar]
- 9.Fisher S L, Kim S-K, Wanner B L, Walsh C T. Kinetic comparison of the specificity of the vancomycin resistance kinase VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- 10.Guan C-D, Wanner B L, Inouye H. Analysis of regulation of phoB expression using a phoB-cat fusion. J Bacteriol. 1983;156:710–717. doi: 10.1128/jb.156.2.710-717.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haldimann A, Fisher S L, Daniels L L, Walsh C T, Wanner B L. Transcriptional regulation of the Enterococcus faecium BM4147 vancomycin resistance gene cluster by the VanS-VanR two-component regulatory system in Escherichia coli. J Bacteriol. 1997;179:5903–5913. doi: 10.1128/jb.179.18.5903-5913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haldimann A, Prahalad M K, Fisher S L, Kim S-K, Walsh C T, Wanner B L. Altered recognition mutants of the response regulator PhoB: a new genetic strategy for studying protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:14361–14366. doi: 10.1073/pnas.93.25.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haldimann, A., and B. L. Wanner. Conditional replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies in bacteria. Unpublished data. [DOI] [PMC free article] [PubMed]
- 15.Jiang, W., A. Haldimann, and B. L. Wanner. Unpublished data.
- 16.Kim, S.-K., and B. L. Wanner. Unpublished data.
- 17.Metcalf W W, Jiang W, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 18.Metcalf W W, Wanner B L. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation using TnphoA′ elements. J Bacteriol. 1993;175:3430–3442. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muda M, Rao N N, Torriani A. Role of PhoU in phosphate transport and alkaline phosphatase regulation. J Bacteriol. 1992;174:8057–8064. doi: 10.1128/jb.174.24.8057-8064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oehler S, Eismann E R, Krämer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips G J, Silhavy T J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 22.Rod M L, Alam K Y, Cunningham P R, Clark D P. Accumulation of trehalose by Escherichia coli K-12 at high osmotic pressure depends on the presence of amber suppressors. J Bacteriol. 1988;170:3601–3610. doi: 10.1128/jb.170.8.3601-3610.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schleif R. Two positively regulated systems, ara and mal. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1300–1309. [Google Scholar]
- 24.Siegele D A, Hu J C. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci USA. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 26.Steed P M, Wanner B L. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol. 1993;175:6797–6809. doi: 10.1128/jb.175.21.6797-6809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wanner B L. Overlapping and separate controls on the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1983;166:283–308. doi: 10.1016/s0022-2836(83)80086-2. [DOI] [PubMed] [Google Scholar]
- 28.Wanner B L. Gene expression in bacteria using TnphoA and TnphoA′ elements to make and switch phoA gene, lacZ (op), and lacZ (pr) fusions. In: Adolph K W, editor. Methods in molecular genetics. Vol. 3. Orlando, Fla: Academic Press; 1994. pp. 291–310. [Google Scholar]
- 29.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- 30.Wanner B L. Phosphate signaling and the control of gene expression in Escherichia coli. In: Silver S, Walden W, editors. Metal ions in gene regulation. Sterling, Va: Chapman and Hall, Ltd.; 1997. pp. 104–128. [Google Scholar]
- 31.Wanner B L, Boline J A. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J Bacteriol. 1990;172:1186–1196. doi: 10.1128/jb.172.3.1186-1196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wanner B L, Chang B-D. The phoBR operon in Escherichia coli K-12. J Bacteriol. 1987;169:5569–5574. doi: 10.1128/jb.169.12.5569-5574.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wanner B L, Wilmes-Riesenberg M R. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in the control of the phosphate regulon in Escherichia coli. J Bacteriol. 1992;174:2124–2130. doi: 10.1128/jb.174.7.2124-2130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb D C, Rosenberg H, Cox G B. Mutational analysis of the Escherichia coli phosphate-specific transport system, a member of the traffic ATPase (or ABC) family of membrane transporters. A role for proline residues in transmembrane helices. J Biol Chem. 1992;267:24661–24668. [PubMed] [Google Scholar]