Abstract
Escherichia coli cell division protein FtsK is a homolog of Bacillus subtilis SpoIIIE and appears to act late in the septation process. To determine whether FtsK localizes to the septum, we fused three N-terminal segments of FtsK to green fluorescent protein (GFP) and expressed them in E. coli cells. All three segments were sufficient to target GFP to the septum, suggesting that as little as the first 15% of the protein is a septum-targeting domain. Localized fluorescence was detectable only in cells containing a visible midcell constriction, suggesting that FtsK targeting normally occurs only at a late stage of septation. The largest two FtsK-GFP fusions were able at least partially to complement the ftsK44 mutation in trans, suggesting that the N- and C-terminal domains are functionally separable. However, overproduction of FtsK-GFP resulted in a late-septation phenotype similar to that of ftsK44, with fluorescent dots localized at the blocked septa, suggesting that high levels of the N-terminal domain may still localize but also inhibit FtsK activity. Interestingly, under these conditions fluorescence was also sometimes localized as bands at potential division sites, suggesting that FtsK-GFP is capable of targeting very early. In addition, FtsK-GFP localized to potential division sites in cephalexin-induced and ftsI mutant filaments, further supporting the idea that FtsK-GFP can target early, perhaps by recognizing FtsZ directly. This hypothesis was supported by the failure of FtsK-GFP to localize in ftsZ mutant filaments. In ftsK44 mutant filaments, FtsA and FtsZ were usually localized to potential division sites between the blocked septa. When the ftsK44 mutation was incorporated into the FtsK-GFP fusions, localization to midcell ranged between very weak and undetectable, suggesting that the FtsK44 mutant protein is defective in targeting the septum.
Recent genetic and cytological evidence suggests that cell division in bacteria requires a complex of proteins localized to the division site that are both cytoplasmic and span the inner membrane (18). Conditional mutations in the fts genes encoding most of these proteins result in inhibition of cell division at various stages in the process (10). In Escherichia coli, ftsZ mutants yield filamentous cells with no sign of septation, indicating a function early in the septation pathway (13), whereas ftsK mutants gave rise to highly indented filaments, suggesting a defect late in the pathway (5). Mutants in other fts genes appear to be arrested as early as ftsZ, such as ftsW (8, 12), or at a later step, such as ftsI (7, 16, 21), but not as late as ftsK. This genetic evidence has been supplemented by the results of cytological studies showing that FtsZ and FtsA localize to division sites (4, 6, 14) and that FtsZ localizes in cells with other fts mutations but that FtsA fails to localize in ftsZ mutants (1, 4). FtsI, which is required for septal peptidoglycan synthesis, also localizes to the septum (23). Two other proteins isolated by their ability to interact with the cell division apparatus, ZipA and FtsN, also localize to the E. coli septum, with ZipA being recruited early and FtsN at a later stage (2, 11). Confirmation of the localization of other Fts proteins to the division site would lend further support for the idea that a large protein complex functions in septation. Moreover, defining which mutants can or cannot support localization would allow further ordering of the assembly pathway of the putative septation complex.
The ftsK gene appears to be essential for cell division in E. coli because the original ftsK44 mutation that defined the gene resulted in a lethal defect in the late stages of septation at the restrictive temperature (5). However, an insertion mutation in the center of ftsK that leaves the N terminus intact causes chain formation as in ftsK44 but is not lethal (9). FtsK is predicted to be a large polytopic protein, with a hydrophobic N terminus and a C terminus with high sequence similarity to a family of proteins involved in intercellular and intracellular DNA transfer (5). One member of this family, Bacillus subtilis SpoIIIE, can partition chromosomes through the completed sporulation septum by a conjugation-like mechanism (19, 24). SpoIIIE has recently been shown to localize to the sporulation septum; since a mutation in its N-terminal hydrophobic domain interferes with its localization, it is likely that this domain is responsible for targeting (25). However, SpoIIIE is not required for vegetative septation in B. subtilis, whereas FtsK appears to be required in E. coli. FtsK also differs from other members of the SpoIIIE family in having a large internal proline-glutamine-rich stretch which may function as a flexible spacer between the N- and C-terminal domains. A similar type of unique spacer domain is present in FtsZ1, a Rhizobium meliloti homolog of FtsZ (15). The roles of FtsK in E. coli septation and any role in DNA dynamics are not understood. As a first step towards understanding more about FtsK function, and about localization of proteins to the E. coli cell division site, we used green fluorescent protein (GFP) to tag small N-terminal domains of FtsK in order to identify those important for localization to the septum. Here we report that a relatively small N-terminal portion of FtsK is able to localize to the E. coli septum. Normally, this targeting occurs late during the septation process, although under some conditions FtsK-GFP can localize early. We also show that a single amino acid substitution in this domain represented by ftsK44 appears to significantly inhibit FtsK targeting and demonstrate that FtsZ and FtsA generally do not localize to septa blocked at the ftsK stage.
MATERIALS AND METHODS
Bacterial strains and growth media.
E. coli JM105 or BSP853 (W3110 rnc40::Tn5) was used to harbor FtsK-GFP fusion plasmids. Strain TOE44 (AB2497 ftsK44), obtained from K. Begg, contains the temperature-sensitive ftsK44 mutation (5). Strain JFL101 (ftsZ84) was obtained from J. Lutkenhaus. Strains AX621 (ftsA1882) and AX655 (ftsI2158) were from the E. coli Genetic Stock Center. LB medium (10 g of tryptone per liter, 5 g of yeast extract per liter, 5 g of NaCl per liter) was used for most experiments, except when nonpermissive conditions were required for the fts mutants, in which case LBNS (10 g of tryptone per liter, 5 g of yeast extract per liter) was used. Chloramphenicol was added to 20 to 25 μg/ml.
Plasmid construction.
To amplify ftsK fragments by PCR, the following primers were made: FtsK1 (5′TTGAGCTCCCTGGAGAGCCTTTCTTGA3′), FtsK3 (5′AATCTAGAAAAGGTATCTACCGGC3′), FtsK4 (5′TTTCTAGATTTGCCGTGATTTTCATC3′), and FtsK7 (5′TTTCTAGAATCATCGCGACGGGTACG3′). The FtsK1 primer included a SacI site and the region including the FtsK ribosome binding sequence and start codon (underlined), whereas the other 3 primers all included an XbaI site and the desired 3′ end of the FtsK insert. The XbaI site was engineered to be in frame with the GFP gene in pZG, as described previously (14). The FtsK1 primer was designed so that a TGA sequence overlapping the FtsK start codon would be in frame with the lacZ gene of the vector, in order to prevent inadvertent expression of lacZ fusions that might interfere with the expression of FtsK-GFP. To amplify the inserts for cloning by PCR, reaction mixtures included primer pairs FtsK1-FtsK3, FtsK1-FtsK4, and FtsK1-FtsK7. PCR mixtures also included template DNA supplied by a dilute suspension of JM105 cells and a mixture of Taq polymerase (Promega) and Vent polymerase (New England Biolabs). The reaction conditions consisted of denaturation at 95°C for 1 min, annealing at 50°C for 2 min, and polymerization at 72°C for 1 min over 30 cycles. The PCR products of 2.6, 0.71, and 0.64 kb, respectively, were purified, cleaved with SacI and XbaI, and cloned into SacI- and XbaI-cleaved pZG. This procedure replaced the ftsZ gene of pZG, a derivative of pBC SK+ containing GFP between the XbaI and PstI sites and lacIq inserted at the EcoRI site, with the ftsK fragments. Plasmids were named pK3G, pK4G, and pK7G, corresponding to the number of the downstream PCR primer. The fragments containing the ftsK44 mutation were cloned by using cells from strain TOE44 as the DNA template for PCR amplification, with FtsK1-FtsK3 and FtsK1-FtsK4 as primer pairs, and otherwise repeating the above-described protocol. The resulting plasmids were pK3(−)G and pK4(−)G. Expression of the insert genes was dependent on the vector lac promoter and regulated by the lacIq gene. The one exception was pK4G(+), from which the lacIq cassette was deleted by cleavage with EcoRI followed by religation. Strain BSP853 containing this plasmid produced significantly higher levels of FtsK-GFP, and IPTG (isopropyl-β-d-thiogalactopyranoside) was not needed to visualize fluorescence.
Growth conditions for fusion protein expression.
For FtsK-GFP localization in the JM105 or BSP853 strain background, cells were grown in LB medium plus chloramphenicol as standing overnight cultures and then subcultured at 37°C and shaken for 1 to 2 h in the same medium. Standing overnight cultures were used to circumvent a reproducible growth lag in the strains containing the active FtsK-GFP fusions. Subsequently, 0.5 to 1 mM IPTG was added to the growing cultures and incubated one to two additional hours at 37°C before microscopic examination. For FtsK-GFP localization in fts mutants, cells were grown at 28 to 32°C in LBNS plus chloramphenicol, with thymine for TOE44, until early logarithmic phase. The culture was then divided into two, 0.5 mM IPTG was added to both aliquots, and one was shifted to 42°C while the other remained at the lower temperature. Aliquots were taken at various times after induction. To rule out the possibility that FtsK-GFP localization was due to the presence of preformed division sites in ftsI mutants, cells were induced at 42°C for 2 h prior to IPTG induction and then grown for an additional 5 h before aliquots were taken for examination. For FtsK-GFP localization in cephalexin-treated cells, strain BSP853 was grown at 37°C in LB medium plus chloramphenicol for several hours until early logarithmic phase and then 10 μg of cephalexin per ml was added and the cultures were monitored between 2 and 4 h after drug addition. Short induction times at this IPTG concentration were necessary for detection of fluorescence localization of FtsK-GFP. Complementation tests with TOE44 were done with plates containing LBNS plus chloramphenicol plus thymine, with or without IPTG, at 28 and 42°C.
For FtsA-GFP localization in TOE44, plasmid pAG (14) containing lacIq was introduced into TOE44. Expression of FtsA-GFP was induced with 40 to 100 μM IPTG for several hours, concomitant with the shift of the culture from 28 to 42°C. Because TOE44 is a thyA mutant, the growth medium was supplemented with 50 μg of thymine per ml.
Microscopic techniques.
Immunofluorescence localization of FtsZ in TOE44 was performed essentially as described previously (1) with polyclonal antiserum against a preparation of purified E. coli FtsZ, which was verified to be specific by immunoblot analysis. The secondary antibody was conjugated to Oregon Green (Molecular Probes). Propidium iodide was used to stain DNA as described previously (17).
For GFP visualization, live cells were viewed immediately after either immobilization in 1% low-melting-point agarose in LB medium or placement on the surface of a thin agar layer on a microscope slide. All images were viewed with an Olympus BX60 fluorescence microscope equipped with a 100× oil immersion plan fluorite objective (numerical aperture = 1.3), a 100-W mercury lamp, a standard fluorescein isothiocyanate filter set, and an Optronics DEI-750 cooled video camera. Images were digitized with a Scion LG3 video card, manipulated with Adobe Photoshop, and printed on a Tektronix Phaser 400 dye-sublimation printer. The FtsK-GFP signals were often so faint that they could not be viewed with the eye but were easily detectable by the video camera, which was capable of on-chip integration. Exposures were generally between 2 and 8 s.
RESULTS
Construction and characterization of FtsK-GFP fusions.
To determine if FtsK is localized to the cell midpoint, we used a GFP-tagging strategy that was successful in confirming the septal localization of FtsZ and demonstrating that FtsA also targets to the septum (14). Because the N terminus of B. subtilis SpoIIIE has been recently implicated in septal localization (25), we reasoned that the N terminus of FtsK might be a localization domain. Therefore, GFP was fused to the C termini of three N-terminal fragments of FtsK. The GFP portion in the fusion proteins was attached to a cytoplasmic portion of FtsK as predicted by previous hydropathy analysis (5).
Three N-terminal fragments of ftsK, encoding 205, 230, and 859 amino acids, were fused to the gene encoding GFP (239 amino acids), creating pK7G, pK4G, and pK3G, respectively (Fig. 1). All three plasmids have identical transcriptional and translational controls and have the same fusion junction with GFP; only the 3′ end of ftsK differs in each case. Initially, we used versions of these plasmids that contained a copy of lacIq to repress transcription from Plac. However, even with full induction with IPTG, these fusions were expressed at very low levels, as assessed by cellular fluorescence (see below) and by immunoblotting (data not shown). Expression was undetectable without IPTG induction. This low expression level may have been due to the rare TTG initiation codon present in the wild-type ftsK sequence that was preserved in the fusions. The low levels of FtsK-GFP may also have been caused in part by proteolysis, since lower-molecular-weight bands were observed in immunoblots (data not shown).
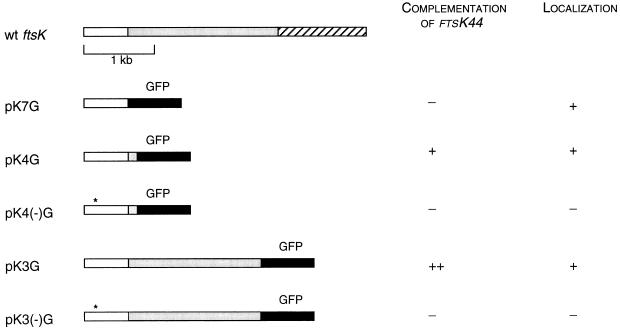
FIG. 1.
Properties of different FtsK-GFP fusions. Structures of the fusion proteins and fusion endpoints are shown at the left. Open and hatched boxes represent the N-terminal and C-terminal domains of FtsK, respectively. Shaded boxes represent portions of the long spacer region of FtsK. Black boxes denote GFP. Asterisks represent the presence and positions of the ftsK44 mutations in the various clones. For complementation, ++ denotes full complementation, + denotes partial complementation, and − denotes no detectable complementation. For localization, + denotes strong midcell localization and − denotes no detectable localization. wt, wild type.
To test if the FtsK-GFP fusions were functional, they were introduced into the ftsK44 mutant strain TOE44. The complementation results are summarized in Fig. 1. At 42°C in the absence of added NaCl in the medium, TOE44 mutant cells gave rise to no colonies. However, pK3G allowed colony formation in this strain under these conditions with a plating efficiency indistinguishable from that at 32°C. Plasmid pK4G also partially complemented the mutant, with a decrease in plating efficiency of about 10-fold. Interestingly, pK7G did not complement at all, suggesting that the residues between 205 and 230 were essential for this activity. The complementation of the ftsK44 mutant by an N-terminal domain of FtsK is similar to the complementation by partial ftsK fragments observed by Begg et al. (5), except that in this case GFP was also attached. These results suggest that FtsK has at least two distinct domains that can function separately, such that a functional N terminus, even when it is attached to GFP, can mix with a functional C terminus to reconstitute normal FtsK activity. The ability of some of the GFP fusions to act as fully functional domains also supports the localization data discussed below.
Localization of FtsK-GFP.
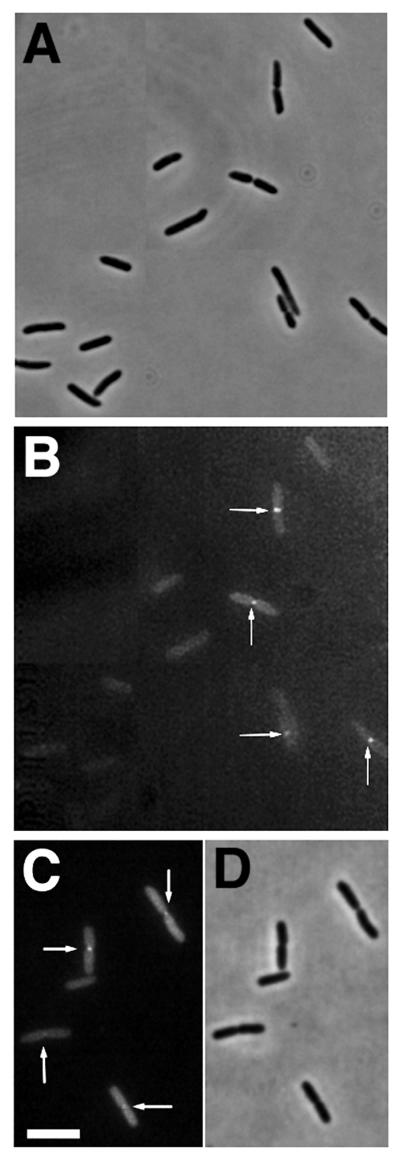
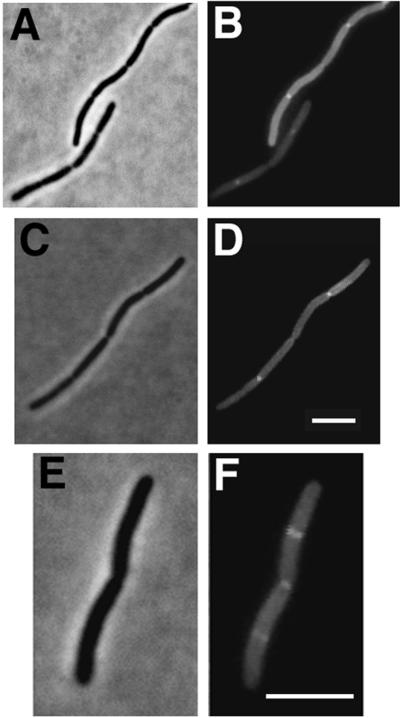
To visualize FtsK-GFP in living cells, the strains containing the three different plasmids were induced for 1 to 2 h with IPTG to express the fusion proteins. Microscopic examination of the cells revealed two populations, one with uniformly distributed dim fluorescence and one with bright fluorescence concentrated at the center (Fig. 2). Detectable fluorescent localization to the septum nearly always occurred in cells with an obvious midcell constriction, whereas cells with no clear constriction had no detectable midcell fluorescence (Fig. 2). This suggested that FtsK-GFP, and also FtsK, might be recruited to the septum later in the cell division process. Occasionally, cells were observed with fluorescent bands instead of dots at midcell, but even these cells contained a slight constriction coincident with the band, suggesting that they represented an initial FtsK localization event. All three FtsK-GFP fusions localized to midcell (Fig. 2 and data not shown), indicating that the inability to complement ftsK44 for colony formation did not prevent the ability to target correctly. These results strongly suggest that the FtsK protein localizes late to the E. coli septum and demonstrate that the first 205 amino acids of this 1,329-amino-acid protein, just the N-terminal 15%, are sufficient to target GFP to the septum. The simplest conclusion that can be drawn is that this segment functions as a midcell targeting domain for FtsK.
FIG. 2.
Late localization of FtsK-GFP to the septum. Cells containing FtsK-GFP fusions on plasmids were grown and examined as described in Materials and Methods. (A and B) Phase-contrast (A) and fluorescence (B) micrographs of a field of JM105 cells containing pKG4 after IPTG induction, with arrows highlighting fluorescence localization at constrictions; (C and D) fluorescence (C) and phase-contrast (D) micrographs of a field of cells with pKG3 in JM105 after IPTG induction. Bar = 5 μm.
The FtsK44 mutant protein domain is defective in localizing to the septum.
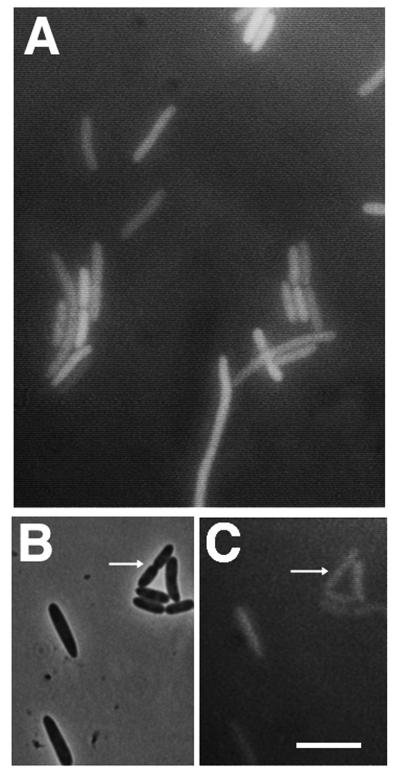
To test if the protein encoded by the temperature-sensitive ftsK44 allele could also localize to the septum, the same 230- and 859-amino-acid segments of FtsK44 present in pK4G and pK3G were used to fuse the mutant protein to GFP, creating pK4(−)G and pK3(−)G. As expected, these plasmids were unable to complement the ftsK44 mutant (Fig. 1). When FtsK44-GFP proteins were expressed from cells in the same experiment with cells expressing FtsK-GFP as a positive control, the FtsK44-GFP cells usually displayed uniform fluorescence (Fig. 3A), suggesting that the FtsK44 mutant protein domain cannot detectably localize to midcell. Over the course of many repeated experiments, we occasionally found very weak localization to constrictions that was barely detectable (Fig. 3B and C), suggesting that the mutant protein may have retained a small amount of targeting activity. This localization defect was particularly apparent in filamentous cells, which facilitated visualization (see below), and was also observed at 28°C, suggesting that the FtsK44 protein was still defective even at temperatures permissive for growth of TOE44.
FIG. 3.
FtsK44-GFP is defective in localization to the septum. (A) Typical fluorescence micrograph showing no fluorescence at constrictions of JM105 cells containing pK4(−)G; (B and C) phase-contrast (B) and fluorescence (C) micrographs of a field of JM105 cells containing pK4(−)G showing very weak fluorescence at constrictions and the brightest localized fluorescence ever observed for this mutant fusion (highlighted by arrows); usually no localization was observed. Bar = 5 μm.
It was possible that the lack of localization of the FtsK44-GFP fusion was due to an additional mutation incurred from PCR amplification, poor expression, or protein degradation. DNA sequencing of the entire inserts containing the GFP fusions to the wild-type and mutant FtsK [pK4G and pK4(−)G] revealed only the single base change corresponding to the ftsK44 mutation, ruling out the possibility of additional mutations. To ascertain whether the FtsK44-GFP fusion protein was expressed, IPTG-induced cells with pK3G, pK3(−)G, pK4G, and pK4(−)G were subjected to immunoblot analysis with anti-GFP antibody. Expression was weak, as expected, but major bands of approximately 160 kDa for pK3G and pK3(−)G and approximately 50 kDa for pK4G and pK4(−)G were observed at similar levels (data not shown). The predicted sizes of the fusions were 122 and 53 kDa, respectively. The discrepancy with the larger fusion may have been due to aberrant migration in sodium dodecyl sulfate-polyacrylamide gel electrophoresis, because of the unusual extended structure of the spacer domain, which is absent in the smaller fusion; similar aberrantly slow migration was observed with R. meliloti FtsZ1, which has a similar type of spacer region (15). In any case, the immunodetection of both FtsK-GFP and FtsK44-GFP fusion proteins at similar levels suggests that the poor FtsK44-GFP localization was not due to the absence of the fusion protein from the cell and instead suggests that FtsK44-GFP is defective in detectably localizing to the septum. These results suggest that FtsK44 is a defective protein that somehow retains enough activity under permissive conditions of high salt or low temperature to complete septation.
Although the ability of the N terminus of FtsK to localize to the septum supports the idea that it is a localization domain, another possibility is that it localizes because it is able to oligomerize with the wild-type FtsK already present at the septum. To address this issue, the localization of FtsK-GFP in the ftsK44 mutant TOE44 was examined under nonpermissive conditions. Under these conditions, ftsK44 cells form moderately long filaments with regular, deep indentations. These indentations represent division sites presumably blocked at the time FtsK normally functions. When it was expressed at low levels, FtsK-GFP fluorescence was often localized to the constricting septa within the short filaments (Fig. 4D and E and data not shown). Presumably, these cells were somewhat filamentous because the pKG4 plasmid was able to only partially complement the ftsK44 phenotype. It is not clear why so many constrictions lacked detectable fluorescence. However, because FtsK44-GFP was defective in localizing to septa (Fig. 3), one explanation for the TOE44 phenotype may be poor localization of the intact FtsK44 protein, which is tolerated by the cell at lower temperatures but not at higher temperatures or low osmolarity. If the assumption is made that FtsK44 in TOE44 cells is poorly localized at the nonpermissive temperature, then the ability of FtsK-GFP to localize in TOE44 under the same conditions supports, but does not prove, the idea that the N-terminal domain is a true localization domain.
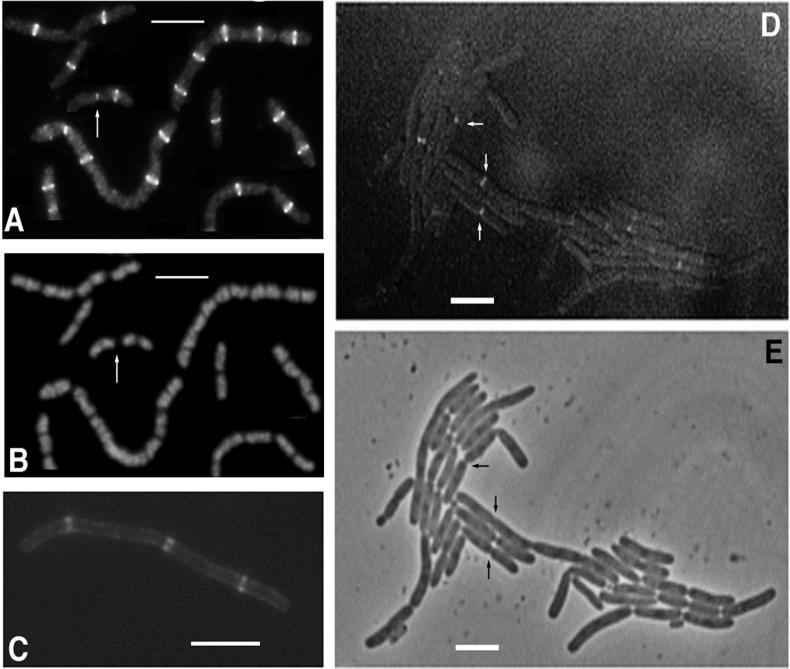
FIG. 4.
Localization of FtsZ and FtsA to new division sites in ftsK filaments. (A and B) IFM of FtsZ in TOE44 filaments showing FtsZ staining (A) and propidium iodide staining of chromosomes (B) from the same cells. Arrows in panels A and B indicate a cell with FtsZ at a nearly constricted septum and at one potential division site. (C) Fluorescence micrograph of FtsA-GFP expressed from pAG in TOE44 (ftsK44) filaments. (D) Fluorescence micrograph showing fluorescent localization at some constrictions of TOE44 filaments containing pKG4. (E) Phase-contrast image of the filaments shown in panel D. Arrows highlight some constrictions that also show fluorescence. Bars = 5 μm.
FtsZ and FtsA localization in an ftsK mutant.
To understand more about the role and placement of FtsK in the cell division pathway, we tested the ability of FtsA and FtsZ to localize in ftsK44 mutant filaments. FtsZ localization was determined by immunofluorescence microscopy (IFM). Surprisingly, FtsZ rings were almost always found at potential, unconstricted division sites and not at the nearly complete septa (Fig. 4A and B). Occasional cells contained a faint dot of fluorescence still visible at the constricting septum, along with one or two FtsZ rings at the potential division sites (Fig. 4A). FtsA-GFP was localized to potential division sites and not blocked septa in a manner similar to that of FtsZ (Fig. 4C). These results suggest two possibilities. One is that the FtsK division block destabilizes the FtsZ ring at the last stage, causing disassembly of the midcell FtsZ ring and new assembly of a ring or rings at unblocked potential division sites. Since FtsA is recruited to the FtsZ ring, it is predicted to localize similarly. The other possibility is that the stage at which the FtsK block occurs is after the FtsZ ring normally disassembles and goes to the potential division sites. Since IFM analysis (17) and monitoring of growing cells with FtsZ-GFP (22) demonstrate that FtsZ persists at the septum until immediately before cell separation, we favor the former idea that the FtsZ ring is destabilized after an FtsK block. Such FtsZ ring instability after a block in constriction has been suggested previously (1, 3, 17).
Overexpressed FtsK-GFP causes an ftsK mutant phenocopy and can localize early.
We made versions of pKG4 in which the lacIq gene was excised, and cells containing the new plasmid, pKG4(+), were viable. Overall expression was high, and in BSP853 no IPTG induction was necessary to observe bright fluorescence. Concentrated dots of fluorescence still localized to many of the constriction points (Fig. 5A to D). However, these cells often formed chains that were very similar to ftsK44 mutant cell chains. This result suggests that although low levels of FtsK-GFP were able to complement ftsK44 at least partially, high levels inhibited septation in wild-type cells and caused a cell division block at a late stage similar to the ftsK44 block. The block was not total, since strains with pKG4(+) and the other FtsK-GFP fusions were able to form colonies on plates containing IPTG. In fact, complementation of TOE44 by pKG3 and pKG4 occurred in the presence of IPTG, suggesting that the observed septation inhibition may have been merely a delay from which the cells could recover. It is possible that this inhibition or delay is due to interference with the proper stoichiometries of the N- and C-terminal domains. For example, at low levels, FtsK N termini might be able to form productive complexes with FtsK C termini, but at high levels, N termini might compete for sites to prevent proper midcell targeting of C termini. Localization of the N-terminal portion of FtsK was probably required for this inhibitory effect, because overexpression of pK(−)G4 resulted in high levels of fluorescence but had no significant effect on cell division (data not shown).
FIG. 5.
FtsK-GFP overproduction correlates with cell division inhibition and early localization. All fields show BSP853 cells overproducing FtsK-GFP from pGK4(+) (no lacIq). Phase-contrast micrographs (A, C, and E) and fluorescence micrographs (B, D, and F) are paired to show localization to constriction points in ftsK44-like filaments (A to D) and an undivided cell with fluorescence at both the midcell constriction and at both potential division sites (E and F). Bars = 5 μm.
Interestingly, some cells overexpressing FtsK-GFP exhibited, in addition to fluorescent dots at constrictions, fluorescent bands at one-quarter and three-quarter positions corresponding to potential division sites. A typical cell with a fluorescent dot at the blocked midcell septum and bands at both potential division sites is shown in Fig. 5E to F. This result indicates that FtsK-GFP is capable of localizing before constriction is visible under some conditions; further evidence for this is presented below.
FtsK-GFP localization in ftsA, ftsI, and ftsZ mutants.
When it is expressed at low levels, FtsK-GFP appears to localize to the septum only after visible constriction begins, implying that it localizes later than FtsZ and perhaps later than some other cell division proteins. One possibility is that the septal localization of FtsK as well as the timing of its targeting requires the proper function and localization of other cell division proteins. To test this idea, FtsK-GFP was expressed in temperature-sensitive mutants of ftsZ, ftsA, and ftsI.
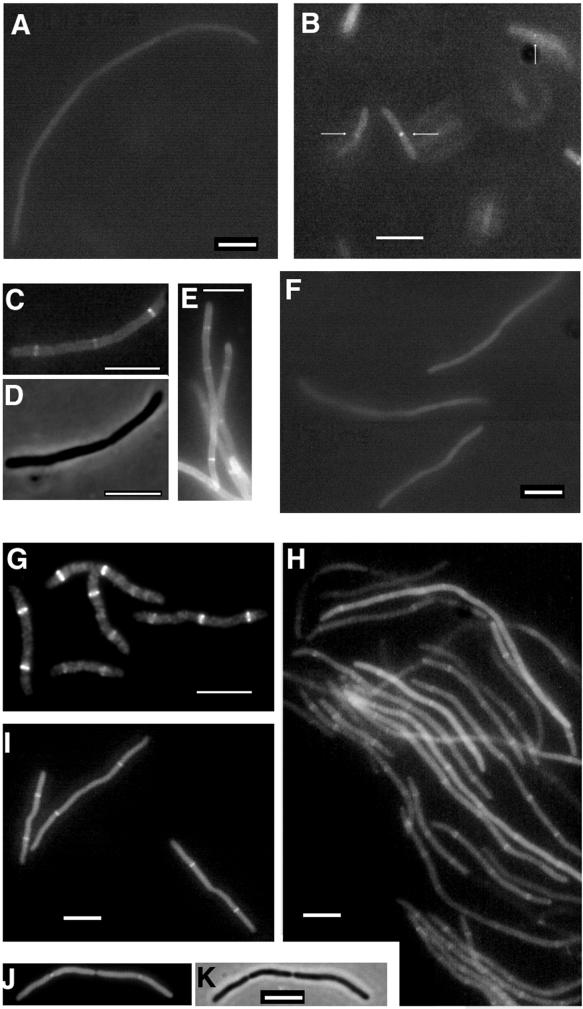
Because FtsK localizes late in wild-type cells, it was expected that FtsK would fail to localize in ftsZ and ftsA mutant filaments. Indeed, FtsK-GFP failed to localize detectably in ftsZ filaments (Fig. 6A), indicating that FtsZ function was required for FtsK localization. Control cells grown at 28°C exhibited localization to constrictions (data not shown). Likewise, FtsK-GFP localization was not detectable in ftsA mutant filaments that were grown for over two generations at the nonpermissive temperature (Fig. 6F), and control cells at 28°C showed localization as expected (Fig. 6B). However, we sometimes observed FtsK-GFP localized to potential division sites between constrictions in short ftsA mutant filaments at early times after shift to the nonpermissive temperature (Fig. 6C to E). Localization to constrictions was rarely observed in these cases. Because repeated searches for FtsK-GFP localization in long ftsA filaments failed, we surmise that the transient localization was probably due to residual activity of FtsA or a factor dependent on FtsA at potential division sites. Taken together, these results suggest that FtsK-GFP localization to the septum requires functional FtsZ and probably also FtsA.
FIG. 6.
FtsK-GFP localization in ftsA, ftsI, and ftsZ mutants and in cephalexin-treated cells. (A) Fluorescence micrograph of pKG4 in a JFL101 (ftsZ84) filament after 3 h at 42°C; (B to E) fluorescence micrographs (B, C, and E) and a phase-contrast micrograph (D) of pKG4 in ftsA1882 cells showing localization to midcell septa (arrows) in cells grown at 28°C (B), and at 60 min after the shift to 42°C (C to E [panel E is of a field different from that shown in panels C and D]); (F) fluorescence micrograph of pKG4 in ftsA1882 filaments 2 h after the shift to 42°C; (G) IFM of FtsZ in ftsI2158 short filaments at 42°C; (H) fluorescence micrograph of pKG4 in ftsI2158 filaments, showing localization; (I to K) fluorescence micrographs (I and J) and a phase-contrast micrograph (K) of BSP853 cells containing pKG7, 4 h after cephalexin addition, showing fluorescent bands at unconstricted midcell sites (I), and 2 h after cephalexin addition, showing fluorescent dots at constrictions (J and K). Bars = 5 μm.
FtsK-GFP localization was then examined under conditions where FtsI, which is responsible for septal peptidoglycan synthesis, is inactivated either by the drug cephalexin (7, 20) or by growth of an ftsI mutant at the nonpermissive temperature (21). Because it is directly involved in septal cell wall biosynthesis, FtsI is thought to act at a later stage than FtsZ or FtsA. FtsK-GFP was able to localize clearly and strongly to potential division sites in filaments generated under both types of conditions (Fig. 6H to K). Targeting to constrictions was observed in short filaments after shorter induction times (Fig. 6J and K) but not at later times when visible constrictions disappeared. The spacing between FtsK-GFP fluorescent bands varied but was at least 6 μm, or approximately four nucleoid lengths. Figure 6I to K clearly shows two FtsK-GFP bands per filament, some of which were over 10 μm apart, implying that the second-generation division sites, but not the first- or third-generation division sites, were recognized. The result shown in Fig. 6I suggested that at this later time point the original midcell FtsZ ring had been disassembled, sometimes with a kink left in the filament, and that targeting to the third-generation sites was delayed. These suggestions are similar to what has been observed for FtsZ in ftsI filaments by us (Fig. 6G) and by others (17). FtsK44-GFP, encoded by pK4(−)G, did not form any detectable bands in cephalexin-induced filaments, supporting the idea that it is defective in localization (data not shown).
The above-described results, from both ftsI mutant filaments and cephalexin-induced filaments, suggest that FtsI function is not strictly necessary for FtsK-GFP localization. Moreover, the presence of a large proportion of FtsK-GFP bands at unconstricted, new division sites supports the idea that FtsK-GFP, when it is expressed at relatively high levels, has the capacity to localize to potential division sites prior to visible constriction.
DISCUSSION
There is a growing body of evidence that the key proteins involved in E. coli cell division may all be localized to the septum region, indicating their direct role in the functioning of the cell division machine. It is already established that FtsZ, FtsA, FtsI, FtsN, and ZipA localize to the septum. Here, we show that the N-terminal 15% of FtsK is sufficient to target GFP to the septum, which suggests that this domain is all that is needed for the wild-type FtsK to recognize the cell midpoint.
FtsK-GFP appears to be targeted late during the septation process. Normally, cells with FtsK-GFP fluorescence at the septum were observed to be in the process of visibly constricting, which is the last step in septation before septum closure and cell separation. The ftsK44 mutant is arrested at a late stage, consistent with the late localization of FtsK. This late localization is similar to that of FtsN (2). Unexpectedly, FtsZ and FtsA were usually not found at septa in cell chains after blockage of FtsK function. This finding might be explained if FtsZ and FtsA did not persist through the entire division process, which FtsK appears to complete. However, evidence from immunofluorescence work and studies of live cells with GFP fusions (22) suggests that FtsZ persists essentially all the way through cell separation. In addition, FtsK-GFP was observed as a band in cells just beginning visible constriction. Therefore, we propose that the presence of FtsK is actually necessary to prevent premature disappearance of FtsZ from the closing septum. FtsZ function along with that of FtsK may still be required at this late stage. Since FtsA seems to follow FtsZ, it is not surprising, then, that FtsA also is absent in the final stages when FtsK function is blocked. As more cell division proteins are characterized, and more dynamic studies of the septation machine are done, it should be possible in the near future to obtain a clearer picture of the pathway of assembly of the cell division apparatus.
What is the function of FtsK? One possibility is that it forms a seal at the last stage of septation, as proposed for SpoIIIE function at the sporulation septum in B. subtilis (25). No obvious perturbation in chromosome segregation has been observed in any ftsK mutant to date (5, 9), indicating that if FtsK has a SpoIIIE-like DNA transport function, it is not essential under the conditions tested. The behavior of an ftsK null allele or a mutation in the C terminus homologous to the DNA-binding region of SpoIIIE will be important to determine in future studies. Perhaps there is functional redundancy with another protein, such as MukB or an uncharacterized segregation protein. One speculation is that FtsK seals the septum closed and simultaneously helps to keep segregated DNA away from the seal. If so, then the ftsK44 mutant would not have a DNA phenotype because the seal would not be complete, obviating a need to remove DNA from the closed septum. The large internal spacer domain of FtsK may impart flexibility to this potentially complicated maneuver.
What does the FtsK N terminus recognize in order to be targeted? Since FtsK has a complex polytopic membrane architecture, particularly in its N-terminal domain, its dependence on FtsZ and FtsA may be via either a direct protein-protein interaction or an indirect interaction with another protein at the division site. The ability of FtsK-GFP, when it is overproduced, to localize to potential division sites in predivisional cells strongly suggests that FtsK has the capacity to recognize FtsZ or another early-acting component of the division apparatus. FtsK-GFP also clearly localized to potential division sites in ftsA mutants at early times after shifting to the nonpermissive temperature but then became diffuse. One explanation for this phenomenon is that FtsK-GFP targeting indeed depends on FtsA, but in this mutant, FtsA was not completely inactivated at early times after the temperature shift. Another possibility is that FtsK-GFP targeting depends on another factor which is FtsA dependent and which takes several generations to be fully inactivated.
In filamentous cells in which FtsI was inactivated, FtsK-GFP clearly localized both to constricting septa and to a subset of potential division sites. One conclusion that can be drawn is that FtsI is not necessary for FtsK-GFP localization. This is significant for two reasons. First, although both FtsK and FtsN appear to be recruited late in septation, FtsN targeting is dependent on FtsI (2), which suggests that the mechanisms of recruitment of FtsN and FtsK may be distinct. Secondly, because the FtsI transpeptidase presumably starts acting well before FtsK does (1, 17), FtsK is likely to recognize an early constituent of the cell division machine such as FtsZ, ZipA, and FtsA. Our observation here that FtsK-GFP often targets new, unconstricted division sites directly supports this model. The apparent contradiction that FtsK-GFP and presumably FtsK are detectable only late in septation in normal cells can be rationalized by a model that couples FtsK levels with timing of localization. In normal dividing cells, FtsK does not attain high-enough levels to assemble the protein at the septum until late in septation, at which point all of the free FtsK becomes bound to the septum. After cell division, the amount of free FtsK presumably returns to the original level and the process continues, ensuring that FtsK never assembles early. In cells overexpressing FtsK-GFP, levels are now significantly higher, such that FtsK can assemble prematurely at potential division sites as well as late during septation. Since FtsZ has generally not been observed to assemble at potential division sites in wild-type cells and yet appears to be required for FtsK targeting, it is likely that the inhibitory effect of high levels of FtsK-GFP on late septation results in a delay in cell division. Such a delay would allow levels of free FtsZ and FtsK-GFP to build up sufficiently to form new FtsZ rings and, hence, new FtsK-GFP targeting sites. In ftsI filaments overexpressing FtsK-GFP, FtsK once again was able to assemble prematurely at potential division sites. However, because a significant amount of FtsK-GFP assembles at unconstricted FtsZ rings and remains there, the level of free FtsK-GFP might be expected to drop. This potential drop may be especially large, because early assembly of FtsK may force it to assemble over the entire cell circumference, perhaps requiring much more protein than for assembly at a constricting septum. Therefore, the concentration of free FtsK-GFP in filamentous cells, even after overproduction, would be too low to assemble at every potential division site. In addition, free FtsZ levels might drop as well, since FtsZ would be tied up in unconstricting rings; previous observations of ftsI filaments with various numbers of FtsZ rings are consistent with this idea (1, 17). Because premature FtsK-GFP localization correlated with a cell division block of some type, it was not possible to determine if the premature targeting per se had any inhibitory effect, for example, on the constriction of the FtsZ ring. Future dynamic studies of cell division in single cells as well as studies of domain interactions and second-site suppressors should prove useful in identifying protein-protein contacts important for FtsK targeting and function.
GFP is a highly useful tool for determining protein localization in living cells and has been used previously to show that the cell division proteins FtsZ, FtsA, and ZipA localize to the septum (11, 14). It usually is much easier to construct and express GFP fusions than to purify proteins and make antibodies, and for some proteins that are hard to purify, GFP fusions may be the only way to perform cytological analysis. Another advantage of GFP, as demonstrated in this paper, is the ability to define localization domains easily. This is especially true for essential cell division proteins, since the studies can be done with tagged domains in a merodiploid. With FtsK, the large N-terminal fusion to GFP was able to complement the ftsK44 mutant, lending greater credence to the results. However, as with any method involving protein fusions, results with GFP must be interpreted with caution. Although the cells are live instead of fixed, a new protein is being expressed. In our study, FtsK-GFP, despite its ability to complement ftsK44, may still have had aberrant activities due to the tag. Keeping these caveats in mind, future studies combining GFP tagging with immunofluorescence and domain swapping should be a fruitful approach toward a deeper understanding of the assembly and function of the bacterial cell division machine.
ACKNOWLEDGMENTS
We thank Ken Begg for the TOE44 strain.
This work was supported by National Science Foundation grant MCB-9410840.
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 3.Addinall S G, Cao C, Lutkenhaus J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–4284. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begg K J, Dewar S J, Donachie W D. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 7.Botta G A, Park J T. Evidence of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 9.Diez A A, Farewell A, Nannmark U, Nystrom T. A mutation in the ftsK gene of Escherichia coli affects cell-cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donachie W D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 11.Hale C A, de Boer P. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 12.Khattar M M, Begg K J, Donachie W D. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol. 1994;176:7140–7147. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993;9:403–410. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 14.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margolin W, Corbo J C, Long S R. Cloning and characterization of a Rhizobium meliloti homolog of the Escherichia coli cell division gene ftsZ. J Bacteriol. 1991;173:5822–5830. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanninga N. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol. 1991;5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 17.Pogliano J, Pogliano K, Weiss D S, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 19.Sharpe M E, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt B G. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 21.Spratt B G. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin binding proteins. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun, Q., and W. Margolin. Real-time tracking of FtsZ dynamics during the bacterial cell cycle. Submitted for publication.
- 23.Weiss D S, Pogliano K, Carson M, Guzman L-M, Fraipont C, Nguyen-Distéche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 24.Wu L J, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 25.Wu L J, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]