Abstract
Gastrointestinal (GI) cancers (gastric cancer, oesophageal cancer, liver cancer, colorectal cancer, etc.) are the most common cancers with the highest morbidity and mortality in the world. The therapy for most GI cancers is difficult and is associated with a poor prognosis. In China, upper GI cancers, mainly gastric cancer (GC) and oesophageal cancer (EC), are very common due to Chinese people’s characteristics, and more than half of patients are diagnosed with distant metastatic or locally advanced disease. Compared to other solid cancers, such as lung cancer and breast cancer, personalized therapies, especially targeted therapy and immunotherapy, in GC and EC are relatively lacking, leading to poor prognosis. For a long time, most studies were carried out by using in vitro cancer cell lines or in vivo cell line-derived xenograft models, which are unable to reproduce the characteristics of tumours derived from patients, leading to the possible misguidance of subsequent clinical validation. The patient-derived models represented by patient-derived organoid (PDO) and xenograft (PDX) models, known for their high preservation of patient tumour features, have emerged as a very popular platform that has been widely used in numerous studies, especially in the research and development of antitumour drugs and personalized medicine. Herein, based on some of the available published literature, we review the research and application status of PDO and PDX models in GC and EC, as well as detail their future challenges and prospects, to promote their use in basic and translational studies or personalized therapy.
Keywords: Upper gastrointestinal cancer, Organoid, PDX, Personalized therapy
Introduction
Gastrointestinal (GI) cancers (including gastric cancer, oesophageal cancer, liver cancer, colorectal cancer, and pancreatic cancer) pose a major challenge to public health and have the highest morbidity and mortality in the world [1, 2]. Upper GI cancers mainly include gastric cancer (GC) and oesophageal cancer (EC), and there were nearly 1.7 million new diagnoses and over 1 million new deaths for GC and EC combined, accounting for 8.7% and 13.2% of the incidence and mortality of all cancers in 2020 [3]. In China, GC and ESCC (oesophageal squamous cell carcinoma, a type of EC that is dominant in China) are very common in people with Chinese characteristics, and more than half of patients are diagnosed with distant metastatic or locally advanced disease and have thus missed the opportunity for radical surgery and have a very poor prognosis [4].
The standards of care for GC and EC patients mainly include surgical resection, drug therapy (chemotherapy, targeted drug therapy, immunotherapy, etc.), and radiotherapy [5–8]. For patients with advanced disease, drug-based comprehensive treatment is the main method [9]. In the era of precision medicine, along with the emergence of many new techniques represented by next-generation sequencing (NGS), significant progress has been made in increasing patient survival with the development of targeted drugs and immunotherapy drugs in a variety of tumours including GC and EC [10, 11]. However, precision therapeutic options for cancer patients are far from being met, especially for GC and EC, compared to other solid cancers, such as lung cancer and breast cancer [12]. Over the past decade, most clinical trials of targeted drugs in GC and EC have failed and produced negative results, and one of the reasons for the failure was the misdirection of preclinical results [13].
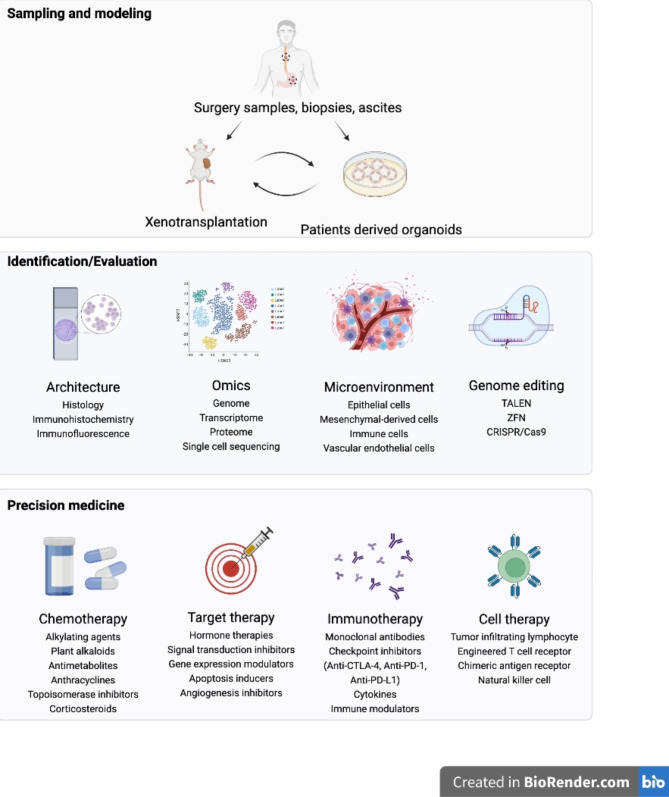
For a long time, most preclinical studies were carried out by using in vitro cancer cell lines or in vivo cell line-derived xenograft (CDX) mouse models, which lack the genetic heterogeneity of original tumors after many passages, are unable to reproduce all the essential characteristics of tumours derived from patients [14–16]. Therefore, cell lines and CDX models have failed to predict human efficacy for most drugs, especially targeted therapeutics [17]. To generate preclinical results that are more reliable, patient-derived organoid (PDO; hereafter referred to as organoids) and xenograft (PDX) models have attracted attention in recent years [18, 19]. Either organoid (ex vivo 3D cell model) or PDX (in vivo animal model; mice are generally used) models are mainly constructed using tumour tissues derived from patients (malignant body fluids can also be used successfully) and preserve the heterogeneity and most of the features of patient tumours [20]. Although organoid and PDX models have different characteristics with advantages and disadvantages (Table 1), due to the high consistency with the features of patient tumours, both models have been widely used in numerous studies, especially in the research and development of antitumour drugs and personalized medicine (Fig. 1).
Table 1.
The differential features of organoid and PDX models
| Characteristics | Organoids | PDX model |
|---|---|---|
| Model type | Ex vivo | in vivo |
| Patient recapitulation | Yes | Yes |
| Stability through passage | Yes | Yes |
| Tumour microenvironment | Rare, few or none | Yes (from host) |
| Maintenance of immune response | No | No |
| Methods to recapitulate immune response | Coculture with immune-related cells | Introduce humanized immune system |
| Scalability | High | Medium |
| Establishment time | Relatively fast, ~weeks | Generally slow, ~months |
| Cost | Relatively low | High |
| Current applications | Preclinical study, translational research, personalized drug screening, etc. | Preclinical study, translational research, personalized drug screening, etc. |
Fig. 1.
Flow diagram of organoid and PDX models derived from patients
It is well known that personalized medicine guided by genotyping is the mainstream precision medicine at present [21]. In addition, personalized functional diagnosis using patient-derived models has emerged as an alternative method to guide personalized therapy [22], which is often referred to as an ‘avatar’ of a patient to predict the therapeutic efficiency of some drug or drug combination. For patients with rare defined driver genes or few actionable variants, personalized therapy guided by functional diagnosis seems to be very important. Herein, based on the available published literature, we review the research and application status of organoid and PDX models in GC and EC, as well as their future challenges and prospects, to promote their use in basic and translational studies or personalized therapy for patients.
Organoid and PDX models in EC
The culture and application of organoids in EC
Based on the few published studies, organoids could be cultured from both the mucosal epithelium and tumour tissues of EC patients using suitable media contianing a variety of growth factors, inhibitors and hormones, using matrigel or basement membrane extract as ECM substitutes [23, 24]. Zheng et al. developed a novel oesophageal minimum essential organoid culture medium (E-MEOM) for culturing murine oesophageal organoids that were immunohistologically and transcriptomically similar to the normal oesophageal epithelium [25]. Vega et al. developed a 3D organotypic culture system and demonstrated that inhibition of Notch signalling promoted the transdifferentiation of normal oesophageal squamous epithelium to a Barrett’s oesophagus (BE)-like metaplasia partially through KLF4 upregulation [26]. Karakasheva et al. developed a protocol that could successfully generate primitive organoids from oesophageal squamous cell samples, allowing the study of the molecular mechanisms underlying oesophageal cancer cell propagation in PDO culture [27]. Kijima et al. established PDOs from both tumour and adjacent normal mucosa biopsies of ESCC patients using medium with exogenous stem cell factors, and the success rate of PDO culture was 68.75% (11/16) for ESCC tumour tissues [28]. They evaluated the growth and structural characteristics of ESCC PDOs treated with 5-FU ex vivo and found that treatment resistance was more conducive to tumour-like organ formation, and the potential treatment-resistant cell population was characterized by high CD44 expression and high autophagy capacity [28]. These organoid culture systems provided a comprehensive experimental platform for studying the molecular mechanisms of oesophageal cancer cell development and drug response. However, at present, the successful rate of esophageal cancer organoids is generally low, and it’s sincerely expected that further technical optimization can overcome this issue.
Organoids have been reported to maintain high accuracy in predicting therapy response in cancer patients (Table 2). Vlachogiannis et al. reported the potential of PDOs to predict therapy response using a living biobank, which included 110 fresh biopsies from 71 patients, with 100% sensitivity and 93% specificity to forecast patients’ responses to chemotherapy or targeted therapy [29]. Li et al. established long-term expansion PDOs from resection tissues of oesophageal adenocarcinoma cancer (EAC) patients using a glandular-preferred protocol which added additional Wnt3A to the growth factors combination of culture medium and tested 24 anticancer compounds in these PDOs [30]. For target agents, the drug sensitivity of PDOs was mostly in accordance with the molecular status, such as TP53 and EGFR mutations. For chemotherapeutic drugs, including 5-fluorouracil, epirubicin, and cisplatin, the resistance of PDOs was consistent with the response in patients [30]. Another study reported that a total of 16 EAC PDOs were successfully grown and characterized using short tandem repeat (STR) analysis, whole-exome sequencing (WES), histology, and immunohistochemistry, indicating recapitulation of the tumour histology and genomic characteristics [31]. Additionally, drug testing using clinically appropriate chemotherapeutics and targeted therapeutics showed an overlap between the patient tumour response and the corresponding organoid response. Together with genomic features, an EAC PDO carrying ERBB2 amplification responded to the targeted HER2 agent mubritinib, while wild-type organoids showed no response, providing insight into personalized treatment using PDO-based drug sensitivity tests. Taken together, due to the capacity of PDOs to recapitulate patient responses, these results verified the feasibility of using organoids for testing drug sensitivity.
Table 2.
Organoid culture and application in EC
| Reference | Sample source | Tissue type | Sample number | Success rate of transplantation | Evaluation |
|---|---|---|---|---|---|
| [30] | EAC | Surgical resections | 32 | 10/32 (31.25%) | H&E, immunohistochemistry, immunofluorescence, whole-exome sequencing, therapy response evaluation |
| [27] | ESCC | Endoscopic biopsies | 16 | 11/16 (68.75%) | H&E, immunohistochemistry, immunofluorescence, therapy response evaluation and resistance mechanism exploration |
| [29] | CRC and GI cancers | Endoscopic biopsies and needle biopsies | 110 | 70% | H&E, immunohistochemistry, next-generation sequencing, therapy response evaluation |
| [31] | EAC | Endoscopic biopsies | 28 | 16/28 (57.2%) | H&E, immunohistochemistry, short tandem repeat analysis, whole-exome sequencing, |
[i]ESCC: oesophageal squamous cell carcinoma; EAC: oesophageal adenocarcinoma cancer; CRC: colorectal cancer; GI: gastrointestinal; H&E: haematoxylin-eosin staining.
The establishment and application of PDX models in EC
Among reported studies establishing preclinical models of EC, PDX models derived from ESCC patients were more frequently observed than those derived from EAC patients (Table 3). A study established 18 EAC PDX models and confirmed their clinicopathological features [32]. A previous study established 4 ESCC and 13 EAC PDX models from a tumour tissue bank [33]. Zhu et al. reported a 61-PDX sequence, one of the largest ESCC PDX cohorts ever reported, and revealed that EGFR may function as a predictive biomarker for cetuximab response [34]. Dodbiba et al. established 21 PDX models from oesophageal/gastroesophageal junction cancers with a success rate of 38%, and among 7 xenografts with drug tests, only the chemosensitivity of 2 xenografts correlated with the patients’ clinical responses [35]. Zou et al. completed the modelling of 25 PDXs derived from 188 fresh endoscopic biopsy tissues of ESCC patients and established PDX models that retained the histologic and genomic characterizations. Tumour growth inhibition from 5 xenografts exposed to paclitaxel and platinum correlated well with the clinical response of patients [36]. Zhang et al. established 37 ESCC PDX models for preclinical drug discovery, in which models carrying HER2 expression had no response to 5-FU/cisplatin [37]. By applying a humanized ESCC PDX model, Liu et al. reported that indomethacin exerted antitumour activity and enhanced cancer immune responses [38]. Ma et al. analysed the heterogeneity of ESCC across 10 cell lines, 80 TCGA tissues and 2 PDX models at the DNA, RNA and protein levels and characterized various novel TP53 mutations, ECM-receptor interactions, focal adhesion, and olfactory transduction pathways (CNGB1) as indicators for accurate research and precision therapeutic development [39]. These observations highlight that EC PDXs has been established as a reliable preclinical model system with histology, genomic variation, and gene expression patterns consistent to the primary tumor, and have proved to be of great application value in oesophageal cancer drug screening.
Table 3.
The establishment and application of PDX models in EC
| Reference | Sample source | Tissue type | Sample number | Success rate of transplantation | Evaluation |
|---|---|---|---|---|---|
| [32] | EAC | Surgical resections | 54 | 18/54 (33.3%) | H&E, short tandem repeat analysis |
| [33] | EAC/ESCC | Surgical resections, endoscopic biopsies and needle biopsies | 61 | 17/61 (27.9%) | H&E, immunohistochemistry |
| [34] | ESCC | Surgical resections | 110 | 61/110 (55.5%) | Immunohistochemistry, gene copy number and mutation analysis |
| [35] | EC/GEJ cancer | Surgical resections | 55 | 21/55 (38.2%) | Immunohistochemistry |
| [36] | ESCC | Endoscopic biopsies | 188 | 25/188 (13.3%) | Immunohistochemistry, DNA sequencing, therapy response evaluation |
| [37] | ESCC | Surgical resections | 96 | 37/96 (38.5%) | Immunohistochemistry |
| [38] | ESCC | NA | 3 | NA | NA |
| [39] | ESCC | NA | 2 | NA | Proteomics analysis |
[i]ESCC: oesophageal squamous cell carcinoma; EAC: oesophageal adenocarcinoma cancer; EC: oesophageal cancer; GEJ: gastroesophageal junction; H&E: haematoxylin-eosin staining; NA: not available
Organoid and PDX models in GC
The culture and application of organoids in GC
PDOs of GC could be cultured from both tissue specimens and malignant ascites (Table 4) [40–43]. Gao et al. developed 15 GC PDOs from 5 patients and identified similar KRAS alterations and drug sensitivity in primary tumours and paired organoids [41]. Malignant ascites from advanced GC with peritoneal metastasis could be collected to generate organoids, showing divergence between individuals but comparability between ascites and PDOs in histological and genomic landscapes. Additionally, ascites-derived PDOs could be used to evaluate the response of chemotherapy regimens and showed similar drug sensitivity to that of patients [40]. Steele et al. reported that GC PDOs from individuals exhibited divergent morphological features and therapeutic regimen efficiency [42]. One patient exhibited a complete response clinically in accordance with the high sensitivity of their corresponding organoids, while another patient did not show a response to therapy agents even though the derived organoids partially responded. Overall, PDO culture techniques allow the generation of preclinical models from metastatic ascites or primary cancer sites, which representing the molecular characteristics and corresponding medical responses similar to parental tumour.
Table 4.
Organoid culture and application in GC
| Reference | Sample source | Tissue type | Sample number | Success rate of transplantation | Evaluation |
|---|---|---|---|---|---|
| [41] | GC | Surgical resections and endoscopic biopsies | 15 | 14/15 (93.3%) | H&E, immunohistochemistry, immunofluorescence, whole-exome sequencing, next-generation sequencing, therapy response evaluation |
| [40] | GC | Malignant ascites | 12 | 11/12 (91.7%) | H&E, immunohistochemistry, whole-exome sequencing, therapy response evaluation |
| [46] | GC | NA | 63 | > 90% | H&E, immunohistochemistry, whole-exome sequencing and transcriptome analysis, therapy response evaluation |
| [53] | GEP-NEN | Fresh clinical samples | 16 GEP-NET and 22 NEC lines | NA | H&E, immunohistochemistry, whole-exome sequencing and transcriptome analysis |
[i]GC: gastric cancer; GEJ: gastroesophageal junction; H&E: haematoxylin-eosin staining; NA: not available; GEP-NEN: gastroenteropancreatic neuroendocrine neoplasm; NEC: neuroendocrine carcinoma
GC PDOs could be expanded long term and subjected to whole-genome sequencing, which can reveal the characteristic mutation style in specific subtypes of GC, such as TP53 mutation in the CIN (chromosomal instability) group, PIK3CA alteration in EB virus, MSI (microsatellite instability), and GS (genomically stable) subtypes. Different types of ERBB2 alterations including amplification and Ser310Phe showed similar regulatory patterns involving the c-MYC-mediated genes CCND2, CDKN1A and THBS1 [44]. On the basis of biomarker detection, ex vivo targeted therapy tests were set up, showing that PDOs harbouring HER2 mutations responded to trastuzumab alone or with 5-fluorouracil. The divergent response to classic chemotherapeutics was investigated, and the IC50 was compared with several cell lines showing a resistance tendency [45]. Therefore, PDO may be a powerful tool for the investigation of molecular pathogenesis and the discovery of biomarkers and targeted medicine.
GC organoid biobanks have been established and comprise an assortment of histological and molecular subtypes, and these normal and cancerous organoid lines recapitulate the morphological, histological, genetic, and transcriptomic characterization of corresponding tumour tissues [46]. Whole-exome sequencing in the GC PDOs revealed the well-documented driver mutations previously reported in GC, such as frequent alterations of CDH1 in diffuse type, TP53 in intestinal type and some other mutations involving RHOA, ERBB2, FGFR2, and MYC, and the similarities in the CIN and GS status to those previously reported in GC were also demonstrated. Organoid-based drug sensitivity ex vivo correlated well with clinical response. For instance, two patients who benefited from 5-fluorouracil and cisplatin after gastrectomy with this combined treatment had sensitive organoids, and another organoid derived from a patient with progressive disease showed no response to capecitabine. High-throughput drug screening was performed in PDOs from 7 patients, and the heterogeneity of agent response was assessed under the conditions of the same patient being given an array of drugs, the same drug being given to various individual patients or spatially different tumour regions from same patient being assessed [46]. Five characteristic organoids derived from gastroesophageal cancer patients in a gastrointestinal cancer cohort were established that captured the histological and genomic features of the parent tissues, such as the intestinal type, diffuse type, ERBB2 amplification type, and temporal intratumor heterogeneity from the same patient (from baseline and posttreatment) [29]. The drug sensitivity of organoids correlated well with clinical treatment response, and the transformation of PDXs from sensitive to resistant to paclitaxel was sequentially generated before and after treatment [29].
Currently, immunotherapy has become a major therapeutic option in the clinic for most cancers, including gastroesophageal cancers. PDOs are being developed to explore the potential mechanisms of immune therapy resistance/response in gastroesophageal cancers by using coculture or air-liquid interface (ALI) systems [47–50]. A study presented a system for the coculture of mouse-derived gastric cancer organoids with immune cells, allowing the identification of a subgroup of gastric cancer patients who would potentially benefit from immunotherapy [49]. Chakrabarti et al. cocultured human gastric cancer organoids (huTGOs) generated from biopsied or resected tissues with cytotoxic T lymphocytes (CTLs) and myeloid-derived suppressor cells (MDSCs) and suggested that HER2-targeted therapy could inhibit CTL effector functions and PD-L1 expression [51]. In ALI system, tumor tissue containing stromal cells and immune cells are separated physically or enzymatically, following by seeded in the collagen gel in a upper surface which is exposed to air-conditions with a porous membrane underneath for nutrient diffusion occurring, so that oxygen can be transported in a more efficient manner [52]. By using ALI technology, Neal and colleagues successfully established co-culture PDOs containing immune cells or fibroblasts from 100 patients representing 28 different tumour types with the success rate of 73% after culture for one-month. These co-culture models maintained the diversity of T cell clones in patients for several weeks [50]. With the rapid development of organoid coculture technology, it provides an valuable platform for further research of personalized immunotherapy. However, the coculture PDOs still need to be more validated.
The establishment and application of PDX models in GC
GC PDX models have been established so that the correlations compared to parent tumours, characterized by histology, genetics and clinical responses, can be evaluated (Table 5) [54–57]. Wang et al. constructed 9 PDX models from 32 GC patients (28.1% success rate) harbouring molecular heterogeneity, including HER2 positivity, c-Met overexpression, and FGFR2 amplification, that responded to molecular targeted therapeutic agents [54]. Gastroscopic biopsies of GC patients were obtained to establish the PDX model, and the overall success rate was 34.1%, in which samples obtained before chemotherapy showed a higher transplantation rate. In addition to the concordance of histopathology and HER2 expression, chemosensitivity between parent tumour tissues and xenografts was investigated, revealing comparable therapeutic responses of corresponding regimens used in clinical treatment [55]. Within these cases, histological transformation from intestinal to diffuse type occurred in case 144, displaying no correlation between PDX-based drug sensitivity and clinically stable disease status [55]. Wang et al. developed mini PDX models for 4 GC patients to achieve personalized screening of chemotherapy or targeted therapy agents [58].
Table 5.
The establishment and application of PDX models in GC
| Reference | Sample source | Tissue type | Sample number | Success rate of transplantation | Evaluation |
|---|---|---|---|---|---|
| [59] | GC/EC | Surgical resections, endoscopic biopsies and needle biopsies | 276 | 98/276 (35.5%) | DNA sequencing, therapy response evaluation |
| [60] | GC | NA | 3 | NA | Karyotyping, whole-exome sequencing, RNA sequencing, and functional studies |
| [61] | GC | Surgical resections | 232 | 35/232 (15.1%) | Immunohistochemistry |
| [62] | GC | Surgical resections, endoscopic biopsies, needle biopsies, pleural fluid and ascites | 54 | 9/54 (16.7%) | Immunohistochemistry |
| [58] | GC | Endoscopic biopsies | 4 | NA | Therapy response evaluation |
| [54] | GC | Surgical resections | 32 | 9/32 (28.1%) | Immunohistochemistry, fluorescent in situ hybridization, therapy response evaluation |
| [64] | GC | NA | 50 | NA | Immunohistochemistry, fluorescent in situ hybridization, next-generation sequencing, therapy response evaluation |
[i]GC: gastric cancer; EC: oesophageal cancer; NA: not available
Notably, PDXs had become a successful tool for drug discovery in GC cancer. Ryan et al. constructed a comprehensive PDX collection of gastroesophageal cancer, including 46 (47%) GC adenocarcinomas, 25 gastroesophageal junction adenocarcinomas (26%), 21 oesophageal adenocarcinomas (32%), and three squamous cell carcinomas (3%), and then evaluated the antitumour activity of rational combination strategies [59]. Song et al. established patient-derived cell lines with peritoneal carcinomatosis, transformed them into orthotopic mouse models, identified major expression and activation traits, and then recapitulated the molecular and phenotypical features of donors [60]. Kuwata et al. successfully established 35 gastric cancer PDX models from 232 engrafted tissues and compared the clinicopathological factors associated with the establishment of PDX and CDX models [61]. Yagishita et al. built a large-scale Japanese patient-derived xenograft library (J-PDX) composed of 298 cross-cancer PDXs, in which 9 PDXs were gastric cancer, with a success rate of 16.7% (9/54) for engraftment [62]. Corso et al. established a comprehensive collection of gastric cancer preclinical models composed of 100 PDX and derivative cell lines or organoids, which included all the major gastric cancer histologic and molecular types identified by The Cancer Genome Atlas [63]. Chen et al. provided a 50-case PDX cohort of gastric cancer, characterized each of their individual histopathological and molecular features, and then evaluated anticancer agents targeting MET, EGFR, HER2 and CDKs in these models [64]. The broad application of these PDX accelerate the development of individualized combination therapies and guide the design of future clinical trials.
Future prospects and challenges of PDO and PDX models in upper GI cancers
Future prospects and challenges of PDO
Precision medicine therapies generally require genomics based on targeted therapies and immunotherapies instead of “one-size-fits-all” chemotherapies [65]. Organoids recapture key characteristics of their corresponding normal or diseased organs and are amenable to current experimental technology. Inspired by cutting edge organoid technology, a deeper understanding of developmental biology and cancer biology could be achieved, as well as the filling of the gap between bench and bedside [19, 66–69]. Great progress has been accomplished in the establishment of PDO models and their application as a predictive preclinical model to evaluate therapeutic responses in vitro, involving lung cancer [70–72], colorectal cancer [29, 73–75], breast cancer [76, 77], ovarian cancer [78, 79], pancreatic cancer [80, 81], liver cancer [82, 83], neuroendocrine neoplasms [53], oesophageal cancer and gastric cancer. However, the following practical challenges still need to be addressed to completely maximize the potential value of upper gastrointestinal PDOs in predicting clinical response:
Optimization of the culture system. With regard to the flexible and diversified application in different scenarios such as with a large cohort for drug screening, omics sequencing (RNA sequencing, single-cell sequencing, etc.), and biobanking of organoids, sustainable expansion of patient-derived tumours and healthy organoids are greatly needed. However, the success rates were comparable for different types and subtypes of cancers. As shown in Tables 2 and 4, the success rate of organoid establishment in gastric cancer was more than 90%, which was significantly higher than that of esophageal cancer, which has a success rates ranging of 31.25-70%. Interestingly, in the existing research, EAC shows a lower success rate than ESCC, even if both of them originate from the esophagus. One of the reason may be different cancer need different culture condition. It has been indicated that multifarious protocols involving Wnt, epidermal growth factor, Noggin, and R-spondin1 showed dramatically divergent effects on organoid outgrowth. Establishment of a standard (basic) protocol for the main type of cancer is a prerequisite, and individualized adjustments based on the characteristics of tumour or healthy tissues are of great value. Except for inhibitors/agonists of specific pathways, the extracellular matrix has an important influence on the formation of organoids.
Quality control. The establishment of organoids depends on the complexity of resection or biopsy tissues or malignant ascites, which might comprise different contents, including cancer cells, normal epithelial cells, mesenchymal-derived cells, or immune cells. To some degree, the existence of varied types of cells reflects the complex tumour microenvironment, while the overexpansion of nonneoplastic portions would mislead the application of organoids in preclinical response to different therapeutic agents. A balance between the presence or absence of normal tissues should be defined to improve the precision of organoids in modelling and clinical prediction.
Drug sensitivity evaluation. The clinical application of organoids has been well developed, either in retrospective observation studies of the consistency between clinical responses and organoid-based drug sensitivity or in intervention studies to predict the individual probable efficacy of potential therapies. Several evaluation indices have been introduced to evaluate the efficiency of different anticancer drugs, including image-based indices such as the size, area, and deep neural network-developed quantification using bright field- or fluorescent field-derived images or cell viability-based indices such as ATP-based AUC or IC50 and flow cytometry-based cell viability. Multidimensional and diversified evaluation approaches should be developed to diagnose drug-induced cell viability to further analyse the relationship between sensitivity results and treatment outcomes. Furthermore, the inhibitors and growth factors in medium may affect gene expression and signal pathway of tumour arganoids, thereby have effects on drug sensitivity.
Microenvironment remodelling. Organoids have been established that can reproduce the genomic landscape and structure of parental tissues, but not all aspects of the microenvironment have been recapitulated, such as mesenchymal-derived cells or immune cells. The preservation of immune cells has been found in ALI-derived organoids, at least for a short term of a month, as mentioned above [50]. However, unlike epithelial cell, which can be continuously passaged and cryoppreserved, the immune element of ALI PDO (such as TILs, TAMs and fibroblast stroma) declines over time and are difficult to persist beyond 60 days [50]. Further optimization of the coculture system is necessary in the future. Besides, since the peripheral immune system play a critical role in antitumour immunity, coculture of organoids with immune components from lymph nodes or peripheral blood may create more comprehensive models that mimic the microenvironment of patients. A coculture system involving tumour organoids and T-cell populations derived from either peripheral blood lymphocytes or TILs was used to explore their potential application in practical immune checkpoint inhibitor testing, which was further adapted into cell therapy evaluations, such as CAR-T cells. In the future, improving the practicality remains an issue to broaden the utility of organoids in preclinical prediction.
Large cohorts with paired organoid-related information and clinical follow-up. Efforts have been invested to probe the possibility of using organoids as biomarkers to predict the response to potential regimens. Among a wealth of clinical studies with organoid-based drug sensitivity tests, a vast number of tumour organoids have been established, while censored data either in drug screening or clinical responses have been found, leading to limited data to determine the potential correlation between organoid-derived drug testing results and clinical responses. Prospective and integral cohort studies are urgently needed to provide valuable insights into individualized precision medicine.
Future prospects and challenges of PDX
As we all known, the PDX is widely used in basic research, drug development and clinical practice because it shows significant heterogeneity of primary tumour and is highly consistent with patient response to treatment. Nevertheless, there are still some limitations of PDX models in cancer research:
Time course. The establishment of PDX is time consuming, which is a major limiting factor in the application of PDX models in real-time personalized medicine. In the literature, establishment of PDX model usually takes 4–8 months, much longer than clinically acceptable waiting times to start treatment.
Engraftment rate. The successful rate of developing PDX models varies among tumor types. As described above, the PDX model has a success rate of 13.3-55.5% and 15.1-35.5% in EC and GC, respectively. There is an urgent need to further optimize the PDX culture technology for various tumors.
Microenvironment remodelling. Human tumor stromal cells and extracellular matrix are gradually replaced by murine counterparts after transplanting into immunodeficient mice [84], and the exact effect of these murine stroma in PDX models remains unclear. Besides, immunodeficient mice lacks components of human immune system, which makes PDX models difficult to study the tumor immune environment and develop immunotherapy strategies. Fortunately, humanized mice, which are immunodeficient mice co-engrafted with human tumours and human hematopoietic stem cells or immune components to reconstitute human immune system, are being investigated [85]. These humanized PDX model offers a potential platform for studying immunotherapies, despite the lack of HLA molecule and lesser functionality of immune cells.
Acknowledgements
Not applicable.
Abbreviations
- PDO
patient-derived organoid
- PDX
patient-derived xenograft
- GI
gastrointestinal
- GC
gastric cancer
- EC
oesophageal cancer
- ESCC
oesophageal squamous cell carcinoma
- EAC
oesophageal adenocarcinoma cancer
- CRC
colorectal cancer
- NGS
next-generation sequencing
- CDX
cell line-derived xenograft
- E-MEOM
oesophageal minimum essential organoid culture medium
- BE
Barrett’s oesophagus
- STR
short tandem repeat
- WES
whole-exome sequencing
- CIN
chromosomal instability
- MSI
microsatellite instability
- GS
genomically stable
- ALI
air-liquid interface
- CTLs
cytotoxic T-lymphocytes
- MDSCs
myeloid-derived suppressor cells
- GEP-NEN
gastroenteropancreatic neuroendocrine neoplasm
- NEC
neuroendocrine carcinoma
Authors’ contributions
Conceptualization, Y.Z., J.G. and C.Z.; Resources, J.L., H.L.,F.Y., P.Q., F.J., S.W. and L.S.; Writing—Original Draft, J.G., J.L., H.L. and C.Z.; Writing—Review and Editing, Y.Z., C.Z., J.G., J.L., and H.L.; Funding Acquisition, J.G.; Supervision, Y.Z., C.Z., and J.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82072728).
Data Availability
All the information generated and analysed is included in the manuscript and relevant tables and figures. Since this is a systematic review of the available literature, no raw data were collected.
Declarations
Competing interests
The authors declare no conflicts of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Gao, Jianqiang Lan, and Haiyan Liao contributed equally to this work.
Contributor Information
Tengfei Chao, Email: turnface@126.com.
Cheng Zhang, Email: qenya@163.com.
Yu Zhu, Email: zhuyu@accibio.com.
References
- 1.Dizdar Ö, Kılıçkap S. Global epidemiology of gastrointestinal cancers. Textbook of Gastrointestinal Oncology, 2019: p. 1–12.
- 2.Arnold M, et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159(1):335–49. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Li M, et al. Time trends of esophageal and gastric cancer mortality in China, 1991–2009: an age-period-cohort analysis. Sci Rep. 2017;7(1):1–8. doi: 10.1038/s41598-017-07071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham D, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355(1):11–20. doi: 10.1056/NEJMoa055531. [DOI] [PubMed] [Google Scholar]
- 6.Al-Batran S-E, et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. The Lancet. 2019;393(10184):1948–57. doi: 10.1016/S0140-6736(18)32557-1. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds J, et al. ICORG 10–14: NEOadjuvant trial in Adenocarcinoma of the oEsophagus and oesophagoGastric junction International Study (Neo-AEGIS) BMC Cancer. 2017;17(1):1–10. doi: 10.1186/s12885-017-3386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro J, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol. 2015;16(9):1090–8. doi: 10.1016/S1470-2045(15)00040-6. [DOI] [PubMed] [Google Scholar]
- 9.Sitarz R, et al. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manage Res. 2018;10:239. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Meltzer SJ. Gastric cancer in the era of precision medicine. Cell Mol Gastroenterol Hepatol. 2017;3(3):348–58. doi: 10.1016/j.jcmgh.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y-M, et al. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Therapy. 2020;5(1):1–11. doi: 10.1038/s41392-020-00323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka T, Yashiro M. Precision medicine for gastrointestinal cancer: recent progress and future perspective. World J Gastrointest Oncol. 2020;12(1):1. doi: 10.4251/wjgo.v12.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranga A, Gjorevski N, Lutolf MP. Drug discovery through stem cell-based organoid models. Adv Drug Deliv Rev. 2014;69:19–28. doi: 10.1016/j.addr.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Li A, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly Representative of Primary Human Gliomas. Mol Cancer Res. 2008;6(1):21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 15.Gillet JP, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A. 2011;108(46):18708–13. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, et al. Microfluidic device for primary tumor spheroid isolation. Exp Hematol Oncol. 2017;6:22. doi: 10.1186/s40164-017-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–54. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 18.Tentler JJ, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Reviews Clin Oncol. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18(7):407–18. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol. 2020;13(1):1–16. doi: 10.1186/s13045-019-0829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer. 2016;16(8):525–37. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauli C, et al. Vitro and in vivo Cancer models to Guide Precision MedicinePersonalized Cancer Models to Guide Precision Medicine. Cancer Discov. 2017;7(5):462–77. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T, et al. Long-term expansion of epithelial organoids from human Colon, Adenoma, Adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 24.Whelan KA, Muir AB, Nakagawa H. Esophageal 3D Culture Systems as modeling tools in esophageal epithelial pathobiology and Personalized Medicine. Cell Mol Gastroenterol Hepatol. 2018;5(4):461–78. doi: 10.1016/j.jcmgh.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng B, et al. A new murine esophageal organoid culture method and organoid-based model of esophageal squamous cell neoplasia. Iscience. 2021;24(12):103440. doi: 10.1016/j.isci.2021.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vega ME, et al. Inhibition of notch signaling enhances transdifferentiation of the esophageal squamous epithelium towards a Barrett’s-like metaplasia via KLF4. Cell Cycle. 2014;13(24):3857–66. doi: 10.4161/15384101.2014.972875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakasheva TA, et al. Generation and characterization of patient-derived head and neck, oral, and esophageal cancer organoids. Curr Protoc Stem Cell Biol. 2020;53(1):e109. doi: 10.1002/cpsc.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kijima T, et al. Three-Dimensional Organoids reveal Therapy Resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell Mol Gastroenterol Hepatol. 2019;7(1):73–91. doi: 10.1016/j.jcmgh.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–6. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat Commun. 2018;9(1):1–13. doi: 10.1038/s41467-018-05190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derouet MF, et al. Towards personalized induction therapy for esophageal adenocarcinoma: organoids derived from endoscopic biopsy recapitulate the pre-treatment tumor. Sci Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-71589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dodbiba L, et al. Primary esophageal and gastro-esophageal junction cancer xenograft models: clinicopathological features and engraftment. Lab Invest. 2013;93(4):397–407. doi: 10.1038/labinvest.2013.8. [DOI] [PubMed] [Google Scholar]
- 33.Damhofer H, et al. Establishment of patient-derived xenograft models and cell lines for malignancies of the upper gastrointestinal tract. J Translational Med. 2015;13(1):1–14. doi: 10.1186/s12967-015-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu H, et al. A subset of esophageal squamous cell carcinoma patient-derived xenografts respond to cetuximab, which is predicted by high EGFR expression and amplification. J Thorac Disease. 2018;10(9):5328. doi: 10.21037/jtd.2018.09.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodbiba L, et al. Appropriateness of using patient-derived xenograft models for pharmacologic evaluation of novel therapies for esophageal/gastro-esophageal junction cancers. PLoS ONE. 2015;10(3):e0121872. doi: 10.1371/journal.pone.0121872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou J, et al. Establishment and genomic characterizations of patient-derived esophageal squamous cell carcinoma xenograft models using biopsies for treatment optimization. J Translational Med. 2018;16(1):1–11. doi: 10.1186/s12967-018-1379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Establishment and characterization of esophageal squamous cell carcinoma patient-derived xenograft mouse models for preclinical drug discovery. Lab Invest. 2014;94(8):917–26. doi: 10.1038/labinvest.2014.77. [DOI] [PubMed] [Google Scholar]
- 38.Liu F, et al. Targeting integrin αvβ3 with indomethacin inhibits patient-derived xenograft tumour growth and recurrence in oesophageal squamous cell carcinoma. Clin Translational Med. 2021;11(10):e548. doi: 10.1002/ctm2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma F, et al. Heterogeneity analysis of esophageal squamous cell carcinoma in cell lines, tumor tissues and patient-derived xenografts. J Cancer. 2021;12(13):3930. doi: 10.7150/jca.52286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, et al. Malignant ascites-derived organoid (MADO) cultures for gastric cancer in vitro modelling and drug screening. J Cancer Res Clin Oncol. 2019;145(11):2637–47. doi: 10.1007/s00432-019-03004-z. [DOI] [PubMed] [Google Scholar]
- 41.Gao M, et al. Development of patient-derived gastric cancer organoids from endoscopic biopsies and surgical tissues. Ann Surg Oncol. 2018;25(9):2767–75. doi: 10.1245/s10434-018-6662-8. [DOI] [PubMed] [Google Scholar]
- 42.Steele NG, et al. An organoid-based preclinical model of human gastric cancer. Cell Mol Gastroenterol Hepatol. 2019;7(1):161–84. doi: 10.1016/j.jcmgh.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S, et al. Three-dimensional Ex vivo culture for drug responses of patient-derived gastric Cancer tissue. Front Oncol. 2021;10:614096. doi: 10.3389/fonc.2020.614096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckers J, et al. Identification and validation of novel ERBB2 (HER2, NEU) targets including genes involved in angiogenesis. Int J Cancer. 2005;114(4):590–7. doi: 10.1002/ijc.20798. [DOI] [PubMed] [Google Scholar]
- 45.Seidlitz T, et al. Human gastric cancer modelling using organoids. Gut. 2019;68(2):207–17. doi: 10.1136/gutjnl-2017-314549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan HH, et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23(6):882–97. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Ootani A, Kuo C. An air–liquid interface culture system for 3D organoid culture of diverse primary gastrointestinal tissues, in Gastrointestinal Physiology and Diseases. 2016, Springer. p. 33–40. [DOI] [PubMed]
- 48.Li X, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20(7):769–77. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakrabarti J et al. Mouse-derived gastric organoid and immune cell co-culture for the study of the tumor microenvironment, in Epithelial Cell Culture. 2018, Springer. p. 157–168. [DOI] [PubMed]
- 50.Neal JT, et al. Organoid modeling of the Tumor Immune Microenvironment. Cell. 2018;175(7):1972. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakrabarti J, et al. Disruption of Her2-induced PD-L1 inhibits tumor cell immune evasion in patient-derived gastric cancer organoids. Cancers. 2021;13(24):6158. doi: 10.3390/cancers13246158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X. Calvin Kuo < an air–liquid Interface Culture System for 3D Organoid Culture of Diverse primary gastrointestinal Tissues.pdf>. Methods Mol Biol. 2016;1422:33–40. doi: 10.1007/978-1-4939-3603-8_4. [DOI] [PubMed] [Google Scholar]
- 53.Kawasaki K, et al. An organoid biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. Cell. 2020;183(5):1420–35. doi: 10.1016/j.cell.2020.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, et al. Establishment of patient-derived gastric cancer xenografts: a useful tool for preclinical evaluation of targeted therapies involving alterations in HER-2, MET and FGFR2 signaling pathways. BMC Cancer. 2017;17(1):1–11. doi: 10.1186/s12885-017-3177-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Y, et al. Establishment and characterization of patient-derived tumor xenograft using gastroscopic biopsies in gastric cancer. Sci Rep. 2015;5(1):1–8. doi: 10.1038/srep08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi YY, et al. Establishment and characterisation of patient-derived xenografts as paraclinical models for gastric cancer. Sci Rep. 2016;6(1):1–12. doi: 10.1038/srep22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang T, et al. Patient-derived gastric carcinoma xenograft mouse models faithfully represent human tumor molecular diversity. PLoS ONE. 2015;10(7):e0134493. doi: 10.1371/journal.pone.0134493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J et al. Personalized Treatment of Advanced Gastric Cancer Guided by the MiniPDX Model. Journal of Oncology, 2022. 2022. [DOI] [PMC free article] [PubMed]
- 59.Moy RH, et al. Defining and targeting Esophagogastric Cancer genomic subsets with patient-derived xenografts. JCO Precision Oncology. 2022;6:e2100242. doi: 10.1200/PO.21.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song S, et al. Patient-derived cell lines and orthotopic mouse model of peritoneal carcinomatosis recapitulate molecular and phenotypic features of human gastric adenocarcinoma. J Experimental Clin Cancer Res. 2021;40(1):1–15. doi: 10.1186/s13046-021-02003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuwata T, et al. Establishment of novel gastric cancer patient-derived xenografts and cell lines: pathological comparison between primary tumor, patient-derived, and cell-line derived xenografts. Cells. 2019;8(6):585. doi: 10.3390/cells8060585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yagishita S, et al. Characterization of the large-scale japanese patient‐derived xenograft (J‐PDX) library. Cancer Sci. 2021;112(6):2454–66. doi: 10.1111/cas.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corso S, et al. A Comprehensive PDX Gastric Cancer Collection captures Cancer cell–intrinsic transcriptional MSI TraitsMSI signature derived from a gastric Cancer PDX platform. Cancer Res. 2019;79(22):5884–96. doi: 10.1158/0008-5472.CAN-19-1166. [DOI] [PubMed] [Google Scholar]
- 64.Chen Z, et al. Characterization and validation of potential therapeutic targets based on the molecular signature of patient-derived xenografts in gastric cancer. J Hematol Oncol. 2018;11(1):1–12. doi: 10.1186/s13045-018-0563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dugger SA, Platt A, Goldstein DB. Drug development in the era of precision medicine. Nat Rev Drug Discovery. 2018;17(3):183–96. doi: 10.1038/nrd.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wensink GE, et al. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precision Oncology. 2021;5(1):1–13. doi: 10.1038/s41698-021-00168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in immunological research. Nat Rev Immunol. 2020;20(5):279–93. doi: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- 68.Bock C, et al. The organoid cell atlas. Nat Biotechnol. 2021;39(1):13–7. doi: 10.1038/s41587-020-00762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schutgens F, Clevers H. Human organoids: tools for understanding biology and treating diseases. Annu Rev Pathol. 2020;15:211–34. doi: 10.1146/annurev-pathmechdis-012419-032611. [DOI] [PubMed] [Google Scholar]
- 70.Hu Y, et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat Commun. 2021;12(1):1–14. doi: 10.1038/s41467-021-22676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi R, et al. Organoid cultures as Preclinical Models of non–small cell lung CancerNon–Small cell Lung Cancer Organoids. Clin Cancer Res. 2020;26(5):1162–74. doi: 10.1158/1078-0432.CCR-19-1376. [DOI] [PubMed] [Google Scholar]
- 72.Kim S-Y, et al. Modeling clinical responses to targeted therapies by patient-derived organoids of Advanced Lung AdenocarcinomaClinical Relevance of Lung Cancer patient-derived Organoids. Clin Cancer Res. 2021;27(15):4397–409. doi: 10.1158/1078-0432.CCR-20-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ooft SN, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11(513):eaay2574. doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 74.Yao Y, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26(1):17–26. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 75.Bruun J, et al. Patient-derived organoids from multiple colorectal Cancer Liver Metastases Reveal Moderate Intra-patient Pharmacotranscriptomic HeterogeneityPharmacological Heterogeneity of Colorectal Liver Metastases. Clin Cancer Res. 2020;26(15):4107–19. doi: 10.1158/1078-0432.CCR-19-3637. [DOI] [PubMed] [Google Scholar]
- 76.Sachs N, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1–2):373–86. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 77.Chen P, et al. Patient-derived Organoids can Guide Personalized‐Therapies for patients with advanced breast Cancer. Adv Sci. 2021;8(22):2101176. doi: 10.1002/advs.202101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Witte CJ, et al. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter-and intrapatient drug responses. Cell Rep. 2020;31(11):107762. doi: 10.1016/j.celrep.2020.107762. [DOI] [PubMed] [Google Scholar]
- 79.Kopper O, et al. An organoid platform for ovarian cancer captures intra-and interpatient heterogeneity. Nat Med. 2019;25(5):838–49. doi: 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 80.Driehuis E, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci. 2019;116(52):26580–90. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tiriac H, et al. Organoid Profiling identifies common responders to Chemotherapy in Pancreatic CancerPancreatic Cancer Organoids parallel patient response. Cancer Discov. 2018;8(9):1112–29. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Broutier L, et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23(12):1424–35. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao W, et al. Modeling liver cancer and therapy responsiveness using organoids derived from primary mouse liver tumors. Carcinogenesis. 2019;40(1):145–54. doi: 10.1093/carcin/bgy129. [DOI] [PubMed] [Google Scholar]
- 84.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 85.Chuprin J, et al. Humanized mouse models for immuno-oncology research. Nat Reviews Clin Oncol. 2023;20(3):192–206. doi: 10.1038/s41571-022-00721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the information generated and analysed is included in the manuscript and relevant tables and figures. Since this is a systematic review of the available literature, no raw data were collected.