Abstract
The Aspergillus nidulans xylanase genes xlnA and xlnB are subject to regulation by ambient pH via the zinc finger transcription factor PacC. In the presence of d-xylose, xlnA is expressed under conditions of alkaline ambient pH while xlnB is expressed at acidic ambient pH. These data have been confirmed for acidity- and alkalinity-mimicking A. nidulans mutants.
In nature, many microbes are exposed to large variations in ambient pH and thus require both an efficient pH homeostatic system and a regulatory mechanism which ensures that those molecules exposed to the environment (such as certain permeases, small metabolites, and extracellular enzymes) are only produced under appropriate pH conditions. The ascomycete Aspergillus nidulans is able to grow over a wide pH range (14). The regulatory circuit controlling pH regulation in this microorganism has been analyzed both genetically and molecularly. As an example, this system regulates the secretion of alkaline phosphatase in alkaline growth conditions and acid phosphatase in acidic environments (2).
pH regulation of gene expression in A. nidulans is mediated by the wide-domain zinc finger transcription factor PacC (16). In alkaline culture conditions, this factor is converted to its truncated functional form in response to the ambient pH signal transduced by the products of six genes (palA, -B, -C, -F, -H, and -I) and is able to activate the expression of those genes whose products are appropriate at alkaline ambient pH and repress those whose expression is suited to acidic pH (1, 3, 10, 11, 16). Activation is exercised by binding to the consensus target sequence 5′-GCCARG-3′ (5). We have previously shown that A. nidulans produces three xylanases when grown on d-xylose as the sole carbon source: one minor xylanase (X24), encoded by xlnB, and two major xylanases (X22 and X34), encoded by xlnA and xlnC, respectively (6, 7, 9, 12). In this article we demonstrate pH regulation of the expression of the xlnA and xlnB genes.
The nucleotide sequences of the upstream regions of the xlnA and xlnB genes have been determined and reveal the presence of two and one consensus PacC binding sites, respectively (Fig. 1). The occurrence of such binding targets suggests that the transcription of these genes might, at least in part, be regulated by this transcription factor.
FIG. 1.
DNA sequences of the xlnA and xlnB gene promoters (EMBL accession no. Z49892 and Z49893, respectively). The translational initiation codon is shown in bold, and consensus PacC binding sites are shown in bold and underlined.
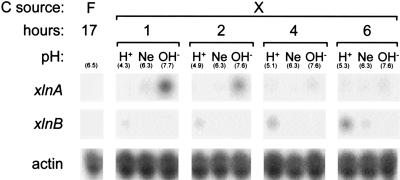
Transcription of the xylanase genes was analyzed in mycelia grown under acidic (pH ∼4.5), neutral (pH ∼6.5), and alkaline (pH ∼7.5) growth conditions. Wild-type conidia were inoculated in minimal medium (MM) (13) supplemented with 0.5% (wt/vol) Casamino Acids and 1% (wt/vol) d-fructose as the sole carbon source. Incubation was done for 17 h at 37°C with orbital shaking, and the mycelia were subsequently transferred to buffered media containing 1% (wt/vol) d-xylose as the sole carbon source for 1, 2, 4, and 6 h. Total RNA was prepared and analyzed by Northern blotting. Figure 2 shows that the xlnA transcript appears shortly after transfer and only in alkaline growth conditions. In contrast, the xlnB transcript is detected after 4 h and mainly in acidic growth conditions. These data suggest pH regulation of the expression of xlnA and xlnB.
FIG. 2.
Expression of xlnA and xlnB genes in A. nidulans wild-type mycelium in response to external pH. Carbon (C) sources present were d-fructose (F) and d-xylose (X). pH conditions are indicated as follows: H+, acidic; Ne, neutral; and OH−, alkaline. The actual pHs of the culture media at the time of recovery of mycelia for RNA isolation are indicated in parentheses. Actin gene expression is shown as a loading control.
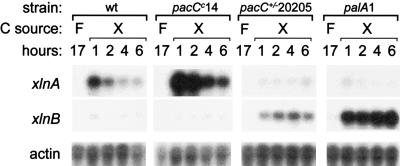
In order to investigate whether this response to ambient pH is mediated by PacC, we examined the transcript levels of the two genes in different pH mutant genetic backgrounds. Wild type, pacCc14 (an extreme alkalinity-mimicking mutant) (2, 16), pacC+/−20205 (an acidity-mimicking pacC mutant [17]), and palA1 (a mutant in the signal transduction pathway mimicking acidic conditions) (1, 2, 10) mycelia were grown in d-fructose MM as noted above and transferred to MM (pH 6.5) containing 1% (wt/vol) d-xylose as the sole carbon source. At different time points after transfer (1, 2, 4, and 6 h) total RNA was isolated and analyzed by Northern blotting. Figure 3 shows that the alkalinity-mimicking pacCc14 mutation simultaneously results in elevated levels of the xlnA transcript and reduced levels of the xlnB messenger. In contrast, the acidity-mimicking pacC+/−20205 and palA1 mutations yield the opposite pattern of expression. The highest level of expression of xlnA occurs in the pacCc14 mutant background, whereas that of xlnB occurs in the palA background.
FIG. 3.
xlnA and xlnB expression in A. nidulans pH-regulatory mutants. Carbon (C) sources present were d-fructose (F) and d-xylose (X). Measurements at the time of recovery of mycelia for RNA isolation showed that the pH of all culture fluids fell within the range of 5.9 to 6.5. Actin gene expression is shown as a loading control.
The analyses described above show an absolute correspondence between the patterns of transcription of the xylanase genes under different ambient pH conditions and those observed in the pH mutant backgrounds. Specific transcription of the xlnA and xlnB genes requires both the presence of an inducer (d-xylose) and optimal ambient pH. In Aspergillus niger the regulation by pH of the two major acidic extracellular proteases was investigated by Northern analysis (8). Due to the absence of pH-regulatory mutants of A. niger, this study was carried out by varying the pH of the culture medium. In the present study, pH regulation of the expression of two A. nidulans xylanase-encoding genes was investigated under different conditions of ambient pH and for different alkalinity- and acidity-mimicking mutants. This constitutes the first demonstration of the control of xylanase gene expression by ambient pH via the wide-domain pH regulator PacC. It is interesting to note the noncoincident, reversed patterns of expression of xlnA and xlnB with respect to pH and the time course of induction. The former might be related to the fact that xlnB codes for an acidic xylanase stable at acidic pH and xlnA encodes a neutral xylanase that is less stable at acidic pH (6, 7).
Including the data presented in this report, four A. nidulans alkaline-expressed genes, ipnA, pacC, prtA, and xlnA, have been studied with respect to pH expression (4, 16). In agreement with the proposed model for PacC as a transcriptional activator (11, 16), each of these genes contains several consensus PacC binding sites in the promoter. Only in the case of ipnA has the physiological relevance of the PacC sites been determined (5). The mechanism for the negative action of PacC in the regulation of acid-expressed genes is poorly understood. Interestingly, pacA, the only acid-expressed A. nidulans gene studied so far, contains an upstream PacC site, but it lies well within the transcribed region and quite close to the putative initiator codon (15, 15a). In contrast, the PacC site in xlnB lies much more nearly in the region where PacC sites occur in alkaline-expressed promoters.
Acknowledgments
This work was supported by the EC-BIOTECH project BIO2-CT93-0174. A.P.M. was the recipient of EC Biotechnology Programme fellowship BIO2-CT94-8136, and M.O. is the recipient of a CSIC postdoctoral contract.
Thanks are due to Herb N. Arst, Jr., for kindly providing us with the A. nidulans mutants used in this work.
REFERENCES
- 1.Arst H N, Jr, Bignell E, Tilburn J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol Gen Genet. 1994;245:787–790. doi: 10.1007/BF00297286. [DOI] [PubMed] [Google Scholar]
- 2.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 3.Denison S H, Orejas M, Arst H N., Jr Signaling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem. 1995;270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 4.Espeso E A, Tilburn J, Arst H N, Jr, Peñalva M A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993;12:3947–3956. doi: 10.1002/j.1460-2075.1993.tb06072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espeso E A, Peñalva M A. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J Biol Chem. 1996;271:28825–28830. doi: 10.1074/jbc.271.46.28825. [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Espinar M T, Piñaga F, Sanz P, Ramón D, Vallés S. Construction of an Aspergillus nidulans multicopy transformant for the xlnB gene and its use to purify the minor X24 xylanase. FEMS Microbiol Lett. 1993;113:223–228. [Google Scholar]
- 7.Fernández-Espinar M T, Piñaga F, de Graaff L H, Visser J, Ramón D, Vallés S. Purification, characterisation and regulation of the synthesis of an Aspergillus nidulans acidic xylanase. Appl Microbiol Biotechnol. 1994;42:555–562. [Google Scholar]
- 8.Jarai G, Buxton F. Nitrogen, carbon, and pH regulation of extracellular acidic proteases of Aspergillus niger. Curr Genet. 1994;26:238–244. doi: 10.1007/BF00309554. [DOI] [PubMed] [Google Scholar]
- 9.MacCabe A P, Fernández-Espinar M T, de Graaff L H, Visser J, Ramón D. Identification, cloning and isolation of the Aspergillus nidulans xlnC gene encoding the 34-kDa xylanase. Gene. 1996;175:29–33. doi: 10.1016/0378-1119(96)00116-3. [DOI] [PubMed] [Google Scholar]
- 10.Negrete-Urtasun S, Denison S H, Arst H N., Jr Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 12.Pérez-González J A, De Graaff L H, Visser J, Ramón D. Molecular cloning and expression in Saccharomyces cerevisiae of two Aspergillus nidulans xylanase genes. Appl Environ Microbiol. 1996;62:2179–2182. doi: 10.1128/aem.62.6.2179-2182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pontecorvo G. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 14.Rossi A, Arst H N., Jr Mutants of Aspergillus nidulans able to grow at extremely acidic pH acidify the medium less than wild type when grown at more moderate pH. FEMS Microbiol Lett. 1990;66:51–54. doi: 10.1016/0378-1097(90)90257-q. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar S, Caddick M X, Bignell E, Tilburn J, Arst H N., Jr Regulation of gene expression by ambient pH in Aspergillus: genes expressed at acid pH. Biochem Soc Trans. 1996;24:360–363. doi: 10.1042/bst0240360. [DOI] [PubMed] [Google Scholar]
- 15a.Sarkar, S., E. A. Espeso, J. Tilburn, and H. N. Arst, Jr. Personal communication.
- 16.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widdick, D. A., and H. N. Arst, Jr. Unpublished data.