Abstract
Survivors of childhood central nervous system (CNS) tumors experience early-onset aging-related phenotypes. DNA methylation (DNAm) age is an emerging epigenetic biomarker of physiologic age and may be predictive of chronic health conditions in long-term survivors. This report describes the course of epigenetic age acceleration using post-diagnosis blood samples (median: 3.9 years post-diagnosis; range: 0.04–15.96) from 83 survivors of pediatric CNS tumors. Epigenetic age acceleration was detected in 72% of patients, with an average difference between chronologic and dnam age of 2.58 years (95% Ci: 1.75–3.41, p < 0.001). Time from diagnosis to sample collection correlated with the magnitude of epigenetic age acceleration.
Keywords: Epigenetics, aging, survivorship, PNET, medulloblastoma
Introduction
Medulloblastoma and primitive neuroectodermal tumor (PNET) are the most common malignant central nervous system (CNS) tumors diagnosed in children.1 Current treatment strategies, consisting of surgery, radiotherapy, and chemotherapy, are associated with long-term survival rates as high as 85%.2 However, these lifesaving therapies can profoundly compromise the long-term function of otherwise healthy organs and tissues. Specifically, long-term survivors of childhood CNS tumors have an elevated risk of multiple chronic health conditions (CHCs), including subsequent neoplasms, neurologic and neurocognitive impairment, endocrine dysfunction, and premature frailty.3–5 For this reason, there is a suspicion that childhood cancer treatment predisposes survivors to an accelerated aging phenotype.4
Aging is a progressive process resulting in increased susceptibility to both morbidity and mortality.6 The aging process also results in changes at the molecular level, including altered DNA methylation (DNAm).7 Several tools are now available to evaluate DNAm as an epigenetic biomarker of physiologic age, even in children and adolescents.8 Recent studies have reported that long-term survivors of childhood cancers experience significant epigenetic age acceleration (EAA).9 However, the onset of a detectable departure between chronologic and epigenetic age following a childhood cancer diagnosis is not yet clear. Therefore, the objective of this study was to characterize the course of EAA among early and late off-therapy individuals treated for pediatric CNS tumors.
Methods
Study participants were diagnosed with medulloblastoma or PNET and treated at Texas Children’s Cancer Center (Houston, TX; 1994–2011). All patients were enrolled in or treated according to St. Jude Medulloblastoma (SJMB) 96 or 03 clinical trial protocols, which generally included surgical resection followed by radiation therapy at 23.4 Gy craniospinal irradiation (CSI) for average-risk patients, and, for high-risk patients, surgical resection, 36.0–39.6 Gy CSI with high-dose chemotherapy with stem cell rescue.10,11 Clinical and demographic factors including sex, race and clinical risk category were abstracted from the electronic medical record. Where appropriate, written informed consent or assent was obtained from participants and/or legal guardians. The study protocol was approved by the institutional review board at Baylor College of Medicine.
Germline DNA was extracted from post-diagnosis whole blood samples and bisulfite converted using the EZ DNA Methylation Kit (Zymo Research) following manufacturer’s instructions. DNA methylation at CpG sites was evaluated using the Illumina Infinium Human Methylation450k Beadchip Array (Illumina Inc, San Diego, CA). Methylation data underwent standard quality control using minifi. Methylation beta-values were normalized using NOOB normalization prior to DNAm calculation.
Descriptive statistics were calculated for cohort demographics and clinical factors. DNAm age was calculated using the Horvath DNA Methylation Age calculator.8 Briefly, the calculator constructs a DNAm age estimate using penalized regression from DNAm measured at 363 CpG sites. Epigenetic age acceleration (EAA) was calculated by taking the difference between the Horvath estimated DNAm age and chronological age at the time of sample collection. A two-sided, paired t-test for difference in means was used to test for a significant difference between DNAm age and chronological age. Next, linear regression models were used to test for associations between EAA and clinical/demographic risk factors, including time from diagnosis to sample collection, sex, race, diagnosis age, and National Cancer Institute risk stratification. Statistical significance was set at a two-sided p < 0.05.
Results
A total of 83 survivors of pediatric CNS tumors were included in the analysis (Table 1). Participants were predominantly male (72.3%), white (45.8%), and had a mean age of diagnosis at 6.8 years. All were diagnosed with medulloblastoma (71.1%) or PNET (28.9%).
Table 1.
demographic characteristics of survivors of medulloblastoma & primitive neuroectodermal tumor (n = 83).
| Characteristic | n(%) |
|---|---|
| Sex | |
| Male | 60 (72.3) |
| Female | 23 (27.7) |
| Race/Ethnicity | |
| Asian | 3 (3.6) |
| Black | 11 (13.3) |
| Hispanic | 27 (32.5) |
| Other/Unknown | 4 (4.8) |
| White | 38 (45.8) |
| Primary Diagnosis | |
| Medulloblastoma | 59 (71.1) |
| pneta | 24 (28.9) |
| Mean(SDb) | |
| Age (years) | |
| Age at Diagnosis | 6.8 (4.0) |
| Age at Sample Collection | 11.6 (4.9) |
| Epigenetic Age | 14.2 (7.0) |
PNET, Primitive neuroectodermal tumor.
SD, Standard deviation.
Epigenetic age was estimated using DNA methylation profiling of samples collected an average of 4.7 years after diagnosis (median: 3.9 years; range: 0.04–15.96 years). On average, the difference between chronologic age and calculated DNAm age at the time of sample collection was 2.58 years (95% CI: 1.75–3.41, p < 0.001) (Figure 1A). Approximately 72% of participants had a DNAm age that was greater than their chronological age. Time from diagnosis to sample collection showed a significant positive association with EAA (β coefficient = 0.46, p < 0.001, R2 = 0.25), such that the mean difference between DNAm age and chronological age at sample collection was 6.9 years (95% CI: 0.70 to 13.12) at 10 years post-diagnosis, compared to 0.5 years (95% CI: −1.52 to 2.55) at one-year post-diagnosis (Figure 1B). No other factors evaluated were significantly associated with EAA (p > 0.05).
Figure 1.
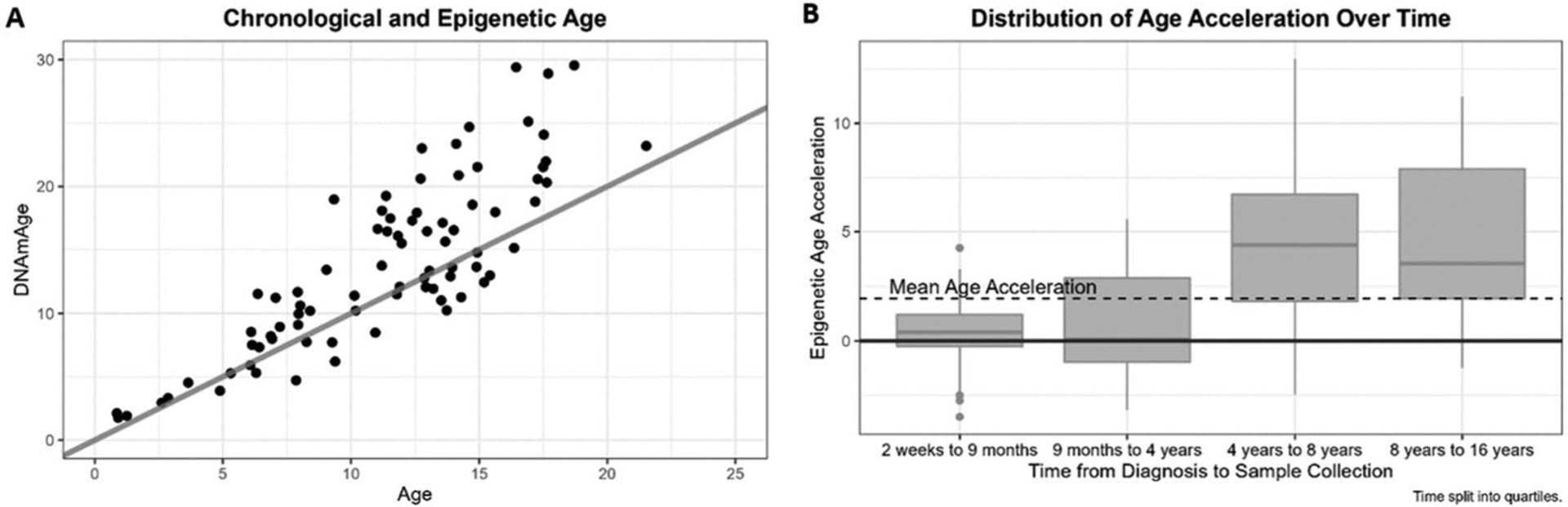
(A) DNAm age and chronological age of all participants with linear trend line. (B) Boxplots showing distribution of epigenetic age acceleration over time since diagnosis.
Discussion
EAA represents an emerging biomarker of physiologic age. EAA has been associated with the incidence of cancer-related, cardiovascular disease-related, and all-cause mortality in elderly populations.12 Because childhood cancer therapy predisposes survivors to excess morbidity and mortality due to aging-related phenotypes, the objective of this study was to determine whether EAA was detectable in the early years following diagnosis. Specifically, we observed that, on average, epigenetic age significantly exceeds chronological age and that the magnitude of this difference is amplified as time from diagnosis increases among survivors of pediatric CNS tumors treated with radiation and/or chemotherapy.
This study adds to the growing body of evidence suggesting childhood cancer treatment is associated with accelerated biologic aging. Recent studies conducted in survivors of childhood cancer have identified other biomarkers of biologic aging, including telomere attrition, associated with late effects of childhood cancer therapy.13,14 Our observation that survivors of pediatric CNS tumors experience accelerated epigenetic aging is consistent with a prospective cohort study of more than 2,000 > 5-year survivors of childhood cancer, including 59 survivors of pediatric medulloblastoma and PNET.9 This study reported an association between the incidence of seven age-related CHCs and EAA among survivors. Notably, the investigators evaluated DNAm age in samples collected an average of approximately two decades after diagnosis. The results of our study expand on this prior work and demonstrate that significant departures between DNAm and chronologic age may not be detectable immediately after diagnosis. However, we observed that the gap between DNAm and chronologic age widened over time, suggesting a potential progressive effect of treatment for pediatric CNS tumors on epigenetic aging. Future studies, including longitudinal studies with serial sample collection, are needed to confirm this observation and explore dynamic changes in DNAm age following childhood cancer treatment.
The mechanisms by which childhood cancer therapy promotes accelerated biologic aging remain unknown. Previous studies have demonstrated that exposure to radiation can alter DNA methylation profiles.1,9 Epigenetic damage may impact pathways involved in cell cycle, DNA repair, and apoptosis; all of which are key molecular pathways involved in the aging process.15 Identifying the molecular processes responsible for the observed link between childhood cancer therapy, altered physiologic aging, and the premature development of CHCs as well as potentially modifiable factors should be the subject of future investigations. Some evidence suggests lifestyle factors, such as tobacco smoking, alcohol consumption, diet, and physical activity, may contribute to advanced epigenetic aging in survivors of childhood cancer.9 Additional work is needed to develop interventions that may improve the physiologic aging profiles of survivors and ultimately reduce the risk of adverse age-related conditions.
Funding
This work was supported in part by funding from the National Institutes of Health National Cancer Institute (K07CA218362), the Cancer Prevention & Research Institute of Texas (RP170668), and the Pablove Foundation Childhood Cancer Research Seed Grant.
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to disclose.
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 2.Packer RJ, Vezina G. Management of and prognosis with medulloblastoma: therapy at a crossroads. Arch Neurol. 2008;65(11):1419–1424. doi: 10.1001/archneur.65.11.1419. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong GT, Liu Q, Yasui Y, et al. Long-term outcomes among adult survivors of childhood central nervous system malignancies in the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2009;101(13):946–958. doi: 10.1093/jnci/djp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ness KK, Krull KR, Jones KE, et al. Physiologic frailty as a sign of accelerated aging among adult survivors of childhood cancer: a report from the St Jude Lifetime cohort study. J Clin Oncol. 2013;31(36):4496–4503. doi: 10.1200/JCO.2013.52.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oeffinger KC, Mertens AC, Sklar CA, et al. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006;355(15):1572–1582. doi: 10.1056/NEJMsa060185. [DOI] [PubMed] [Google Scholar]
- 6.Burger S The aging process. Sciences (New York). 1963;3(8):10–13. doi: 10.1002/j.2326-1951.1963.tb00730.x. [DOI] [Google Scholar]
- 7.Campisi J, Vijg J. Does damage to DNA and other macromolecules play a role in aging? If so, how? J Gerontol A Biol Sci Med Sci. 2009;64(2):175–178. doi: 10.1093/gerona/gln065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath S DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin N, Li Z, Song N, et al. Epigenetic age acceleration and chronic health conditions among adult survivors of childhood cancer. J Natl Cancer Inst. 2021;113(5):597–605. doi: 10.1093/jnci/djaa147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gajjar A, Chintagumpala M, Ashley D, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multi-centre trial. Lancet Oncol. 2006;7(10):813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 11.Gajjar A, Robinson GW, Smith KS, et al. Outcomes by clinical and molecular features in children with medulloblastoma treated with risk-adapted therapy: results of an international phase III trial (SJMB03). J Clin Oncol. 2021;39(7):822–835. doi: 10.1200/JCO.20.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenet. 2016;8(1):1–7. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Man T-K, Aubert G, Richard MA, et al. Short NK- and naïve T-cell telomere length is associated with thyroid cancer in childhood cancer survivors: a report from the childhood cancer survivor study. Cancer Epidemiol Biomarkers Prev. 2022;31(2):453–460. doi: 10.1158/1055-9965.EPI-21-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song N, Li Z, Qin N, et al. Shortened leukocyte telomere length associates with an increased prevalence of chronic health conditions among survivors of childhood cancer: a report from the St. Jude Lifetime Cohort. Clin Cancer Res. 2020;26(10):2362–2371. doi: 10.1158/1078-0432.CCR-19-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics. 2013;8(8):839–848. doi: 10.4161/epi.25498. [DOI] [PMC free article] [PubMed] [Google Scholar]


